Abstract
Biofilm dispersion by Pseudomonas aeruginosa in response to environmental cues is dependent on the cytoplasmic BdlA protein harboring two sensory PAS domains and a chemoreceptor domain, TarH. The closest known and previously characterized BdlA homolog is the flavin adenine dinucleotide (FAD)-binding Aer, the redox potential sensor and aerotaxis transducer in Escherichia coli. Here, we made use of alanine replacement mutagenesis of the BdlA PAS domain residues previously demonstrated to be essential for aerotaxis in Aer to determine whether BdlA is a potential sensory protein. Five substitutions (D14A, N23A, W60A, I109A, and W182A) resulted in a null phenotype for dispersion. One protein, the BdlA protein with the G31A mutation (BdlA-G31A), transmitted a constant signal-on bias as it rendered P. aeruginosa biofilms hyperdispersive. The hyperdispersive phenotype correlated with increased interaction of BdlA-G31A with the phosphodiesterase DipA under biofilm growth conditions, resulting in increased phosphodiesterase activity and reduced biofilm biomass accumulation. We furthermore demonstrate that BdlA is a heme-binding protein. None of the BdlA protein variants analyzed led to a loss of the heme prosthetic group. The N-terminal PASa domain was identified as the heme-binding domain of BdlA, with BdlA-dependent nutrient-induced dispersion requiring the PASa domain. The findings suggest that BdlA plays a role in intracellular sensing of dispersion-inducing conditions and together with DipA forms a regulatory network that modulates an intracellular cyclic d-GMP (c-di-GMP) pool to enable dispersion.
INTRODUCTION
Pseudomonas aeruginosa is one of the principal pathogens associated with cystic fibrosis (CF) pulmonary infection and ranks second among the most common pathogens isolated from chronic and burn wounds (15). The root cause of chronic and persistent infections is the formation of biofilms, which are comprised of microorganisms enmeshed in a hydrated polymer matrix attached to a solid surface (11). Biofilm formation by P. aeruginosa has been shown to progress through multiple developmental stages, beginning with reversible attachment to a surface, followed by irreversible attachment and the development of microcolonies (36, 39, 40). The biofilm developmental cycle comes full circle when biofilms disperse (39, 40). Biofilm dispersion is a process in which sessile, surface-attached organisms liberate themselves from the biofilm and transition to the motile lifestyle. The transition is induced when bacteria sense a myriad of environmental cues that include changes in growth medium composition, pH, oxygen and carbon substrate concentrations, and exposure to heavy metals and nitric oxide (3, 4, 18, 22, 33, 40, 47). Several proteins have been demonstrated to play a role in this process (5, 18, 33, 38, 40, 47). These proteins have been shown to modulate levels of the intracellular signaling molecule cyclic di-GMP (c-di-GMP). c-di-GMP has been associated with controlling the transition between motile and sessile lifestyles, with high concentrations of this molecule correlating with biofilm formation, while low concentrations favor motility (e.g., twitching and swarming) and the free-swimming lifestyle (12). The involvement of c-di-GMP-modulating proteins in biofilm dispersion suggests that this process coincides with alteration of c-di-GMP levels. In addition to the c-di-GMP-modulating proteins, the P. aeruginosa chemotaxis transducer protein BdlA has been demonstrated to be essential for biofilm dispersion. BdlA was identified in a mutant screen, with inactivation of bdlA rendering P. aeruginosa biofilms dispersion deficient in response to various environmental cues (33) and nitric oxide (5). However, the protein lacks the typical domains required for cyclic di-GMP modulation. Instead, BdlA harbors a TarH/methyl-accepting chemotaxis protein (MCP) domain and two PAS domains (Fig. 1A).
Fig 1.
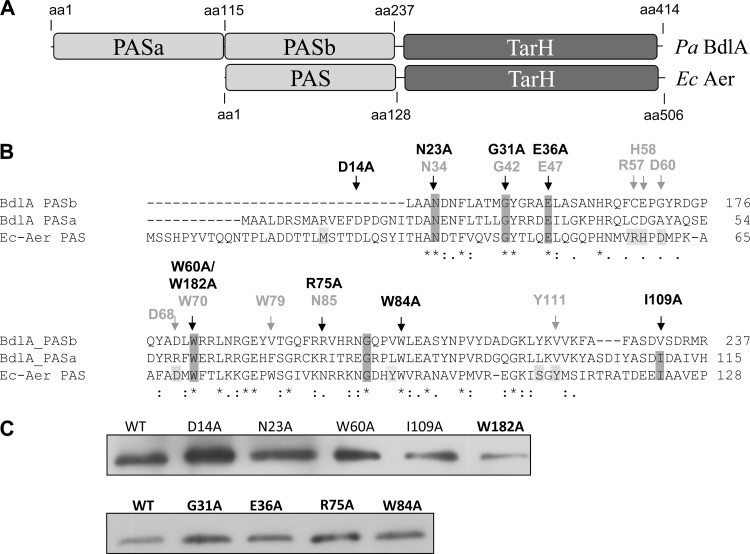
Similarity of BdlA to the aerotaxis transducer protein Aer. (A) Domain structure of BdlA and Aer. PAS, Per-Arnt-Sim sensory domain; MCP/TarH, homolog of Tar (taxis toward aspartate and related amino acids) chemoreceptor domain. aa, amino acid. (B) Alignment of the PASa and PASb amino acid sequences of BdlA with the Aer PAS domain. Aer amino acid residues essential for chemotaxis (dark gray box), but not FAD binding (light gray box), are conserved in BdlA. Residues subjected to alanine substitution mutagenesis in BdlA are indicated by black arrows. Equivalent Aer residues are indicated in light gray. (C) BdlA site-directed mutants are soluble, as indicated by the detection of His-tagged BdlA in cell extracts by immunoblot analysis. Cell extracts were obtained following cell lysis and removal of cell debris by centrifugation at 21,000 × g. WT, wild type.
PAS domains are sensory input domains and protein-protein interaction sites that have been identified in a large family of signal transduction proteins that spans the three divisions of cellular life: archaea, bacteria, and eukarya. Stimuli recognized by PAS domains, which share a characteristic three-dimensional fold, include, but are not limited to, light, oxygen, and redox potential (46). Biological processes in which PAS domains have been implicated are diverse and include global regulation of metabolism, nitrogen fixation, aerotaxis, circadian clocks, hypoxia responses, ion channel function, and developmental processes in prokaryotes and eukaryotes (46). While the response depends on the activity of the modular effector domains, the specificity in sensing arises from cofactors and prosthetic groups that are associated with PAS domains: p-hydroxycinnamic acid, heme, flavin adenine dinucleotide (FAD), and flavin mononucleotide (FMN) (32). Although PAS domains are widespread, the detailed mechanisms of signal transduction have been elucidated only for the bacterial photosensor photoactive yellow protein (PYP), oxygen sensor FixL, and Aer (37). Aer is a membrane-associated protein that mediates the aerotactic response in Escherichia coli by detecting oxygen-related cellular redox changes with its PAS-associated FAD prosthetic group (14, 45). The PAS domain in the Aer aerotaxis transducer has been identified as the site of sensory specificity by comparison of similar domains in other oxygen/redox-sensing proteins (6, 50) and by the generation of a chimera between the PAS domain of Aer and the C terminus of the serine chemoreceptor Tsr, which fully restored chemotaxis in a Δaer Δtsr strain (BT3354) (37).
The sensory protein Aer is the closest known homologue of BdlA, and similar to Aer, BdlA harbors PAS and TarH/MCP-like domains (although BdlA harbors two PAS domains, PASa and PASb [PASa/b], while Aer harbors only one) (Fig. 1A). Furthermore, BdlA also lacks orthodox methylation sites based on sequence alignments with the methylation-independent receptor Aer, which is in agreement with the previous findings that dispersion is independent of CheB (7, 33). Despite the similarity in sequence and domain structure, little is known about BdlA and its role in nutrient-induced biofilm dispersion. Is BdlA a sensory protein? If so, what determines its specificity? In this study, we made use of alignments between the PAS domains of BdlA and Aer to identify amino acid residues essential for dispersion. We demonstrate the association of a heme prosthetic group with the first BdlA PAS domain, PASa, and identify amino acid residues that are critical for dispersion of P. aeruginosa biofilms and protein interaction with the previously identified phosphodiesterases DipA and RbdA. Based on the similarity of BdlA to Aer, we propose that BdlA plays a sensory role in dispersion.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
All bacterial strains and plasmids used in this study are listed in Table 1. P. aeruginosa strain PAO1 was used as the parental strain. P. aeruginosa planktonic cultures were grown in Luria-Bertani (LB) minimal medium or Vogel-Bonner minimal medium (VBMM) (41) in shake flasks at 220 rpm. Antibiotics were used at the following concentrations: 50 to 75 μg/ml gentamicin and 200 to 250 μg/ml carbenicillin for P. aeruginosa and 20 μg/ml gentamicin and 50 μg/ml ampicillin for E. coli.
Table 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant genotype or description | Source or reference |
|---|---|---|
| Strains | ||
| Escherichia coli | ||
| DH5α | λ− ϕ80dlacZΔM15 Δ(lacZYA-argF)U169 recA1 endA hsdR17(rK− mK−) supE44 thi-1 gyrA relA1 | Invitrogen Corp. |
| BL21 | F− ompT hsdSB (rB− mB−) gal dcm (DE3) | Invitrogen Corp. |
| P. aeruginosa | ||
| PAO1 | Wild type | B.H. Holloway |
| ΔbdlA strain | ΔbdlA in PAO1, Kmr | 27, 33 |
| Plasmids | ||
| pCR2.1-TOPO | TA cloning vector; Kmr Apr | Invitrogen Corp. |
| pRK2013 | Helper plasmid for triparental mating; mob tra, Kmr | 16 |
| pJN105 | Arabinose-inducible gene expression vector; pBRR-1 multiple cloning site; araC-PBAD, Gmr | 34 |
| pET101D | Vector for directional cloning and high level V5/6×His fusion protein expression, Ampr | Invitrogen Corp. |
| pMJT1 | araC-PBAD cassette of pJN105 cloned into pUCP18, Ampr (Carbr) | 23 |
| pET-bdlA-V5/6×His | bdlA cloned into pET101D | This study |
| pJN-bdlA-V5/6×His | C-terminal V5/6×His-tagged bdlA cloned into pJN105 at EcoRI/SpeI | This study |
| pJN-bdlA-PASbTarH-V5/6×His | C-terminal V5/6×His-tagged PASbTar of bdlA cloned into pJN105 at NheI/SpeI | This study |
| pJN-bdlA-TarH-V5/6×His | C-terminal V5/6×His-tagged Tar of bdlA cloned into pJN105 at NheI/SpeI | This study |
| pJN-bdlA-W60A | bdlA-V5/His with W60A mutation in pJN105 | This study |
| pJN-bdlA-W182A | bdlA-V5/His with W182A mutation in pJN105 | This study |
| pUC-bdlA | bdlA in pUCP20, Carbr | D. Hassett |
| pUC-bdlA-D14A | bdlA with D14A mutation in pUCP20 | D. Hassett |
| pUC-bdlA-N23A | bdlA with N23A mutation in pUCP20 | D. Hassett |
| pUC-bdlA-G31A | bdlA with G31A mutation in pUCP20 | D. Hassett |
| pUC-bdlA-E36A | bdlA with E36A mutation in pUCP20 | D. Hassett |
| pUC-bdlA-R75A | bdlA with R75A mutation in pUCP20 | D. Hassett |
| pUC-bdlA-W84A | bdlA with W84A mutation in pUCP20 | D. Hassett |
| pUC-bdlA-I109A | bdlA with I109 mutation in pUCP20 | D. Hassett |
| pJN-bdlA-D14A-His | C-terminal 6×His-tagged bdlA with D14A mutation cloned into pJN105 at NheI/SacI | This study |
| pJN-bdlA-N23A-His | C-terminal 6×His-tagged bdlA with N23A mutation cloned into pJN105 at NheI/SacI | This study |
| pJN-bdlA-G31A-His | C-terminal 6×His-tagged bdlA with G31A mutation cloned into pJN105 at NheI/SacI | This study |
| pJN-bdlA-E36A-His | C-terminal 6×His-tagged bdlA with E36A mutation cloned into pJN105 at NheI/SacI | This study |
| pJN-bdlA-R75A-His | C-terminal 6×His-tagged bdlA with R75A mutation cloned into pJN105 at NheI/SacI | This study |
| pJN-bdlA-W84A-His | C-terminal 6×His-tagged bdlA with W84A mutation cloned into pJN105 at NheI/SacI | This study |
| pJN-bdlA-I109A-His | C-terminal 6×His-tagged bdlA with I109 mutation cloned into pJN105 at NheI/SacI | This study |
| pJN-bdlA-W60A-His | C-terminal 6×His-tagged bdlA with W60A mutation cloned into pJN105 at NheI/SacI | This study |
| pJN-bdlA-W182A-His | C-terminal 6×His-tagged bdlA with W182A mutation cloned into pJN105 at NheI/SacI | This study |
Strain construction.
Complementation of a ΔbdlA strain was accomplished by placing the respective genes under the control of an arabinose-inducible promoter in the pJN105 (34) or pMJT-1 (23) vector or an isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible promoter in a pUCP20 vector. C-terminal V5/6×His tagging was accomplished by subcloning into pET101D (Invitrogen) while C-terminal 6×His tagging was accomplished by introducing the tag via PCR (using the sequence CATCATCACCATCACCAT). The tagged constructs were introduced into pJN105 and pMJT-1. Site-directed mutagenesis of bdlA was accomplished by using a GeneArt site-directed mutagenesis kit (Invitrogen) according to the manufacturer's protocol. The identity of vector inserts was confirmed by sequencing. Plasmids were introduced into P. aeruginosa via conjugation or electroporation. Primers used for strain construction are listed in Table 2.
Table 2.
Oligonucleotides
| Oligonucleotide function and name | Sequencea |
|---|---|
| General cloning | |
| bdlA_pET_for | CACCATGGCGGCCCTGGACC |
| bdlA_pET_rev | GAGATCGGCGTTGAGGGTGC |
| PASab_pET_rev | CATGCGGTCGCTGACATCGC |
| PASa_pET_rev | CATCTCGTGTTCCTGGTGGAC |
| pETHis_rev_EcoRI | GCGCGCGAATTCTCAATGGTGATGGTGATG |
| pETHis_XbaI_rev | GCGCGCTCTAGATCAATGGTGATGGTGATG |
| bdlAf_His_Eco | GAATTCATGGCGGCCCTGGACCG |
| bdlA_NheI_for | GCGCGCGCGCTAGCATGGCGGCCCTGGACCG |
| bdlA_NoPAS1_NheI_for | GCGCGCGCTAGCATGGAGCAAGCTGGATGCCC |
| bdlA_TarH_only_NheI_for | GCGCGCGCGCTAGCATGAGCGATGTCAGCGACCGC |
| bdlAHis_SacI_rev | GCGCGCGCGAGCTCCTAATGGTGATGGTGATGATGGAGATCGGCGTTGAGGGTGCGCG |
| Site-directed mutagenesis | |
| W60Afor | CGGAAGACTACCGGCGCTTCGCGGAACGCCTGCGGCGCGGCGAAC |
| W60rev | GTTCGCCGCGCCGCAGGCGTTCCGCGAAGCGCCGGTAGTCTTCCG |
| W182for | GCCCACAGTACGCCGACCTCGCGCGCCGCCTGAACCGCGGCG |
| W182rev | CGCCGCGGTTCAGGCGGCGCGCGAGGTCGGCGTACTGTGGGC |
Restriction sites are underlined.
Biofilm formation.
Biofilms were grown using a once-through continuous-flow tube reactor system for dispersion assays and to obtain protein samples; biofilms were grown in flow cells to view the biofilm architecture prior to and after induction of dispersion as previously described (1, 2, 36, 39, 43). Biofilms were grown at 22°C in 1/20 diluted LB minimal medium or 1/5 diluted VBMM in the absence or presence of 0.1% arabinose. Antibiotics were used in tube reactors at the following concentrations: 2 μg/ml gentamicin and 10 μg/ml carbenicillin. To determine biofilm biomass accumulation over the course of biofilm formation, biofilms were harvested, homogenized, serially diluted, and spread plated onto LB agar. Biofilm biomass was determined via CFU counts. The number of bacterial cells present in biofilm effluents was similarly determined via CFU counts, with biofilm effluents collected daily (for 30 min on ice) over the course of 5 days following initiation of biofilm growth. Quantitative analysis of confocal laser scanning microscopy (CLSM) images of flow cell-grown biofilms was performed using COMSTAT (19).
Biofilm dispersion.
Biofilm dispersion assays were performed as previously described (40). Briefly, following 5 days of biofilm growth in continuous-flow tube reactors, biofilm dispersion was induced by a sudden increase of the carbon concentration in the growth medium by the addition of 20 mM glutamate. Subsequently, the optical density at 600 nm of the biofilm effluent, collected at 1-min intervals, was measured, with dispersion events indicated by an increase in the turbidity of the effluent. In addition, dispersion was visualized by confocal microscopy, and the response was subsequently quantitated by COMSTAT analysis as previously described (33).
FAD and heme detection for BdlA cofactor determination.
Cofactor binding to BdlA or truncated peptides was tested in P. aeruginosa with arabinose-inducible V5- or 6×His-tagged constructs, respectively, using UV-visible light (Vis) spectroscopy, high-performance liquid chromatography (HPLC) analysis, and luminometric FAD determination. Strains expressing the intact or truncated BdlA were grown in the presence or absence of 25 μM hemin, protoporphyrin IX, or FAD (Sigma). The use of exogenous hemin is based on the finding that Burkholderia cepacia was shown to take up hemin (42) and that P. aeruginosa possesses a heme uptake system (28, 29). In addition, protein extracts were preincubated in the absence or presence of 3 μM FAD or 5 μM hemin and subsequently purified again (17, 28, 44). The full-length BdlA and truncated constructs were purified using a Qiagen Ni-nitrilotriacetic acid (NTA) spin kit and an Invitrogen Ni-NTA purification kit according to manufacturers' protocols. The UV-Vis spectra (200 to 800 nm) of the purified constructs were obtained using a DU 700 series Beckman-Coulter UV-Vis spectrophotometer. HPLC analysis was performed on an Agilent 1100 HPLC system equipped with an autosampler, degasser, and detector, with the samples separated using a reverse-phase C18 Targa column (2.1 by 40 mm; 5-μm particle size) at a flow rate of 0.2 ml/min. Purified BdlA was analyzed for the presence of potential cofactors by HPLC following heat denaturation. Commercially available c-di-GMP, NAD, FAD, riboflavin, FMN, NADP, protoporphyrin IX, and hemin were used as standards. Separation and detection of c-di-GMP, NAD, FAD, riboflavin, FMN, and NADP was accomplished by monitoring absorbance at 253 nm using the following gradient of buffer B (methanol plus 10 mM ammonium acetate), with buffer A (10 mM ammonium acetate) making up the total volume: 0 to 9 min, 1% buffer B; 9 to 14 min, 15% B; 14 to 19 min, 25% B; 19 to 24 min, 37.5% B; 24 to 29 min, 50% B; 29 to 40 min, 90% B; and 40 to 50 min, 1% B. Protoporphyrin IX and hemin were detected at 415 nm using the same buffers and the following gradient, with buffer A again making up the total volume: 0 to 6 min, 1% B; 6 to 10 min, 10% B; 10 to 12 min, 20% B; 12 to 14 min, 25% B; 14 to 16 min, 30% B; 16 to 18 min, 35% B; 18 to 20 min, 40% B; 20 to 22 min, 45% B; 22 to 28 min, 50% B; 28 to 30 min, 60% B; 30 to 32 min, 70% B; 32 to 34 min, 80% B, 32 to 42 min, 90% B; 42 to 55 min, 10% B.
In addition to UV-Vis spectroscopy and HPLC analysis, FAD content of purified BdlA constructs and total cellular extracts following perchloric acid precipitation (25) was analyzed by a luminometric assay as previously described (20, 48, 49), using 250 ng/ml apo-d-amino acid oxidase (apo-DAAO) in a prereaction mixture of 23 mM d-alanine, 7.5 μg of horseradish peroxidase, and 25 μM luminol in a total reaction volume of 200 μl. Stock solutions of 0.8 mg/ml horseradish peroxidase (Sigma), 1.12 M d-alanine (Sigma), and 1 mg/ml apo-DAAO (Calzyme, San Luis Obispo, CA) in 0.1 M sodium phosphate, pH 7.0, and 3.75 mM luminol (Sigma) in the reaction buffer of 0.1 M sodium bicarbonate–0.1 M NaCl, pH 9.2, were used. Luminescence was counted for 0.1 s every 60 s for 60 min in a microtiter plate reader (Beckman-Coulter) at 37°C. Peak luminometric readings were used to determine FAD levels using a standard curve established with FAD amounts ranging from 100 to 50,000 fmol.
Determination of BdlA protein interactions using pulldown assays.
In vivo pulldown assays were used to determine whether wild-type and mutant BdlA proteins interact with the phosphodiesterases DipA and RbdA. Briefly, 200 μg of cellular extracts obtained from cells grown planktonically or as biofilms was immunoprecipitated with V5-tagged proteins used as bait and His-tagged proteins used as prey. The resulting immunoprecipitation eluates were separated by SDS/PAGE and assessed by immunoblot analysis for the presence of BdlA or DipA using anti-His or anti-V5 antibodies. Mouse anti-His (Invitrogen) and anti-V5 (Invitrogen) antibodies were used for immunoprecipitation at 1 μg/ml and for immunoblotting at 0.1 μg/ml.
Phosphodiesterase activity determination of cell extracts.
Phosphodiesterase activity of total cell extracts was determined using the synthetic chromogenic substrate bis(p-nitrophenyl) phosphate (bis-pNPP) (Sigma-Aldrich) essentially as previously described (8, 26, 38) by measuring the release of p-nitrophenol (pNP) at 405 nm. An extinction coefficient for p-nitrophenol of 1.78 × 104 M−1 cm−1 was used.
Statistical analysis.
Student's t test was performed for pairwise comparisons of groups, and multivariate analyses were performed using a one-way analysis of variance (ANOVA) followed by a postpriori test using Sigma Stat software. All experiments were carried out in triplicate.
RESULTS
To start elucidating the role of BdlA in dispersion, we focused on the PAS sensory input domains. The closest known homologue of the P. aeruginosa BdlA was found to be the aerotaxis receptor Aer of Escherichia coli that regulates the motile behavior of bacteria in response to gradients of oxygen, redox potential, and certain nutrients (6, 37, 46). Alignment of the two BdlA PAS domains, PASa/b, with the sensory PAS domain of Aer not only revealed similarities between PASa and PASb (44%) but also indicated the presence of several conserved amino acids (Fig. 1A), some of which have been demonstrated in Aer to be essential for the aerotactic response in E. coli (9, 37, 46). For example, amino acid substitutions of N34, F66, and N85 diminished the aerotaxis response and increased the tumbling frequency, while substitutions of G42, E47, R57, H58, D60, D68, W79, and G90 abolished the aerotaxis response, indicating that these residues were essential for aerotaxis and signaling (9, 37, 46). In contrast, mutation of Y111 reversed the aerotaxis response. Of these, amino acid residues N34, G42, E47, and W70 were conserved in both BdlA PAS domains (Fig. 1B).
PAS domain residues important for dispersion of P. aeruginosa biofilms.
To determine whether these conserved amino acids play a role in BdlA function and dispersion, the respective BdlA amino acid residues (D14, N23, G31, E36, W60, R75, W84, I109, and W182) were substituted by site-directed mutagenesis (Fig. 1B). The resulting site-directed BdlA mutants generated by alanine substitution were tested for their capability to restore biofilm dispersion by ΔbdlA mutant biofilms. Biofilm dispersion assays were carried out using tube reactor-grown biofilms. Dispersion was indicated by an increase in the absorbance of biofilm effluents following exposure to dispersion-inducing conditions. Dispersion was induced by a sudden increase in the glutamate concentration. Under the conditions tested, ΔbdlA mutant biofilms were dispersion deficient, while a ΔbdlA mutant complemented with wild-type bdlA dispersed (Table 3) as previously described (33). Likewise, complementation of a ΔbdlA mutant with BdlA harboring the substitutions R75A and W84A fully restored the dispersion phenotype of the ΔbdlA mutant to wild-type levels (Table 3), indicating that these amino acid residues, which are conserved in both BdlA and Aer, do not play a role in BdlA-dependent dispersion. Similarly, alanine substitution of E36 did not alter BdlA function as this mutant biofilm dispersed (Table 3), while the equivalent Aer residue was found to be essential for aerotaxis (9). In contrast, substitutions of alanines for D14, N23, W60, I109, and W182 rendered the proteins nonfunctional, as indicated by the nondispersing phenotype of the complemented ΔbdlA mutant biofilms (Table 3). With the exception of BdlA with the mutation D14A (BdlA-D14A), alanine substitutions of homologue amino acids N23, W60, I109, and W182 were shown in Aer to be associated with an aerotaxis null phenotype (9). Similarly, alanine substitution of BdlA G31 rendered biofilms dispersion deficient (Table 3). Dispersion deficiency was not due to the absence or improper folding of BdlA site-directed mutant variants as all proteins were found to be soluble, as determined by immunoblot analysis (Fig. 1C). Moreover, no significant differences in predicted secondary structures were noted for BdlA protein variants compared to wild-type BdlA, as indicated by the PELE program (SDSC Biology Workbench [http://workbench.sdsc.edu/]) (data not shown).
Table 3.
Identification of BdlA residues essential for dispersion
| P. aeruginosa strainsa | Dispersion responseb | Biofilm biomass (μm3/μm2 [SD])c |
|
|---|---|---|---|
| Prior to dispersion | After dispersion | ||
| PAO1 | + | 12.8 (8.1) | 1.2 (1.6) |
| ΔbdlA strain | - | 11.7 (5.4) | 11.3 (6.9) |
| ΔbdlA/bdlA strain | + | 11.7 (6.9) | 2.8 (1.9) |
| ΔbdlA/bdlA-D14A strain | − | 9.6 (1.4) | 9.9 (1.2) |
| ΔbdlA/bdlA-N23A strain | − | 15.8 (0.2) | 15.8 (0.3) |
| ΔbdlA/bdlA-G31A strain | (−) | 1.7 (0.3) | 2.0 (0.17) |
| ΔbdlA/bdlA-E36A strain | + | 7.3 (0.02) | 3.7 (0.24) |
| ΔbdlA/bdlA-W60A strain | − | 14.3 (10.2) | 13.4 (11.0) |
| ΔbdlA/bdlA-R75A strain | + | 14.8 (3.0) | 2.4 (1.3) |
| ΔbdlA/bdlA-W84A strain | + | 15.2 (0.3) | 1.1 (0.09) |
| ΔbdlA/bdlA-I109A strain | − | 17.5 (2.5) | 18.6 (1.2) |
| ΔbdlA/bdlA-W182A strain | − | 9.9 (6.7) | 8.9 (6.1) |
bdlA constructs were expressed using pUCP20 with the exception of bdlA-W60A and bdlA-W182A, which were expressed using pJN105.
Dispersion response in response to glutamate was determined using the biofilm tube reactor dispersion method. +, dispersion detected; −, no dispersion detected; (−), lack of dispersion probably due to hyperdispersion phenotype.
Visualization and quantitation of the biofilm biomass prior to and following induction of dispersion by glutamate were done by confocal microscopy and subsequent COMSTAT analysis.
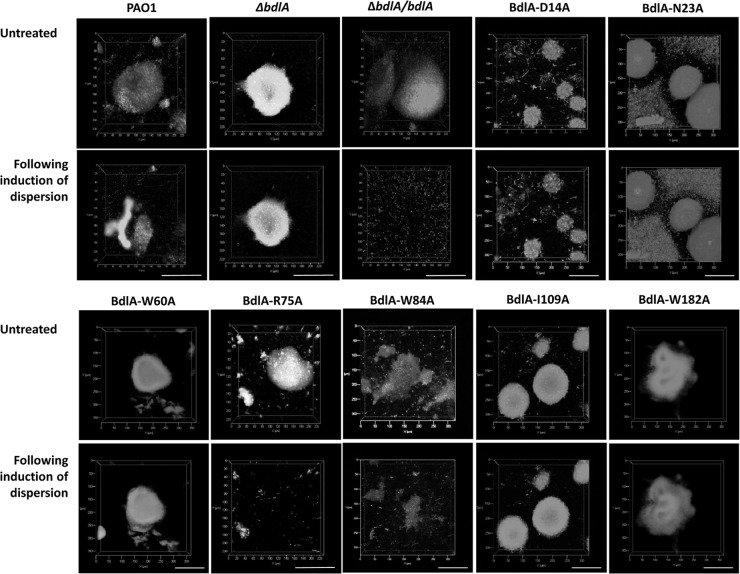
To further confirm our findings of site-directed BdlA protein variants rendering P. aeruginosa dispersion deficient, the dispersion to glutamate was visualized by confocal laser scanning microscopy (CLSM) combined with COMSTAT analysis. Dispersion-deficient ΔbdlA mutant biofilms were used as negative controls while wild-type and complemented ΔbdlA mutant biofilms (ΔbdlA/bdlA strain), which were found to disperse in response to glutamate in our tube reactor dispersion screen (Table 3), were used as positive controls. While all complemented mutant strains formed biofilms on glass, the ΔbdlA/bdlA-R75A and ΔbdlA/bdlA-W84A strains dispersed (Fig. 2 and Table 3), confirming our dispersion results obtained using the absorbance of medium effluent as an indicator. Our findings indicate that selected BdlA PAS domain residues that are conserved in the Aer PAS domain and are essential for the aerotactic response are also essential for dispersion. However, not all residues essential for aerotaxis were found to play a role in dispersion (Fig. 2 and Table 3) or even to be conserved in BdlA (Fig. 1B).
Fig 2.
Biofilm dispersion is impaired following alanine substitutions of D14, N23, W60, I109, and W182 in BdlA. Confocal images of ΔbdlA mutant biofilms complemented with pUC-bdlA harboring site-directed mutations in D14, N23, R75, W84, and I109, and ΔbdlA mutant biofilms complemented with pJN-bdlA harboring site-directed mutations in W60 and W182 prior to and 30 min following addition of glutamate. P. aeruginosa PAO1, the ΔbdlA mutant, and a complemented bdlA strain (ΔbdlA/pUC-bdlA strain) were used as controls. Biofilms were grown in flow cells for 5 days before induction of dispersion. Biofilms were stained with the Live/Dead BacLight viability stain (Invitrogen Corp.). Scale bar, 100 μm.
BdlA does not bind FAD.
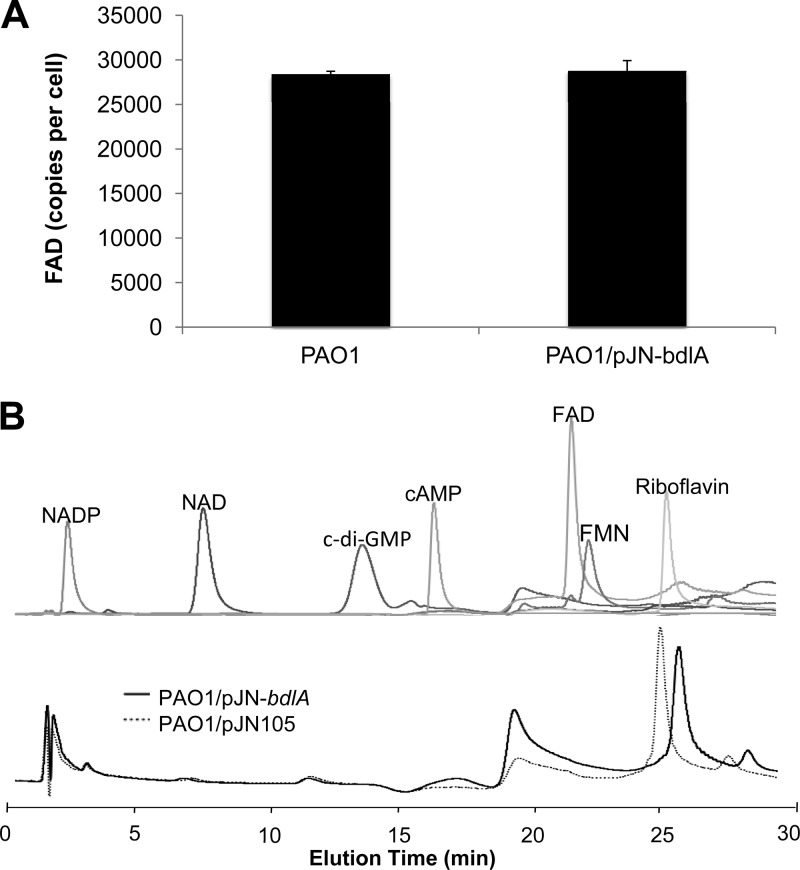
Sensory proteins harboring PAS domains can recognize and respond to a variety of signals, with the specificity of sensing often defined via associations with cofactors of prosthetic groups (32). Aer contains a flavin adenine dinucleotide (FAD) cofactor that affords the protein redox sensing capabilities (37). The Aer amino acid W70 (equivalents in BdlA are W60 in PASa and W182 in PASb) is conserved in all Aer-type proteins, and its presence is now considered diagnostic for predicting whether a given PAS domain binds FAD (Fig. 1B). Similarly, I109 was shown in Aer to reduce FAD binding (9). Additional amino acids involved in FAD binding in Aer (Fig. 1B, R57, H58, D60, and D68) (37) were not conserved in BdlA PASa/b domains (Fig. 1B). The presence of the W60, I109, and W182 residues nevertheless suggested binding of the BdlA PAS domains to an FAD prosthetic group. Further support was derived from alanine substitutions of BdlA, with BdlA-W60A, BdlA-W182A, and BdlA-I109A not being able to restore the biofilm dispersion-deficient phenotype of the ΔbdlA mutant to wild-type levels (Fig. 1C and Table 3). However, no difference in total FAD content was noted in cells lacking or overexpressing bdlA (Fig. 3), nor did purified BdlA bind FAD, as determined by HPLC analysis (Fig. 3) and UV-Vis spectroscopy (data not shown). Preincubation of cells overexpressing bdlA with FAD prior to protein purification also did not result in detectable levels of FAD bound to BdlA (Fig. 3), suggesting that a cofactor other than FAD is associated with the BdlA PAS domains. Additional cofactors and prosthetic groups have been reported to be associated with PAS domains (32). Under the conditions tested, we were unable to confirm riboflavin, FMN, cyclic di-GMP, NAD, and NADP as potential cofactors/prosthetic groups of BdlA (Fig. 3).
Fig 3.
BdlA does not bind FAD, FMN, NAD+, NADP+, or riboflavin. (A) Total FAD content of P. aeruginosa PAO1/pJN105 or PAO1 overexpressing bdlA. (B) Elution profile of FAD, FMN, NAD+, NADP+, or riboflavin by HPLC and supernatants of purified BdlA. Detection limit for each compound tested was <1 pmol. BdlA was purified by Ni-NTA affinity chromatography, and supernatants of denatured BdlA protein were analyzed by HPLC. Ni-NTA eluates obtained from PAO1 cells harboring the empty vector pJN105 were used as controls.
BdlA harbors a heme prosthetic group bound to the PASa domain.
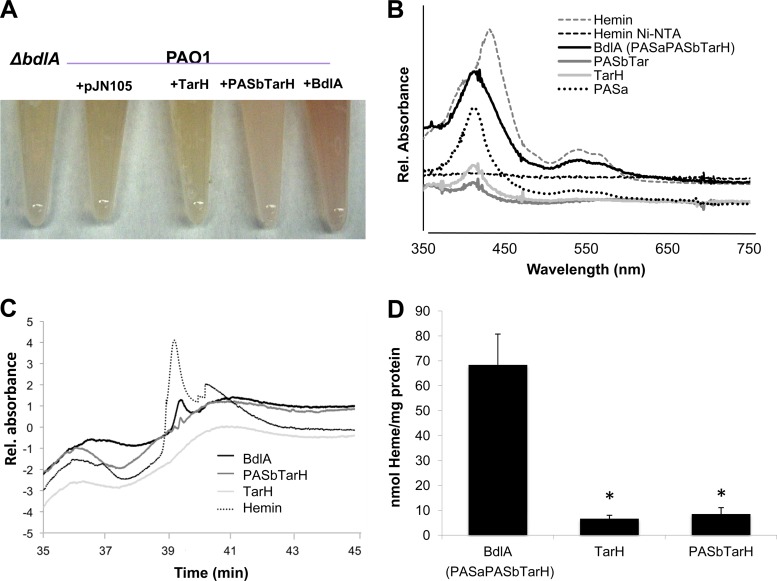
During purification of BdlA, we noticed that cells overexpressing bdlA were reddish while cells lacking bdlA were not (Fig. 4A). In addition, the UV-Vis spectrum of purified BdlA showed a characteristic absorption maximum at 420 nm (referred to as a Soret band) and a broad shoulder at 550 nm, which is characteristic of heme (Fig. 4B). Since heme is structurally diverse, we were unable to properly detect the respective heme by HPLC analysis as we were unable to locate a suitable heme standard. To get around this problem, we made use of previous observations that bacterial cells can take up hemin, which is subsequently incorporated into proteins (29, 42). BdlA was purified using Ni-NTA affinity chromatography from cells grown in the presence of an exogenously added hemin, denatured by heat, and the resulting supernatant was analyzed by HPLC, which confirmed the presence of hemin. The obtained eluates revealed a peak with a retention time identical to hemin (Fig. 4C). To ensure that the hemin was associated with BdlA, hemin alone was subjected to Ni-NTA affinity chromatography, and the eluates were analyzed by HPLC. No peak was detected for hemin alone following purification, indicating that hemin was bound to BdlA (Fig. 4C). It is of interest that when protoporphyrin IX instead of hemin was used, no protoporphyrin IX was detected in supernatants of purified and denatured BdlA by HPLC analysis, nor did the UV-Vis spectrum of purified BdlA show absorption maxima characteristic for protoporphyrin IX (not shown).
Fig 4.
BdlA is a heme-binding protein. (A) Color of extracts of cells overexpressing intact and truncated BdlA tagged with V5/6×His. Cell extracts of the ΔbdlA mutant and PAO1 harboring the empty vector pJN1005 were used as controls. (B) UV-Vis spectra of purified intact and truncated V5/6×His-tagged BdlA proteins. Hemin was used as a control. (C) HPLC elution profile of purified intact and truncated V5/6×His-tagged BdlA proteins. Hemin was used as a control. Representative results are shown in panels A to C. (D) Heme content of intact BdlA and truncated BdlA composed only of the TarH domain or lacking the PASa domain (PASbTarH). *, significantly different from intact BdlA (P < 0.05), as determined by ANOVA and Sigma Stat. Error bars indicate standard deviations. Experiments were carried out in triplicate. Rel, relative.
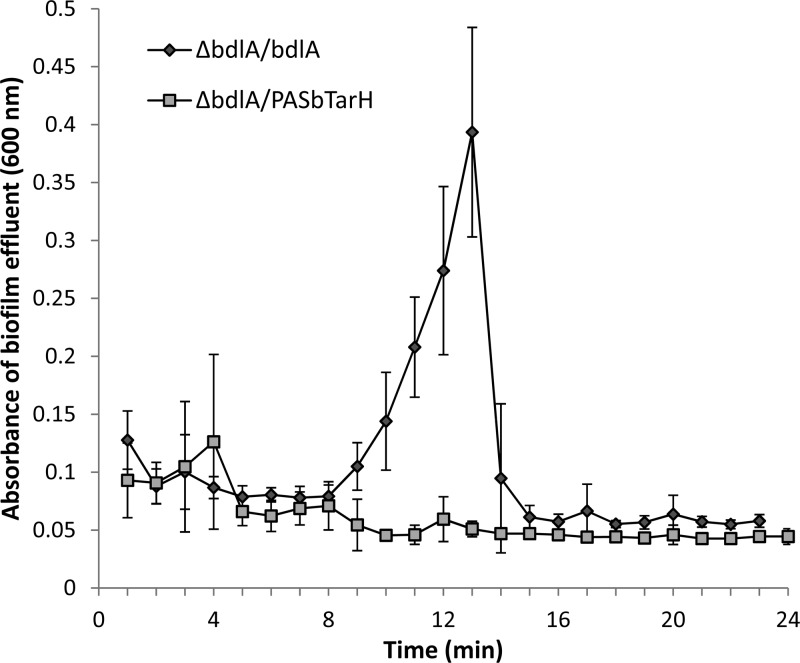
Purified BdlA was subsequently determined to harbor 70 μmol of heme per mg of protein using a heme assay (Fig. 4D). To determine which BdlA domain is associated with heme, two C-terminal V5/6×His-tagged truncated BdlA constructs were generated, a truncated BdlA lacking the first PAS domain (PASbTarH) and one only containing the C-terminal TarH domain (TarH). The resulting proteins were soluble (data not shown). Compared to intact BdlA, both truncated BdlA proteins contained little heme (<10 μmol of heme/mg of protein), as determined using a heme assay (Fig. 4D). Similarly, UV-Vis spectra of truncated BdlA lacked the characteristic absorption maxima observed for intact BdlA and lacked the respective HPLC peak indicative of hemin (Fig. 4B). The findings suggested that the heme group is associated with PASa, with BdlA PASbTarH and BdlA TarH containing only residual heme (Fig. 4).
PASa domain of BdlA is important for dispersion.
We next asked whether the PASa domain associated with heme binding is essential for BdlA to induce dispersion. We therefore made use of a truncated BdlA construct lacking the PASa domain (PASbTarH). The truncated protein was found to be soluble, indicating proper folding (data not shown). We subsequently tested ΔbdlA mutant biofilms complemented with intact or truncated BdlA for dispersion in response to glutamate. While the ΔbdlA/bdlA strain biofilms dispersed, complementation of the ΔbdlA mutant with BdlA lacking PASa did not restore the biofilm dispersion phenotype to wild-type levels (Fig. 5). The observations indicated that truncated BdlA is inactive, with the PASa domain being required for BdlA function.
Fig 5.
The heme-binding BdlA PASa domain is essential for nutrient-induced dispersion. BdlA lacking the first PASa domain does not restore the dispersion-deficient phenotype of the ΔbdlA mutant to wild-type levels. Error bars indicate standard deviation. Experiments were carried out in triplicate.
PAS domain residues important for dispersion do not impair hemin binding of BdlA.
To determine whether site-directed BdlA mutants that were unable to restore the dispersion-deficient phenotype of the ΔbdlA mutant to wild-type levels (Fig. 2 and Table 3) are impaired in heme binding, cell extracts overexpressing the respective proteins were analyzed for heme content. No difference in heme-binding levels was noted among the complemented strains using a heme assay (Fig. 6A). Cell extracts obtained from ΔbdlA strains contained significantly smaller amounts of heme (Fig. 6A). To ensure that the detection of heme was not due to residual hemin, V5/6×His-tagged BdlA-W60A and BdlA-W182A proteins were purified from P. aeruginosa and analyzed for the presence of hemin. Compared to intact BdlA, no significant difference in heme levels was detected (Fig. 6B). The findings strongly suggest that none of the amino acid residues essential for BdlA-dependent dispersion are involved in hemin binding. Moreover, the results strongly suggest that impaired hemin binding does not contribute to the dispersion-deficient phenotype.
Fig 6.
Heme binding is unaltered in site-directed BdlA protein variants. (A) Heme content in cell extracts obtained from planktonic ΔbdlA cells expressing bdlA or bdlA harboring site-directed mutations. (B) Heme content of purified P. aeruginosa V5/6×His-tagged BdlA and V5/6×His-tagged BdlA harboring site-directed mutations in W60 and W182. Error bars indicate standard deviations. Experiments were repeated three times.
Mutation of G31 results in a hyperdispersive phenotype.
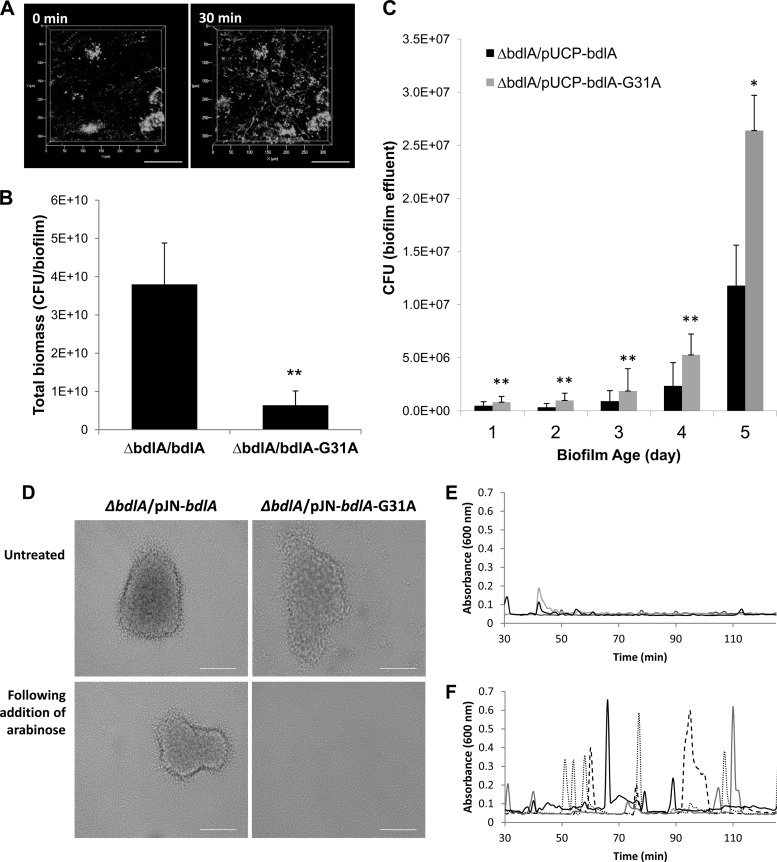
To further elucidate the contribution of the BdlA amino acid sequence to its function in triggering the dispersion response, we analyzed the phenotype associated with the BdlA-G31A substitution. While alanine substitution of BdlA-G31A rendered biofilms dispersion deficient when they were tested via a nutrient-induced dispersion assay (Table 3), ΔbdlA/bdlA-G31A strain biofilms were characterized by little biofilm biomass accumulation. The finding suggested that lack of dispersion may be due to insufficient biofilm biomass accumulation. However, during the microscopic analysis of ΔbdlA/bdlA-G31A biofilms, we noticed P. aeruginosa cells dispersing before the induction of dispersion when the same position of the biofilm was viewed over time (Fig. 7A). The observation implied that the mutated BdlA is in a signal-on mode, even in the absence of an external signal, and suggested that the dispersion-deficient phenotype observed using the tube reactor assay was instead due to ΔbdlA/bdlA-G31A biofilms hyperdispersing. This was further supported by the significantly reduced biofilm biomass accumulation of ΔbdlA/bdlA-G31A strain biofilms compared to that of a ΔbdlA/bdlA strain (Fig. 7B). Moreover, ΔbdlA/bdlA-G31A biofilms were characterized by a 2- to 3-fold increased detection of bacteria present in biofilm effluents compared to wild-type and complemented biofilms over the course of 5 days of biofilm growth (Fig. 7C).
Fig 7.
Alanine substitution of BdlA residue G31 renders P. aeruginosa biofilms hyperdispersive. (A) Confocal image of the ΔbdlA/pUC-bdlA-G31A strain dispersing in the absence of dispersion-inducing conditions. Images were acquired at the same position over the course of 30 min. Scale bar, 100 μm. (B) Total biomass of ΔbdlA/pUC-bdlA and ΔbdlA/pUC-bdlA-G31A strain biofilms following 5 days of growth under flowing conditions, as determined by CFU counts (number of CFU/biofilm). **, significantly different (P < 0.05) compared to the ΔbdlA/bdlA strain. (C) Bacterial cells detached from ΔbdlA/pUC-bdlA and ΔbdlA/pUC-bdlA-G31A strain biofilms over the course of biofilm formation, as determined by viability counts of cells present in biofilm effluents. Significant difference compared to the ΔbdlA/pUC-bdlA strain on the same day is indicated as follows: **, P < 0.05; *, P < 0.1. (D) ΔbdlA mutant biofilms complemented with pJN-bdlA-His or pJN-bdlA-G31A-His were grown in flow cells diluted 5-fold with VBMM for 6 days, and bright-field images were acquired prior to and 12 h following the addition of 0.5% arabinose. A total of 20 images per growth condition were acquired in triplicate. Representative images are shown. Scale bar, 100 μm. (E and F) Detection of dispersion following induction of bdlA gene expression by addition of arabinose to the growth medium. Dispersion experiments were carried out in tube reactor-grown biofilms. Dispersion was indicated by an increase in the turbidity of biofilm tube reactor effluents. Graphs shown are representative of three independent biofilm replicates, indicated by dotted, dashed, and solid lines. Dispersion was detected upon addition of arabinose to the ΔbdlA/pJN-bdlA-G31A-His strain (F) but not to the ΔbdlA/pJN-bdlA-His strain (E).
To further determine whether alanine substitution of G31 locks BdlA in the signal-on mode and, thus, renders P. aeruginosa hyperdispersive, we made use of a complemented mutant strain (the ΔbdlA/pJN-bdlA-G31A strain), which harbored the bdlA-G31A mutant under the control of the arabinose-inducible PBAD promoter. This allowed mutant biofilms to form wild-type-like biofilms within 144 h of growth in the absence of arabinose (Fig. 7D), after which time arabinose was added to the growth medium to induce the transcription of bdlA-G31A. Following arabinose supplementation, the biofilm architecture was monitored by bright-field microscopy for signs of dispersion. Within 30 min following arabinose addition, increased motility and dislodging cells were observed in ΔbdlA/pJN-bdlA-G31A biofilms, ultimately resulting in biofilms composed of a thin layer of cells devoid of microcolonies and clusters within 12 h following arabinose induction (Fig. 7D). In contrast, no increased motility or loss of biofilm biomass was observed in ΔbdlA/pJN-bdlA strain biofilms. The finding strongly suggested that induced expression of bdlA-G31A but not wild-type bdlA resulted in a hyperdispersive phenotype. To quantitatively monitor for dispersion events immediately following induction of bdlA gene expression, we made use of tube reactor-grown biofilms. Addition of arabinose to the growth medium resulted in repeated dispersion events of the ΔbdlA/pJN-bdlA-G31A strain over the course of more than 2 h (Fig. 7F), a response that was absent in ΔbdlA/pJN-bdlA biofilms used as controls (Fig. 7E).
Site-directed BdlA protein variants differentially interact with the phosphodiesterases DipA and RbdA required for dispersion.
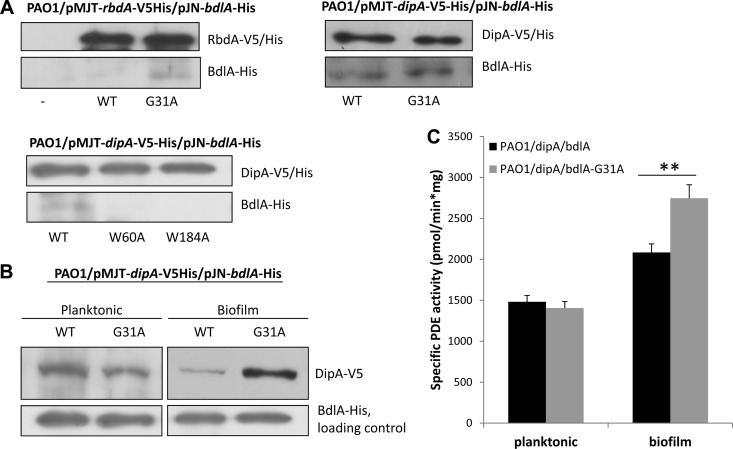
Our findings strongly suggested that the alanine substitution of G31 causes BdlA to transmit a constant signal-on bias as it rendered biofilms hyperdispersive. Recent evidence suggested that dispersion by P. aeruginosa requires two phosphodiesterases (RbdA and DipA) and coincides with increased DipA phosphodiesterase activity and reduced c-di-GMP levels compared to biofilm and planktonic cells (38). In contrast, ΔbdlA mutant biofilms were characterized by increased c-di-GMP levels, suggesting a link between BdlA and the modulation of c-di-GMP levels (33). We therefore asked whether BdlA interacts with DipA and RbdA. In vivo pulldowns indicated that BdlA interacts with DipA under planktonic conditions, while interactions with RbdA appeared to be limited, if not absent (Fig. 8A). Moreover, in vivo pulldowns were also used to determine whether the impaired dispersion response of the ΔbdlA/bdlA-W60A and ΔbdlA/bdlA-W182A strains versus the hyperdispersive phenotype of the ΔbdlA/pJN-bdlA-G31A strain was due to differential protein interactions. Alanine substitution of the G31 residue promoted interaction of BdlA with RbdA, while interactions with DipA appeared to be comparable to those observed for wild-type BdlA (Fig. 8A). In contrast, alanine substitutions of W60 and W182 significantly reduced the interaction with DipA (Fig. 8A). The findings suggested a link between dispersion capabilities of P. aeruginosa and protein interactions of BdlA with phosphodiesterases.
Fig 8.
Wild-type BdlA and site-directed BdlA protein variants differentially interact with the phosphodiesterases RbdA and DipA. (A) In vivo pulldowns were obtained from P. aeruginosa grown planktonically and coexpressing V5/6×His-tagged RbdA or DipA and C-terminally His-tagged BdlA wild type or protein variants (BdlA-G31A, BdlA-W60A, and BdlA-W182A). BdlA-DipA and BdlA-RbdA interactions were assessed via anti-V5 pulldowns, followed by immunoblot analysis for the presence of His-tagged BdlA using anti-His antibodies. Anti-His probing simultaneously visualized the presence of V5/6×His-tagged RbdA and DipA (top panels) used as internal controls. −, negative control using PAO1/pJN-bdlA-His. (B) Immunoblot assays demonstrating a biofilm-specific increase in DipA levels in cells producing BdlA-G31A compared to those harboring wild-type BdlA. Anti-V5 antibodies were used to probe for DipA. BdlA proteins (loading control) were detected using anti-His antibodies. (C) Specific phosphodiesterase activity was determined using cell extracts obtained from P. aeruginosa PAO1 biofilms coexpressing dipA and bdlA or bdlA-G31A and measuring the release of p-nitrophenol (pNP) at 405 nm. WT, G31A, W60A, and W182A indicate the BdlA variants used.
Considering that DipA has previously been demonstrated to be upregulated in dispersing cells (38), we asked whether the hyperdispersive biofilms bearing the BdlA-G31A variant would exhibit changes in DipA levels. While little difference was noted under planktonic conditions, biofilm cells producing BdlA-G31A demonstrated significantly increased DipA abundance compared to cells producing wild-type BdlA (Fig. 8B). The findings suggested that BdlA G31 either contributes to DipA protein stabilization or promotes higher DipA levels specifically under biofilm growth conditions. We therefore determined whether the G31A substitution-specific changes in DipA protein interactions and levels correspond to changes in c-di-GMP turnover rates. This was accomplished by analyzing overall phosphodiesterase activity in planktonic and biofilm cells of PAO1/pMJT-dipA-V5His/pJN-bdlA-His and PAO1/pMJT-dipA-V5His/pJN-bdlA-G31A-His using the synthetic chromogenic substrate bis-pNPP. Under the conditions tested, the phosphodiesterase activity of cell extracts obtained from biofilms coexpressing dipA and bdlA-G31A was 2,746 pmol/min mg−1 and 32% higher than the phosphodiesterase activity of cell extracts obtained from biofilms coexpressing dipA and wild-type bdlA (Fig. 8C). Increased phosphodiesterase activity upon coexpression of dipA and bdlA-G31A was further supported by the finding of decreased biofilm biomass accumulation compared to cells coexpressing dipA and bdlA (data not shown). No difference in the total cellular phosphodiesterase activity was observed when the two strains were grown planktonically (Fig. 8C), suggesting biofilm-specific modulation of DipA levels as well as phosphodiesterase activity by BdlA.
DISCUSSION
BdlA is essential for dispersion by the pathogen P. aeruginosa. However, no specific function for BdlA has been identified. The hypothesis that BdlA is a sensory protein arose from its domain organization, a comparison to similar domains in oxygen/redox-sensing proteins including Aer, and the presence of conserved and essential PAS residues. When aligned with Aer, BdlA harbored several conserved amino acid residues whose substitutions were previously demonstrated to diminish or abolish the aerotaxis response in E. coli (9, 37, 46).
Here, we demonstrate that dispersion induced in response to environmental conditions requires five BdlA PAS domain residues, D14, N23, W60, W182, and I109, that, if mutated, render BdlA inactive by abolishing dispersion. In contrast, mutation of G31 rendered P. aeruginosa biofilms hyperdispersive, indicating that residue G31 is required for the modulation of BdlA activity, with mutation of G31 rendering BdlA constitutively signal-on. Moreover, with the exception of residue I109, the respective amino acids are located in the PAS core region and the EF loop of the PAS domain. The PAS core has been previously demonstrated to contribute to protein-protein interactions and signaling, while the EF loop is part of a linker connecting the PAS core to the β-scaffold that maintains the structural integrity of the PAS domain (35, 37). In Aer, residues involved in FAD cofactor binding and signaling were similarly located within the EF loop and PAS core region, respectively (37). Considering that BdlA PAS residues N23 and G31 are highly conserved within the PAS superfamily (Fig. 1B) (35, 46) and that mutation of D14, N23, G31, W60/W182, and I109 affected neither heme binding nor solubility of BdlA, our findings strongly suggest an involvement of these PAS residues in dispersion in response to environmental conditions, likely by contributing to the active site of BdlA.
The Aer FAD binding residues are variable and only conserved in a subset of PAS domain proteins (37). With the exception of one amino acid residue (Aer W70; BdlA W60/W182), none of the FAD binding residues in Aer are conserved in BdlA (Fig. 1B). This is consistent with our finding of BdlA not binding FAD but, instead, a heme prosthetic group. Considering that the specificity of PAS sensory domains is dependent on the presence of a cofactor or prosthetic group, the findings suggest that if BdlA is indeed a sensory protein, it performs this function in a manner dissimilar from Aer as the initial signal transduction events, as well as the signal output, differ between these two proteins. The membrane-bound Aer senses environmental oxygen levels indirectly by sensing changes in the redox state of its FAD cofactor (14). In the case of BdlA, which is located in the cytoplasm, sensing specificity likely arises from its heme prosthetic group. Proteins harboring a heme PAS domain include membrane-bound FixL proteins which are biological oxygen sensors and the E. coli Dos, a membrane-bound phosphodiesterase that degrades cyclic AMP in a redox-dependent manner (10). The Dos heme-binding region is 60% homologous to FixL PAS domains and capable of binding O2, CO, and NO (13). While the signals for BdlA are not known at this time, it is likely that BdlA is involved in sensing and enabling dispersion in a redox-dependent manner, potentially by sensing the internal energy status of the cells.
In addition to signaling and ligand and/or cofactor binding, PAS domains mediate protein-protein interactions (21, 24). PAS-containing proteins have been demonstrated to either homodimerize or heterodimerize with other proteins through their PAS domains (21, 30, 31, 35). The present study suggests a possible PAS domain site for protein-protein interaction. One site surrounds G31/Y32, which corresponds to PYP residue Y52, with residues located within 10 Å of Y52 forming part of a dimerization interface in PYP (35). Here, we demonstrate that the likely interaction partners of BdlA include the recently identified phosphodiesterases DipA and RbdA required for dispersion by P. aeruginosa biofilms (38). The interactions between the phosphodiesterases and BdlA were increased upon alanine substitution of G31, which also resulted in significantly increased DipA levels and increased phosphodiesterase activity under biofilm growth conditions. Similar to BdlA, both DipA and RbdA harbor PAS domains and are essential for dispersion in response to a variety of dispersion-inducing conditions (33, 38). It is thus likely that they do not represent functionally redundant activities but, instead, are part of a pathway or protein complex capable of modulating an intracellular c-di-GMP pool to enable dispersion. A contribution of BdlA to the modulation of c-di-GMP level is further supported by the finding that site-directed mutation of BdlA W60 and W182 not only impaired the dispersion response but also resulted in reduced DipA-BdlA protein interactions. While future research will determine whether BdlA is directly involved in dispersion signal propagation as well as modulation of the intracellular c-di-GMP pool via protein-protein interaction, our findings indicate that the BdlA PAS domain(s) plays an important role in regulating dispersion.
ACKNOWLEDGMENTS
We thank D. Hassett for providing pUCP20-derived plasmids.
This work was supported by a grant from the NIH (RO1 A107525701).
Footnotes
Published ahead of print 24 August 2012
REFERENCES
- 1. Allegrucci M, et al. 2006. Phenotypic characterization of Streptococcus pneumoniae biofilm development. J. Bacteriol. 188: 2325–2335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Allegrucci M, Sauer K. 2007. Characterization of colony morphology variants isolated from Streptococcus pneumoniae biofilms. J. Bacteriol. 189: 2030–2038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Applegate DH, Bryers JD. 1991. Effects on carbon and oxygen limitations and calcium concentrations on biofilm removal processes Biotechnol. Bioeng. 37: 17–25 [DOI] [PubMed] [Google Scholar]
- 4. Barraud N, et al. 2006. Involvement of nitric oxide in biofilm dispersal of Pseudomonas aeruginosa. J. Bacteriol. 188: 7344–7353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Barraud N, et al. 2009. Nitric oxide signaling in Pseudomonas aeruginosa biofilms mediates phosphodiesterase activity, decreased cyclic di-GMP levels, and enhanced dispersal. J. Bacteriol. 191: 7333–7342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bibikov SI, Biran R, Rudd KE, Parkinson JS. 1997. A signal transducer for aerotaxis in Escherichia coli. J. Bacteriol. 179: 4075–4079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bibikov SI, Miller AC, Gosink KK, Parkinson JS. 2004. Methylation-independent aerotaxis mediated by the Escherichia coli Aer protein. J. Bacteriol. 186: 3730–3737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bobrov AG, Kirillina O, Perry PD. 2005. The phosphodiesterase activity of the HmsP EAL domain is required for negative regulation of biofilm formation in Yersinia pestis FEMS Microbiol. Lett. 247: 123–130 [DOI] [PubMed] [Google Scholar]
- 9. Campbell AJ, Watts KJ, Johnson MS, Taylor BL. 2010. Gain-of-function mutations cluster in distinct regions associated with the signalling pathway in the PAS domain of the aerotaxis receptor, Aer. Mol. Microbiol. 77: 575–586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chan MK. 2001. Recent advances in heme-protein sensors. Curr. Opin. Chem. Biol. 5: 216–222 [DOI] [PubMed] [Google Scholar]
- 11. Costerton JW, Stewart PS, Greenberg EP. 1999. Bacterial biofilms: a common cause of persistent infections. Science 284: 1318–1322 [DOI] [PubMed] [Google Scholar]
- 12. D'Argenio DA, Miller SI. 2004. Cyclic di-GMP as a bacterial second messenger. Microbiology 150: 2497–2502 [DOI] [PubMed] [Google Scholar]
- 13. Delgado-Nixon VM, Gonzalez G, Gilles-Gonzalez M-A. 2000. Dos, a heme-binding PAS protein from Escherichia coli, is a direct oxygen sensor. Biochemistry 39: 2685–2691 [DOI] [PubMed] [Google Scholar]
- 14. Edwards JC, Johnson MS, Taylor BL. 2006. Differentiation between electron transport sensing and proton motive force sensing by the Aer and Tsr receptors for aerotaxis. Mol. Microbiol. 62: 823–837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Emori TG, Gaynes RP. 1993. An overview of nosocomial infections, including the role of the microbiology laboratory. Clin. Microbiol. Rev. 6: 428–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Figurski DH, Helinski DR. 1979. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc. Natl. Acad. Sci. U. S. A. 76: 1648–1652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Giardina G, et al. 2008. NO sensing in Pseudomonas aeruginosa: structure of the transcriptional regulator DNR. J. Mol. Biol. 378: 1002–1015 [DOI] [PubMed] [Google Scholar]
- 18. Gjermansen M, Ragas P, Sternberg C, Molin S, Tolker-Nielsen T. 2005. Characterization of starvation-induced dispersion in Pseudomonas putida biofilms. Environ. Microbiol. 7: 894–904 [DOI] [PubMed] [Google Scholar]
- 19. Heydorn A, et al. 2000. Quantification of biofilm structures by the novel computer program COMSTAT. Microbiology 146: 2395–2407 [DOI] [PubMed] [Google Scholar]
- 20. Hinkkanen A, Decker K. 1983. Luminometric determination of FAD in subpicomole quantities. Anal. Biochem. 132: 202–208 [DOI] [PubMed] [Google Scholar]
- 21. Huang ZJ, Edery I, Rosbash M. 1993. PAS is a dimerization domain common to Drosophila Period and several transcription factors. Nature 364: 259–262 [DOI] [PubMed] [Google Scholar]
- 22. James GA, Korber DR, Caldwell DE, Costerton JW. 1995. Digital image analysis of growth and starvation responses of a surface-colonizing Acinetobacter sp. J. Bacteriol. 177: 907–915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kaneko Y, Thoendel M, Olakanmi O, Britigan BE, Singh PK. 2007. The transition metal gallium disrupts Pseudomonas aeruginosa iron metabolism and has antimicrobial and antibiofilm activity. J. Clin. Invest. 117: 877–888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kay SA. 1997. PAS, present, and future: clues to the origins of circadian clocks. Science 276: 753–754 [DOI] [PubMed] [Google Scholar]
- 25. Keppler D, Rudigier J, Decker K. 1970. Enzymic determination of uracil nucleotides in tissues. Analyt. Biochem. 38: 105–114 [DOI] [PubMed] [Google Scholar]
- 26. Kuchma SL, et al. 2007. BifA, a cyclic-Di-GMP phosphodiesterase, inversely regulates biofilm formation and swarming motility by Pseudomonas aeruginosa PA14. J. Bacteriol. 189: 8165–8178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kulasakara H, et al. 2006. Analysis of Pseudomonas aeruginosa diguanylate cyclases and phosphodiesterases reveals a role for bis-(3′-5′)-cyclic-GMP in virulence. Proc. Natl. Acad. Sci. U. S. A. 103: 2839–2844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lansky IB, et al. 2006. The cytoplasmic heme-binding protein (PhuS) from the heme uptake system of Pseudomonas aeruginosa is an intracellular heme-trafficking protein to the δ-regioselective heme oxygenase. J. Biol. Chem. 281: 13652–13662 [DOI] [PubMed] [Google Scholar]
- 29. Létoffé S, Redeker V, Wandersman C. 1998. Isolation and characterization of an extracellular haem-binding protein from Pseudomonas aeruginosa that shares function and sequence similarities with the Serratia marcescens HasA haemophore. Mol. Microbiol. 28: 1223–1234 [DOI] [PubMed] [Google Scholar]
- 30. Lindebro MC, Poellinger L, Whitelaw ML. 1995. Protein-protein interaction via PAS domains: role of the PAS domain in positive and negative regulation of the bHLH/PAS dioxin receptor-Arnt transcription factor complex. EMBO J. 14: 3528–3539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. McGuire J, Coumailleau P, Whitelaw ML, Gustafsson JA, Poellinger L. 1995. The basic helix-loop-helix/PAS factor Sim is associated with hsp90. J. Biol. Chem. 270: 31353–31357 [DOI] [PubMed] [Google Scholar]
- 32. Möglich A, Ayers RA, Moffat K. 2009. Structure and signaling mechanism of Per-ARNT-Sim domains. Structure 17: 1282–1294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Morgan R, Kohn S, Hwang S-H, Hassett DJ, Sauer K. 2006. BdlA, a chemotaxis regulator essential for biofilm dispersion in Pseudomonas aeruginosa. J. Bacteriol. 188: 7335–7343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Newman JR, Fuqua C. 1999. Broad-host-range expression vectors that carry the arabinose-inducible Escherichia coli araBAD promoter and the araC regulator. Gene 227: 197–203 [DOI] [PubMed] [Google Scholar]
- 35. Pellequer J-L, Wager-Smith KA, Kay SA, Getzoff ED. 1998. Photoactive yellow protein: a structural prototype for the three-dimensional fold of the PAS domain superfamily. Proc. Natl. Acad. Sci. U. S. A. 95: 5884–5890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Petrova OE, Sauer K. 2009. A novel signaling network essential for regulating Pseudomonas aeruginosa biofilm development. PLoS Pathog. 5: e1000668 doi:10.1371/journal.ppat.1000668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Repik A, et al. 2000. PAS domain residues involved in signal transduction by the Aer redox sensor of Escherichia coli. Mol. Microbiol. 36: 806–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Roy AB, Petrova OE, Sauer K. 2012. The phosphodiesterase DipA (PA5017) is essential for Pseudomonas aeruginosa biofilm dispersion. J. Bacteriol. 194: 2904–2915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sauer K, Camper AK, Ehrlich GD, Costerton JW, Davies DG. 2002. Pseudomonas aeruginosa displays multiple phenotypes during development as a biofilm. J. Bacteriol. 184: 1140–1154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sauer K, et al. 2004. Characterization of nutrient-induced dispersion in Pseudomonas aeruginosa PAO1 biofilm. J. Bacteriol. 186: 7312–7326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Schweizer HP. 1991. The agmR gene, an environmentally responsive gene, complements defective glpR, which encodes the putative activator for glycerol metabolism in Pseudomonas aeruginosa. J. Bacteriol. 173: 6798–6806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Smalley JW, Charalabous P, Birss AJ, Hart CA. 2001. Detection of heme-binding proteins in epidemic strains of Burkholderia cepacia. Clin. Diagn. Lab. Immunol. 8: 509–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Southey-Pillig CJ, Davies DG, Sauer K. 2005. Characterization of temporal protein production in Pseudomonas aeruginosa biofilms. J. Bacteriol. 187: 8114–8126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Suske WA, et al. 1997. Purification and characterization of 2-hydroxybiphenyl 3-monooxygenase, a novel NADH-dependent, FAD-containing aromatic hydroxylase from Pseudomonas azelaica HBP1. J. Biol. Chem. 272: 24257–24265 [DOI] [PubMed] [Google Scholar]
- 45. Taylor BL, Rebbapragada A, Johnson MS. 2001. The FAD-PAS domain as a sensor for behavioral responses in Escherichia coli. Antioxid. Redox Signal. 3: 867–879 [DOI] [PubMed] [Google Scholar]
- 46. Taylor BL, Zhulin IB. 1999. PAS domains: internal sensors of oxygen, redox potential, and light. Microbiol. Mol. Biol. Rev. 63: 479–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Thormann KM, Saville RM, Shukla S, Spormann AM. 2005. Induction of rapid detachment in Shewanella oneidensis MR-1 biofilms. J. Bacteriol. 187: 1014–1021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Watts KJ, Sommer K, Fry SL, Johnson MS, Taylor BL. 2006. Function of the N-terminal cap of the PAS domain in signaling by the aerotaxis receptor Aer. J. Bacteriol. 188: 2154–2162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Xie Z, Ulrich LE, Zhulin IB, Alexandre G. 2010. PAS domain containing chemoreceptor couples dynamic changes in metabolism with chemotaxis. Proc. Natl. Acad. Sci. U. S. A. 107: 2235–2240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Zhulin IB, Taylor BL, Dixon R. 1997. PAS domain S-boxes in archaea, bacteria and sensors for oxygen and redox. Trends Biochem. Sci. 22: 331–333 [DOI] [PubMed] [Google Scholar]