Abstract
Adrenocorticotropin (ACTH) signaling increases glucocorticoid production by promoting the interaction of transcription factors and coactivator proteins with the promoter of steroidogenic genes. The nuclear receptor steroidogenic factor 1 (SF-1) is essential for steroidogenic gene transcription. Sphingosine (SPH) is a ligand for SF-1. Moreover, suppression of expression of acid ceramidase (ASAH1), an enzyme that produces SPH, increases the transcription of multiple steroidogenic genes. Given that SF-1 is a nuclear protein, we sought to define the molecular mechanisms by which ASAH1 regulates SF-1 function. We show that ASAH1 is localized in the nuclei of H295R adrenocortical cells and that cyclic AMP (cAMP) signaling promotes nuclear sphingolipid metabolism in an ASAH1-dependent manner. ASAH1 suppresses SF-1 activity by directly interacting with the receptor. Chromatin immunoprecipitation (ChIP) assays revealed that ASAH1 is recruited to the promoter of various SF-1 target genes and that ASAH1 and SF-1 colocalize on the same promoter region of the CYP17A1 and steroidogenic acute regulatory protein (StAR) genes. Taken together, these results demonstrate that ASAH1 is a novel coregulatory protein that represses SF-1 function by directly binding to the receptor on SF-1 target gene promoters and identify a key role for nuclear lipid metabolism in regulating gene transcription.
INTRODUCTION
In the human adrenal cortex, adrenocorticotropin (ACTH) regulates cortisol biosynthesis by activating a cyclic AMP/protein kinase A (cAMP/PKA)-dependent signaling pathway that leads to rapid cholesterol import and transport, as well as the transcriptional activation of genes required for steroid hormone production (51, 62). The transcription of most steroidogenic genes is regulated by steroidogenic factor 1 (SF-1; NR5A1), which in response to ACTH signaling binds to target gene promoters and facilitates the recruitment of coactivator proteins and RNA polymerase II (6, 21, 30, 50, 62). SF-1 is a member of the nuclear receptor superfamily of transcription factors (34) whose structure can be divided into functional domains: an amino-terminal conserved DNA binding domain (DBD) consisting of two zinc-binding modules, an intervening hinge region that contains a ligand-independent activation domain (activation domain 1 [AF-1]), and a carboxy-terminal ligand-binding domain (LBD) that contains a conserved AF-2 hexamer domain (LLIEML) that is critical for receptor activation (43). The hinge region and LBD both participate in transcriptional repression and ligand-dependent activation. These domains serve as the interface for interactions between SF-1 and numerous coregulatory proteins, including steroid hormone coactivator 1 (SRC-1) (10, 29) and silencing mediator for retinoid and thyroid hormone receptors (SMRT) (21). Generally, coregulators bind to the AF-1 and/or AF-2 domains of nuclear receptors through LXXLL motifs (nuclear receptor [NR] boxes), where X is any amino acid and L is a leucine (22, 44). Additional LXXLL-related motifs where L is substituted for an isoleucine, phenylalanine, or methionine have also been reported (14, 81).
The ability of SF-1 to bind to target promoters is regulated by posttranslational modifications, including phosphorylation (13, 21, 63, 64), sumoylation (7, 31), and acetylation (6, 27, 30), as well as protein-protein interactions (5, 12, 37, 45, 61, 79, 82). More recently, ligand binding has also been implicated in the regulation of SF-1 activity (32, 36, 38, 75, 77). Crystallographic studies using bacterially expressed SF-1 have demonstrated that phospholipids are present in the ligand-binding pocket and that ligand binding is required for maximal activity of the receptor (32, 38, 58, 77). We previously identified phosphatidic acid (PA) and sphingosine (SPH) as endogenous ligands for SF-1 (75). SPH is an antagonist that binds to SF-1 under basal conditions and prevents receptor binding to the CYP17A1 promoter, thus decreasing cAMP-stimulated CYP17A1 mRNA expression and steroid hormone biosynthesis (75).
SPH is produced by the hydrolysis of ceramide (67, 76) in a reaction catalyzed by ceramidases (N-acylsphingosine amidohydrolase) (11, 26, 53). Five human ceramidases have been cloned and are categorized by their optimal pH for in vitro activity as acid (ASAH1) and neutral (ASAH2) as well as three isoforms of alkaline (ACER1 to ACER3) (26, 53, 78). ASAH1 is a glycoprotein processed from a 55-kDa precursor into a heterodimeric protein formed by α (14-kDa) and β (40-kDa) subunits via autoproteolytic cleavage (3, 66). In vivo studies demonstrated that ASAH1 requires sphingolipid activator proteins (SAP; saposin), mainly SAP-D, as cofactors for maximal activity (39). ASAH1 has been reported to localize to lysosomes (17) and to be secreted extracellularly from murine endothelial cells, macrophages, and human fibroblasts (56). This ceramidase is required for development because targeted disruption of the gene in mice leads to an early, embryonic lethal phenotype (16). In addition, a genetic deficiency in ASAH1 resulting in reduced enzymatic activity causes Farber's disease, a lysosomal sphingolipid storage disorder (49). In adrenocortical cells, we have recently characterized a novel role for ASAH1 as a negative regulator of steroidogenic gene transcription and steroidogenic capacity (41). ASAH1 knockdown induces the expression of multiple SF-1 target genes, including those for steroidogenic acute regulatory protein (StAR), melanocortin 2 receptor (MC2R), and CYP17A1, and results in increased cortisol and dehydroepiandrosterone (DHEA) secretion (41), supporting a role for ASAH1 in regulating the transactivation potential of SF-1.
Based on the role of SPH as an SF-1 antagonist (75) and the fact that SPH-mediated repression of CYP17A1 transcription is dependent on ASAH1 expression (75), as well as the role of ASAH1 in global steroidogenic gene expression and steroid hormone secretion (41), we sought to define the mechanism by which ASAH1 regulates SF-1 function in H295R adrenocortical cells. We show that ASAH1 and SAP-D are localized in the nuclei of H295R cells and that ACTH/cAMP signaling acutely increases ASAH1 activity concomitant with nuclear ceramide metabolism. Significantly, ASAH1 represses SF-1 activity by directly binding to the receptor on DNA through an LXXLL motif in the α subunit of the enzyme. The ability of ASAH1 to repress SF-1 function requires a direct interaction with the receptor as well as a catalytically competent enzyme. Finally, cAMP modulates the recruitment of ASAH1 to the promoters of various SF-1 target genes, suggesting a global coregulatory role for ASAH1 in adrenocortical cells.
MATERIALS AND METHODS
Reagents.
Dibutyryl cAMP (Bt2cAMP) was obtained from Sigma (St. Louis, MO). N-[12-(7-Nitro-2-1,3-benzoxadiazol-4-yl-amino) dodecanoyl]-d-erythro-sphingosine (NBD-12-ceramide) was purchased from Avanti Polar Lipids, Inc. (Alabaster, AL), and 12-[N-(7-nitro-2-1,3-benzoxadiazol-4-yl-amino) amino]-dodecanoic acid from Invitrogen (Carlsbad, CA).
Cell culture.
H295R human adrenocortical cells (55, 68) were obtained from William E. Rainey (Georgia Health Sciences University, Augusta, GA) and cultured in Dulbecco's modified Eagle's medium (DMEM)-F12 medium (Mediatech Inc., Manassas, VA) supplemented with 10% Nu-Serum I (BD Biosciences, Palo Alto, CA), 1% ITS Plus (BD Biosciences), antibiotics, and antimycotics. CV-1 monkey kidney cells were obtained from the American Type Culture Collection (ATCC) (Manassas, VA) and cultured in Eagle's minimum essential medium (MEM) (Mediatech, Inc.) supplemented with 10% fetal bovine serum (Mediatech, Inc.), antibiotics, and antimycotics. H295R ASAH1 knockdown (ASAH1KD) cells were generated as described previously (44) and cultured in DMEM-F12 medium (Mediatech, Inc.) supplemented with 10% Nu-Serum I (BD Biosciences), 1% ITS Plus (BD Biosciences), antibiotics, antimycotics, 1 μg/ml blasticidin (Sigma), 50 μg/ml zeocin (Invitrogen), and 5 μg/ml tetracycline (Invitrogen) to induce ASAH1 short hairpin RNA (shRNA) production. Y-1 mouse adrenocortical cells were obtained from the ATCC and cultured in DMEM supplemented with 10% fetal bovine serum (Mediatech, Inc.), antibiotics, and antimycotics. MCF-7 human breast cancer cells were obtained from the ATCC and cultured in phenol red-free MEM (Mediatech, Inc.) supplemented with 10% fetal bovine serum (Mediatech, Inc.), 11 mM sodium pyruvate (Sigma), 0.01 mg/ml bovine insulin (Sigma), antibiotics, and antimycotics.
Nuclear extract purification and WB.
H295R cells were subcultured into 100-mm dishes, and cytoplasmic, nuclear envelope, and nucleoplasmic fractions were purified using the Nuclei Pure isolation kit (Sigma) by following the manufacturer's instructions. Aliquots (30 μg protein) of cytoplasmic, nuclear envelope, and nucleoplasmic fractions were separated by SDS-PAGE and transferred to polyvinylidene difluoride (PVDF) membranes (Millipore, Temecula, CA). Blots were probed with antibodies against SF-1 (1:7,500; Millipore), ASAH1 (1:2,000; HPA005468; Sigma), ASAH2 (1:1,000; PRS4743; Sigma), ACER3 (1:3,000; sc-101848; Santa Cruz, Santa Cruz, CA), SAP-D (1:5,000; sc-27024-R; Santa Cruz), lysosome-associated membrane protein 1 (LAMP1; 1:2,000; sc-18822; Santa Cruz), P450c17α (1:1,000; sc-66849; Santa Cruz), or StAR (1:2,000; sc-25806; Santa Cruz). Expression was detected using an ECF Western blotting (WB) kit (Amersham Bioscience, Piscataway, NJ) and visualized by scanning blots on a VersaDoc 4000 imager (Bio-Rad, Hercules, CA).
Immunostaining and confocal microscopy.
Immunostaining and confocal microscopy were performed as described previously (36). Briefly, H295R cells were plated on glass coverslips, fixed, permeabilized, blocked with 1% bovine serum albumin (BSA) for 1 h, and then incubated with anti-ASAH1 or anti-SAP-D antibodies (diluted 1:200 in 1% BSA) overnight, followed by incubation with secondary goat anti-rabbit rhodamine-conjugated IgG (Thermo Scientific) for 1 h. Coverslips were stained with 4,6′-diamino-2-phenylindole dihydrochloride (DAPI; Invitrogen) for 2 min and then mounted onto slides using Fluoromount G (Southern Biotech, Birmingham, AL). Confocal images were captured using a laser-scanning microscope (LSM 510; Zeiss, Thornwood, NY) equipped with argon and helium-neon lasers, with excitation wavelengths of 436 and 542 nm for DAPI and rhodamine, respectively.
ASAH1 activity assay.
H295R wild-type (WT) or ASAH1KD (pretreated for 96 h with 5 μg/ml tetracycline) cells were subcultured into 100-mm dishes and treated for 30 min to 6 h with 0.4 mM Bt2cAMP. In vitro ASAH1 activity assay was performed as previously described (40). Thin-layer chromatography (TLC) plates were visualized by fluorescence scanning on a VersaDoc 4000 imager (Bio-Rad). NBD-dodecanoic acid formation was quantified by densitometric analysis and normalized to the protein content of each sample.
Analysis of sphingolipid species.
Sphingolipid concentrations in the nuclei of H295R WT and ASAH1KD (pretreated for 96 h with 5 μg/ml tetracycline) cells were analyzed by liquid chromatography, electrospray ionization, and tandem mass spectrometry (LC-ESI-MS-MS) as described previously (69, 70).
Site-directed mutagenesis.
Mutagenesis was carried out using a Stratagene QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA) and confirmed by sequencing. To mutate putative nuclear receptor (NR) binding motifs in ASAH1, the first and fourth leucine (L) residues within each motif were mutated to alanine (A) using the following primers: ASAH1-NR1 (5′-TAC CCA AGT GCC AGC GCG GCT TTC ACC GAG-3′) and ASAH1-NR5 (5′-GTG GGC ATG GCT ACA GGA GCT AAA CCA GGA-3′). SF-1 pocket mutants (G341K, Y436A, and K440E) have been previously described (36, 75).
Transient transfection and reporter gene analysis.
H295R cells were transfected with 100 ng pGL3-CYP17A1-2x57 reporter plasmid (61), 25 ng wild-type (WT) or mutant (G341W or Y436A) pCMV6myc/FLAG-hSF1 (RC207577; Origene, Rockville, MD), 50 ng WT (pCMV6myc/FLAG-ASAH1) or mutant (pCMV6myc/FLAG-ASAH1-NR1 or pCMV6myc/FLAG-ASAH1-NR5) plasmid, 25 ng pBKCMV-SRC-1 (B. O'Malley, Baylor College of Medicine, Houston, TX), and/or 25 ng pCMX-SMRT (Y. Nakatani, Harvard Medical School, Boston, MA) using GeneJuice (EMD Biosciences). Cells were cotransfected with 1.5 ng Renilla luciferase plasmid under the control of the thymidine kinase (TK) promoter (pRL-TK; Promega, Madison, WI). Twenty-four hours after transfection, cells were treated with 0.4 mM Bt2cAMP for 16 h. In some experiments, cells were pretreated for 1 h with 0.5 μM or 5 μM D-NMAPPD {N-[(1R,2R)-2-hydroxy-1-(hydroxymethyl)-2-(4-nitrophenyl)-ethyl]-tetradecanamide} before treatment with Bt2cAMP. Firefly and Renilla luciferase activities were determined using a dual-luciferase assay (Promega), and CYP17A1 reporter gene activity (pGL3; firefly) was normalized to Renilla activity.
coIP.
For endogenous protein interaction assays, H295R cells were treated with 0.4 mM Bt2cAMP for 30 min. Coimmunoprecipitation (coIP) assays were also performed with tagged proteins by transfecting CV-1 cells with 5 μg pCMV6-GFP-hSF-1 (RG207577; Origene) and pCMV6myc/FLAG-ASAH1 WT or pCMV6myc/FLAG-ASAH1-NR1 or pCMV6myc/FLAG-ASAH1-NR5 mutant expression plasmid for 72 h. Cells were washed twice with phosphate-buffered saline (PBS) and harvested into radioimmunoprecipitation assay (RIPA) buffer (1× PBS, 1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% SDS) containing protease inhibitors (EMD Chemicals). Lysates were then sonicated 6 times for 5 s, followed by centrifugation at 12,000 rpm for 15 min at 4°C. Samples were precleared with 20 μl BSA (20 mg/ml) and immunoprecipitated overnight at 4°C on a tube rotator using protein A/G Plus-agarose (Santa Cruz) and anti-SF-1 (Millipore, for H295R lysates) or anti-FLAG (for CV-1 lysates; Sigma) antibodies. The immobilized protein complexes were washed twice in RIPA buffer and twice in PBS and then separated by SDS-PAGE. Western blots were probed with anti-ASAH1, anti-SAP-D, anti-SF-1, or anti-green fluorescent protein (anti-GFP; Invitrogen) antibodies, and expression was detected using an ECF Western blotting kit (GE Biosciences) and visualized using a VersaDoc 4000 imager (Bio-Rad).
In vitro binding assays.
His-tagged SF-1 was expressed and purified from Escherichia coli as previously described (74). ASAH1 was in vitro translated from the pCMV6-XL5-ASAH1 plasmid (RC212434; Origene) using the TNT Quick Coupled transcription/translation system kit (Promega). ASAH1 expression was confirmed by subjecting samples to SDS-PAGE and Coomassie staining and exposing the gel to a radioactive-imager screen. ASAH1 expression was visualized by radioactive-imager scanning (FLA 7000; Fuji, Japan). Twenty microliters of in vitro-translated ASAH1 was incubated with 50 μl His-tagged immobilized SF-1, 10 μl BSA (10 mg/ml), and 1 ml buffer (4 M NaCl, 160 nM Tris-HCl, 40 mM imidazole [pH 7.9]) in a tube rotator for 24 h at 4°C. Complexes were washed 3 times and subjected to SDS-PAGE and Coomassie staining. The gel was dried, exposed to a radioactive-imager screen, and visualized by radioactive-imager scanning (Fuji, Japan).
Transient transfection and real-time RT-PCR.
H295R ASAH1KD cells were transfected with 5 μg of pCMV6FLAG/myc-ASAH1 wild type or pCMV6FLAG/myc-ASAH1 NR1 for 72 h using Lipofectamine (Invitrogen). Cells were concurrently treated with 5 μg/ml tetracycline to induce ASAH1 knockdown. Untransfected groups that were untreated or treated with 5 μg/ml tetracycline were also included as controls. Total RNA was extracted using Isol-RNA lysis reagent (5 Prime, Inc., Gaithersburg, MD) and amplified using a One-Step SYBR green reverse transcription-PCR (RT-PCR) kit (Quanta Biosciences, Gaithersburg, MD) and primers for DAX-1 (forward, 5′-CCA AAT GCT GGA GTC TGA ACA TC-3′, and reverse, 5′-CCA AAT GCT GGA GTC TGA ACA TA-3′) and MC2R (forward, 5′-GTG GTG CTT ACG GTC ATC TGG A-3′, and reverse, 5′-AGG CAC AGG ATG AAG ACC AGC A-3′) genes. Gene expression was normalized to β-actin mRNA content and calculated using the delta-delta cycle threshold (ΔΔCT) method.
Mammalian 2-hybrid assay.
ASAH1 and SF-1 genes (full-length and truncation mutants) were cloned into the SalI and EcoRV sites of pBIND and/or pACT vectors (Promega). H295R cells were subcultured into 24-well plates and transfected with the pG5 firefly luciferase reporter in combination with pBIND and pACT vectors expressing fusions of the Gal4 DNA binding domain and VP16 activation domain, respectively, with SF-1 or ASAH1. In some experiments, cells were also cotransfected with 50 ng SRC-1 or SMRT expression plasmid. The ratio of pG5 to pBIND to pACT in transient transfections was 50 ng to 50 ng to 50 ng. Cells were treated 24 h later with 0.4 mM Bt2cAMP for 16 h before being harvested and assayed for dual-luciferase activity.
ChIP.
A chromatin immunoprecipitation (ChIP) assay was performed as previously described (12). Briefly, H295R cells were subcultured into 150-mm dishes and treated with 0.4 mM Bt2cAMP for 1 h. After cross-linking with 1% formaldehyde, cells were harvested into RIPA buffer and lysates sonicated to obtain optimal DNA fragment lengths of 100 to 1,000 bp (5 times for 10 s), followed by centrifugation at 12,000 rpm for 15 min at 4°C. The purified chromatin was precleared with 1 μg IgG and immunoprecipitated overnight at 4°C on a tube rotator using 5 μg primary antibody [anti-trimethyl (Lys20) histone H4 (Millipore), anti-RNA polymerase II CTD (Millipore), anti-SF-1 (07-618; Millipore), anti-ASAH1 (HPA005468; Sigma), anti-ASAH2 (PRS4743; Sigma), anti-ACER3 (sc-18822; Santa Cruz), anti-SRC-1 (Millipore), and anti-HDAC1 (Millipore)] and 30 μl protein A/G Plus agarose (Santa Cruz). Real-time PCR was carried out using 20% output, 5% input (diluted 1:4), ABsolute qPCR SYBR green fluorescein mix (Thermo Scientific), and primer sets for the following: CYP17A1 (forward, 5′-GGC TGG GCT CCA GGA GAA TCT TTC TTC CAC-3′, and reverse, 5′-CGG CAG GCA AGA TAG ACA GCA GTG GAG TAG-3′), StAR (forward, 5′-GCA GTG TGA GGC AAT CGC TCT-3′, and reverse, 5′-TGT TTC CTG GCA AAT GTG GCA-3′), MC2R (forward, 5′-TTG CCC AGA AAG TTC CTG CTT-3′, and reverse, 5′-TTC TCC TGC TTG TGG TTA AGG-3′), DAX-1 (forward, 5′-TCC TGC TTT TAA AGA GCA CCC GCC CCT-3′, and reverse, 5′-CGG CGC CCG TAG CCC AGT TCT-3′), and NR4A2 (forward, 5′-GAT CAG CTT GGA CTC CCT TGA AAG-3′, and reverse, 5′-CAT TCC AGA AAT TGG CAT CTT TTG-3′). For negative controls, primers for β-actin (forward, 5′-TGC ACT GTG CGG CGA AGC-3′, and reverse, 5′-TCG AGC CAT AAA AGG CAA-3′) or CYP17A1 noncoding region (forward, 5′-GAC AAG GGC ACA GAA GTT ATC ATC-3′, and reverse, 5′-CAG GGA GGG CAG CTG CCC ATC ATC-3′) were used. PCRs were carried out as follows: (i) 1 cycle of 95°C for 15 min; (ii) 60 cycles of 95°C for 15 s, 55°C for 30 s, and 72°C for 30 s; (iii) 1 cycle of 95°C for 30 s and 60°C for 30 s; (iv) 80 cycles of 60°C for 10 s; and (v) cooling to 4°C. Output ΔCT values were normalized to input values. Some PCRs were resolved by 2% agarose gel electrophoresis.
reChIP.
For sequential ChIP (reChIP), after the first IP using anti-ASAH1 or anti-SF-1 antibodies (as described for ChIP), immobilized protein/DNA complexes were eluted from agarose beads using 50 μl Tris-EDTA (TE) buffer containing 10 mM dithiothreitol (DTT) (37°C for 30 min). Chromatin was diluted 20-fold in RIPA buffer and submitted to another round of immunoprecipitation using 5 μg primary antibody (anti-SF-1 [07-618; Millipore] or anti-ASAH1 [HPA005468; Sigma]) and 30 μl protein A/G Plus agarose (Santa Cruz) overnight at 4°C on a tube rotator. The immobilized protein/DNA complexes were subjected to a series of 5-min washes as described above. The cross-links were reversed and protein was digested using proteinase K (100 μg/ml). DNA was purified by phenol-chloroform extraction and ethanol precipitation. Real-time PCR was carried out using 20% output, 5% input (diluted 1:4), ABsolute qPCR SYBR green fluorescein mix (Thermo Scientific), and primer sets for the following: CYP17A1 (forward, 5′-GGC TGG GCT CCA GGA GAA TCT TTC TTC CAC-3′, and reverse, 5′-CGG CAG GCA AGA TAG ACA GCA GTG GAG TAG-3′) and StAR (forward, 5′-GCA GTG TGA GGC AAT CGC TCT-3′, and reverse, 5′-TGT TTC CTG GCA AAT GTG GCA-3′), which amplify the proximal (∼300 bp) promoter region of each gene. For negative controls, primers for β-actin (forward, 5′-TGC ACT GTG CGG CGA AGC-3′, and reverse, 5′-TCG AGC CAT AAA AGG CAA-3′) or CYP17A1 noncoding region (forward, 5′-GAC AAG GGC ACA GAA GTT ATC ATC-3′, and reverse, 5′-CAG GGA GGG CAG CTG CCC ATC ATC-3′) were used. PCRs were carried out as follows: (i) 1 cycle of 95°C for 15 min; (ii) 40 cycles of 95°C for 15 s, 52°C for 30 s, and 72°C for 30 s; (iii) 1 cycle of 95°C for 30 s and 60°C for 30 s; (iv) 80 cycles of 60°C for 10 s; and (v) cooling to 4°C. Output ΔCT values were normalized to input values. Some PCRs were resolved by 2% agarose gel electrophoresis.
Statistical analysis.
One-way analysis of variance and Tukey's multiple comparison tests were performed using GraphPad InStat software (GraphPad Software Inc., San Diego, CA). Significant differences were defined as a P value of <0.05 and are indicated in the figures by asterisks, carets, or pound signs.
RESULTS
ASAH1 and SAP-D are expressed in the nuclei of H295R cells.
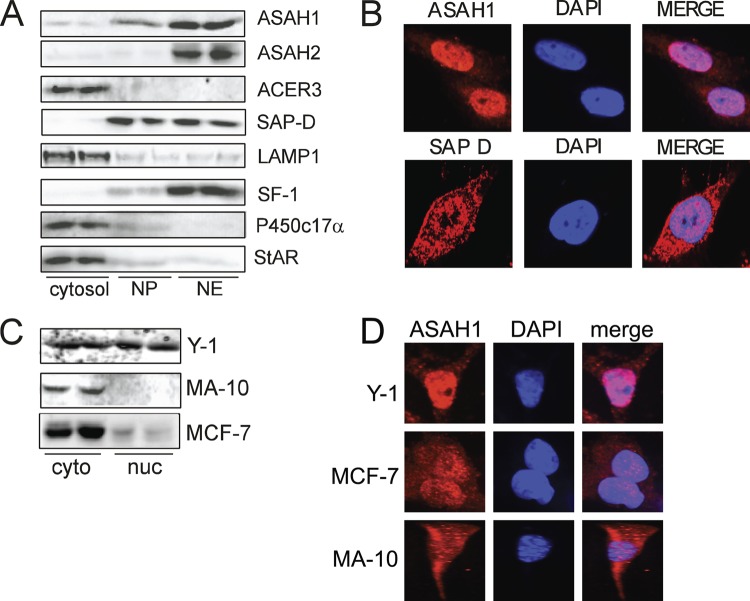
We have previously shown that SPH is an antagonist for SF-1 and represses the transcription of CYP17A1 (75). Moreover, ASAH1 suppression confers increased steroidogenic gene expression and steroid hormone secretion (41). However, the mechanism by which SPH production is controlled and the specific parameters governing ligand-dependent SF-1 activity are unknown. Because ceramidase activity is the only source of SPH (67, 76) and SF-1 is a nuclear protein, we used confocal microscopy and Western blotting to determine the subcellular localization of ceramidases in H295R adrenocortical cells. As shown in Fig. 1A, ASAH1 and its coactivator protein SAP-D are expressed in the nuclear envelope and nucleoplasmic fractions. Interestingly, ASAH2 is also expressed in the nuclei of these cells, whereas ACER3 is cytoplasmic. Because ASAH1 has been shown to have an optimal in vitro activity at acidic pH, it was characterized as a lysosomal protein (17). Therefore, to validate the purity of the nuclear extracts, we assessed the expression of LAMP1 (lysosomal), P450c17α (microsomal), and StAR (mitochondrial), all of which were found in their predicted subcellular fractions (Fig. 1A). As expected, SF-1 is primarily localized in the nuclear envelope fraction, with slight expression in the nucleoplasmic fraction. Confocal analysis confirmed that ASAH1 and SAP-D are expressed in the nuclei of H295R cells (Fig. 1B). Interestingly, probing the subcellular localization of ASAH1 in other cell lines revealed that ASAH1 is localized in both compartments in Y-1 mouse adrenocortical cells, with predominantly and cytoplasmic localization in MA-10 Leydig cells and MCF-7 breast cancer cells (Fig. 1C and D). However, ASAH1 was detected at low levels in the nuclear compartment of MCF-7 cells (Fig. 1C).
Fig 1.
ASAH1 and SAP-D are expressed in the nuclei of H295R cells. (A) Cytosolic, nuclear envelope (NE), and nucleoplasmic (NP) fractions were isolated from H295R cells as described in Materials and Methods. Lysates (30 μg protein) were separated by SDS-PAGE, followed by Western blotting using ASAH1, ASAH2, ACER3, SF-1, SAP-D, LAMP1, P450c17α, or StAR antibodies. (B) H295R cells were plated onto glass coverslips, fixed, permeabilized, and incubated with anti-ASAH1 or anti-SAP-D antibodies overnight, followed by a 2-min incubation with the nuclear marker DAPI. Coverslips were washed and incubated with antirhodamine and mounted, and immunofluorescence was detected by confocal microscopy. (C) Nuclear and cytoplasmic extracts from Y-1, MA-10, and MCF-7 cells were separated by SDS-PAGE, followed by Western blotting using an anti-ASAH1 antibody. (D) Y-1, MA-10, or MCF-7 cells were plated onto glass coverslips, fixed, permeabilized, and incubated overnight with anti-ASAH1, followed by a 2-min incubation with the nuclear marker DAPI. Coverslips were washed and incubated with antirhodamine and mounted, and immunofluorescence was detected by confocal microscopy.
ACTH/cAMP signaling promotes nuclear sphingolipid metabolism in an ASAH1-dependent manner.
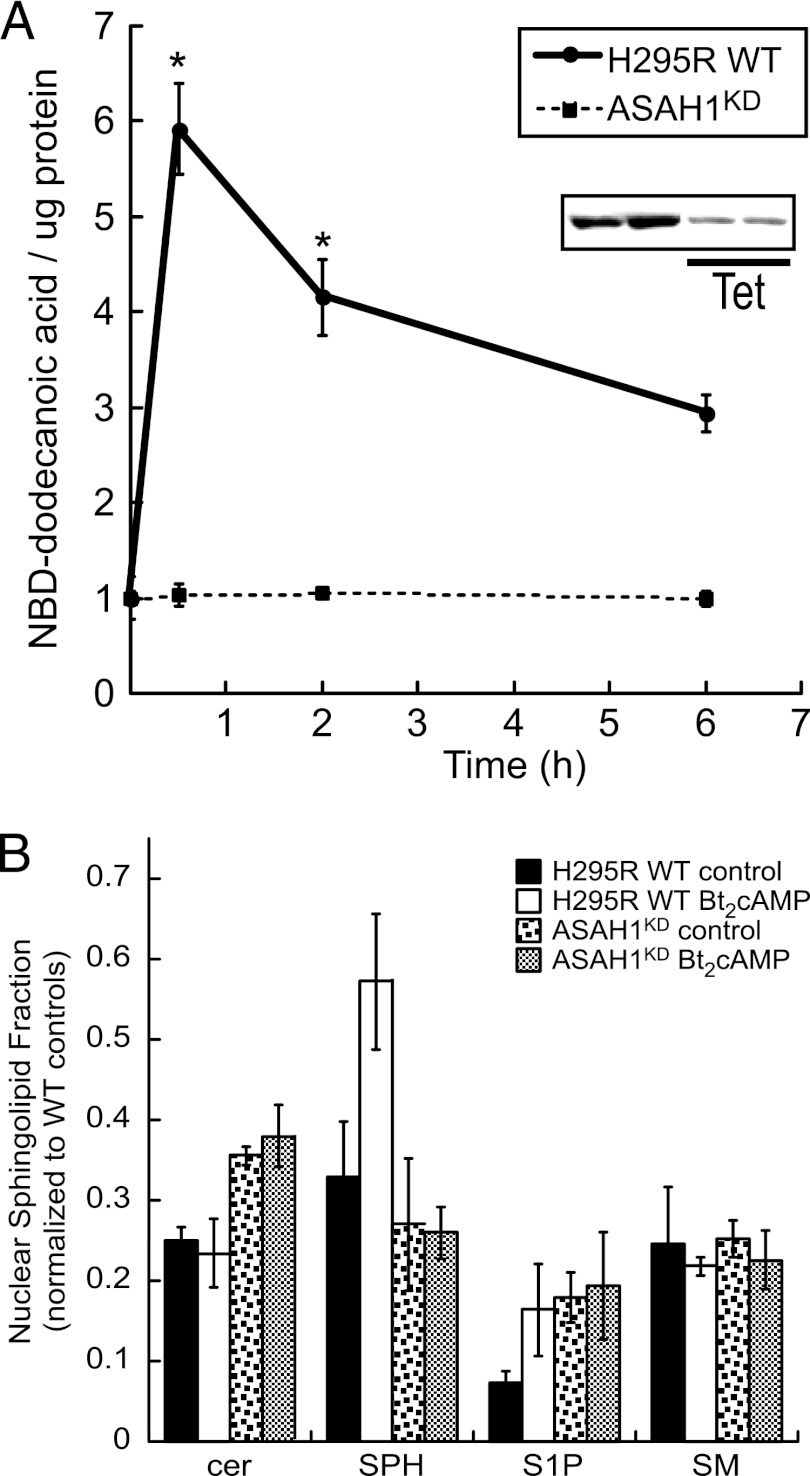
Because cortisol biosynthesis is primarily regulated by the ACTH/cAMP pathway and ACTH signaling induces ceramide turnover in H295R cells (47), we next investigated the effect of ACTH/cAMP signaling on nuclear ceramidase activity. H295R wild-type (WT) cells were treated with 0.4 mM Bt2cAMP and isolated nuclei were purified for in vitro activity assays. As shown in Fig. 2A, 30 min of Bt2cAMP stimulation increased ceramidase activity 6-fold. Given that ASAH2 is also expressed in the nuclei of H295R cells (Fig. 1A), we sought to define the relative contribution of ASAH1 to nuclear ceramidase activity, by performing in vitro activity assays using nuclei that were purified from an H295R stable cell line that expresses a tetracycline-regulated short hairpin RNA (shRNA) against ASAH1 (ASAH1KD cells) to selectively knock down ASAH1 expression (41). As shown in Fig. 2A, no increase in ceramidase activity was observed in response to Bt2cAMP in ASAH1KD cells. To determine the relative fraction of cellular versus nuclear ceramide, we used mass spectrometry to assess the sphingolipid content of whole cells and nuclei that were isolated from wild-type or ASAH1KD cells that were treated with Bt2cAMP for 30 min. We have previously found that Bt2cAMP rapidly stimulates the catabolism of complex sphingolipids, thereby resulting in decreases in the cellular amounts of ceramide and sphingomyelin and increased SPH and sphingosine-1-phosphate (S1P) (47, 48). Consistent with our previous findings, 30 min of stimulation of wild-type cells with Bt2cAMP increased the nuclear fraction of SPH and S1P 1.6- and 2.2-fold, respectively (Fig. 2B). Moreover, suppressing ASAH1 expression increased the nuclear fraction of ceramide from 25% to 35.5%, suggesting that silencing ASAH1 reduces the nuclear conversion of ceramide to SPH. The increase in the nuclear fraction of ceramide was mirrored by a reduction of nuclear SPH from 33% in wild-type control cells to 27% in untreated ASAH1KD cells. Finally, the increase in the nuclear fraction of SPH observed in response to Bt2cAMP stimulation of wild-type cells was attenuated in ASAH1KD cells (Fig. 2B).
Fig 2.
ACTH/cAMP signaling promotes nuclear sphingolipid metabolism in an ASAH1-dependent manner. (A) H295R wild-type (WT) and ASAH1 knockdown (ASAH1KD) (pretreated for 96 h with 5 μg/ml tetracycline [Tet]) cells were treated for 30 min or 2 or 6 h with 0.4 mM Bt2cAMP, and nuclear lysates were isolated as described in Materials and Methods. H295R WT or ASAH1KD nuclear extracts (150 μl) were incubated with 2 μl 1 mM NBD-12-ceramide in acetate buffer (0.5 M, pH 4.5) at 37°C for 2 h. Reactions were stopped, and lipids were extracted and spotted on TLC plates. Plates were developed and visualized by fluorescent scanning. NBD-dodecanoic acid formation was quantified and normalized to the protein content of each sample. Data represent means ± standard errors of the means (SEMs) from 3 separate experiments, each performed in duplicate. (Inset) Representative Western blot of controls and Tet-treated H295RKD cells demonstrating decreased ASAH1 protein levels. *, statistically different from value for untreated control group (P < 0.05). (B) H295R WT or ASAH1KD (pretreated for 96 h with 5 μg/ml Tet) cells were subcultured into 100-mm dishes and treated with 0.4 mM Bt2cAMP for 30 min, and sphingosine (SPH), ceramide (cer), sphingosine-1-phosphate (S1P), and sphingomyelin (SM) concentrations in purified whole cells or nuclei were quantified by LC-ESI-MS-MS using appropriate standards and normalized to total cellular protein concentrations. Data are graphed as the means ± standard deviations of two experiments, each performed in duplicate, and are plotted as the nuclear fraction of each (nuclear/cellular) sphingolipid species with normalization to the amounts for wild-type untreated whole-cell sphingolipid.
ASAH1 overexpression attenuates SF-1-stimulated CYP17A1 reporter gene activity.
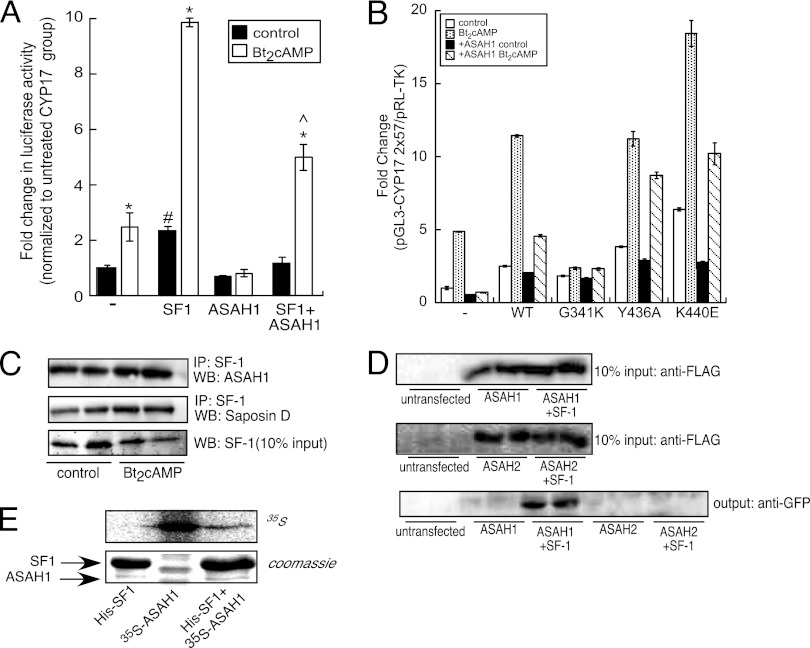
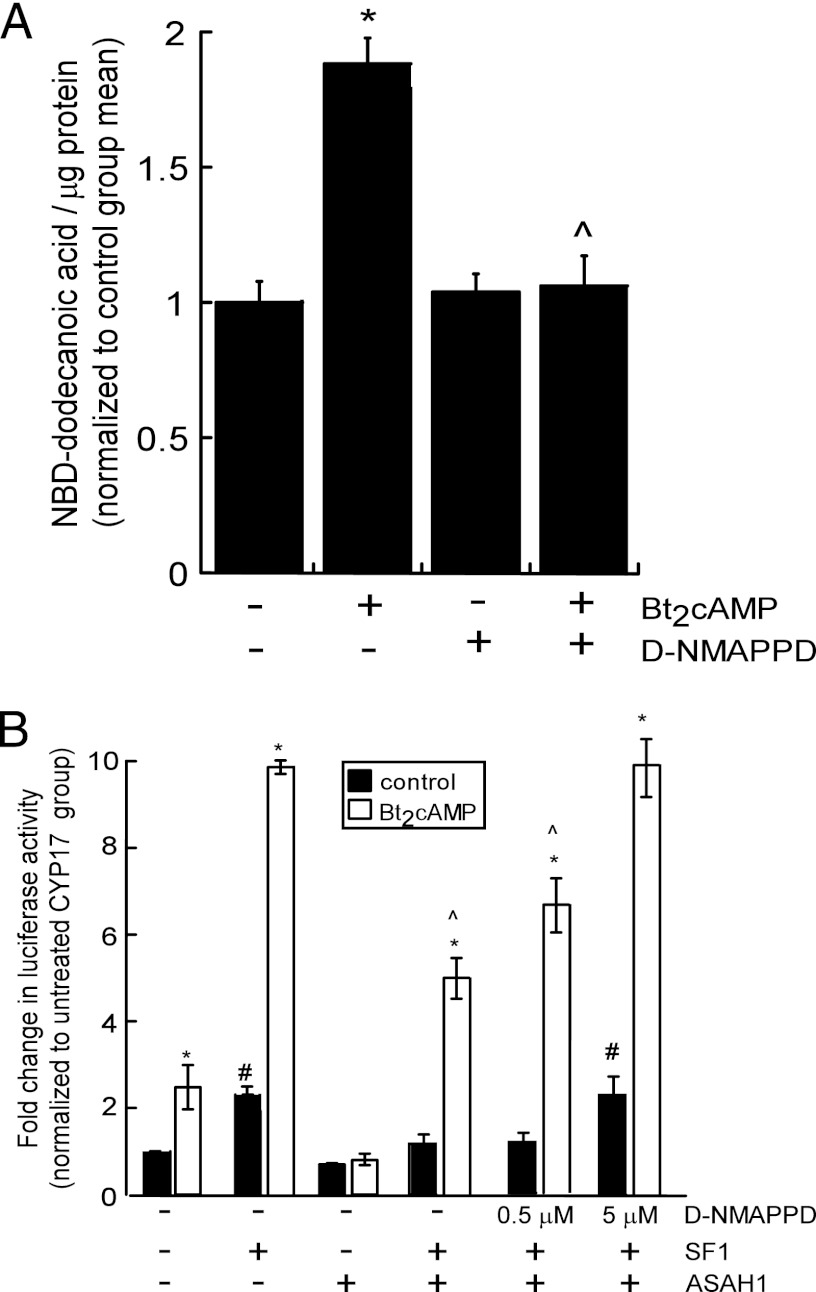
Next, we determined the role of ASAH1 in modulating CYP17A1 gene expression by carrying out reporter gene assays. Consistent with previous results (61), SF-1 increased basal and Bt2cAMP-stimulated CYP17A1 reporter gene activity 2.4- and 9.8-fold, respectively (Fig. 3A). ASAH1 not only abrogated Bt2cAMP-dependent CYP17A1 reporter gene activity but also inhibited SF-1-stimulated CYP17A1 reporter gene activity in both untreated and Bt2cAMP-stimulated cells (Fig. 3A).
Fig 3.
ASAH1 binds to SF-1. (A) H295R cells were transfected with 100 ng pGL3-CYP17A1-2x57, 25 ng pCMV6myc/FLAG-hSF1, 50 ng pCMV6myc/FLAG-ASAH1, and 1.5 ng pRL-TK. Twenty-four hours after transfection, cells were treated with 0.4 mM Bt2cAMP for 16 h and harvested for dual-luciferase assay. Data are expressed as fold increase in pGL3-CYP17A1 reporter gene activity and represent the means ± SEMs of 3 separate experiments, each performed in triplicate. Asterisks, carets, and pound signs indicate statistically significant differences (P < 0.05) from values for the untreated control group within each transfection group, Bt2cAMP-treated SF-1-transfected group, and untreated CYP17A1 group, respectively. (B) Cells were transfected with pGL3-CYP17A1-2x27 (100 ng), wild-type or mutant pCMV6GFP-SF1 (25 ng), or pCMV6mycFLAG-ASAH1 (50 ng) and pRL-TK (1.5 ng) using Gene Juice. Cells were treated with 0.4 mM Bt2cAMP for 16 h and harvested for dual-luciferase assay. Data are expressed as fold increase in pGL3-CYP17A1 firefly luciferase activity normalized to pRL-TK Renilla luciferase activity and represent the means ± SEMs of 3 separate experiments, each performed in triplicate. Asterisks indicate statistically significant differences (P < 0.05) from the values for the untreated control group. (C) H295R cells were treated with 0.4 mM Bt2cAMP for 30 min, and lysates were subjected to IP using an anti-SF-1 antibody and protein A/G agarose. Immobilized proteins were washed, separated by SDS-PAGE, and analyzed by Western blotting. Blots were hybridized to anti-ASAH1 or anti-SAP-D antibodies (top two blots). Ten percent inputs were subjected to SDS-PAGE and Western blotting using an antibody against SF-1 (bottom blot). (D) H295R cells were transfected with 5 μg pCMV6-GFP-hSF-1 and pCMV6myc/FLAG-ASAH1 or pCMV6myc/FLAG-ASAH2 expression plasmids for 72 h. Lysates were subjected to IP with an anti-FLAG antibody and protein A/G agarose. Immobilized proteins were washed and separated by SDS-PAGE, followed by Western blotting using an anti-GFP antibody (bottom blot). Ten percent inputs were subjected to SDS-PAGE and Western blotting using an antibody against FLAG (top two blots). (E) Hig-tagged SF-1 was bacterially expressed from E. coli and ASAH1 was in vitro transcribed/translated from the pCMV6myc/FLAG-ASAH1 vector. Twenty micrograms of in vitro-translated 35S-ASAH1 was incubated with 50 μl immobilized His-tagged SF-1 in a tube rotator for 24 h at 4°C. SF-1/ASAH1 complexes were then subjected to SDS-PAGE and Coomassie staining. The gel was dried, exposed to a radioactive-imager screen, and visualized by radioactive-imager scanning.
ASAH1 interacts directly with SF-1.
Based on the nuclear localization of ASAH1 (Fig. 1), the effect of ASAH1 on SF-1-stimulated CYP17A1 reporter gene activity (Fig. 3A), and the role of SPH as an antagonist for SF-1 (75), we hypothesized that ASAH1 modulates SF-1 activity by directly binding to the receptor. SF-1 has been shown to bind to phospholipids (4, 32, 36, 38, 77) and sphingolipids (75). Moreover, structural studies have found that G341, Y436, and K440 all coordinate interaction with the phospholipid head group (32, 38), and we have previously shown that mutation of G341 increases the amount of SPH bound to the receptor (75). To determine the effect of ASAH1 on the transactivation potential of SF-1 pocket mutants, we performed reporter gene assays. As shown in Fig. 3B, transfection of ASAH1 attenuated cAMP-stimulated reporter gene activity and reduced basal luciferase activity by 33%. Overexpression of wild-type SF-1 increased CYP17A1 reporter gene activity 2.5- and 11.4-fold in control and Bt2cAMP-treated cells, respectively, while ASAH1 reduced the stimulatory effect of the wild-type receptor by 50% in cells stimulated with Bt2cAMP (Fig. 3B). Consistent with our previous findings (36, 75), the G341K mutant exhibited a markedly reduced increase in reporter gene activity in response to Bt2cAMP in comparison to the wild-type receptor. Moreover, expression of ASAH1 had no effect on basal or Bt2cAMP-stimulated reporter gene activity of the G341K mutant. In contrast, ASAH1 inhibited the ability of the Y436A and K440E mutants to stimulate reporter gene activity by 22 and 45% in Bt2cAMP-treated cells, respectively (Fig. 3B). Although the K440E mutant has impaired phospholipid binding capabilities (32, 38, 77), this mutant is still repressed by SPH (75), suggesting that ASAH1 acts to repress the transactivation potential of the K440E by facilitating the delivery of SPH to the ligand binding pocket.
To determine if ASAH1 and SF-1 interact, coIP assays were carried out; these revealed that endogenous ASAH1 and SAP-D coimmunoprecipitate with SF-1 (Fig. 3C). Additionally, GFP-tagged SF-1 coimmunoprecipitated with FLAG-tagged ASAH1 (Fig. 3D). Importantly, ASAH2, which is also localized in the nuclei of H295R cells (Fig. 1A), failed to interact with SF-1 (Fig. 3D). The interaction between SF-1 and ASAH1 was further probed in in vitro pulldown assays using bacterially expressed His-tagged SF-1 and in vitro-translated 35S-ASAH1. As shown in Fig. 3E (bottom), immobilized SF-1 copurified with radiolabeled ASAH1 (lane 3), indicating a direct interaction between the two proteins.
Mapping the ASAH1 region that interacts with SF-1.
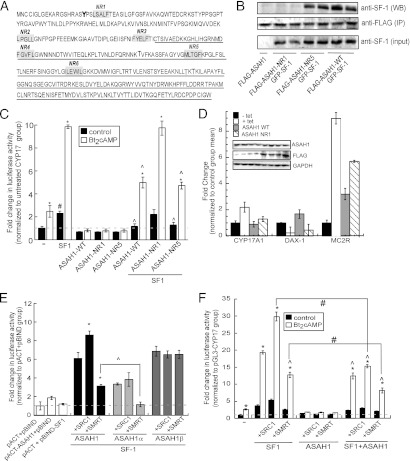
As mentioned earlier, nuclear receptors interact with coregulatory proteins via NR boxes. In silico analysis of ASAH1 revealed 6 putative nuclear receptor-binding (NR) motifs (Fig. 4A). To test whether ASAH1 binds to SF-1 through one of these motifs, we carried out site-directed mutagenesis as described in Materials and Methods. As shown in Fig. 4B, ASAH1 harboring a mutation of NR1 (located in the α subunit of ASAH1 [Fig. 4A]) was unable to interact with the receptor, indicating that this region is required for mediating the binding of ASAH1 to SF-1. Conversely, mutation of NR5 (located in the β subunit of ASAH1 [Fig. 4A]) had no effect on the ability of ASAH1 to coimmunoprecipitate with SF-1 (Fig. 4B). Mutation of all other putative NR interaction motifs did not affect the ability of ASAH1 to repress SF-1-dependent CYP17A1 promoter activity (data not shown). Consistent with a role for NR1 in conferring SF-1 repression, reporter gene assays revealed that mutation of NR1 rendered ASAH1 incapable of repressing SF-1/Bt2cAMP-dependent CYP17A1 promoter activity, whereas mutation of NR5 in ASAH1 had no effect on the ability of the ceramidase to suppress SF-1 activity (Fig. 4C).
Fig 4.
(A) Schematic representation of the putative NR motifs (numbered NR1 to NR6) in ASAH1. The β subunit sequence is underlined. NR motifs were mutated as described in Materials and Methods and used for panel B. (B) H295R cells were transfected with 5 μg pCMV6-GFP-hSF-1 and pCMV6myc/FLAG-ASAH1-wild type (WT) or pCMV6myc/FLAG-ASAH1-NR1 or pCMV6myc/FLAG-ASAH1-NR5 expression plasmids for 72 h. Lysates were subjected to IP with an anti-FLAG antibody and protein A/G agarose. Immobilized proteins were washed and separated by SDS-PAGE, followed by Western blotting using an anti-SF-1 antibody (top blot). Ten percent inputs were subjected to SDS-PAGE and Western blotting using antibodies against FLAG (middle blot) or SF-1 (bottom blot). (C) H295R cells were transfected with 100 ng pGL3-CYP17A1-2x57, 25 ng pCMV6myc/FLAG-hSF1, 1.5 ng pRL-TK, and 50 ng pCMV6myc/FLAG-ASAH1-wild type (WT), pCMV6myc/FLAG-ASAH1-NR1, or pCMV6myc/FLAG-ASAH1-NR5. Twenty-four hours after transfection, cells were treated with 0.4 mM Bt2cAMP for 16 h and harvested for dual-luciferase assay. Data are expressed as fold increase in pGL3-CYP17A1 reporter gene activity and represent the means ± SEMs of 3 separate experiments, each performed in triplicate. Asterisks, carets, or pound signs indicate statistically significant differences (P < 0.05) from values for the untreated controls within each transfection group, Bt2cAMP-treated SF-1-transfected group, or untreated CYP17A1 group, respectively. (D) H295R ASAH1KD cells were transfected with expression plasmids for ASAH1 wild type (WT) or NR1 mutant for 72 h in the presence of tetracycline (tet, 5 μg/ml, 96 h). Untransfected groups that were left untreated (− tet) or treated with 5 μg/ml tetracycline for 96 h (+ tet) were used as controls. Total RNA was isolated, and the mRNA levels of CYP17, DAX-1, and MC2R were measured by real-time RT-PCR. Data are expressed as the means ± SEMs of 3 separate experiments, each performed in triplicate, and normalized to the − tet control group. Asterisks and carets indicate statistically significant differences (P < 0.05) from values for the untranfected − tet and + tet control groups, respectively. (Inset) Representative Western blots of ASAH1 and FLAG-tagged ASAH1 WT or NR1 protein levels. GAPDH, glyceraldehyde-3-phosphate dehydrogenase. (E) H295R cells were transfected with pGL5luc, pACT, and pBIND (empty vectors) or pGL5luc, pBIND-SF-1 and pACT-ASAH1, pACT-ASAH1Δα, or pACT-ASAH1Δβ plasmid in the presence or absence of pBKCMV-SRC-1 or pCMX-SMRT expression plasmids (50 ng each plasmid). Transfected cells were harvested after 48 h, and luciferase activity was quantified by luminometry. Data represent the means ± SEMs of at least 3 separate experiments, each performed in triplicate. Asterisks or carets denote statistically significant differences (P < 0.05) from values for the coregulator-untransfected group or as indicated in the graph, respectively. (F) H295R cells were transfected with 100 ng pGL3-CYP17A1-2x57, 25 ng pCMV6myc/FLAG-hSF1, 50 ng pCMV6myc/FLAG-ASAH1, 1.5 ng pRL-TK, and 25 ng pBKCMV-SRC-1 or pCMX-SMRT. Twenty-four hours after transfection, cells were treated with 0.4 mM Bt2cAMP for 16 h and harvested for dual-luciferase assay. Data are expressed as fold increase in pGL3-CYP17A1 reporter gene activity and represent the means ± SEMs of 3 separate experiments, each performed in triplicate. Asterisks, carets, or pound signs indicate statistically significant differences (P < 0.05) from the untreated control group within each transfection group, the Bt2cAMP-treated SF-1-transfected group, or as indicated in the figure, respectively.
To investigate the role of ASAH1 in endogenous SF-1-dependent gene expression, we tested the effect of overexpressing ASAH1 NR1 on the transcriptional levels of three SF-1 target genes, those for CYP17A1, DAX-1 (dosage-sensitive sex reversal, adrenal hypoplasia critical region, on chromosome X, gene 1) and MC2R, in H295R ASAH1KD cells. Consistent with our previous results (41), tetracycline-induced ASAH1 knockdown resulted in a 2.2-fold increase in CYP17A1 gene expression and a 2.8-fold decrease and a 9-fold increase in DAX-1 and MC2R mRNA expression, respectively, compared to those of the control (tetracycline minus) (Fig. 4D). Transfecting the knockdown cells with wild-type ASAH1 reduced the expression of CYP17A1 to 89% of that observed for cells not treated with tetracycline, whereas transfection with the ASAH1 NR1 mutant resulted in a 1.3-fold increase in CYP17A1 gene expression compared to that in tetracycline-minus cells. In contrast, overexpressing wild-type ASAH1 led to a 1.7-fold increase in DAX-1 transcription compared to that in control cells, which is consistent with our previous report of a role for ASAH1 as a positive regulator of DAX-1 gene expression (41). Consistent with this finding, overexpression of the ASAH1 NR1 mutant was unable to increase DAX-1 transcript levels as observed for the ASAH1 WT-transfected group (Fig. 4D). Unexpectedly, overexpressing either wild-type or the NR1 mutant ASAH1 resulted in an increase in MC2R mRNA expression compared to that in cells that were not treated with tetracycline (Fig. 4D). Although the induction in MC2R mRNA expression observed in cells transfected with the NR1 mutant is greater than that observed for cells expressing the wild-type enzyme, it is unclear why reintroduction of wild-type ASAH1 into the tetracycline-inducible cell line resulted in increased MC2R expression.
To further investigate the interaction between ASAH1 and SF-1, GAL4 chimeric mutants of ASAH1 were generated by cloning the primary genomic sequence of the α or β subunit of ASAH1 into the pACT vector. Mammalian two-hybrid assays in H295R cells were then carried out to test the interaction between GAL4-fused, full-length ASAH1, GAL4-ASAH1 α, or GAL4-ASAH1 β and VP16–SF-1. Consistent with coIP studies (Fig. 4B), ASAH1α interacted with SF-1, but surprisingly, the β subunit of ASAH1 also interacted with the receptor (Fig. 4E), suggesting that additional regions independent of the NR motifs within the β subunit of ASAH1 are also involved in mediating the binding of ASAH1 to SF-1. Because the transcriptional potential of SF-1 is modulated by its interaction with various coregulatory proteins (12), the ability of coregulators to alter the interaction between ASAH1 and SF-1 was tested using mammalian 2-hybrid assays. Unexpectedly, SMRT significantly reduced the interaction between full-length ASAH1 and SF-1 and completely prevented the interaction between ASAH1α and the receptor (Fig. 4E). While SRC-1 facilitated the interaction between full-length ASAH1 and SF-1 1.4-fold, neither coregulator affected the interaction between ASAH1β and SF-1 (Fig. 4E). Furthermore, as shown in Fig. 4F, ASAH1 not only inhibited the ability of SRC-1 to potentiate SF-1-dependent promoter activity but also potentiated the repressive activity of SMRT. Collectively, these interaction studies demonstrate that ASAH1 suppresses SF-1 activity by binding to the receptor through at least one NR motif and suggest that the repressive function of ASAH1 is mediated, at least in part, by its ability to abrogate SRC-1 coactivator activity while potentiating the repressive function of SMRT.
The catalytic activity of ASAH1 is required for its corepressive function.
To determine whether ceramidase activity (SPH production) is required for the repressive effect of ASAH1 on SF-1 function, H295R cells were pretreated with the ceramidase inhibitor D-NMAPPD (60) and the ability of ASAH1 to inhibit SF-1 activity was assessed in reporter gene assays. Administration of D-NMAPPD prevented the Bt2cAMP-stimulated increase in ceramidase activity (Fig. 5A) and rendered ASAH1 unable to suppress SF-1/Bt2cAMP-induced CYP17A1 reporter gene activity (Fig. 5B).
Fig 5.
Catalytically active ASAH1 is required for suppression of SF-1 activity. (A) H295R cells were pretreated for 1 h with 5 μM D-NMAPPD, followed by treatment with 0.4 mM Bt2cAMP for 30 min. Lysates were isolated and then incubated with 2 μl 1 mM NBD-12-ceramide in acetate buffer (0.5 M, pH 4.5) at 37°C for 2 h. Reactions were stopped, and lipids were extracted and spotted on TLC plates. Plates were developed and visualized by fluorescent scanning. NBD-dodecanoic acid formation was quantified and normalized to the protein content of each sample. Data represents means ± SEMs of 3 separate experiments, each performed in triplicate. (B) H295R cells were transfected with pGL3-CYP17A1-2x57 and expression plasmids for SF-1 and ASAH1. Cells were pretreated for 1 h with 0.5 μM or 5 μM D-NMAPPD, followed by treatment with 0.4 mM Bt2cAMP for 16 h. Cells were then harvested for dual-luciferase assays. Data are expressed as fold increase in pGL3-CYP17A1 reporter gene activity and represent the means ± SEMs of 3 separate experiments, each performed in triplicate. Asterisks, carets, or pound signs indicate statistically significant differences (P < 0.05) from the values for the untreated controls within each group, treated SF-1-transfected group, or untreated pGL3-CYP17A1 group, respectively.
ACTH/cAMP signaling promotes the recruitment of ASAH1 to multiple steroidogenic gene promoters.
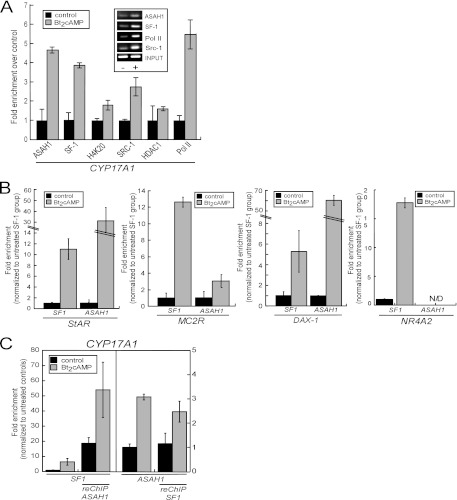
Given that ASAH1 binds to SF-1 (Fig. 3 and 4A), we next sought to determine if ASAH1 acts in a coregulator protein manner by binding to the nuclear receptor in a chromatin context. ChIP assays were performed using chromatin isolated from H295R cells treated with 0.4 mM Bt2cAMP for 1 h. The primer set used amplifies the promoter region −104/+43 of the CYP17A1 promoter, which contains an ACTH/cAMP-responsive SF-1 binding site at −57/−37 (12). Elevated ASAH1 occupancy at the promoter was observed after Bt2cAMP stimulation, which coincided with the enrichment of SF-1 at the CYP17A1 promoter (Fig. 6A). Consistent with transcriptional initiation, Bt2cAMP also promoted the recruitment of RNA polymerase II and SRC-1 and increased the levels of trimethylated histone H4 at lysine 20 (H4K20). Importantly, neither ASAH2 nor ACER3 was detected in this region of the CYP17A1 promoter under these conditions (data not shown). Next, we investigated whether ASAH1 and SF-1 can occupy the same SF-1 target gene promoter together as part of a complex by performing reChIP assays. As shown in Fig. 6B, there was an enrichment of ASAH1 at SF-1-bound promoters in unstimulated cells. While Bt2cAMP increased the recruitment of SF-1 to the CYP17 promoter approximately 8-fold, there was only a 2.5-fold increase in the enrichment of ASAH1 at SF-1-bound promoters in response to stimulation with Bt2cAMP (Fig. 6B).
Fig 6.
Bt2cAMP promotes the recruitment of ASAH1 to multiple steroidogenic gene promoters. (A) H295R cells were treated with 0.4 mM Bt2cAMP for 1 h and cross-linked with 1% formaldehyde, and the sheared chromatin was immunoprecipitated (IP) using antibodies against SF-1, ASAH1, SRC-1, trimethyl-H4K20, RNA polymerase II, and HDAC 1. The relative amount of IP promoter fragments was analyzed by quantitative PCR with a primer set that amplifies region −104/+43 of the CYP17A1 promoter and a primer set for β-actin as a negative control. IP chromatin fragments were normalized to their respective levels in the input DNA. (Inset) Representative agarose gels of the PCR products obtained for output and input DNA in control (−) and Bt2cAMP-treated (+) samples. Data represent the means ± standard deviations of at least three experiments, each performed in duplicate. (B) H295R cells were treated with 0.4 mM Bt2cAMP for 1 h and cross-linked with 1% formaldehyde, and reChIP assays were carried out as described in Materials and Methods. The IP chromatin fragments were analyzed by quantitative PCR using primers that amplify the proximal promoter of the CYP17A1 gene. The relative amount of IP promoter fragments was normalized to their respective levels in the input DNA. (C) H295R cells were treated with 0.4 mM Bt2cAMP for 1 h and cross-linked with 1% formaldehyde, and the sheared chromatin was immunoprecipitated using antibodies against SF-1 or ASAH1. The IP DNA fragments were analyzed by quantitative PCR using primers that amplify the proximal promoter regions of the StAR, MC2R, DAX-1, and NR4A2 genes as well as a primer set for β-actin as a negative control. The relative amount of IP promoter fragments was normalized to their respective levels in the input DNA and is graphed as fold enrichment over untreated SF-1-immunoprecipitated DNA. Data represent the means ± standard deviations of two experiments, each performed in duplicate.
Finally, to examine the recruitment of ASAH1 to additional SF-1-regulated promoters, ChIP assays were carried out using primers that amplify the proximal promoter region of the StAR, DAX-1, and MC2R genes, which encompass at least one SF-1 binding site. We have recently shown that silencing ASAH1 increases the acetylation of histone H3 at these promoters (41). As shown in Fig. 6C, Bt2cAMP stimulation promoted the recruitment of SF-1 to all promoters. Interestingly, Bt2cAMP also promoted the recruitment of SF-1, but not ASAH1, to the NR4A2 promoter, which encodes the orphan nuclear receptor Nurr1 (Fig. 6C).
DISCUSSION
A growing body of literature supports the role of sphingolipid metabolism in the regulation of steroid hormone production (42). Our group has previously identified the role of SPH as an antagonist for SF-1 (75). However, because the mechanisms that control ligand availability are unknown, we sought to determine the role of ASAH1, an SPH-generating enzyme, in regulating SF-1 function. We demonstrate that ASAH1 is expressed in the nuclei of H295R cells and its activity is acutely regulated by ACTH/cAMP signaling. ASAH1 represses the transcriptional potential of SF-1 by directly binding to the receptor on the promoter of multiple steroidogenic genes. Significantly, the interaction between ASAH1 and SF-1 occurs through a functional NR motif, which is essential for conferring ASAH1-dependent repression of SF-1.
Mounting evidence has linked sphingolipids to various nuclear processes (reviewed in reference 35). Nuclear sphingosine kinase 2 (SPHK2), which converts SPH into sphingosine-1-phosphate, has recently been shown to modulate gene expression by regulating histone acetylation (20). In addition to SPHK2, other sphingolipid-metabolizing enzymes, such as sphingomyelinases and sphingomyelin synthases, have also been detected in nuclear extracts from different cell lines (1, 2, 46, 72). Due to the hydrophobic nature of most sphingolipids, the nuclear expression of sphingolipid enzymes suggests that there is dynamic, localized production of bioactive lipids, which may have unique roles in nuclear processes independently from their cytoplasmic functions. The present study reveals that ASAH1 and its coactivator protein SAP-D are expressed in the nuclei of H295R cells (Fig. 1), providing further evidence for the functional significance of localized sphingolipid metabolism. Consistent with this premise, mass spectrometric analysis revealed that ACTH/cAMP signaling promotes rapid nuclear sphingomyelin and ceramide turnover with a concomitant accumulation of SPH and S1P in an ASAH1-dependent manner (Fig. 2B).
Our findings identify a novel mechanism by which nuclear localization of ASAH1 acts to regulate gene expression by modulating the transcriptional potential of a nuclear receptor. Nuclear import of proteins is usually based on the presence of a basic amino acid sequence designated the nuclear localization signal (NLS), which mediates binding to importins and translocation through the nuclear pore complex (NPC) (19). In addition to this “classical” mechanism of protein nuclear import, other pathways for the transport of non-NLS-containing proteins into the nucleus have been described. One such mechanism is the nuclear import of glycoproteins via a mechanism that utilizes sugar-binding carrier proteins (e.g., lectins) (15). Given that ASAH1 contains no NLS sequences but is glycosylated at multiple residues during processing (17), it is possible that sugar-binding proteins mediate its nuclear import in H295R cells. Alternatively, it was recently shown that nuclear import of extracellular-regulated kinase (ERK), which does not contain a canonical NLS, is mediated by casein kinase II-dependent phosphorylation of three residues (54), which promote ERK binding to importin-7 and translocation through NPCs (8). Notably, probing the subcellular localization of ASAH1 in other cell lines revealed that this ceramidase is nuclear in murine Y-1 adrenocortical cells, whereas it is cytoplasmic in MCF-7 breast cancer and MA-10 Leydig cells (Fig. 1C), suggesting that the nuclear expression of ASAH1 is cell type specific.
The transcriptional activity of nuclear receptors is influenced by their ability to interact with numerous coregulator proteins. Conserved sequences containing an NR motif of LXXLL (14, 23, 24, 81), LXX-(I/H)-IXXX-(I/L) helix (52), or (L/I)-XX-(I/V)-I (CoRNR) box (25) have been implicated in receptor-corepressor interaction. Our study demonstrates that ASAH1 is a novel SF-1 coregulator that represses receptor activity by a direct protein-protein interaction involving an NR motif located in the α subunit of ASAH1. Site-directed mutagenesis revealed that this motif is required not only for interaction with the receptor (Fig. 4B) but also for the enzyme to repress SF-1 activity (Fig. 4C). However, it is unclear why overexpression of both the wild type and NR1 mutant increased the endogenous mRNA expression of the MC2R gene (Fig. 4D). The fact that ASAH1 colocalizes with SF-1 on the promoter of the CYP17A1 and StAR genes (Fig. 6B) and is recruited to the promoter of multiple SF-1 target genes, including MC2R and DAX-1 (Fig. 6C), is consistent with a coregulator function. Coregulators are recruited to DNA by nuclear receptors and either have intrinsic catalytic properties or recruit enzymatic protein complexes (e.g., histone deacetylases [HDAC], methyltransferases, or acetyltransferases) to mediate chromatin and/or nuclear receptor modifications that influence gene transcription. Coactivators enhance nuclear receptor-mediated transcription, while corepressors inhibit gene expression, primarily through their interaction with unliganded or antagonist-bound nuclear receptor (18). We have recently reported that ASAH1 negatively regulates histone H3 acetylation at SF-1 target promoters (41). Furthermore, Hait et al. (20) have recently reported that sphingosine kinase 2-produced nuclear S1P modulates gene transcription by binding and suppressing the activity of HDACs 1 and 2. Thus, it is tempting to speculate that upon binding to SF-1, ASAH1 mediates the recruitment of histone-modifying enzymes (e.g., HDAC) to target DNA. Our data presented in Fig. 3B, showing that ASAH1 is able to repress the transactivation potential of the K440E SF-1 pocket mutant that is unable to bind to phospholipids (32, 38, 77) but still repressed by SPH (75), suggest that the factors that govern phospholipid binding and SPH binding to the receptor may be different. Significantly, a novel role for another nuclear lipid-metabolizing enzyme, inositol phosphate multikinase (IMPK), in regulating the transactivation potential of SF-1 has been recently described (4). In an elegant study, Blind and colleagues show that activation of SF-1 occurs by IMPK-catalyzed phosphorylation of the phospholipid ligand when the substrate phosphoinositol phosphate is bound to the receptor (4). These studies provide further evidence for the pivotal role that localized lipid metabolism plays in regulating SF-1 function by controlling the nuclear concentrations of ligands.
The molecular determinants of SF-1 interaction with various coregulators, such as DAX-1 (9, 28, 71), SRC-1 (10), SRC2 (21), peroxisome proliferator-activated receptor γ coactivator 1α (80), and diacylglycerol kinase θ (DGKθ) (36), have been described. Suzuki et al. (71) and Zhang et al. (81) proposed a model for DAX-1-dependent SF-1 and estrogen receptor (ER) α transcriptional repression, respectively, where DAX-1 binding to nuclear receptor competes with coactivators (e.g., SRC-1) as well as recruits corepressors (e.g., NCoR). Because ASAH1 expression prevented SRC-1 from inducing SF-1-dependent CYP17A1 promoter activity while potentiating the repressive activity of SMRT (Fig. 4C), it is possible that the underlying mechanism by which ASAH1 suppresses SF-1 function is similar to that for DAX-1. Significantly, pharmacological inhibition of ASAH1 rendered the enzyme incapable of repressing SF-1 (Fig. 5B). Collectively, these findings suggest that the corepressive function of ASAH1 requires not only an interaction with SF-1 but also a catalytically competent enzyme. This premise is consistent with the role of ASAH1 in controlling the nuclear levels of the SF-1 antagonist SPH (75).
Our group has previously reported an NR-dependent interaction between SF-1 and DGKθ (36), the enzyme that produces the SF-1 agonist PA. DGKθ activity and binding to SF-1 are required events for cAMP-dependent CYP17A1 mRNA expression (36). The findings described within support a regulatory mechanism in which the interaction of ASAH1 with SF-1 represses receptor activity possibly by facilitating ligand exchange (i.e., the exchange of PA for SPH) and/or promoting corepressor recruitment (e.g., SMRT and HDAC). It is counterintuitive to think that activation of the ACTH/cAMP signaling cascade, which is the main activator of steroidogenic gene transcription, would stimulate ASAH1 DNA recruitment and SF-1 binding. However, because SF-1-containing transcriptional complex assembly at target promoters is dynamic and cyclical, as demonstrated by Winnay and Hammer (79) and our group (12), it is probable that ACTH evokes spatiotemporal transcriptional regulation where a cyclic, reciprocal exchange of coactivators and corepressors ensures optimal target gene transcription. Furthermore, several transcriptional studies support the coordinate localization of coactivators and corepressors in the same complex (57), which has been proposed as a possible feedback mechanism to avoid dysregulated transcriptional activation (33). Even though ASAH1 acts as a repressor of SF-1, it is provocative to speculate that ASAH1 recruitment to DNA in response to ACTH/cAMP signaling is important for fine-tuning steroid hormone output. This premise is consistent with recent work by our group demonstrating that suppressing ASAH1 expression leads to increased steroidogenic gene transcription and the overproduction of steroid hormones in H295R cells (41). Additionally, because SPH activates DGK activity (59), it is possible that ASAH1 may also play a role in regulating PA availability, thus indirectly mediating SF-1 activation. Future reChIP experiments to identify additional ASAH1 target genes will provide further insight into a global role of ASAH1 as a transcriptional regulator.
Notably, the ability of various nuclear receptors, including ERβ (73), PPARγ (65), and SF-1 (21), to interact with coregulatory proteins is regulated by phosphorylation. Therefore, it is possible that posttranslational modifications of SF-1 may be involved in temporally regulating the receptor's affinity for ASAH1 in response to ACTH stimulation. Phosphorylation of SF-1 at Ser203 enhances the receptor's ability to interact with SRC2 and SMRT (21). It is equally likely that phosphorylation of ASAH1 may regulate the ability of the ceramidase to interact with binding partners. Indeed, we have preliminary data supporting a role for phosphorylation of ASAH1 in controlling binding to SF-1 (N. C. Lucki et al., unpublished observation). In summary, we show that ASAH1 is a nuclear protein that acts via a novel coregulatory mechanism to control steroidogenic gene transcription by binding to SF-1 at the promoter of target genes in H295R cells. Moreover, our data lend support to the emerging role of localized nuclear lipid metabolism in the regulation of gene transcription.
ACKNOWLEDGMENTS
This work is supported by National Institutes of Health grants DK084178 (to M.B.S.) and GM069388 (to A.H.M.) and by a predoctoral fellowship from the American Heart Association (10PRE3230019; to N.C.L.).
Footnotes
Published ahead of print 27 July 2012
REFERENCES
- 1.Albi E, Magni MP. 1997. Chromatin neutral sphingomyelinase and its role in hepatic regeneration. Biochem. Biophys. Res. Commun. 236:29–33 [DOI] [PubMed] [Google Scholar]
- 2.Albi E, Magni MV. 1999. Sphingomyelin synthase in rat liver nuclear membrane and chromatin. FEBS Lett. 460:369–372 [DOI] [PubMed] [Google Scholar]
- 3.Bernardo K, et al. 1995. Purification, characterization, and biosynthesis of human acid ceramidase. J. Biol. Chem. 270:11098–11102 [DOI] [PubMed] [Google Scholar]
- 4.Blind RD, Suzawa M, Ingraham HA. 2012. Direct modification and activation of a nuclear receptor-PIP2 complex by the inositol lipid kinase IPMK. Sci. Signal. 5:ra44 doi:10.1126/scisignal.2003111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Børud B, et al. 2002. The nuclear receptor coactivators p300/CBP/cointegrator-associated protein (p/CIP) and transcription intermediary factor 2 (TIF2) differentially regulate PKA-stimulated transcriptional activity of steroidogenic factor 1. Mol. Endocrinol. 16:757–773 [DOI] [PubMed] [Google Scholar]
- 6.Chen WY, Juan LJ, Chung BC. 2005. SF-1 (nuclear receptor 5A1) activity is activated by cyclic AMP via p300-mediated recruitment to active foci, acetylation, and increased DNA binding. Mol. Cell. Biol. 25:10442–10453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen WY, Lee WC, Hsu NC, Huang F, Chung BC. 2004. SUMO modification of repression domains modulates function of nuclear receptor 5A1 (steroidogenic factor-1). J. Biol. Chem. 279:38730–38735 [DOI] [PubMed] [Google Scholar]
- 8.Chuderland D, Konson A, Seger R. 2008. Identification and characterization of a general nuclear translocation signal in signaling proteins. Mol. Cell 31:850–861 [DOI] [PubMed] [Google Scholar]
- 9.Crawford PA, Dorn C, Sadovsky Y, Milbrandt J. 1998. Nuclear receptor DAX-1 recruits nuclear receptor corepressor N-CoR to steroidogenic factor 1. Mol. Cell. Biol. 18:2949–2956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Crawford PA, Polish JA, Ganpule G, Sadovsky Y. 1997. The activation function-2 hexamer of steroidogenic factor-1 is required, but not sufficient for potentiation by SRC-1. Mol. Endocrinol. 11:1626–1635 [DOI] [PubMed] [Google Scholar]
- 11.Cuvillier O. 2002. Sphingosine in apoptosis signaling. Biochim. Biophys. Acta 1585:153–162 [DOI] [PubMed] [Google Scholar]
- 12.Dammer EB, Leon A, Sewer MB. 2007. Coregulator exchange and sphingosine-sensitive cooperativity of steroidogenic factor-1, general control nonderepressed 5, p54, and p160 coactivators regulate cyclic adenosine 3′,5′-monophosphate-dependent cytochrome P450c17 transcription rate. Mol. Endocrinol. 21:415–438 [DOI] [PubMed] [Google Scholar]
- 13.Desclozeaux M, Krylova IN, Horn F, Fletterick RJ, Ingraham HA. 2002. Phosphorylation and intramolecular stabilization of the ligand binding domain in the nuclear receptor steroidogenic factor 1. Mol. Cell. Biol. 22:7193–7203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dubbink HJ, et al. 2006. Androgen receptor ligand-binding domain interaction and nuclear receptor specificity of FXXLF and LXXLL motifs as determined by L/F swapping. Mol. Endocrinol. 20:1742–1755 [DOI] [PubMed] [Google Scholar]
- 15.Duverger E, Pellerin-Mendes C, Mayer R, Roche AC, Monsigny M. 1995. Nuclear import of glycoconjugates is distinct from the classical NLS pathway. J. Cell Sci. 108(Part 4):1325–1332 [DOI] [PubMed] [Google Scholar]
- 16.Eliyahu E, Park JH, Shtraizent N, He X, Schuchman EH. 2007. Acid ceramidase is a novel factor required for early embryo survival. FASEB J. 21:1403–1409 [DOI] [PubMed] [Google Scholar]
- 17.Ferlinz K, et al. 2001. Human acid ceramidase: processing, glycosylation, and lysosomal targeting. J. Biol. Chem. 276:35352–35360 [DOI] [PubMed] [Google Scholar]
- 18.Glass CK, Rosenfeld MG. 2000. The coregulator exchange in transcriptional functions of nuclear receptors. Genes Dev. 14:121–141 [PubMed] [Google Scholar]
- 19.Görlich D. 1997. Nuclear protein import. Curr. Opin. Cell Biol. 9:412–419 [DOI] [PubMed] [Google Scholar]
- 20.Hait NC, et al. 2009. Regulation of histone acetylation in the nucleus by sphingosine-1-phosphate. Science 325:1254–1257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hammer GD, et al. 1999. Phosphorylation of the nuclear receptor SF-1 modulates cofactor recruitment: integration of hormone signaling in reproduction and stress. Mol. Cell 3:521–526 [DOI] [PubMed] [Google Scholar]
- 22.Heery DM, Kalkhoven E, Hoare S, Parker MG. 1997. A signature motif in transcriptional co-activators mediates binding to nuclear receptors. Nature 387:733–736 [DOI] [PubMed] [Google Scholar]
- 23.Heldring N, et al. 2007. Structural insights into corepressor recognition by antagonist-bound estrogen receptors. J. Biol. Chem. 282:10449–10455 [DOI] [PubMed] [Google Scholar]
- 24.Hsia EY, Goodson ML, Zou JX, Privalsky ML, Chen HW. 2010. Nuclear receptor coregulators as a new paradigm for therapeutic targeting. Adv. Drug Deliv. Rev. 62:1227–1237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hu X, Lazar MA. 1999. The CoRNR motif controls the recruitment of corepressors by nuclear hormone receptors. Nature 402:93–96 [DOI] [PubMed] [Google Scholar]
- 26.Huwiler A, Kolter T, Pfeilschifter J, Sandhoff K. 2000. Physiology and pathophysiology of sphingolipid metabolism and signaling. Biochim. Biophys. Acta 1485:63–99 [DOI] [PubMed] [Google Scholar]
- 27.Ishihara SL, Morohashi K. 2005. A boundary for histone acetylation allows distinct expression patterns of the Ad4BP/SF-1 and GCNF loci in adrenal cortex cells. Biochem. Biophys. Res. Commun. 329:554–562 [DOI] [PubMed] [Google Scholar]
- 28.Ito M, Yu R, Jameson JL. 1997. DAX-1 inhibits SF-1-mediated transactivation via a carboxy-terminal domain that is deleted in adrenal hypoplasia congenita. Mol. Cell. Biol. 17:1476–1483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ito M, Yu RN, Jameson JL. 1998. Steroidogenic factor-1 contains a carboxy-terminal transcriptional activation domain that interacts with steroid receptor coactivator-1. Mol. Endocrinol. 12:290–301 [DOI] [PubMed] [Google Scholar]
- 30.Jacob AL, Lund J, Martinez P, Hedin L. 2001. Acetylation of steroidogenic factor 1 protein regulates its transcriptional activity and recruits the coactivator GCN5. J. Biol. Chem. 276:37659–37664 [DOI] [PubMed] [Google Scholar]
- 31.Komatsu T, et al. 2004. Small ubiquitin-like modifier 1 (SUMO-1) modification of the synergy control motif of Ad4 binding protein/steroidogenic factor 1 (Ad4BP/SF-1) regulates synergistic transcription between Ad4BP/SF-1 and Sox9. Mol. Endocrinol. 18:2451–2462 [DOI] [PubMed] [Google Scholar]
- 32.Krylova IN, et al. 2005. Structural analyses reveal phosphatidyl inositols as ligands for the NR5 orphan receptors SF-1 and LRH-1. Cell 120:343–355 [DOI] [PubMed] [Google Scholar]
- 33.Kumar R, Gururaj AE, Vadlamudi RK, Rayala SK. 2005. The clinical relevance of steroid hormone receptor corepressors. Clin. Cancer Res. 11:2822–2831 [DOI] [PubMed] [Google Scholar]
- 34.Lala DS, Rice DA, Parker KL. 1992. Steroidogenic factor I, a key regulator of steroidogenic enzyme expression, is the mouse homolog of fushi tarazu-factor I. Mol. Endocrinol. 6:1249–1258 [DOI] [PubMed] [Google Scholar]
- 35.Ledeen RW, Wu G. 2008. Nuclear sphingolipids: metabolism and signaling. J. Lipid Res. 49:1176–1186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li D, et al. 2007. Cyclic AMP-stimulated interaction between steroidogenic factor 1 and diacylglycerol kinase θ facilitates induction of CYP17. Mol. Cell. Biol. 27:6669–6685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li LA, et al. 1999. Function of steroidogenic factor 1 domains in nuclear localization, transactivation, and interaction with transcription factor TFIIB and c-Jun. Mol. Endocrinol. 13:1588–1598 [DOI] [PubMed] [Google Scholar]
- 38.Li Y, et al. 2005. Crystallographic identification and functional characterization of phospholipids as ligands for the orphan nuclear receptor steroidogenic factor-1. Mol. Cell 17:491–502 [DOI] [PubMed] [Google Scholar]
- 39.Linke T, et al. 2001. Interfacial regulation of acid ceramidase activity. Stimulation of ceramide degradation by lysosomal lipids and sphingolipid activator proteins. J. Biol. Chem. 276:5760–5768 [DOI] [PubMed] [Google Scholar]
- 40.Lucki N, Sewer MB. 2009. The cAMP-responsive element binding protein (CREB) regulates the expression of acid ceramidase (ASAH1) in H295R human adrenocortical cells. Biochim. Biophys. Acta 1791:706–713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lucki NC, Bandyopadhyay S, Wang E, Merrill AH, Sewer MB. 2012. Acid ceramidase (ASAH1) is a global regulator of steroidogenic capacity and adrenocortical gene expression. Mol. Endocrinol. 26:228–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lucki NC, Sewer MB. 2008. Multiple roles for sphingolipids in steroid hormone biosynthesis. Subcell. Biochem. 49:387–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mangelsdorf DJ, et al. 1995. The nuclear receptor superfamily: the second decade. Cell 83:835–839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McInerney EM, et al. 1998. Determinants of coactivator LXXLL motif specificity in nuclear receptor transcriptional activation. Genes Dev. 12:3357–3368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Monté D, DeWitte F, Hum DW. 1998. Regulation of the human P450scc gene by steroidogenic factor 1 is mediated by CBP/p300. J. Biol. Chem. 273:4585–4591 [DOI] [PubMed] [Google Scholar]
- 46.Neitcheva T, Peeva D. 1995. Phospholipid composition, phospholipase A2 and sphingomyelinase activities in rat liver nuclear membrane and matrix. Int. J. Biochem. Cell Biol. 27:995–1001 [DOI] [PubMed] [Google Scholar]
- 47.Ozbay T, Merrill AH, Jr, Sewer MB. 2004. ACTH regulates steroidogenic gene expression and cortisol biosynthesis in the human adrenal cortex via sphingolipid metabolism. Endocr. Res. 30:787–794 [DOI] [PubMed] [Google Scholar]
- 48.Ozbay T, Rowan A, Leon A, Patel P, Sewer MB. 2006. Cyclic adenosine 5′-monophosphate-dependent sphingosine-1-phosphate biosynthesis induces human CYP17 gene transcription by activating cleavage of sterol regulatory element binding protein 1. Endocrinology 147:1427–1437 [DOI] [PubMed] [Google Scholar]
- 49.Park J, Schuchuman E. 2006. Acid ceramidase and human disease. Biochim. Biophys. Acta 1758:2133–2138 [DOI] [PubMed] [Google Scholar]
- 50.Parker KL, et al. 2002. Steroidogenic factor 1: an essential mediator of endocrine development. Recent Prog. Horm. Res. 57:19–36 [DOI] [PubMed] [Google Scholar]
- 51.Payne AH, Hales DB. 2004. Overview of steroidogenic enzymes in the pathway from cholesterol to active steroid hormones. Endocr. Rev. 25:947–970 [DOI] [PubMed] [Google Scholar]
- 52.Perissi V, et al. 1999. Molecular determinants of nuclear receptor-corepressor interaction. Genes Dev. 13:3198–3208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pettus BJ, Chalfant CE, Hannun YA. 2002. Ceramide in apoptosis: an overview and current perspectives. Biochim. Biophys. Acta 1585:114–125 [DOI] [PubMed] [Google Scholar]
- 54.Plotnikov A, Chuderland D, Karamansha Y, Livnah O, Seger R. 2011. Nuclear extracellular signal-regulated kinase 1 and 2 translocation is mediated by casein kinase 2 and accelerated by autophosphorylation. Mol. Cell. Biol. 31:3515–3530 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 55.Rainey WE, Bird IM, Mason JI. 1994. The NCI-H295 cell line: a pluripotent model for human adrenocortical studies. Mol. Cell. Endocrinol. 100:45–50 [DOI] [PubMed] [Google Scholar]
- 56.Romiti E, et al. 2000. Neutral/alkaline and acid ceramidase activities are actively released by murine endothelial cells. Biochem. Biophys. Res. Commun. 275:746–751 [DOI] [PubMed] [Google Scholar]
- 57.Rosenfeld MG, Lunyak VV, Glass CK. 2006. Sensors and signals: a coactivator/corepressor/epigenetic code for integrating signal-dependent programs of transcriptional response. Genes Dev. 20:1405–1428 [DOI] [PubMed] [Google Scholar]
- 58.Sablin EP, et al. 2009. Structure of SF-1 bound by different phospholipids: evidence for regulatory ligands. Mol. Endocrinol. 23:25–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sakane F, Yamada K, Kanoh H. 1989. Different effects of sphingosine, R59022 and anionic amphiphiles on two diacylglycerol kinase isozymes purified from porcine thymus cytosol. FEBS Lett. 255:409–413 [DOI] [PubMed] [Google Scholar]
- 60.Selzner M, et al. 2001. Induction of apoptotic cell death and prevention of tumor growth by ceramide analogues in metastatic human colon cancer. Cancer Res. 61:1233–1240 [PubMed] [Google Scholar]
- 61.Sewer MB, et al. 2002. Transcriptional activation of human CYP17 in H295R adrenocortical cells depends on complex formation among p54(nrb)/NonO, protein-associated splicing factor, and SF-1, a complex that also participates in repression of transcription. Endocrinology 143:1280–1290 [DOI] [PubMed] [Google Scholar]
- 62.Sewer MB, Waterman MR. 2003. ACTH modulation of transcription factors responsible for steroid hydroxylase gene expression in the adrenal cortex. Microsc. Res. Tech. 61:300–307 [DOI] [PubMed] [Google Scholar]
- 63.Sewer MB, Waterman MR. 2002. Adrenocorticotropin/cyclic adenosine 3′,5′-monophosphate-mediated transcription of the human CYP17 gene in the adrenal cortex is dependent on phosphatase activity. Endocrinology 143:1769–1777 [DOI] [PubMed] [Google Scholar]
- 64.Sewer MB, Waterman MR. 2002. cAMP-dependent transcription of steroidogenic genes in the human adrenal cortex requires a dual-specificity phosphatase in addition to protein kinase A. J. Mol. Endocrinol. 29:163–174 [DOI] [PubMed] [Google Scholar]
- 65.Shao D, et al. 1998. Interdomain communication regulating ligand binding by PPAR-gamma. Nature 396:377–380 [DOI] [PubMed] [Google Scholar]
- 66.Shtraizent N, et al. 2008. Autoproteolytic cleavage and activation of human acid ceramidase. J. Biol. Chem. 283:11253–11259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Spiegel S, Milstien S. 2002. Sphingosine 1-phosphate, a key cell signaling molecule. J. Biol. Chem. 277:25851–25854 [DOI] [PubMed] [Google Scholar]
- 68.Staels B, Hum DW, Miller WL. 1993. Regulation of steroidogenesis in NCI-H295 cells: a cellular model of the human fetal adrenal. Mol. Endocrinol. 7:423–433 [DOI] [PubMed] [Google Scholar]
- 69.Sullards MC. 2000. Analysis of sphingomyelin, glucosylceramide, ceramide, sphingosine, and sphingosine 1-phosphate by tandem mass spectrometry. Methods Enzymol. 312:32–45 [DOI] [PubMed] [Google Scholar]
- 70.Sullards MC, Merrill AH., Jr 2001. Analysis of sphingosine 1-phosphate, ceramides, and other bioactive sphingolipids by high-performance liquid chromatography-tandem mass spectrometry. Sci. STKE 2001:pl1. [DOI] [PubMed] [Google Scholar]
- 71.Suzuki T, Kasahara M, Yoshioka H, Morohashi K, Umesono K. 2003. LXXLL-related motifs in Dax-1 have target specificity for the orphan nuclear receptors Ad4BP/SF-1 and LRH-1. Mol. Cell. Biol. 23:238–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tamiya-Koizumi K, Umekawa H, Yoshida S, Kojima K. 1989. Existence of Mg2+-dependent, neutral sphingomyelinase in nuclei of rat ascites hepatoma cells. J. Biochem. 106:593–598 [DOI] [PubMed] [Google Scholar]
- 73.Tremblay A, Tremblay GB, Labrie F, Giguere V. 1999. Ligand-independent recruitment of SRC-1 to estrogen receptor beta through phosphorylation of activation function AF-1. Mol. Cell 3:513–519 [DOI] [PubMed] [Google Scholar]
- 74.Urs AN, et al. 2007. Steroidogenic factor-1 is a sphingolipid binding protein. Mol. Cell. Endocrinol. 265–266:174–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Urs AN, Dammer E, Sewer M. 2006. Sphingosine regulates the transcription of CYP17 by binding to steroidogenic factor-1. Endocrinology 147:5249–5258 [DOI] [PubMed] [Google Scholar]
- 76.Wang E, Norred WP, Bacon CW, Riley RT, Merrill AH., Jr 1991. Inhibition of sphingolipid biosynthesis by fumonisins. Implications for diseases associated with Fusarium moniliforme. J. Biol. Chem. 266:14486–14490 [PubMed] [Google Scholar]
- 77.Wang W, et al. 2005. The crystal structures of human steroidogenic factor-1 and liver receptor homologue-1. Proc. Natl. Acad. Sci. U. S. A. 102:7505–7510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wattenberg BW, Pitson SM, Raben DM. 2006. The sphingosine and diacylglycerol kinase superfamily of signaling kinases: localization as a key to signaling function. J. Lipid Res. 47:1128–1139 [DOI] [PubMed] [Google Scholar]
- 79.Winnay JN, Hammer GD. 2006. Adrenocorticotropic hormone-mediated signaling cascades coordinate a cyclic pattern of steroidogenic factor 1-dependent transcriptional activation. Mol. Endocrinol. 20:147–166 [DOI] [PubMed] [Google Scholar]
- 80.Yazawa T, et al. 2010. PPAR-gamma coactivator-1alpha regulates progesterone production in ovarian granulosa cells with SF-1 and LRH-1. Mol. Endocrinol. 24:485–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhang H, Thomsen JS, Johansson L, Gustafsson JA, Treuter E. 2000. DAX-1 functions as an LXXLL-containing corepressor for activated estrogen receptors. J. Biol. Chem. 275:39855–39859 [DOI] [PubMed] [Google Scholar]
- 82.Zhou D, et al. 2000. PNRC: a proline-rich nuclear receptor coregulatory protein that modulates transcriptional activation of multiple nuclear receptors, including orphan receptors SF1 (steroidogenic factor 1) and ERRalpha1 (estrogen related receptor alpha-1). Mol. Endocrinol. 14:986–998 [DOI] [PubMed] [Google Scholar]