Abstract
Poly(A) binding proteins (PABPs) specifically bind the polyadenosine tail of mRNA and have been shown to be important for RNA polyadenylation, translation initiation, and mRNA stability. Using a modified L1 retrotransposition vector, we examined the effects of two PABPs (encoded by PABPN1 and PABPC1) on the retrotransposition activity of the L1 non-long-terminal-repeat (non-LTR) retrotransposon in both HeLa and HEK293T cells. We demonstrated that knockdown of these two genes by RNA interference (RNAi) effectively reduced L1 retrotransposition by 70 to 80% without significantly changing L1 transcription or translation or the status of the poly(A) tail. We identified that both poly(A) binding proteins were associated with the L1 ribonucleoprotein complex, presumably through L1 mRNA. Depletion of PABPC1 caused a defect in L1 RNP formation. Knockdown of the PABPC1 inhibitor PAIP2 increased L1 retrotransposition up to 2-fold. Low levels of exogenous overexpression of PABPN1 and PABPC1 increased L1 retrotransposition, whereas unregulated overexpression of these two proteins caused pleiotropic effects, such as hypersensitivity to puromycin and decreased L1 activity. Our data suggest that PABPC1 is essential for the formation of L1 RNA-protein complexes and may play a role in L1 RNP translocation in the host cell.
INTRODUCTION
Long interspersed element 1s (L1s or LINE-1s) are the most abundant autonomous non-long-terminal-repeat (non-LTR) human retrotransposons, accounting for ∼17% of human DNA (45). Although most L1 copies are functionally inactive, there are ∼80 to 100 retrotransposition-competent L1s in the human genome (9). L1s have had a great impact on shaping the human genome by their own retrotransposition and by mobilization of nonautonomous elements (Alu, SVA, and processed pseudogenes) in trans (17, 18, 21, 32, 60, 64). Full-length L1 elements are ∼6 kb long and contain a 5′ untranslated region (5′UTR) and two nonoverlapping open reading frames (ORF1 and ORF2), followed by a short 3′UTR ending in a polyadenosine tail encoded, at least in part, in the DNA (5, 20, 23, 24, 31, 66). The product of ORF1 is a 40-kDa protein (ORF1p) with nucleic acid binding and chaperone activities (35, 36, 39, 40, 51). ORF2 encodes an ∼150-kDa multifunctional protein (ORF2p) with endonuclease (EN) (26) and reverse transcriptase (RT) activities (53, 62) and a cysteine-rich domain of unknown function (22).
L1 proliferates through a complex life cycle beginning with RNA polymerase II (Pol II) transcription of its mRNA, which is then exported to the cytoplasm for translation. Both ORFs are translated in the cytoplasm and exhibit a strong cis preference, presumably via direct association with the mRNA molecule encoding them (21, 44, 72). They form a ribonucleoprotein (RNP) particle likely to be a direct retrotransposition intermediate that is then presumed to enter the nucleus in some form (34, 43, 50). Several lines of evidence suggest that L1 transposes via the mechanism known as target-primed reverse transcription (TPRT) (13, 26), which is utilized by other retrotransposons, such as the distantly related R2 elements of invertebrates (46) and the even more distantly related mobile group II introns (80).
L1 activity is often measured by use of a cell culture-based retrotransposition assay (56). Typically, an L1 element is cloned into a pCEP-based vector (Invitrogen) behind the cytomegalovirus (CMV) promoter and tagged with a retrotransposition indication marker in the 3′UTR. This marker (also called a retrotransposition indicator gene) contains its own promoter, open reading frame, and poly(A) signal and is placed in the reverse complementary orientation relative to L1 transcription. This marker is interrupted by a gamma globin intron which is in the same orientation as L1 transcription, i.e., it is designed to be spliced out of the retrotransposition transcript but not the marker transcript. Thus, the marker is inactive on the plasmid and becomes active only once the intron is spliced and the whole reverse-transcribed cassette becomes part of a chromosome. Depending on the type of marker, L1 activity can be calculated by applying a colony formation assay (antibiotic resistance marker) or fluorescence cell sorting (enhanced green fluorescent protein [EGFP] marker).
As a streamlined parasitic element, L1 requires host functions, in addition to the L1-encoded proteins, to complete its life cycle. For example, several tissue-specific transcription factors can bind the internal promoter in the L1 5′UTR and promote L1 transcription by host-encoded Pol II (4, 25, 70, 76). Recent studies suggest that proteins involved in DNA repair can affect L1 retrotransposition, most likely through affecting the TPRT process (69). On the other hand, the host cell has evolved mechanisms to restrict L1 expression and activity to minimize the harmful effects of L1 retrotransposition. These machineries include DNA methylation (33), various RNA interference pathways (68, 75), and the APOBEC3 family of cytidine deaminases (8, 11, 38, 57, 59). However, the involvement of host functions in L1 expression and retrotransposition is far from completely understood.
A common property of functional L1 elements is the DNA-encoded poly(A) sequence located at the 3′ end of the RNA, which is a candidate binding substrate for poly(A) binding proteins (PABPs). Three classes of PABPs are found in the human genome: the cytoplasmic PABPs, i.e., PABPC1, PABPC3, and iPABP; the nuclear PABP (PABPN1); and the X-linked PABP (PABPC5) (48). They all contain one or more RNA recognition motifs (RRMs) that interact with poly(A) tails. By binding nascent tracts of poly(A) sequence synthesized by poly(A) polymerase (PAP), PABPN1 together with the cleavage and polyadenylation specificity factor (CPSF) stimulates processive synthesis of poly(A) tails by PAP (41, 42). When a poly(A) tail reaches a critical length (∼250 nucleotides [nt]), binding of PABPN1 no longer promotes or may even disrupt the interaction between CPSF and PAP and thereby terminates processive elongation of poly(A) tails (41, 42). Once a poly(A)+ mRNA enters the cytoplasm, PABPC1 binds the poly(A) tail and promotes efficient translation initiation by interacting with the cap-binding factor eIF4G and forming a “closed loop” mRNA structure (27, 42, 48). PABPC1 also plays an important role in mRNA stability, although the mechanism for this is less well defined. Several lines of evidence suggest that PABPC1 is involved in a deadenylation-dependent decapping pathway (5′-to-3′ exoribonuclease XRN1 and/or exosome-mediated 3′-to-5′ degradation) (15). Although the proteins are referred to as PABPN1 and PABPC1, several studies indicate that both proteins can shuttle between the nucleus and the cytoplasm, suggesting that they may promote nucleocytoplasmic RNA trafficking (1, 10, 37). PABPC1 was found to bind to mouse BC1 SINE element RNPs, but there is no prior evidence that PABPs are associated with L1 RNP complexes (58, 73).
In this study, we modified an L1 retrotransposition construct to allow facile incorporation of short hairpin RNA (shRNA) cassettes from the TRC (The RNAi Consortium) collection of ∼75,000 anti-human gene constructs (54) and evaluated candidate genes that may affect the L1 retrotransposition pathway by RNA interference (RNAi) screening. Knockdown of either PABPN1 or PABPC1 reduced L1 activity by 70 to 80% without significantly affecting L1 RNA abundance and translation levels. The reduced PABPC1 levels in these experiments did not detectably affect cell viability, poly(A) tail length, intron splicing ability, or the activity of the retrotransposition reporter gene used to measure transposition efficiency. Knockdown studies indicated that PABPC1 was responsible for efficient formation of the L1 RNP complex, providing an explanation for a specific role in retrotransposition. Low-level exogenous expression of both poly(A) binding proteins increased L1 retrotransposition, but overexpression of PABPs had a negative effect. Our data suggest that in addition to their known “generic” functions, PABPs may have novel properties important for retrotransposon proliferation.
MATERIALS AND METHODS
Plasmids.
The pCEP-puro plasmid (61) was first modified by inserting a ligation-independent cloning (LIC) adaptor cassette into the SalI site, between the puro gene and the CMV promoter. A synthetic human L1 element (ORFeus-Hs) (2) tagged with either the neoAI or eGFPAI retrotransposition reporter was cloned between the NotI and BamHI sites to generate pLD108 or pLD190, respectively. pCEP-puro GFP was made by cloning the EGFP gene into the NheI and BamHI sites of pCEP-puro. To construct plasmids expressing the human PABPN1 and PABPC1 genes, PCRs were performed to amplify cDNA copies of these two genes by using primers JB13474 (5′-CTAGCTAGCCGGGAATTCGGCCATTACGGCCGGG-3′) and JB13469 (5′-ATAAGAATGCGGCCGCTTAGTAAGGGGAATACCATGATGTC-3′) for PABPN1 and primers JB13470 (5′-CTAGCTAGCATGAACCCCAGTGCCCCCAGCTACCC-3′) and JB13471 (5′-ATAAGAATGCGGCCGCTTAGCCCATCTGCTGGCCGCCGGTCATGCTGGCCATAACAGTTGGAACACCGGTGGCA-3′) for PABPC1. The templates for these were from the Ultimate ORF collection of sequence-verified human ORFs (Invitrogen, Carlsbad, CA). The amplicons were digested with NheI and NotI and cloned between the NheI and NotI sites of pcDNA3.1 to generate plasmids pLD141 (pcDNA-PABPN1) and pLD142 (pcDNA-PABPC1), respectively. pLD289 was generated from pLD190 by replacing the CMV promoter and wild-type ORF1 with the TRE promoter and ORF1-FLAG sequence. The detailed behavior of these Tet-L1 plasmids will be described in detail elsewhere (K. O'Donnell, W. An, C. T. Schrum, E. S. Davis, S. J. Wheelan, and J. D. Boeke, unpublished data). pLD437 was generated by inserting a hammerhead ribozyme-aptamer after the eGFPAI marker in pLD289. The pLKO.1 plasmid, all shRNA clones, and lentiviruses containing shRNA were purchased from Sigma-Aldrich (St. Louis, MO).
Antibodies.
Full-length human L1 ORF1p (tagged with an N-terminal 6×His tag) was overexpressed in Escherichia coli BL21 and purified under denaturing conditions. Four hens were immunized using purified ORF1 protein as antigen, with eggs collected 3 months later, by Invitrogen (Carlsbad, CA). Chicken anti-ORF1 IgY antibody was isolated and purified from egg yolks by Gallus Immunotech Inc. (Ontario, Canada; http://www.gallusimmunotech.com/pages.php?page=instruction) and was then resuspended in phosphate-buffered saline (PBS) at ∼10 mg/ml. Anti-human PABPN1, anti-human PABPC1, and anti-p53 were purchased from Abcam (Cambridge, MA). Anti-human P62 (nucleoporin 62) antibody was purchased from Covance (Denver, PA). Monoclonal antitubulin and anti-FLAG antibodies were purchased from Sigma-Aldrich (St. Louis, MO). Polyclonal rabbit anti-human ORF1 antibody was a gift from Gerald Schumann (Paul Ehrlich Institut, Germany).
Ligation-independent cloning.
A modified L1-shRNA plasmid (pLD108 or pLD190) was first digested with BstZ17I at 37°C overnight and then purified by phenol extraction and ethanol precipitation. About 3 μg of digested plasmid was treated with T4 DNA polymerase (10 units; NEB, Ipswich, MA) in the presence of 1× NEB buffer 2 and 1 mM dCTP at room temperature for 1 h in a volume of 100 μl. The T4 polymerase was heat inactivated by incubation at 75°C for 20 min. The treated plasmid was diluted with water to a final concentration of 3 ng/μl. PCR was performed to amplify the U6-shRNA cassette from individual TRC shRNA clones, using primers JB13303 (5′-ATTGGATTGGAAGTAGTACGCGGCCGCCCCCTTCACCGAGGGCCTATTTC-3′) and JB13304 (5′-AATAGGTGAAGGTTAGTACGCGGCCGCAAAATTGTGGATGAATACTGCCATTTGTCT-3′). The PCR products were first gel purified using a QIAquick gel extraction kit (Qiagen, Carlsbad, CA) and then treated with T4 DNA polymerase (3 units) for 1 h at room temperature in a total volume of 20 μl with 1 mM dGTP added. The T4 polymerase was then heat inactivated. Each insert was diluted with water to a final volume of 100 μl. For each cloning reaction, 2 μl each of vector (∼6 ng) and insert (∼1 to 10 ng) was combined and incubated at room temperature for 1 h. The entire 4-μl mixture was used for transformation.
Infection with lentiviruses expressing shRNAs.
HeLa cells were seeded in 24-well plates (10,000 cells/well) the day before infection. On day 1, 5-μl lentivirus aliquots at ∼3 × 108 transduction units/ml (Sigma-Aldrich, St. Louis, MO) were added to cells in growth medium containing Polybrene (8 μg/ml). The next day, the cells were transferred to 10-cm plates with fresh growth medium containing puromycin (2.5 μg/ml) and selected for 2 weeks until stable Puror cell pools were acquired.
Cell culture and L1 retrotransposition assay.
HeLa and HEK293T cells were maintained in Dulbecco's modified Eagle medium (DMEM) (high glucose) supplemented with 10% fetal bovine serum, 4 mM l-glutamine, and 100 U/ml penicillin-streptomycin (Invitrogen, Carlsbad, CA).
L1 retrotransposition assays were conducted in either HeLa or HEK293T cells as described previously (56, 72). Briefly, HeLa or HEK293T cells were seeded into 6-well plates (2 × 105 cells/well) the day before transfection. The next day, each well was transfected with 1 μg plasmid by use of Fugene 6 (Roche Applied Science, Indianapolis, IN) according to the manufacturer's protocol. The day after transfection, cells were trypsinized and transferred to a 6-cm plate with 4 ml DMEM containing puromycin (2.5 μg/ml for HeLa cells and 1 μg/ml for HEK293T cells). After 3 days of puromycin selection, the L1 retrotransposition frequency of constructs with the eGFPAI reporter (pLD190 backbone) was evaluated by fluorescence-activated cell sorter (FACS) analysis using a Becton, Dickinson LSR II flow cytometer. For plasmids containing the neoAI reporter (pLD108 backbone), puromycin-resistant cells were trypsinized and seeded in 10-cm plates with G418 (0.5 mg/ml). Two weeks later, G418r colonies were fixed and stained with crystal violet. For retrotransposition assays with the cell pools generated by lentivirus infection, plasmid JM101/L1.3 (with a Hygr gene in the vector and the neoAI retrotransposition marker) (56) was used. L1 retrotransposition assays using plasmids pLD289 and pLD437 were done in HEK293T-tet-onLD cells, generated from HEK293T cells by integration of a linearized vector carrying the tet transactivator. The day after transfection, puromycin (1 μg/ml), doxycycline (500 ng/ml), and theophylline (various final concentrations) were added. Five days after transfection, retrotransposition efficiency was measured by FACS analysis.
Real-time RT-PCR.
Three days after puromycin selection, total RNA was extracted using an RNeasy minikit (Qiagen, Valencia, CA) according to the manufacturer's protocol. One microgram of RNA was used for cDNA synthesis in a 20-μl reaction mix, using a Superscript III reverse transcription kit (Invitrogen, Carlsbad, CA). Real-time PCR was performed using a 1-μl cDNA sample as the template in a 20-μl reaction mix on a Step One Plus instrument (Applied Biosystems, Carlsbad, CA). Primers used for real-time PCR were as follows: for the beta-actin gene, JB12931 (5′-GCTCGTCGTCGACAACGGCT-3′) and JB12932 (5′-CAAACATGATCTGGGTCATCTTCTC-3′); for the puro gene, JB13421 (5′-GACATCGGCAAGGTGTGG-3′) and JB13422 (5′-AGGCCTTCCATCTGTTGCT-3′); for ORFeus-Hs L1 ORF1, JB13415 (5′-GCTGGATGGAGAACGACTTC-3′) and JB13416 (5′-TTCAGCTCCATCAGCTCCTT-3′); for ORFeus-Hs L1 ORF2, JB13417 (5′-CTGATCAGCCGCATCTACAA-3′) and JB13418 (5′-TGGTCTTGATCTGCATCTCG-3′); for PABPN1, JB13371 (5′-CTCGAGTCAGGGAGATGGAG-3′) and JB13372 (5′-ACACGGTTGACTGAACCACA-3′); for PABPC1, JB13377 (5′-TTGGAGCTAGGGCAAAAGAA-3′) and JB13378 (5′-TTTGCGCTTAAGTTCCGTCT-3′); and for PAIP2, JB13892 (5′-AAGATCCAAGTCGCAGCAGT-3′) and JB13893 (5′-CCTCTTCCAGCATTTCTTGG-3′). The gene knockdown efficiency was calculated by the 2−ΔΔCT method after normalization to the beta-actin mRNA level. A minimum of three biological and three technical replicates for each gene was included, and the standard error was calculated.
Poly(A) tail length assay.
The poly(A) tail length assay was performed using a poly(A) tail length assay kit (Affymetrix, Cleveland, OH) according to the manufacturer's protocol. G/I tailing was performed with 1 μg total RNA, 4 μl 5× tail buffer mix, 2 μl 10× tail enzyme mix, and water to a total volume of 20 μl, and the reaction mix was incubated at 37°C for 60 min. Next, 2 μl 10× tail stop solution was added to stop the G/I tailing reaction. For reverse transcription, 2 μl G/I-tailed RNA sample, 4 μl 5× RT buffer mix, and 2 μl RT enzyme mix were mixed in a total of 20 μl and incubated at 44°C for 60 min and 92°C for 10 min, after which the mix was held at 4°C. The reverse transcription sample was diluted with water to 40 μl, and then 2 μl diluted RT sample was used in a 25-μl PCR mixture containing 5 μl 5× PCR buffer mix, 1 μl 10 μM universal PCR reverse primer, 1 μl 10 μM gene-specific forward primer, and 1.25 units HotStart-IT Taq DNA polymerase. PCR conditions were as follows: 94°C for 2 min; 35 cycles of 92°C for 10 s, 58°C for 30 s, and 72°C for 30 s; 72°C for 5 min; and a hold at 4°C. Five microliters of PCR product was resolved in a 2% agarose gel and stained with ethidium bromide.
The primers used were as follows: for L1 RNA, the gene-specific forward primer JB14067 (5′-GGATCCAGACATGATAAGATACATTGATGA-3′); for actin RNA, the actin forward primer included in the kit; for glyceraldehyde-3-phosphate dehydrogenase (GAPDH) RNA, JB11565 (5′-GACCCTCACTGCTGGGGAGTCC-3′); and for keratin RNA, JB11568 (5′-CCGCCGGATAGTGGATGGCAAAGTG-3′). Additional primers included the following: A0 primer for L1 RNA, JB14398 (5′-GTTGTTAACTTGTTTATTGCAGCTT-3′); A0 primer for actin, JB15084 (5′-AAGGTGTGCACTTTTATTCAACTGGTCTCAA-3′); A0 primer for GAPDH, JB14399 (5′-GTTGAGCACAGGGTACTTTATTGAT-3′); and A0 primer for keratin RNA, JB14401 (5′-GAACTCTGAACTTTTTATTGGCCTC-3′).
Coimmunoprecipitation.
HEK293T cells were transfected with 1 μg pLD108 by use of Fugene HD (Roche, Indianapolis, IN) according to the manufacturer's manual. The day after transfection, cells were trypsinized, transferred to a 6-cm plate, and selected with puromycin (1 μg/ml) for 3 days. After the puromycin selection, ∼106 cells were collected and lysed in 0.5 ml M-PER buffer (Thermo Scientific, Minneapolis, MN). Cell lysate was obtained by centrifugation for 15 min at 13,000 × g. One hundred microliters of cell lysate was diluted to 500 μl with PBS, and then 5 μl anti-ORF1 IgY, 20 μl protease inhibitor (25×) (Roche, Indianapolis, IN), and 10 μl donkey anti-IgY agarose beads (Gallus Immunotech Inc., Ontario, Canada) were added. The immunoprecipitation (IP) mix was incubated at 4°C overnight with rotation. The next day, the beads were washed 3 times with PBS and resuspended in 30 μl SDS-PAGE loading buffer. The sample was boiled for 10 min, and 5 μl was loaded on the gel. For treatment with nuclease, nuclease was first added to the diluted cell lysate and incubated at 37°C for 30 min before adding the antibody and agarose beads.
Immunoblot assays.
After a 3-day puromycin selection, transfected HEK293T cells were lysed in M-PER buffer (Thermo Scientific, Rockford, IL) and centrifuged for 15 min at 13,000 × g to clarify the cell lysate. Clarified cell lysate (10 μl) was mixed with 10 μl 2× loading buffer (0.1 M Tris-HCl, pH 6.8, with 4.0% SDS, 20% glycerol, 5% β-mercaptoethanol, and 0.2% bromophenol blue), and samples were separated in 4% to 20% SDS-PAGE gels. After transfer to nitrocellulose membranes, membranes were probed with anti-ORF1p IgY antibody. Western blots were developed with ECL-Plus detection reagent (GE Healthcare, Piscataway, NJ), which was detected using an imaging system (LAS3000 instrument [Fujifilm] and Image Reader LAS-3000 software) on the “high” setting. The signals in the electronic file were quantified using Multi-Gauge software (Fujifilm), a program based on band density. Only nonsaturated signals were quantified, and the background was subtracted. Results were normalized by using the tubulin controls as a reference and are presented as fold differences relative to the control. A minimum of two Western blots were performed, and a representative blot is shown.
LEAP assay.
The L1 element amplification protocol (LEAP) was performed according to the method of Kulpa and Moran (44). Briefly, HEK293T cells were transfected with L1 expression plasmid (1 μg/400,000 cells) by use of Fugene HD (Roche Applied Science, Indianapolis, IN) according to the manufacturer's protocol. The day after transfection, cells were trypsinized and then selected on puromycin at 1 μg/ml for 2 weeks. On the day of harvest, ∼800 million cells were washed with PBS three times and then resuspended in 10 ml cold PBS. Cells were pelleted at 3,000 × g for 5 min in a swinging-bucket rotor and then lysed with 1 ml buffer (1.5 mM KCl, 2.5 mM MgCl2, 5 mM Tris-HCl, pH 7.5, 1% deoxycholic acid, 1% Triton X-100, 1× protease inhibitor cocktail) for 5 min on ice. The lysate was clarified by centrifugation at 3,000 × g for 5 min at 4°C, and the supernatant was transferred to an 8.5% to 17% sucrose cushion (4 and 6 ml, respectively). The gradient was spun at 39,000 rpm (SW40.1 rotor; 178,000 × g) for 2 h at 4°C. The pellet was resuspended in 5 mM Tris-HCl (pH 7.5) with 1× protease inhibitor, with glycerol added to a final concentration of 50%. RNP preparations were stored at −80°C.
For the LEAP reaction, various amounts of RNP were added to 50-μl reaction mixes containing 50 mM Tris-HCl, pH 7.5, 50 mM KCl, 5 mM MgCl2, 10 mM dithiothreitol, 0.4 μM 3′ LEAP primer (JB11560 [5′-GCGA2GCACAGAATTAATACGACTCACTATAGGTTTTTTTTTTTT-3′]), 20 units RNasin, a 0.2 mM concentration of each deoxynucleoside triphosphate (dNTP), and 0.05% Tween 20 and incubated at 37°C for 1 h. The LEAP reaction product (1 μl) was used as the template in a 50-μl PCR mix with 5 μl 10× PCR buffer (Roche Applied Science, Indianapolis, IN), 5 μl dNTPs (2.5 mM concentration of each), 1 μl each of primers JB11564 (5 μM) (5′-GCGAGCACAGAATTAATACGACTC-3′) and JB14067 (5 μM) (5′-GGATCCAGACATGATAAGATACATTGATGA-3′), and 1 μl FastStart Taq polymerase (Roche Applied Science, Indianapolis, IN). The reaction was carried out with an initial denaturation at 94°C for 5 min, followed by 35 cycles of 94°C for 30 s, 56°C for 30 s, and 72°C for 30 s and a final extension at 72°C for 7 min. An aliquot (10 μl) of each PCR product was loaded onto a 1.5% agarose gel. Band density was quantified using Multi-Gauge software.
Puromycin sensitivity assay.
HEK293T cells were cotransfected with L1 expression plasmid and PABP expression plasmid in six-well plates (1 μg L1 plasmid plus 1 μg PABP plasmid plus 10 ng pCAG-eGFP/40,000 cells). The following day, cells were treated with trypsin, and an equal number of cells was plated in a 96-well plate (∼10,000 cells/well) with puromycin at 2 μg/ml or without puromycin selection. Another portion of the cells was analyzed by FACS analysis to determine the transfection efficiency. Three days later, 20 μl cell viability solution (CellTiter-Blue; Promega, Madison, WI) was added to each well. The plate was incubated at 37°C for 2 h and then read using a microplate reader with an excitation wavelength of 550 nm and an emission wavelength of 600 nm. The values obtained from coexpression of L1 and empty pcDNA were set as 100%. Four independent transfections were performed for each construct, and all values were normalized to the transfection efficiency acquired by FACS analysis.
Cell fractionation.
Nuclear and cytoplasmic fractions were prepared using NE-PER reagent (Thermo Scientific, Minneapolis, MN) according to the manufacturer's instructions.
RESULTS
shRNA screening for host functions involved in L1 retrotransposition.
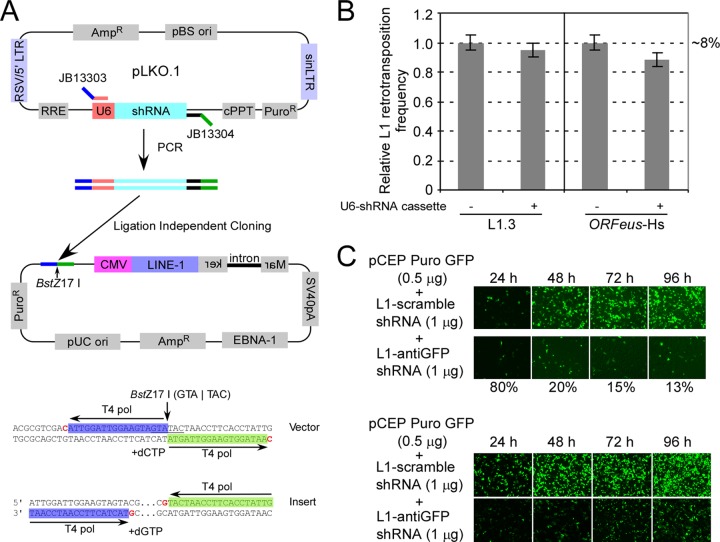
To evaluate host functions involved in the L1 life cycle, we knocked down individual genes by using shRNA in an established cell culture-based L1 retrotransposition assay. To make such screening efficient, we first engineered our pCEP4-based L1 retrotransposition vector by cloning an LIC adaptor sequence between the puro gene and the CMV promoter (3).
As illustrated in Fig. 1A, a unique BstZ17I restriction site is located in the middle of this adaptor fragment, containing two “C-less” 18-bp sequences (blue and green). These two sequences can be removed upon BstZ17I digestion and the action of the (3′-to-5′) exonuclease activity of T4 DNA polymerase in the presence of dCTP. Similarly, two 18-bp overhangs can be produced on the PCR insert in the presence of T4 DNA polymerase and dGTP. We PCR amplified the U6 promoter and shRNA cassette from various TRC shRNA clones by using a pair of universal primers (Fig. 1A). The U6-shRNA fragments were subcloned into a modified L1 retrotransposition vector by ligation-independent cloning, with a >95% success rate (see Materials and Methods for details). Using this strategy, the L1 element and shRNA are delivered in a single plasmid throughout the L1 retrotransposition assay, and the cells maintain a constant ratio of shRNA to L1 expression cassette (and thus, presumably, gene products) in every cell. The insertion of a control empty U6 cassette (no shRNA cloned between U6 and the terminator) had no or minimal effects on the activity of either native L1 (L1.3) or a synthetic human L1 element (ORFeus-Hs) (2) (Fig. 1B). shRNA cassettes cloned in this manner were capable of efficiently knocking down the eGFP gene shortly after transfection into both HeLa and HEK293T cells (Fig. 1C).
Fig 1.
(A) (Top) Strategy of subcloning U6-shRNA cassette from TRC shRNA plasmid into L1 retrotransposition vector. Blue and green lines represent the adaptor sequences for LIC cloning. (Bottom) Sequence of LIC adaptor sequence used in this study. The adaptor includes sequences highlighted with blue and green and a BstZ17I site (underlined). Residues in red represent the boundaries of overhangs after T4 polymerase treatment. (B) Relative L1 retrotransposition efficiencies, with and without U6-shRNA cassette, for native human L1.3 and synthetic human L1 (ORFeus-Hs). The absolute value of L1 activity from the vector without the U6-shRNA cassette was 8% and was arbitrarily set at 1 for comparison. Six independent experiments were done, and standard errors are shown. (C) (Top) Subcloned anti-eGFP shRNA efficiently knocked down eGFP expression in HeLa cells. (Bottom) Subcloned anti-eGFP shRNA efficiently knocked down eGFP expression in HEK293T cells. Cells were cotransfected with pCEP-puro GFP (0.5 μg) and pLD108-shRNA (1.0 μg) plasmids, and eGFP expression was monitored at different time points until 96 h posttransfection. The numbers at the bottom represent relative fluorescence signals of the bottom row (with anti-eGFP shRNA) compared to the top row (with scrambled shRNA).
In this study, we used the synthetic human L1 element ORFeus-Hs in all subsequent analyses, because its elevated expression of RNA and proteins compared to that with native L1 gave a broader dynamic signal range for investigation (2). Since the ORFeus-Hs sequence is comprehensively recoded (40% different from L1RP at the DNA sequence level), it eliminates the background signal from endogenous L1 RNA. We also performed immunoblots to probe L1 ORF1p in cell lysates collected from untransfected and transfected HEK293T cells. Cells transfected with the L1 expression plasmid produced >100-fold more ORF1p than that in untransfected cells, a background easily subtracted from our strong signal (data not shown).
Two poly(A) binding proteins affect LINE-1 retrotransposition.
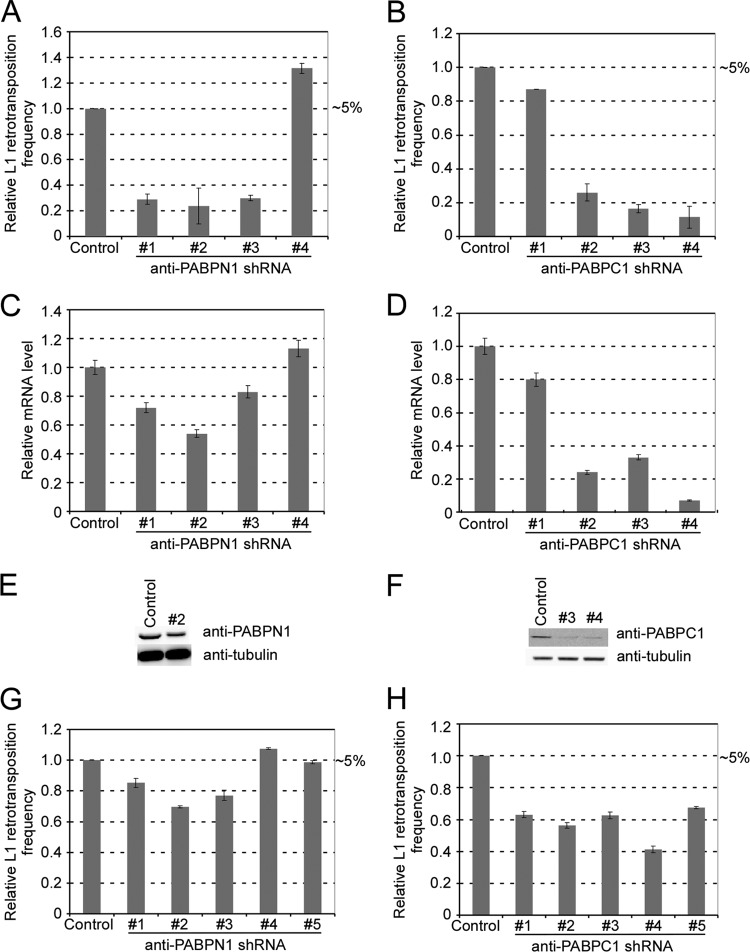
In the process of screening a number of candidate genes, we identified two poly(A) binding proteins, “nuclear” poly(A) binding protein 1 (PABPN1) and “cytoplasmic” poly(A) binding protein 1 (PABPC1), as important for L1 retrotransposition. As shown in Fig. 2A, three of four shRNA clones targeting PABPN1 effectively reduced L1 retrotransposition efficiency (in the neoAI reporter assay) to 20 to 30% of that for the control sample (pLD108-anti-eGFP), which contains an shRNA targeted against eGFP. Similarly, knocking down the PABPC1 gene decreased L1 activity to 15 to 25% of the control level (Fig. 2B). Similar results were obtained using pLD190 (eGFPAI marker) in HEK293T cells (data not shown) when pLD190-scramble shRNA was used as a control. To evaluate the knockdown efficiency of shRNA, we extracted RNAs from cells after 3 days of puromycin selection and performed real-time RT-PCR to measure endogenous mRNA levels of PABPN1 and PABPC1 in the transfected cells. As seen by comparing Fig. 2C and D to Fig. 2A and B, respectively, L1 transposition activity was consistently well correlated with the mRNA levels of these two genes. We chose shRNA 2 (TRCN0000000121) for PABPN1 (N1#2) and shRNAs 3 (TRCN0000074640) and 4 (TRCN0000074641) for PABPC1 (C1#3 and C1#4) for further analyses, since they most effectively knocked down their targets. Immunoblot results from the same batch of cells indicated that cells transfected with these three constructs had reduced levels of PABPN1 (69% of wild-type level) and PABPC1 (19% of wild-type level for C1#3 and 24% of wild-type level for C1#4) (Fig. 2E and F).
Fig 2.
PABPN1 and PABPC1 affect L1 retrotransposition in HeLa cells. (A) Relative L1 retrotransposition with the PABPN1 gene knocked down. Control, pLD108-anti-eGFP shRNA. shRNAs 1 to 4, pLD108-antiPABPN1 shRNAs. The absolute value of the L1 retrotransposition level of the control was 5% and was arbitrarily set at 1 for comparison. (B) Relative L1 retrotransposition with the PABPC1 gene knocked down. Control, pLD108-anti-eGFP shRNA. shRNAs 1 to 4 are vector pLD108-antiPABPC1 shRNAs. The absolute value of the L1 retrotransposition level of the control was 5% and was arbitrarily set at 1 for comparison. (C) Relative PABPN1 mRNA levels in cells transfected with the constructs used for panel A. After 3 days of puromycin selection, RNAs were extracted and PABPN1 mRNA levels were measured by real-time RT-PCR and normalized by the simultaneous measurement of the beta-actin gene. (D) Relative PABPC1 mRNA levels in cells transfected with the constructs used for panel B. After 3 days of puromycin selection, RNAs were extracted and PABPC1 mRNA levels were measured by real-time RT-PCR. The mRNA levels were normalized by the simultaneous measurement of the beta-actin transcript. (E) PABPN1 protein level in cells transfected with pLD108-antiPABPN1#2. Cell lysates were obtained after 3 days of puromycin selection and probed with anti-PABPN1 antibody. (F) PABPC1 protein levels in cells transfected with pLD108-antiPABPC1#3 and pLD108-antiPABPC1#4. Cell lysates were obtained after 3 days of puromycin selection and probed with anti-PABPN1 antibody. (G) L1 retrotransposition efficiencies in HeLa cell pools preinfected with lentiviruses containing anti-PABPN1 shRNAs 1 to 5. The absolute value of the L1 retrotransposition level in the control was 5% and was arbitrarily set at 1 for comparison. (H) L1 retrotransposition efficiencies in HeLa cell pools preinfected with lentiviruses containing anti-PABPC1 shRNAs 1 to 5. The absolute value of the L1 retrotransposition level in the control was 5% and was arbitrarily set at 1 for comparison. All data represent the averaged results for at least three independent experiments. Error bars represent standard deviations.
For validation, we also preinfected HeLa cells with lentivirus preparations containing the same set of shRNA clones against PABPN1 and PABPC1 and then obtained individual cell pools. Transductants were selected in the presence of puromycin, which tags the pLKO.1 vector used. Similar L1 retrotransposition assays were done using individual preinfected cell pools, except that a hygromycin marker was used in the L1 retrotransposition construct (JM101/L1.3) (56). L1 retrotransposition was inhibited in these transductants, and the extent of knockdown was correlated with the data acquired in the plasmid assay (compare Fig. 2G and H to Fig. 2A and B, respectively). Interestingly, overall, the shRNA cassettes cloned into the plasmid produced a substantially larger impact on the L1 retrotransposition activity than the lentivirally transduced/integrated shRNA copies, presumably due to the fact that the pCEP4-based plasmid maintains multiple copies in each cell (71).
Since PABPs may be associated with a large number of cellular mRNAs and potentially affect their expression, we evaluated whether knocking down these two genes had any effect on cell viability, intron splicing, or integrated marker expression rather than L1 activity. We transfected the cells with L1-shRNA coexpression constructs and selected transfectants with puromycin. At 3 days posttransfection, we seeded the same number of Puror cells in puromycin-containing medium and counted the colonies 2 weeks later (Table 1). With various numbers of starting Puror cells seeded, no defect in efficiency of plating (viability) was found when the two PABP genes were knocked down.
Table 1.
HeLa cell colony formation ability with PABPs knocked down
| shRNA | Mean no. of colonies ± SEb |
||
|---|---|---|---|
| 1,000 cells | 500 cells | 200 cells | |
| Controla | 373 ± 31 | 215 ± 10 | 99 ± 5 |
| Anti-PABPN1 shRNA#2 | 368 ± 20 | 236 ± 11 | 95 ± 6 |
| Anti-PABPC1 shRNA#3 | 397 ± 25 | 212 ± 14 | 107 ± 12 |
| Anti-PABPC1 shRNA#4 | 487 ± 10 | 362 ± 38 | 174 ± 17 |
pLD108-anti-eGFP shRNA.
For the indicated total number of Puror cells seeded on the plate. Data are for three independent experiments.
To examine whether knockdown of PABPs had any effect on the capability of the cells to generate neomycin-resistant colonies, we first generated a neomycin-resistant cell pool by an L1-neoAI retrotransposition assay using JM101/L1.3 (56), which is hygromycin resistant but carries no shRNA. This pool was amplified and subsequently transfected with L1-shRNA coexpression plasmids. After a 3-day puromycin selection, the same number of Puror cells were seeded for colony formation in G418 medium (Table 2). The same analysis was done using a cell pool pretransfected with an L1 eGFPAI reporter (L1RP/eGFPAI) in HEK293T cells (Fig. 3A). We found that knockdown of PABPN1 had a negative effect on the ability of the cells to generate Neor colonies (∼1/3 of the control level) but not eGFP expression. A decreased level of PABPC1 did not interfere with either retrotransposition reporter gene postintegration.
Table 2.
neo marker gene expression with PABPs knocked down
| shRNA | Mean no. of colonies ± SEb |
|
|---|---|---|
| 1,000 cells | 500 cells | |
| Controla | 306 ± 14 | 163 ± 10 |
| Anti-PABPN1 shRNA#2 | 113 ± 8 | 66 ± 15 |
| Anti-PABPC1 shRNA#3 | 315 ± 14 | 158 ± 14 |
| Anti-PABPC1 shRNA#4 | 387 ± 19 | 244 ± 18 |
pLD108-anti-eGFP shRNA.
For the indicated total number of Neor cells seeded on the plate. Data are for three independent experiments.
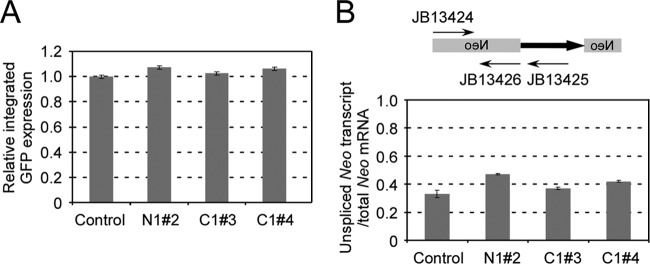
Fig 3.
(A) Effect of shRNA cassette on expression of integrated eGFP marker. A pool of HEK293T cells that had an integrated eGFP marker was first acquired by L1 retrotransposition using L1RP-eGFP. The cells were then transfected with pLD108 containing different shRNA cassettes and analyzed by FACS analysis 1 week after transfection. eGFP expression of the control (pLD108-scramble) was set at 100%. A total of six independent experiments were done, and standard errors are shown. (B) Effect of shRNA on the splicing ability of the globin intron within the neo marker. Total RNAs were extracted from HeLa cells transfected with different vectors after 3 days of puromycin selection. Real-time RT-PCR was performed using two exon primers (JB13424 and JB13426) or exon-intron primers (JB13424 and JB13425) for the neoAI reporter. The ratio of unspliced RNA and total RNA was calculated. Three experiments were performed, and standard errors are shown.
The gamma globin intron interrupting the L1 reporter genes ensures that L1 retrotransposition events occur only through an RNA intermediate (56). Polyadenylation and splicing are known to be correlated processes in some cases and are both thought to occur on the C-terminal domain of Pol II immediately cotranscriptionally (63). We therefore examined whether poly(A) binding proteins might have negative effects on intron splicing efficiency, which could decrease the number of observed G418r colonies as an artifact. We designed a real-time RT-PCR strategy to evaluate the splicing efficiency of this intron when the PABPN1 or PABPC1 gene was knocked down. As shown in Fig. 3B, the splicing ability was decreased by ∼30% when PABPN1 was knocked down and ∼5 to 15% when PABPC1 was knocked down. In summary, knockdown of PABPN1 had negative effects on colony formation ability (neo) and intron splicing ability that may account for the decreased “apparent” L1 retrotransposition activity. PABPC1 knockdown had no effect on cell viability and marker expression. Although intron splicing ability was affected by PABPC1 knockdown, this effect (5 to 15%) cannot account for a 4-fold change of retrotransposition activity, suggesting a more specific involvement of this factor in the L1 life cycle.
PABP knockdown has minimal effects on L1 poly(A) tail length or expression.
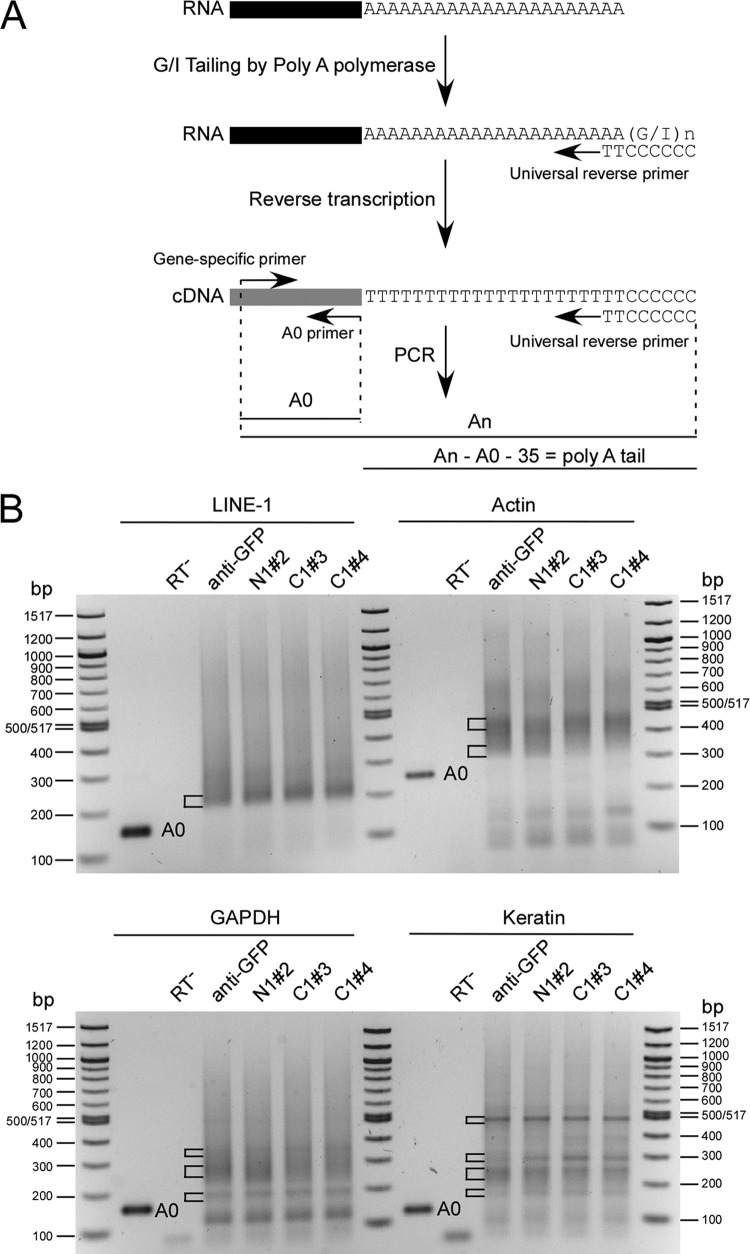
We extracted total RNA from Puror (5 days) cells and measured the L1 mRNA poly(A) tail length by using a poly(A) tail length assay kit. As shown in Fig. 4A, the poly(A) tail lengths of the genes of interest were determined as the sizes of PCR-amplified poly(A) products minus the calculated length from the gene-specific forward primer to the putative polyadenylation start site (A0) and the length of the universal primer (35 nt). For L1 mRNA, all samples tested showed a minimum poly(A) tail length of ∼28 to 40 nt, seen as a strong band in each lane of Fig. 4B. This could account for mRNA that has gone through the translation and deadenylation processes, making it especially favorable for PCR amplification. The smears above the major band represent L1 mRNAs with a longer poly(A) tail. No obvious shortening of the poly(A) tail was observed when either PABP was knocked down in the cell. We also evaluated the poly(A) tails of three housekeeping genes (actin, GAPDH, and keratin), using the same set of RNA samples. Most beta-actin mRNA transcripts contained poly(A) tails of ∼35 nt (bottom bracket) to ∼112 nt (top bracket), with a peak at ∼112 nt. For GAPDH, there was a distinct population of mRNAs that contained an ∼13-nt poly(A) tail (bottom bracket), and the majority of transcripts contained a poly(A) tail with a length of ∼80 nt (middle bracket) to ∼200 nt (top bracket). The poly(A) tail length for keratin mRNA started at ∼32 nt (bottom bracket) and displayed a strong band at ∼317 nt (top bracket). There were also two populations of mRNAs containing poly(A) tails of ∼115 nt (second bracket from bottom) and ∼161 nt (third bracket from bottom). Importantly, we did not observe any change in the poly(A) tail lengths of any of these genes when PABPs were knocked down (Fig. 4B).
Fig 4.
Effects of knocking down PABPs on the poly(A) tail length of L1 and other genes. (A) Scheme of poly(A) tail length assay, redrawn from the user protocol of a poly(A) tail length assay kit (Affymetrix). The A0 primer represents the reverse primer located just before the poly(A) tail for each gene of interest. (B) Poly(A) tail lengths measured for L1, actin, GAPDH, and keratin mRNAs. Total RNAs were extracted from HeLa cells (after 5 days of puromycin selection) transfected with pLD108-anti-eGFP, pLD108-antiPABPN1#2, pLD108-antiPABPC1#3, and pLD108-antiPABPC1#4, and poly(A) tail lengths were measured by use of a poly(A) tail length assay kit. A0 products are unique-sequence PCR products which correspond to the appropriate mRNA and lack any 3′ poly(A) tail. The exact sizes of A0 products are as follows: for L1, 142 bp; for actin, 223 bp; for GAPDH, 153 bp; and for keratin, 122 bp. “An” represents PCR products containing mRNAs with different poly(A) tail lengths. The inferred length of the poly(A) tail was determined as the length difference between An and the sum of the lengths of A0 and the universal reverse primer (35 nt).
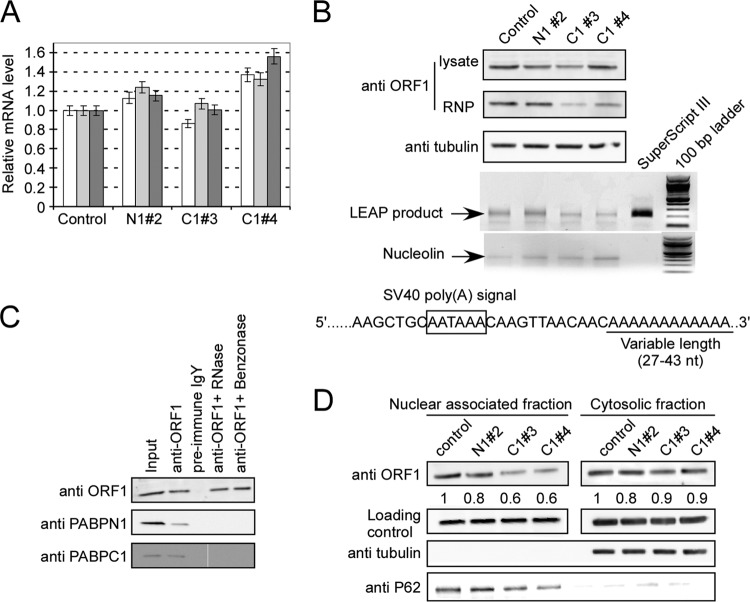
Next, we examined whether knocking down PABPs affected L1 expression. We measured the steady-state RNA levels of puro, ORF1, and ORF2 expressed from the plasmid by using real-time RT-PCR. No defect in L1 transcription was observed in cells with PABPN1 or -C1 knocked down (Fig. 5A). Given the fact that PABPC1 binds the poly(A) tail in the cytoplasm and is important for translation initiation, we probed for ORF1p in whole-cell lysates prepared from Puror cells transfected with various L1-shRNA coexpression plasmids. We found that knockdown of both PABPs decreased L1 ORF1p expression by 13 to 30% (for N1#2, ∼20%; for C1#3, ∼30%; and for C1#4, ∼13%) (Fig. 5B, top panel). For PABPC1, the level of ORF1p produced in the cell was lower than its own knockdown efficiency (81% and 76%, respectively) (Fig. 2F).
Fig 5.
Effects of knocking down PABPs on expression of L1. (A) Relative mRNA levels of Puro transcript; L1 ORF1 and ORF2 levels were measured by real-time RT-PCR. White bars, Puro; light gray bars, ORF1; dark gray bars, ORF2. The mRNA level of pLD108-anti-eGFP was used as a control and set at 1. (B) Expression of L1 ORF1p. Whole-cell lysates prepared from HEK293T cells (after 5 days of puromycin selection) transfected with different L1-shRNA coexpression vectors were probed with anti-ORF1 antibody. The RNP samples were prepared from the same amount (500 μl) of whole-cell lysate, and the pellets were resuspended in 50 μl PBS. Ten-microliter RNP samples were loaded and probed with the anti-ORF1 antibody. The same amount of RNP sample (2 μg) was used in the LEAP assay, and 10 μl of PCR product from each reaction was loaded onto a 1.5% agarose gel. The nucleolin mRNA panel represents the SuperScript RT-PCR product of this cellular mRNA marker for all RNP preparations. The LEAP signal is shown as a semiquantitative readout of the ability of the L1 RNP to produce its own cDNA. Sequences of LEAP products indicated that the cloned products contained poly(A) tails of 27 to 43 nt. (C) Both PABPN1 and PABPC1 bind to ORF1p through L1 RNA. Cell lysate prepared from HEK293T cells transfected with pLD108 was immunoprecipitated with anti-ORF1 IgY and probed with rabbit anti-ORF1p, anti-PABPN1, and anti-PABPC1 antibodies. Lane 1, input; lane 2, anti-ORF1 IgY was used for co-IP; lane 3, preimmune IgY antibody was used for co-IP; lane 4, cell lysate was pretreated with RNase before co-IP with anti-ORF1 IgY; lane 5, cell lysate was pretreated with Benzonase before co-IP with anti-ORF1 IgY. (D) Localization of ORF1 protein. Nucleus-associated and cytosolic fractions were prepared from HEK293T cells (after 5 days of puromycin selection) transfected with different L1-shRNA coexpression plasmids and probed for anti-ORF1 antibody. Alpha-tubulin and p62 (nucleoporin 62) were used as quality control markers for cell fractionations, and p53 was used as a loading control. The numbers under the top panels represent the relative ORF1p abundance in each lane compared to the control. All experiments were performed at least twice, and representative gels are shown.
PABPC1 is important for L1 RNP formation.
We prepared L1 RNP complexes from PABP knockdown cells and evaluated whether PABPs were important for RNP formation. As shown in Fig. 5B (middle panel), the relative level of ORF1p in the RNP complex was not changed by PABPN1 knockdown but was diminished by a greater degree than in the whole-cell lysate (∼64% compared to ∼30% for C1#3 and 38% compared to ∼13% for C1#4) when the PABPC1 level was knocked down. We could not obtain quantifiable data for ORF2p by immunoblotting. To assess ORF2's endogenous reverse transcription function, we conducted LEAP assays using the same RNP preps. As shown in Fig. 5B, RNPs prepared from HEK293T cells with PABPC1 knocked down produced about 50% of the signal seen with the control when equal amounts of RNP (2 μg) were added to the reaction mixture (for PANPN1#2, 114%; for PABPC1#3, 53%; and for PABPC1#4, 47%). We cloned and sequenced 5 clones of LEAP products from each lane. Sequencing results revealed that they had the expected structure (i.e., they were derived from L1 RNA) and contained a poly(A) tail of 27 to 43 A's.
These results prompted us to investigate the subcellular localization of ORF1p. We separated the nucleus-associated and soluble cytosolic fractions and detected the distribution of ORF1p. As shown in Fig. 5D, the ORF1 protein was distributed about equally in both fractions of the PABPN1 knockdown cells, just like the wild-type control. Thus, PABN1 levels do not appear to affect ORF1p intracellular localization. Conversely, in the PABC1 knockdown cells, ORF1p was underrepresented in the nucleus-associated fraction, possibly because its association with the RNP complex is required for translocation to or across the nuclear envelope. The ORF1p level in the cytosolic fraction was not noticeably changed when PABPC1 was knocked down.
We performed a co-IP experiment using anti-ORF1p antibody and identified that both poly(A) binding proteins are associated with the L1 RNP complex. This binding appears to be mediated by RNA, presumably L1 RNA, as it was reversed by prior addition of RNase to the IP reaction mix (Fig. 5C). These findings strongly support the notions that both PABPs are components of L1 RNP complexes and that the PABPC1 protein is critical for RNP formation and may be involved in its nuclear import.
Poly(A) tail length is important for L1 RNP formation.
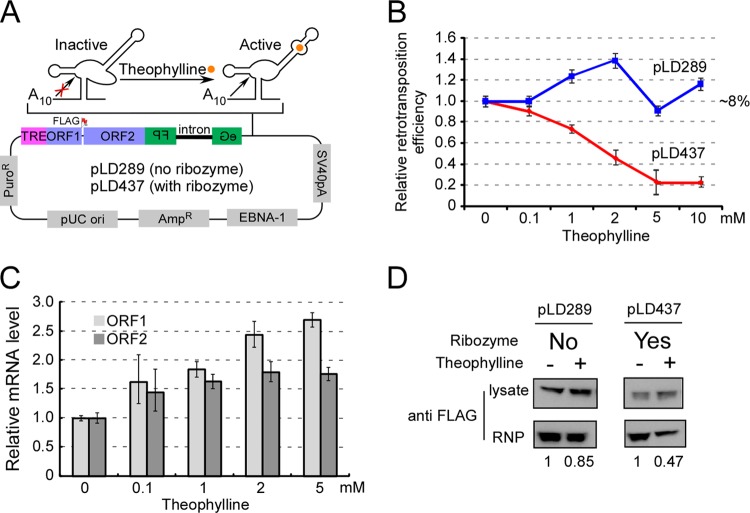
To validate the results mentioned above, we introduced an aptamer-controlled self-cleaving ribozyme into an L1 retrotransposition construct (Fig. 6A). As shown by Win and Smolke, this ribozyme becomes active and cleaves at a specific location when the ligand (theophylline) is present (74). We included 10 A's before the cleavage site so that the cleaved L1 mRNA would contain an oligo(A) tail that was ∼1/4 to 1/3 as long as the wild-type poly(A) tail determined by our LEAP assay data (Fig. 5B). While the retrotransposition ability of the control construct (pLD289 without the ribozyme) was largely unchanged by theophylline treatment, the L1 construct with the ribozyme cassette (pLD437) showed decreased activity with increasing concentrations of theophylline (Fig. 6B). We collected the cells (transfected with pLD437) at various theophylline concentrations and measured the L1 mRNA level by real-time PCR. The L1 mRNA level was not decreased using either an oligo(dT) primer (Fig. 6C) or a random hexamer primer (not shown) for reverse transcription.
Fig 6.
Reduced poly(A) tail length affects L1 RNP formation. (A) Diagram of an L1 retrotransposition construct with a hammerhead ribozyme-aptamer controlled by theophylline. A self-cleavage ribozyme-aptamer (74) was inserted between the eGFPAI marker and the simian virus 40 (SV40) poly(A) signal that normally specifies polyadenylation in this construct. Upon adding theophylline to the medium, the ribozyme cleaves off the SV40 poly(A) signal and therefore leaves only a short oligo(A) tail (A10). Orange circle, theophylline; TRE, tetracycline-regulated promoter; FLAG, FLAG epitope tag at C terminus of ORF1p; pLD289, L1 construct without the ribozyme cassette inserted; pLD437, L1 construct with the ribozyme cassette inserted. (B) Retrotransposition ability of pLD289 and pLD437 in HEK293T-tet-on cells with different concentrations of theophylline. The absolute activity of pLD289 (∼8%) was set at 1. (C) Relative mRNA levels of L1 ORF1 and ORF2 in Tet-on HEK293T cells (transfected with pLD437) with various concentrations of theophylline. Tet-on HEK293T cells were transfected with pLD437. The next day, doxycycline (final concentration, 500 ng/ml) and theophylline were added to the medium. At 5 days posttransfection, total RNA was prepared and real-time RT-PCR was performed. Both the beta-actin and puromycin genes were used as internal controls. All data represent averaged results for at least three independent experiments. Error bars represent standard deviations. (D) A shorter poly(A) tail affects L1 RNP formation. Tet-on HEK293T cells were transfected with pLD289 and pLD437. Transfected cells were selected in DMEM containing 1 μg/ml puromycin and 500 ng/ml doxycycline for 5 days. Theophylline was added to a final concentration of 10 mM and incubated for 2 more days. L1 RNPs were prepared from each cell pool and probed with anti-FLAG antibody. The signals were quantified using Multi-Gauge software (Fujifilm), based on band densitometry. For each sample pair, the signal with theophylline was compared to the signal without theophylline (assigned as 1), with normalization to signals from the total cell lysate (upper panel). The experiment was done twice from two independent transfections and cell cultures.
We then evaluated the RNP formation ability of L1 RNA with a shorter poly(A) tail. Two cell pools were first generated by transfecting pLD289 and pLD437 into Tet-on HEK293T cells. Both plasmids contained ORFeus-Hs with a FLAG tag at the C terminus of ORF1, which does not interfere with retrotransposition (data not shown). These two pools were amplified for 5 days in the presence of puromycin (1 μg/ml) and doxycycline (500 ng/ml), which induced L1 repression. Fresh medium containing theophylline only (10 mM), with no doxycycline, was then added to induce ribozyme cleavage and shut down L1 transcription. Two days later, cell lysate and RNPs were prepared according to the method of Kulpa and Moran (44) and probed with anti-FLAG antibody. As shown in Fig. 6D, the ability of L1 without ribozyme (pLD289) to form an effective RNP complex was decreased by 15% by adding theophylline (presumably this modest decrease was due to a nonspecific effect of the theophylline). In contrast, in the presence of both ribozyme and theophylline [pLD437, resulting in L1 mRNA with a shorter poly(A) tail], the RNP formation ability was reduced to 48% of the no-theophylline control level.
Effect of PABP overexpression on L1 retrotransposition.
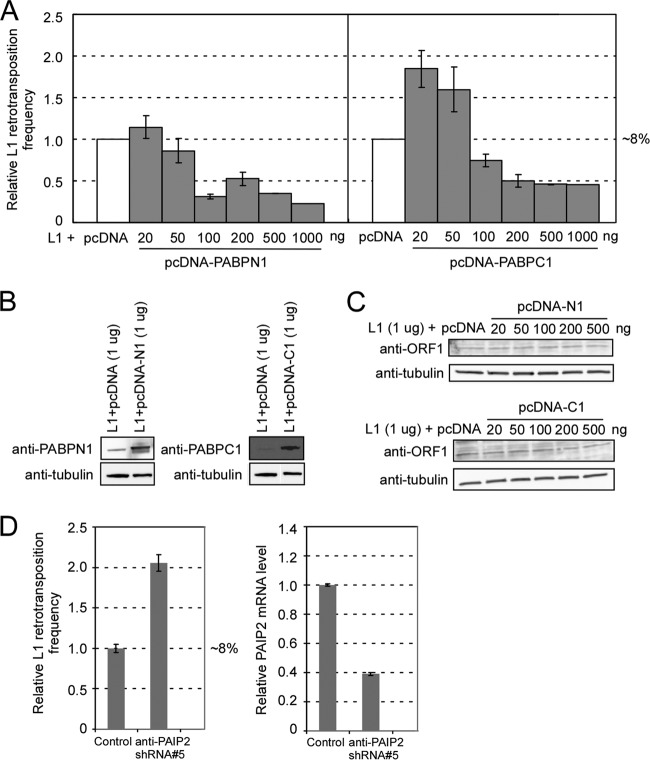
To further probe interactions between the L1 element and poly(A) binding proteins, we cloned PABPN1 and PABPC1 cDNAs into the pcDNA3.1 vector and cotransfected the resulting plasmids with an L1 construct (1 μg:1 μg per 200,000 cells) into HEK293T cells. We found that the protein levels of PABPN1 and -C1 increased by 5- and 25-fold, respectively (Fig. 7B), but overexpression of either protein dramatically and paradoxically decreased the L1 retrotransposition ability, to ∼20% to 50% (Fig. 7A). We then titrated the amounts of PABP expression plasmids from 20 to 500 ng, with a fixed amount of L1 plasmid (1 μg). As shown in Fig. 7A, moderate exogenous expression of the PABP gene (20 ng of PABPN1 or 20 to 50 ng of PABPC1) increased L1 retrotransposition, and this effect was reversed with increased amounts of PABP DNAs (Fig. 7A). The ORF1p expression remained unchanged in response to increased amounts of poly(A) binding proteins (Fig. 7C).
Fig 7.
Effects of overexpression of PABPN1 and PABPC1 on L1 retrotransposition. (A) pLD190 (L1-eGFPAI) (1 μg) was cotransfected with different amounts of pcDNA-PABPN1 (pLD141) or pcDNA-PABPC1 (pLD142), and the retrotransposition efficiency was compared with the result of cotransfection of pLD190 and the same amount of pcDNA3.1 empty vector in HEK293T cells, which was set at 100%. (B) Overexpression of PABPs in HEK293T cells transfected with pcDNA-PABPN1 and pcDNA-PABPC1. Cell lysates were prepared at 3 days posttransfection and probed with anti-PABPN1 and anti-PABPC1 antibodies. Alpha-tubulin was used as an internal control. (C) ORF1p expression was unchanged with PABP overexpression. Cell lysates were prepared at 3 days posttransfection and probed with anti-ORF1 IgY. Alpha-tubulin was used as an internal control. (D) Effect of knocking down PAIP2 on L1 retrotransposition. Control, pLD190-scramble shRNA; anti-PAIP2 shRNA#5, pLD190-antiPAIP2 shRNA#5 (TRCN0000153678). (Left) Relative L1 retrotransposition frequency; (right) relative mRNA level of PAIP2 in the cells. mRNA levels were normalized by the simultaneous measurement of the beta-actin gene.
In high-level overexpression experiments, we observed what we interpret to be a pleiotropic effect, namely, extensive cell death upon puromycin selection for coexpression of poly(A) binding proteins (>200 ng) and L1, a behavior not seen when L1 was expressed alone. Thus, the observed reduction in transposition efficiency with a high level of PABPs was presumably a result of this nonspecific hypersensitivity to puromycin. To investigate this phenomenon, we cotransfected PABP expression plasmids and various L1 vectors, including wild-type L1 and mutant forms, followed by assessment of cell viability under puromycin selection. As shown in Table 3, cells containing retrotransposition-defective L1 mutants and PABPs had the same sensitivity to puromycin, indicating that the effect we observed above was due to unregulated expression of poly(A) binding proteins instead of the combination of L1 and PABPs.
Table 3.
Effects of coexpression of L1 and PABPs on cell sensitivity to puromycin
| L1 construct | Cell viability under puromycin selection relative to control levela |
|
|---|---|---|
| pcDNA-PABPN1 | pcDNA-PABPC1 | |
| ORFeus-Hsb | 0.62 ± 0.07 | 0.62 ± 0.08 |
| ORFeus-Hs EN−c | 0.61 ± 0.07 | 0.63 ± 0.07 |
| ORFeus-Hs RT−d | 0.60 ± 0.03 | 0.52 ± 0.04 |
| ORFeus-Hs EN− RT−e | 0.60 ± 0.06 | 0.55 ± 0.08 |
The value for coexpression of L1 and empty pcDNA was set at 1. Data are means ± standard errors for three independent experiments.
Wild-type ORFeus-Hs cloned into pCEP-puro.
ORFeus-Hs H230A mutant cloned into pCEP-puro.
ORFeus-Hs D702A mutant cloned into pCEP-puro.
ORFeus-Hs H230A D702A mutant cloned into pCEP-puro.
We then attempted to increase PABPC1 activity in a more physiological way by knocking down the expression of poly(A) binding protein interacting protein 2 (PAIP2), an inhibitor of PABPC1 (16, 77). Such knockdown of PAIP2 resulted in an elevated level of L1 retrotransposition, to up to 200% of the control level, presumably due to increased activity of PABPC1 (Fig. 7D).
DISCUSSION
Interactions between transposons and the host involve a complicated and dynamic relationship that is just beginning to be understood. Previous studies suggested that L1 retrotransposition may be influenced by host factors such as proteins in DNA repair pathways, cytidine deaminase, and retinoblastoma proteins (11, 28, 29, 55, 57, 69). However, these findings were obtained using different L1 elements within various backgrounds (i.e., cell types). In this study, we developed a system to examine candidate genes affecting L1 retrotransposition by using a modified L1 plasmid that allows consistent coexpression of an L1 element with either of two distinct retrotransposition reporters and an shRNA of interest. Compared to traditional RNAi screening methods, this strategy fits well with the goal of this study. For example, the standard protocol for TRC shRNA delivery involves lentivirus packaging followed by individual cell pool generation by puromycin selection (2 weeks), requiring an extensive amount of work prior to transfection. Transient knockdown using small interfering RNA (siRNA) requires cotransfection of siRNA and an L1 retrotransposition plasmid. Moreover, the short intracellular half-life of siRNAs is limiting, as some retrotransposition assays take more than 2 weeks to carry out. Having the L1 element and shRNA on the same plasmid completely avoids the preinfection step, simplifies the transfection procedure, and minimizes variation between experiments. Given the fact that pCEP4-based plasmids replicate at ∼30 copies per cell (71), the cloned shRNA is expressed continuously at high levels in the cell throughout the L1 retrotransposition assay period (up to 2 to 3 weeks). Also, even if the copy number varies between individual cells, the ratio of shRNA to L1 RNA and proteins should remain constant, providing an intrinsic normalization. Furthermore, our data indicate that shRNAs cloned into the plasmid produce a more efficient knockdown effect than those integrated into the chromosome by lentivirus infection (Fig. 2G and H). All PCR and LIC cloning steps can easily be done in a 96-well plate format, a method we hope to apply to future high-throughput screening.
In an attempt to identify host factors affecting L1 retrotransposition, two poly(A) binding protein genes (PABPN1 and PABPC1) were discovered. We did not observe significant generic growth or expression defects in the cells when these two genes were knocked down by shRNA. Interestingly, the poly(A) tail length of L1 transcripts was not affected by knockdown of either of the PABPs. Knockdown (by 76% to 81%) of PABPC1 resulted in ∼13 to 30% inhibition of L1 ORF1p expression. This finding is consistent with results obtained by Yoshida et al., who observed a 10 to 15% decrease of reporter gene (luciferase) translation when PABPC1 was knocked down by RNAi (77). It is believed that PABPC1 is regulated by the stability of its interacting protein, PAIP2. Depletion of PABPC1 induces PAIP2 ubiquitylation and degradation, and this in turn releases free PABPC1 molecules originally bound by PAIP2. It is apparent from our work that reducing the amount of PABPC1 associated with L1 mRNA causes a severe and specific defect in L1 RNP formation and cellular localization of ORF1p, either directly or (perhaps) indirectly, by recruiting other host factors less efficiently (30, 78).
Increased L1 retrotransposition by moderate expression of exogenous PABPs strengthens our conclusion that poly(A) binding proteins are important for the L1 life cycle. Interestingly, unregulated overexpression of PABPN1 and PABPC1 caused an apparent decrease in L1 retrotransposition activity. Using wild-type and mutant L1 constructs, we demonstrated that excessive overexpression of both PABPs caused hypersensitivity to puromycin. This is consistent with previous studies which showed that the consequences of unregulated overexpression of PABPN1 and PABPC1 included an increased cell doubling time, a reduced cloning efficiency, and a significantly changed expression level for at least 202 different genes (12, 14, 47, 49).
To test whether moderate and presumably more physiologic increases of PABPs (or their activities) could upregulate L1 activity, we knocked down the PABPC1 inhibitor PAIP2 gene (77) or overexpressed modest amounts of PABPs. As a result of this maneuver, we observed increased L1 retrotransposition. On the other hand, knockdown of another PABPC1-interacting protein, PAIP1 (a protein that stimulates translation [52]), decreased L1 activity (data not shown). Thus, at least for PABPC1, we have clear evidence both that low levels of the protein interfere with L1 retrotransposition and that moderate increases in its level can stimulate L1 retrotransposition.
Proposed function of poly(A) binding proteins in L1 life cycle.
Co-IP experiments indicated that both poly(A) binding proteins are associated with L1 RNP complexes, presumably via the ability of both proteins to bind to L1 mRNA. PABPC1 is known to be generically important to translation initiation. But our study revealed unexpected results: knocking down PABPC1 by shRNA can interfere with retrotransposition, with minimal effects on L1 protein abundance, suggesting novel functions of the PABPC1 protein specific to the L1 life cycle. Less functional RNP was assembled with reduced levels of PABPC1, even though excess ORF1p molecules were produced in the cytoplasm. Since PABPC1 can shuttle between the cytoplasm and the nucleus (1, 37), our data suggest that it may provide a direct trafficking function for L1 RNP nuclear translocation. We speculate that the remaining free L1 proteins and RNA might be degraded in the cytoplasm (30, 67, 79).
The mechanism by which PABPN1 affects L1 retrotransposition is not clear. Based on the data we presented, PABPN1 has a negative effect on neo expression and intron splicing. Since PABPN1 knockdown caused no L1-specific defect, PABPN1 possibly affects L1 activity indirectly through influences on the expression of the neo marker and/or other genes.
Nonautonomous retrotransposition Alu elements rely on the L1 machinery for their mobilization. Since no structural similarity between L1 and Alu RNAs is known, we previously proposed that Alu's poly(A) sequence resembles the L1 RNA poly(A) tail and thus hijacks L1 ORF2 for mobilization (7). Studies have shown that poly(A) tail length is very important to the retrotransposition ability of Alu (6), and poly(A) binding proteins are hypothesized to play an important role in Alu retrotransposition (19, 65). Indeed, we saw decreased Alu retrotransposition with both PABPN1 and PABPC1 knocked down (data not shown), suggesting a more general involvement of poly(A) binding proteins in retrotransposon proliferation.
In summary, our study provides a simple and efficient system for evaluating candidate genes involved in L1 retrotransposition by RNAi and has the potential for use in high-throughput screening. Other than the known functions of binding the poly(A) sequence and initiating translation, PABPC1 appears to play a role in L1 RNP formation and cellular localization of ORF1p.
ACKNOWLEDGMENTS
We thank Nick Conrad for a critical reading of the manuscript and for helpful discussions and Jeffrey Coller for helpful advice on the poly(A) tail length assay.
This research was supported by grant P01CA16519 from the National Cancer Institute and grant R01-GM36481 from the National Institutes of Health to J.D.B.
Footnotes
Published ahead of print 20 August 2012
REFERENCES
- 1. Afonina E, Stauber R, Pavlakis GN. 1998. The human poly(A)-binding protein 1 shuttles between the nucleus and the cytoplasm. J. Biol. Chem. 273: 13015–13021 [DOI] [PubMed] [Google Scholar]
- 2. An W, et al. 2011. Characterization of a synthetic human LINE-1 retrotransposon ORFeus-Hs. Mob. DNA 2: 2 doi:10.1186/1759-8753-2-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Aslanidis C, de Jong PJ. 1990. Ligation-independent cloning of PCR products (LIC-PCR). Nucleic Acids Res. 18: 6069–6074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Athanikar JN, Badge RM, Moran JV. 2004. A YY1-binding site is required for accurate human LINE-1 transcription initiation. Nucleic Acids Res. 32: 3846–3855 doi:10.1093/nar/gkh698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Belancio VP, Whelton M, Deininger P. 2007. Requirements for polyadenylation at the 3′ end of LINE-1 elements. Gene 390: 98–107 doi:10.1016/j.gene.2006.07.029 [DOI] [PubMed] [Google Scholar]
- 6. Bennett EA, et al. 2008. Active Alu retrotransposons in the human genome. Genome Res. 18: 1875–1883 doi:10.1101/gr.081737.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Boeke JD. 1997. LINEs and Alus—the polyA connection. Nat. Genet. 16: 6–7 doi:10.1038/ng0597-6 [DOI] [PubMed] [Google Scholar]
- 8. Bogerd HP, et al. 2006. Cellular inhibitors of long interspersed element 1 and Alu retrotransposition. Proc. Natl. Acad. Sci. U. S. A. 103: 8780–8785 doi:10.1073/pnas.0603313103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Brouha B, Jr, et al. 2003. Hot L1s account for the bulk of retrotransposition in the human population. Proc. Natl. Acad. Sci. U. S. A. 100: 5280–5285 doi:10.1073/pnas.0831042100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Calado A, Kutay U, Kuhn U, Wahle E, Carmo-Fonseca M. 2000. Deciphering the cellular pathway for transport of poly(A)-binding protein II. RNA 6: 245–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chen H, et al. 2006. APOBEC3A is a potent inhibitor of adeno-associated virus and retrotransposons. Curr. Biol. 16: 480–485 doi:10.1016/j.cub.2006.01.031 [DOI] [PubMed] [Google Scholar]
- 12. Corbeil-Girard LP, et al. 2005. PABPN1 overexpression leads to upregulation of genes encoding nuclear proteins that are sequestered in oculopharyngeal muscular dystrophy nuclear inclusions. Neurobiol. Dis. 18: 551–567 doi:10.1016/j.nbd.2004.10.019 [DOI] [PubMed] [Google Scholar]
- 13. Cost GJ, Feng Q, Jacquier A, Boeke JD. 2002. Human L1 element target-primed reverse transcription in vitro. EMBO J. 21: 5899–5910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Davies JE, Sarkar S, Rubinsztein DC. 2008. Wild-type PABPN1 is anti-apoptotic and reduces toxicity of the oculopharyngeal muscular dystrophy mutation. Hum. Mol. Genet. 17: 1097–1108 doi:10.1093/hmg/ddm382 [DOI] [PubMed] [Google Scholar]
- 15. Decker CJ, Parker R. 2002. mRNA decay enzymes: decappers conserved between yeast and mammals. Proc. Natl. Acad. Sci. U. S. A. 99: 12512–12514 doi:10.1073/pnas.212518099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Derry MC, Yanagiya A, Martineau Y, Sonenberg N. 2006. Regulation of poly(A)-binding protein through PABP-interacting proteins. Cold Spring Harb. Symp. Quant. Biol. 71: 537–543 doi:10.1101/sqb.2006.71.061 [DOI] [PubMed] [Google Scholar]
- 17. Dewannieux M, Esnault C, Heidmann T. 2003. LINE-mediated retrotransposition of marked Alu sequences. Nat. Genet. 35: 41–48 doi:10.1038/ng1223 [DOI] [PubMed] [Google Scholar]
- 18. Dewannieux M, Heidmann T. 2005. L1-mediated retrotransposition of murine B1 and B2 SINEs recapitulated in cultured cells. J. Mol. Biol. 349: 241–247 doi:10.1016/j.jmb.2005.03.068 [DOI] [PubMed] [Google Scholar]
- 19. Dewannieux M, Heidmann T. 2005. Role of poly(A) tail length in Alu retrotransposition. Genomics 86: 378–381 doi:10.1016/j.ygeno.2005.05.009 [DOI] [PubMed] [Google Scholar]
- 20. Dombroski BA, Mathias SL, Nanthakumar E, Scott AF, Kazazian HH., Jr 1991. Isolation of an active human transposable element. Science 254: 1805–1808 [DOI] [PubMed] [Google Scholar]
- 21. Esnault C, Maestre J, Heidmann T. 2000. Human LINE retrotransposons generate processed pseudogenes. Nat. Genet. 24: 363–367 doi:10.1038/74184 [DOI] [PubMed] [Google Scholar]
- 22. Fanning T, Singer M. 1987. The LINE-1 DNA sequences in four mammalian orders predict proteins that conserve homologies to retrovirus proteins. Nucleic Acids Res. 15: 2251–2260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Fanning TG. 1983. Size and structure of the highly repetitive BAM HI element in mice. Nucleic Acids Res. 11: 5073–5091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Fanning TG, Singer MF. 1987. LINE-1: a mammalian transposable element. Biochim. Biophys. Acta 910: 203–212 [DOI] [PubMed] [Google Scholar]
- 25. Fedorov AV. 2008. Regulation of mammalian LINE1 retrotransposones transcription. Tsitologiia 50: 1011–1022 [PubMed] [Google Scholar]
- 26. Feng Q, Moran JV, Kazazian HH, Jr, Boeke JD. 1996. Human L1 retrotransposon encodes a conserved endonuclease required for retrotransposition. Cell 87: 905–916 doi: S0092-8674(00)81997-2 [DOI] [PubMed] [Google Scholar]
- 27. Fortes P, et al. 2000. The yeast nuclear cap binding complex can interact with translation factor eIF4G and mediate translation initiation. Mol. Cell 6: 191–196 [PubMed] [Google Scholar]
- 28. Gasior SL, Roy-Engel AM, Deininger PL. 2008. ERCC1/XPF limits L1 retrotransposition. DNA Repair (Amst.) 7: 983–989 doi:10.1016/j.dnarep.2008.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gasior SL, Wakeman TP, Xu B, Deininger PL. 2006. The human LINE-1 retrotransposon creates DNA double-strand breaks. J. Mol. Biol. 357: 1383–1393 doi:10.1016/j.jmb.2006.01.089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Goodier JL, Zhang L, Vetter MR, Kazazian HH., Jr 2007. LINE-1 ORF1 protein localizes in stress granules with other RNA-binding proteins, including components of RNA interference RNA-induced silencing complex. Mol. Cell. Biol. 27: 6469–6483 doi:10.1128/MCB.00332-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Grimaldi G, Skowronski J, Singer MF. 1984. Defining the beginning and end of KpnI family segments. EMBO J. 3: 1753–1759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hancks DC, Goodier JL, Mandal PK, Cheung LE, Kazazian HH., Jr 2011. Retrotransposition of marked SVA elements by human L1s in cultured cells. Hum. Mol. Genet. 20: 3386–3400 doi:10.1093/hmg/ddr245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hata K, Sakaki Y. 1997. Identification of critical CpG sites for repression of L1 transcription by DNA methylation. Gene 189: 227–234 [DOI] [PubMed] [Google Scholar]
- 34. Hohjoh H, Singer MF. 1996. Cytoplasmic ribonucleoprotein complexes containing human LINE-1 protein and RNA. EMBO J. 15: 630–639 [PMC free article] [PubMed] [Google Scholar]
- 35. Hohjoh H, Singer MF. 1997. Sequence-specific single-strand RNA binding protein encoded by the human LINE-1 retrotransposon. EMBO J. 16: 6034–6043 doi:10.1093/emboj/16.19.6034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Holmes SE, Singer MF, Swergold GD. 1992. Studies on p40, the leucine zipper motif-containing protein encoded by the first open reading frame of an active human LINE-1 transposable element. J. Biol. Chem. 267: 19765–19768 [PubMed] [Google Scholar]
- 37. Hosoda N, Lejeune F, Maquat LE. 2006. Evidence that poly(A) binding protein C1 binds nuclear pre-mRNA poly(A) tails. Mol. Cell. Biol. 26: 3085–3097 doi:10.1128/MCB.26.8.3085-3097.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kinomoto M, et al. 2007. All APOBEC3 family proteins differentially inhibit LINE-1 retrotransposition. Nucleic Acids Res. 35: 2955–2964 doi:10.1093/nar/gkm181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kolosha VO, Martin SL. 1997. In vitro properties of the first ORF protein from mouse LINE-1 support its role in ribonucleoprotein particle formation during retrotransposition. Proc. Natl. Acad. Sci. U. S. A. 94: 10155–10160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kolosha VO, Martin SL. 2003. High-affinity, non-sequence-specific RNA binding by the open reading frame 1 (ORF1) protein from long interspersed nuclear element 1 (LINE-1). J. Biol. Chem. 278: 8112–8117 doi:10.1074/jbc.M210487200 [DOI] [PubMed] [Google Scholar]
- 41. Kuhn U, et al. 2009. Poly(A) tail length is controlled by the nuclear poly(A)-binding protein regulating the interaction between poly(A) polymerase and the cleavage and polyadenylation specificity factor. J. Biol. Chem. 284: 22803–22814 doi:10.1074/jbc.M109.018226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kuhn U, Wahle E. 2004. Structure and function of poly(A) binding proteins. Biochim. Biophys. Acta 1678: 67–84 doi:10.1016/j.bbaexp.2004.03.008 [DOI] [PubMed] [Google Scholar]
- 43. Kulpa DA, Moran JV. 2005. Ribonucleoprotein particle formation is necessary but not sufficient for LINE-1 retrotransposition. Hum. Mol. Genet. 14: 3237–3248 doi:10.1093/hmg/ddi354 [DOI] [PubMed] [Google Scholar]
- 44. Kulpa DA, Moran JV. 2006. Cis-preferential LINE-1 reverse transcriptase activity in ribonucleoprotein particles. Nat. Struct. Mol. Biol. 13: 655–660 doi:10.1038/nsmb1107 [DOI] [PubMed] [Google Scholar]
- 45. Lander ES, et al. 2001. Initial sequencing and analysis of the human genome. Nature 409: 860–921 doi:10.1038/35057062 [DOI] [PubMed] [Google Scholar]
- 46. Luan DD, Korman MH, Jakubczak JL, Eickbush TH. 1993. Reverse transcription of R2Bm RNA is primed by a nick at the chromosomal target site: a mechanism for non-LTR retrotransposition. Cell 72: 595–605 [DOI] [PubMed] [Google Scholar]
- 47. Ma S, Musa T, Bag J. 2006. Reduced stability of mitogen-activated protein kinase kinase-2 mRNA and phosphorylation of poly(A)-binding protein (PABP) in cells overexpressing PABP. J. Biol. Chem. 281: 3145–3156 [DOI] [PubMed] [Google Scholar]
- 48. Mangus DA, Evans MC, Jacobson A. 2003. Poly(A)-binding proteins: multifunctional scaffolds for the post-transcriptional control of gene expression. Genome Biol. 4: 223 doi:10.1186/gb-2003-4-7-223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Marie-Josee Sasseville A, et al. 2006. The dynamism of PABPN1 nuclear inclusions during the cell cycle. Neurobiol. Dis. 23: 621–629 doi:10.1016/j.nbd.2006.05.015 [DOI] [PubMed] [Google Scholar]
- 50. Martin SL. 1991. Ribonucleoprotein particles with LINE-1 RNA in mouse embryonal carcinoma cells. Mol. Cell. Biol. 11: 4804–4807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Martin SL, Bushman FD. 2001. Nucleic acid chaperone activity of the ORF1 protein from the mouse LINE-1 retrotransposon. Mol. Cell. Biol. 21: 467–475 doi:10.1128/MCB.21.2.467-475.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Martineau Y, et al. 2008. Poly(A)-binding protein-interacting protein 1 binds to eukaryotic translation initiation factor 3 to stimulate translation. Mol. Cell. Biol. 28: 6658–6667 doi:10.1128/MCB.00738-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Mathias SL, Scott AF, Kazazian HH, Jr, Boeke JD, Gabriel A. 1991. Reverse transcriptase encoded by a human transposable element. Science 254: 1808–1810 [DOI] [PubMed] [Google Scholar]
- 54. Moffat J, et al. 2006. A lentiviral RNAi library for human and mouse genes applied to an arrayed viral high-content screen. Cell 124: 1283–1298 doi:10.1016/j.cell.2006.01.040 [DOI] [PubMed] [Google Scholar]
- 55. Montoya-Durango DE, et al. 2009. Epigenetic control of mammalian LINE-1 retrotransposon by retinoblastoma proteins. Mutat. Res. 665: 20–28 doi:10.1016/j.mrfmmm.2009.02.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Moran JV, et al. 1996. High frequency retrotransposition in cultured mammalian cells. Cell 87: 917–927 [DOI] [PubMed] [Google Scholar]
- 57. Muckenfuss H, et al. 2006. APOBEC3 proteins inhibit human LINE-1 retrotransposition. J. Biol. Chem. 281: 22161–22172 doi:10.1074/jbc.M601716200 [DOI] [PubMed] [Google Scholar]
- 58. Muddashetty R, et al. 2002. Poly(A)-binding protein is associated with neuronal BC1 and BC200 ribonucleoprotein particles. J. Mol. Biol. 321: 433–445 [DOI] [PubMed] [Google Scholar]
- 59. Niewiadomska AM, et al. 2007. Differential inhibition of long interspersed element 1 by APOBEC3 does not correlate with high-molecular-mass-complex formation or P-body association. J. Virol. 81: 9577–9583 doi:10.1128/JVI.02800-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Ostertag EM, Kazazian HH., Jr 2001. Biology of mammalian L1 retrotransposons. Annu. Rev. Genet. 35: 501–538 doi:10.1146/annurev.genet.35.102401.091032 [DOI] [PubMed] [Google Scholar]
- 61. Ostertag EM, Prak ET, DeBerardinis RJ, Moran JV, Kazazian HH., Jr 2000. Determination of L1 retrotransposition kinetics in cultured cells. Nucleic Acids Res. 28: 1418–1423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Piskareva O, Denmukhametova S, Schmatchenko V. 2003. Functional reverse transcriptase encoded by the human LINE-1 from baculovirus-infected insect cells. Protein Expr. Purif. 28: 125–130 [DOI] [PubMed] [Google Scholar]
- 63. Proudfoot NJ, Furger A, Dye MJ. 2002. Integrating mRNA processing with transcription. Cell 108: 501–512 [DOI] [PubMed] [Google Scholar]
- 64. Raiz J, et al. 2012. The non-autonomous retrotransposon SVA is trans-mobilized by the human LINE-1 protein machinery. Nucleic Acids Res. 40: 1666–1683 doi:10.1093/nar/gkr863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Roy-Engel AM, et al. 2002. Active Alu element “A-tails”: size does matter. Genome Res. 12: 1333–1344 doi:10.1101/gr.384802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Scott AF, et al. 1987. Origin of the human L1 elements: proposed progenitor genes deduced from a consensus DNA sequence. Genomics 1: 113–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Shyu AB, Wilkinson MF, van Hoof A. 2008. Messenger RNA regulation: to translate or to degrade. EMBO J. 27: 471–481 doi:10.1038/sj.emboj.7601977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Soifer HS, Zaragoza A, Peyvan M, Behlke MA, Rossi JJ. 2005. A potential role for RNA interference in controlling the activity of the human LINE-1 retrotransposon. Nucleic Acids Res. 33: 846–856 doi:10.1093/nar/gki223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Suzuki J, et al. 2009. Genetic evidence that the non-homologous end-joining repair pathway is involved in LINE retrotransposition. PLoS Genet. 5: e1000461 doi:10.1371/journal.pgen.1000461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Tchenio T, Casella JF, Heidmann T. 2000. Members of the SRY family regulate the human LINE retrotransposons. Nucleic Acids Res. 28: 411–415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Vidal M, Wrighton C, Eccles S, Burke J, Grosveld F. 1990. Differences in human cell lines to support stable replication of Epstein-Barr virus-based vectors. Biochim. Biophys. Acta 1048: 171–177 [DOI] [PubMed] [Google Scholar]
- 72. Wei W, et al. 2001. Human L1 retrotransposition: cis preference versus trans complementation. Mol. Cell. Biol. 21: 1429–1439 doi:10.1128/MCB.21.4.1429-1439.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. West N, Roy-Engel AM, Imataka H, Sonenberg N, Deininger P. 2002. Shared protein components of SINE RNPs. J. Mol. Biol. 321: 423–432 [DOI] [PubMed] [Google Scholar]
- 74. Win MN, Smolke CD. 2007. A modular and extensible RNA-based gene-regulatory platform for engineering cellular function. Proc. Natl. Acad. Sci. U. S. A. 104: 14283–14288 doi:10.1073/pnas.0703961104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Yang N, Kazazian HH., Jr 2006. L1 retrotransposition is suppressed by endogenously encoded small interfering RNAs in human cultured cells. Nat. Struct. Mol. Biol. 13: 763–771 doi:10.1038/nsmb1141 [DOI] [PubMed] [Google Scholar]
- 76. Yang N, Zhang L, Zhang Y, Kazazian HH., Jr 2003. An important role for RUNX3 in human L1 transcription and retrotransposition. Nucleic Acids Res. 31: 4929–4940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Yoshida M, et al. 2006. Poly(A) binding protein (PABP) homeostasis is mediated by the stability of its inhibitor, Paip2. EMBO J. 25: 1934–1944 doi:10.1038/sj.emboj.7601079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Zekri L, Huntzinger E, Heimstadt S, Izaurralde E. 2009. The silencing domain of GW182 interacts with PABPC1 to promote translational repression and degradation of microRNA targets and is required for target release. Mol. Cell. Biol. 29: 6220–6231 doi:10.1128/MCB.01081-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Zheng D, et al. 2008. Deadenylation is prerequisite for P-body formation and mRNA decay in mammalian cells. J. Cell Biol. 182: 89–101 doi:10.1083/jcb.200801196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Zimmerly S, Guo H, Perlman PS, Lambowitz AM. 1995. Group II intron mobility occurs by target DNA-primed reverse transcription. Cell 82: 545–554 [DOI] [PubMed] [Google Scholar]