Abstract
Accumulating evidence suggests that HtrA1 (high-temperature requirement A1) is involved in modulating crucial cellular processes and implicated in life-threatening diseases, such as cancer and neuropathological disorders; however, the exact functions of this protease in vivo remain unknown. Here, we show that loss of HtrA1 function increases fibroblast growth factor 8 (FGF8) mRNA levels and triggers activation of FGF signaling, resulting in dorsalization in zebrafish embryos. Notably, HtrA1 directly cleaves FGF8 in the extracellular region, and this cleavage results in decreased activation of FGF signaling, which is essential for many physiological processes. Therefore, HtrA1 is indispensable for dorsoventral patterning in early zebrafish embryogenesis and serves as a key upstream regulator of FGF signaling through the control of FGF levels. Furthermore, this study offers insight into new strategies to control human diseases associated with HtrA1 and FGF signaling.
INTRODUCTION
Fibroblast growth factor (FGF) signaling is a crucial pathway involved in the control of many key physiological and developmental processes (3, 42, 43). These wide-ranging biological roles of FGF signaling suggest that the FGF signaling pathway is tightly regulated within different cell types, and thus, many positive and negative regulators of FGF signaling are likely to be required to orchestrate the various physiological processes (4, 15, 20, 28, 42). Additionally, recent studies have focused on elucidating different ways to regulate FGF signaling, such as removal of FGF ligands via receptor-mediated endocytosis (35, 39, 47) and inhibition of FGF signaling by the extracellular matrix metalloprotease 2 (MMP2) (44) in the extracellular space. However, the regulatory proteins that directly control the levels of FGF ligands and thus regulate FGF signaling remain unknown.
The protease known as high-temperature requirement A1 (HtrA1) belongs to the HtrA serine protease family, which is highly conserved from microorganisms to multicellular organisms, including humans (8, 9, 26) (see Fig. S1 in the supplemental material). Several cellular and molecular studies have suggested that HtrA1 serves as a key player in regulating various cellular processes through cleavage of and/or binding to pivotal factors that participate in cell proliferation, migration, and fate (8, 9). Moreover, dysregulation of HtrA1 expression or its serine protease activity has been implicated in the pathogenesis of important diseases, such as cancer (1, 6–8, 23), arthritis (19), age-related macular degeneration (AMD) (11, 14, 27, 46, 48), and neuropathological disorders (18, 22, 41). The physiological functions of HtrA1 in vivo, however, remain largely unknown. Recent genetic studies have shown that HtrA1-deficient mice do not display serious defects, which may reflect functional redundancy of HtrAs in mice (27, 48). To investigate the physiological functions of HtrA1 in vivo and the molecular mechanism underlying its in vivo functions, we used morpholino (MO)-based knockdown (KD) in the zebrafish system. Here, we demonstrate that HtrA1 is essential for dorsoventral patterning during early zebrafish embryogenesis and that this protease serves as a key regulator of FGF signaling by controlling levels of FGF8, a factor that functions in the initial step to drive FGF signaling.
MATERIALS AND METHODS
Zebrafish maintenance.
Adult wild-type zebrafish (Danio rerio, 3 to 4 months old) were raised under standard laboratory conditions described in a laboratory manual on zebrafish (36). Embryos were obtained by natural mating after the initiation of the light cycle and cultured at 28.5°C for subsequent experimental procedures. Developmental stages were assigned in accordance with those defined by Kimmel et al. (30). The animal experiments were reviewed and approved by the Institutional Animal Care and Use Committees at the College of Medicine, Catholic University of Korea (IACUC no. CUMC-2010-0206-01).
Chemicals and antibodies.
All chemicals were purchased from Sigma Chemical Co. unless otherwise stated. SU5402 was purchased from Calbiochem. Immunological assays were performed with anti-phospho-extracellular signal-regulated kinase (anti-p-ERK) polyclonal antibody (PAb; Cell Signaling Technology), anti-ERK PAb and anti-GFP monoclonal antibodies (MAbs; Santa Cruz Biotechnology), anti-Flag and anti-β-actin MAbs (Sigma), and anti-HtrA1 PAb (Abfrontier Co., South Korea) (29). Horseradish peroxidase (HRP)-conjugated secondary anti-mouse and -rabbit immunoglobulin G (IgG) were purchased from Santa Cruz Biotechnology. Recombinant FGF8 was purchased from Peprotech, Inc.
Injection of morpholino (MO) and mRNA.
Two antisense MOs against the zHtrA1 splice donor site of E2 (zHtrA1 MO, 5′-TGTGAGAAACTTACTTGCGGTAAAG-3′; the exon is in bold) (see Fig. S3C in the supplemental material) and the 3′ downstream region of the zHtrA1 start codon (zHtrA1-ATG MO, 5′-GCAGAATAAAGTGACCAAAATCATG-3′; the start codon is underlined) (see Fig. S4D in the supplemental material) and a standard control MO (5′-CCTCTTACCTCAGTTACAATTTATA-3′) were synthesized by Gene Tools, LLC. All the MOs were resuspended in RNase-free water (Ambion) and injected into the yolks of one- to two-cell-stage embryos. Capped full-length RNAs encoding zHtrA1 and zFGF8 were synthesized with pCS-zHtrA1 or pCS-zFGF8, respectively, as templates using the mMessage mMachine kit (Ambion). These RNAs (0.3 fmol zHtrA1 and 0.1 fmol zFGF8) were injected at the one-cell stage.
In rescue experiments, zHtrA1 MO and the zHtrA1 and zFGF8 RNAs were injected at the one-cell stage in the amounts specified above. An FGF8 signaling inhibitor, SU5402 (Calbiochem), was dissolved in DMSO to a concentration of 40 mM. The zHtrA1 KD embryos were treated with serial doses of SU5402 (2.5 to 20 μM) from 2.5 h postfertilization (hpf; 256-cell stage) to 10 hpf (tail bud stage).
Reverse transcription-PCR (RT-PCR) and quantitative real-time PCR (qRT- PCR).
Total RNA was extracted from human embryonic kidney 293 (HEK293) cells, zebrafish embryos, and adult zebrafish tissues using RNAiso Plus (TaKaRa). cDNA synthesis was carried out with a PrimeScript first-strand cDNA synthesis kit (TaKaRa) using 1.2 μg of total RNA as the template. qRT-PCR was carried out in a reaction mixture containing SYBR premix Ex Taq (TaKaRa) and using a CFX96 real-time PCR detection system (Bio-Rad). Data were normalized to zβ-actin, and quantitative measures were obtained using the ΔΔCT method. The PCR was carried out with primers listed in Tables S2 and S3 in the supplemental material.
Whole-mount in situ hybridization (WISH).
Antisense digoxigenin (DIG)-labeled RNA probes were synthesized by in vitro transcription using T3 (Ambion), T7, or SP6 RNA polymerases (TaKaRa) with a DIG-RNA-labeled mix (Roche, Basel, Switzerland) according to the manufacturer's instructions. WISH was performed according to the method described in reference 36, and the WISH results were visualized by an anti-DIG Fab fragment conjugated with alkaline phosphatase and the NBT-BCIP (nitroblue tetrazolium–5-bromo-4-chloro-3-indolylphosphate) system (Roche).
Plasmid construction.
RT-PCR cloning of zHtrA1 was performed with Pfu DNA polymerase (Stratagene) from the cDNA prepared from 24-hpf zebrafish embryos. The following zHtrA1 primers used for cloning were designed using sequence information from GenBank accession number NM_001002219 (see Table S2 in the supplemental material): zHtrA1-1 and zHtrA1-2 for the coding region corresponding to amino acids (aa) 1 to 259 and zHtrA1-3 and zHtrA1-4 for the coding region corresponding to aa 260 to 479. The PCR product was digested with the appropriate restriction enzymes, and the resulting zHtrA1 product was inserted into the pCS2+ plasmid. The resulting plasmid was designated pCS-zHtrA1. The putative zHtrA1 mature form (aa 149 to 480) was generated by PCR using pCS-zHtrA1 as a template and the zHtrA1-5 and zHtrA1-4 primers. The PCR product was inserted into the pCS2+ and pGEX 4T-1 plasmids and designated pCS-M-zHtrA1 and pGST-zHtrA1, respectively. A QuikChange site-directed mutagenesis kit (Stratagene) was used according to the manufacturer's instructions to generate the proteolytically inactive zHtrA1-S327A mutant. The pCS-zFGF8 construct was generated by inserting the coding region (aa 1 to 210) and 3′ untranslated region of zFGF8 into the pCS2+ vector.
Various expression vectors for HtrA1 were constructed as previously described (29). The putative HtrA1 mature form (aa 149 to 480) was generated by PCR using pCS-HtrA1 as a template. The PCR product of the HtrA1 putative mature form was inserted into the pCS2+ and pGEX 4T-1 plasmids, and the resulting constructs were designated pCS-M-HtrA1 and pGST-HtrA1, respectively. The human FGF8 clone was purchased from Source BioScience imaGenes, and FGF8 was subcloned into the pCS2+ plasmid to yield pCS-FGF8. The sequence integrity of all plasmid constructs was verified by automated DNA sequencing (COSMO Co., South Korea).
Immunoblot (IB) analyses.
MO or mRNA-injected embryos and SU5402-treated embryos and transfected HEK293 cells were lysed in cell lysis buffer (Cell Signaling Technology) and placed on ice for 15 min. After centrifugation at 15,000 × g for 15 min at 4°C, protein concentrations were determined by the Bradford protein assay (Bio-Rad). Protein samples were resolved by 15% (cellular proteins) and 10% (embryonic proteins) sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to a nitrocellulose membrane.
For dot blot analysis, culture media and cells were separated by centrifugation at 1,000 × g for 5 min at 4°C. Cells were washed with ice-cold phosphate-buffered saline (PBS) and lysed in a total volume of 60 μl in cell lysis buffer for preparation of cell lysates. Culture medium was concentrated 10-fold (to 60 μl) by Amicon Ultra-15 PL10K centrifugal filters (Millipore). Culture medium and cell lysates were filtered through nitrocellulose membranes in the dot blot apparatus (Bio-Rad). Membranes were blocked with 5% bovine serum albumin (BSA) in TBST (10 mM Tris-HCl [pH 7.5], 150 mM NaCl, 0.1% Tween 20), incubated with the specific Ab for the indicated time, and washed with TBST buffer three times. Membranes were then incubated with HRP-conjugated secondary Ab for 1 h and washed with TBST buffer three times, and proteins were detected with the enhanced chemiluminescence (ECL) immunoblotting system (Neuronex, South Korea). The band intensities were quantified by densitometry with Multi-Gauge version 3.0 (Fujifilm). The band intensity of p-ERK was normalized to that of ERK to quantify the levels of ERK activation, and the values are presented as fold change relative to the untreated control.
Purification of the recombinant glutathione S-transferase (GST) fusion proteins and in vitro cleavage assays.
The GST-HtrA1 fusion proteins were induced in the BL21 strain (F− ompT hsdSB (rB− mB−) gal dcm) (Stratagene) and purified as described previously (40). Protein concentrations were determined by comparison with bovine serum albumin (BSA) of known concentration by SDS-PAGE followed by staining of the gel with Coomassie brilliant blue.
The [35S]Met-labeled FGF8 protein was prepared with a TNT quick coupled transcription/translation system (Promega) supplemented with [35S]methionine (Perkin Elmer Life Sciences). Cleavage reactions were performed by incubating the [35S]Met-labeled FGF8 protein with 0.6 μM GST-HtrA1 protein in 20 μl cleavage buffer (50 mM Tris-HCl [pH 7.5], 1 mM dithiothreitol) for 3 h at 37°C. The reaction mixtures were analyzed by 15% SDS-PAGE, and dried gels were exposed to X-ray film. To determine the endoproteolytic activity of HtrA1, 10 μM β-casein was used as an exogenous substrate and was incubated with 0.75 μM GST-zHtrA1 protein in a total volume of cleavage buffer of 100 μl for 3 h at 37°C. The reaction mixtures were analyzed by 15% SDS-PAGE, and the cleavage of β-casein was visualized by staining of the gel with Coomassie brilliant blue.
Imaging analyses and statistical analysis.
Live embryos after injection and embryos processed for WISH were mounted in 3% methylcellulose and 80% glycerol in egg water, respectively. The image data were visualized with a dissection microscope (Leica) using a camera system (Leica DFC 490 and Leica application suite, v. 2.8.1).
HEK293 cells were transfected using Fugene HD reagent (Roche) according to the manufacturer's instructions. At 18 h posttransfection, cells were fixed in 4% paraformaldehyde for 5 min at room temperature. Fixed cells were blocked for 1 h with phosphate-buffered saline (PBS) containing 2% BSA-PBST (PBS with 0.1% Tween 20) at room temperature. After being washed with blocking solution, cells were washed with PBST for 5 min and incubated with 2 μg/ml 4′,6-diamidino-2-phenylindole (DAPI) for 5 min. Expression of HtrA1 and the cell death caused by the HtrA1 proteins were analyzed by confocal microscopy (Carl Zeiss).
All data are presented as means ± standard errors of the means (SEM). Levels of statistical significance were assessed by one-way analysis of variance (ANOVA) and Tukey's post hoc comparisons of means using SPSS version 13.0.
RESULTS
Conservation of HtrA1 function in vertebrates.
We identified the putative zebrafish HtrA1 (zHtrA1), which consists of five functional domains common to human HtrA1 (HtrA1), using a series of TBLASTN analyses in the NCBI zebrafish genome resources (see Fig. S1 and S2 in the supplemental material). This zHtrA1 exhibited 68% identity with HtrA1 at the amino acid sequence level and 90% sequence identity in the protease domain (Fig. 1A; also, see Table S1 in the supplemental material). To investigate the functional similarity between HtrA1 and zHtrA1, we examined the serine protease activity of zHtrA1 (Fig. 1B and C). The zHtrA1 cleaved β-casein similarly to HtrA1. Similar to the catalytically inactive mutant HtrA1-S328A, mutation of serine 327 to alanine (S327A) in the protease domain of zHtrA1 abrogated its proteolytic activity. Consistent with these results, the active zHtrA1 induced cell death at a level comparable to that induced by HtrA1, whereas the zHtrA1-S327A did not (Fig. 1D). Taken together, these data demonstrate that zHtrA1 is the true ortholog of HtrA1 and imply that the zebrafish system is a suitable vertebrate model for studying the physiological functions of HtrA1.
Fig 1.
zHtrA1 is the true zebrafish ortholog of human HtrA1. (A) Schematic view of human and zebrafish HtrA1 showing the signal peptide (SP), insulin-like growth factor binding protein-binding (IGFBP), Kazal inhibitor (KI), serine protease, and PDZ domains. The table indicates amino acid identity between the serine protease domains. (B) Rapid affinity purification of the HtrA1 proteins. (C) Endoproteolytic activity of the HtrA1 serine proteases. GST-HtrA1 proteins (0.75 μM) were incubated for the indicated times at 37°C with β-casein (10 μM) as an exogenous substrate. (D) Induction of cell death in an HtrA1 serine protease-dependent manner. The extent of cell death was calculated by dividing the number of round cells with nuclear condensation and fragmentation (dead cells [arrowheads]) by the total number of GFP-positive cells and is expressed as a percentage. “Mock” refers to cells transfected with a control plasmid expressing only GFP. DIC, differential interference contrast; M-HtrA2, mature HtrA2 used as a positive control for cell death. Bar, 10 μm. (E) Stage-specific expression of zHtrA1. Total RNA was extracted from zebrafish embryos at the indicated developmental time points, and RT-PCR was carried out with the zHtrA1 primer set and normalized to β-actin used as an internal control. (F to H) Spatial expression of zHtrA1 at the indicated developmental time points. WISH was performed on zebrafish embryos at different developmental stages using a DIG-labeled zHtrA1 antisense RNA probe, corresponding to nucleotide positions 1 to 784 of zHtrA1. hpf, hour postfertilization; s, somite; ea, embryonic axis; pf, pectoral fin bud.
We investigated the temporal and spatial patterns of zHtrA1 gene expression from the zygote (one-cell stage) to the early larval periods (protruding-mouth stage) (http://zfin.org/action/marker/view/ZDB-GENE-040704-64) (Fig. 1E to H). Maternal zHtrA1 transcripts were maintained at a constant level during early embryogenesis (from the zygote to blastula periods), and zygotic zHtrA1 transcripts were markedly increased at the gastrula period and were maintained for periods exceeding 72 hpf (Fig. 1E). To define the embryonic regions in which zHtrA1 is specifically expressed, we performed whole-mount in situ hybridization (WISH) (Fig. 1F to H). Specifically, zHtrA1 was ubiquitously expressed during early embryogenesis (Fig. 1F). After the blastula period, zHtrA1 was expressed from the anterior to the posterior poles along the dorsal mesoderm (Fig. 1G). From the pharyngula period, zHtrA1 was expressed in the somites and along the axis derived from the mesoderm (Fig. 1H). These results raise the possibility that HtrA1 plays critical roles in embryonic patterning during early zebrafish embryogenesis (38).
Loss of HtrA1 function specially leads to dorsalized phenotypes in early zebrafish embryogenesis.
To study the function of HtrA1 in vivo, we knocked down the endogenous expression of zHtrA1 by microinjecting a 25-mer zHtrA1 splice-blocking morpholino (zHtrA1 MO), which is complementary to the sequence that encompasses the splice junction between exon 2 and intron 2 (see Fig. S3C, E2-I2, in the supplemental material). The zHtrA1 transcripts were barely detectable in the zHtrA1 MO-injected embryos even by RT-PCR (Fig. 2A); nonetheless, we could detect the 1,131-bp aberrant splice variant following a long exposure with UV light (see Fig. S3A in the supplemental material). Sequence analysis revealed deletion of E2 and E3 in the aberrantly spliced transcript, resulting in a premature termination codon at the 3′ end of E1 (see Fig. S3B in the supplemental material). These results indicate that the aberrant zHtrA1 mRNA is eliminated via the nonsense-mediated mRNA decay pathway (see Fig. S3C) (45).
Fig 2.
Knockdown of zHtrA1 causes dorsalization during zebrafish embryogenesis. (A and B) Knockdown of zHtrA1. Zebrafish embryos were injected with the indicated MO at the one-cell stage. Aberrant splicing caused by the zHtrA1 MO in the zHtrA1 pre-mRNA was assessed by RT-PCR (see Fig. S3A and Table S2 in the supplemental material). Un, uninjected; cMO, control MO. (B) Morphological changes visible in the zHtrA1 KD embryos. The dorsal (D) and ventral (V) regions of all embryos were oriented in the same direction. Phenotypic frequencies (percent) were calculated by dividing the number of dorsalized embryos (P) by the total number of embryos. The zHtrA1-ATG MO was designed to prevent translation of the zHtrA1 protein (see Fig. S4D in the supplemental material). Injection with zHtrA1-ATG MO led to dorsalized phenotypes, consistent with phenotypes obtained with zHtrA1 MO (see Fig. S4E and F). (C) Expression levels of the dorsoventral patterning markers. WISH was performed on the shield-stage embryos with probes for chd and gsc. Arrowheads indicate the dorsal territories. (D and E) Expansion of dorsally derived somites in the zHtrA1 KD embryos. WISH was performed on 7-somite-stage embryos with both the myoD (somatic mesoderm) and pax2.1 probes (7 embryos per group) (D). Dorsal view, anterior to the top. (E) The sizes of the fourth somite (double-headed arrow in panel D) are presented as means ± SEM (*, P < 0.0001 compared with control group). One-way ANOVA was used for statistical analyses.
Following knockdown of zHtrA1, we could clearly distinguish the morphological features in the zHtrA1 KD embryos from those in the control MO (cMO)-injected embryos, which displayed a wild-type phenotype (Fig. 2B; also, see Fig. S4A and B in the supplemental material). Notably, at the bud stage, the zHtrA1 KD embryos exhibited elongated phenotypes (Fig. 2B; 146 mutants out of 148 morphants, 99% elongated phenotype), which are characteristic of dorsalized phenotypes reminiscent of FGF gain of function (16, 17). We also identified additional characteristics of the dorsalized phenotypes in the embryos with loss of zHtrA1 function at the pharyngula period (see Fig. S4B in the supplemental material [51 mutants out of 56 morphants, P1 + P2 = 91%]): loss of the ventral caudal fin or a shortened and twisted tail (see Fig. S4C) (16). In parallel, we verified the specificity of the resulting dorsalized phenotypes using a zHtrA1-ATG MO that was designed to specifically inhibit translation of zHtrA1 protein (Fig. 2B; also, see Fig. S4D to F).
To characterize loss-of-zHtrA1 function mutations at the molecular level, we performed WISH on the shield-stage embryos with mesodermal markers (Fig. 2C). In contrast to the dorsally restricted expression pattern seen in the wild type, expression of the dorsal mesodermal markers chordin (chd) and gooscoid (gsc) was remarkably increased and expanded toward the ventral side in the zHtrA1 KD embryos. Using the specific somatic mesodermal marker myoD, we also found additional characteristics of dorsalized phenotypes, such as a dramatic expansion of the somatic region, in the 7-somite-stage (12-hpf) zHtrA1 KD embryos (Fig. 2D and E). These findings suggest that the dorsalized phenotypes caused by loss of zHtrA1 function may be associated with activation of FGF signaling pathways.
Loss of HtrA1 causes the activation of FGF signaling.
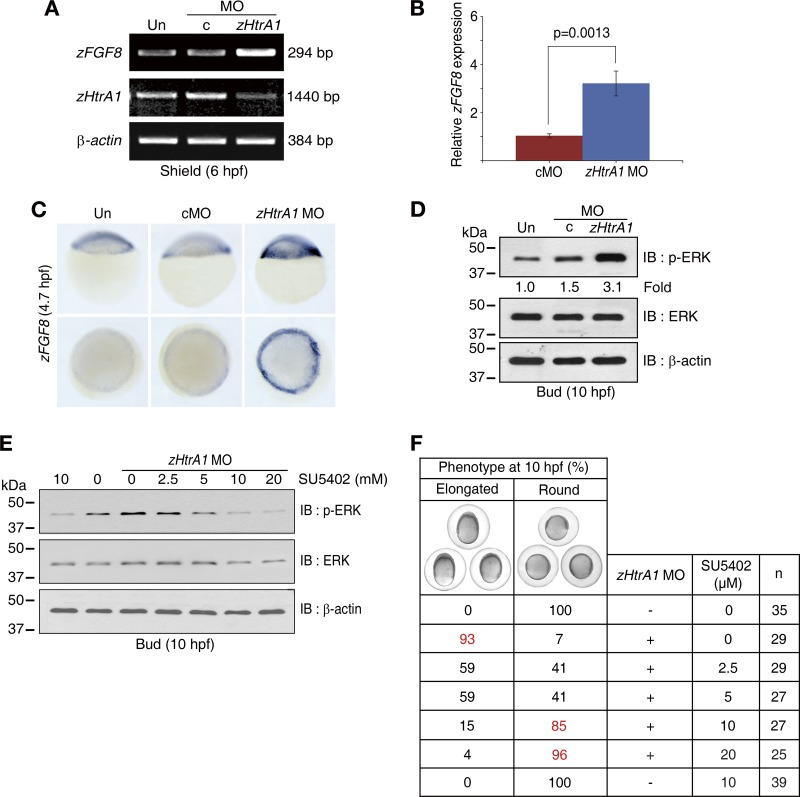
To further investigate these possibilities, we examined the expression levels of several FGF and BMP family genes, which are known to be involved in dorsoventral patterning of the zebrafish gastrula (16, 17, 33) (see Fig. S5A to C in the supplemental material). The expression of zFGF family genes (zFGF3, zFGF8, and zFGF24) was increased by KD of zHtrA1, although no alteration in the expression of BMP family genes was noted in these embryos, suggesting that loss of zHtrA1 function is intimately associated with gain of FGF function rather than loss of BMP function. Notably, the expression of zFGF8 was much higher than that of other zFGFs and, in fact, was 3-fold higher in the zHtrA1 KD embryos than in controls (Fig. 3A and B). Consistent with the dorsalized phenotypes of the embryos injecting with zFGF8 seen at the bud stage (see Fig. S5D in the supplemental material) (16), the increase of zFGF8 transcript in the zHtrA1 KD embryos (Fig. 3A to C) supports their dorsalized phenotypes at the molecular level (Fig. 2).
Fig 3.
Loss of zHtrA1 function activates the FGF signaling pathway. (A to C) The upregulation of zFGF8 in zHtrA1 KD embryos. The levels of zFGF8 mRNA were assessed by RT-PCR with the zFGF8 O primer set (A) and quantitative real-time PCR (qRT-PCR) (7 experiments were performed; more than 8 embryos per group were used for each experiment) with the 5′-U1 primer set (B) (see Table S2 in the supplemental material). Data are means ± SEM. (C) WISH was performed with the zFGF8 probe (11 embryos per group). Top and bottom panels show lateral and animal pole views, respectively. (D) FGF8-induced ERK activation in the zHtrA1 KD embryos. At 10 hpf, embryonic lysates were resolved by 10% SDS-PAGE, and activation of ERK was assessed by IB with anti-p-ERK PAb. The levels of p-ERK were normalized to ERK, and the values are presented as fold change relative to the untreated control. (E and F) Specificity of FGF8-induced ERK activation in the zHtrA1 KD embryos. The zHtrA1 MO was injected at the one-cell stage, and then embryos were treated with SU5402 at 2.5 hpf (initial stage of zFGF8 expression). At 10 hpf, embryonic lysates were resolved by 10% SDS-PAGE, followed by IB for p-ERK (E). Phenotypic rescue of elongated embryos was morphologically analyzed under a stereo dissection microscope (F). Phenotypic frequencies (percent) were calculated by dividing the number of elongated (or round) forms of embryos by the total number of embryos (n).
To assess the mechanistic consequences of upregulation of the zFGF8 signal in the zHtrA1 KD embryos, we analyzed the levels of ERK phosphorylation (p-ERK), which reflects ERK activation, as this protein is a downstream effector of FGF signaling (Fig. 3D). The levels of p-ERK were dramatically increased in the zHtrA1 KD embryos compared to those in uninjected and cMO-injected embryos, indicating activation of FGF signaling in response to an increase in zFGF8 via knockdown of zHtrA1. In parallel, activation of FGF signaling was verified by an increase of the p-ERK following injection of zFGF8 RNA into embryos (see Fig. S5E in the supplemental material) (3, 43). Additionally, pea3 and erm, which are known to be downstream transcription factors in the FGF signaling cascade and are activated via ERK phosphorylation (42), were strikingly expanded on the dorsal region of zHtrA1 KD embryos in the shield stage (see Fig. S5F in the supplemental material).
To substantiate our findings that zHtrA1 KD is involved in the dorsalized phenotypes of embryos via direct activation of FGF signaling, we treated the zHtrA1 KD embryos with SU5402 (32), a molecule that interrupts FGF-dependent signal transduction through inhibition of the tyrosine kinase activity of FGFR1 (Fig. 3E and F). The increase in p-ERK levels (Fig. 3E) and the dorsalized phenotypes (Fig. 3F) in the zHtrA1 KD embryos were restored to nearly wild-type states in a dose-dependent manner following treatment with SU5402. These data indicate that the dorsalized phenotypes elicited by the loss of zHtrA1 function result from activation of FGF signaling and a concomitant increase in the FGF8 signal. Moreover, our findings implicate HtrA1 as a key player in regulation of FGF signaling.
FGF8 positively regulates FGF8 expression through an FGF signaling feedback loop.
Several lines of evidence suggest that FGFs, including FGF8, are involved in increasing the expression of FGF genes through a positive feedback loop of FGF signaling (Fig. 4E) (10, 13). To determinate whether FGF8 expression is regulated via such a positive feedback loop of FGF signaling in the zebrafish system, we injected the zFGF8 RNA, which can be experimentally distinguished from endogenous zFGF8 mRNA by quantitative real-time PCR (qRT-PCR) (Fig. 4A), into one-cell-stage embryos (Fig. 4B). Activation of FGF signaling occurred in the zFGF8 RNA-injected embryos, as increases in transcript levels of the FGF signaling targets pea3 and erm were noted (Fig. 4B; also, see Fig. S6A in the supplemental material). This finding is consistent with results demonstrating that FGF signaling is activated by increased zFGF8 as a consequence of loss of zHtrA1 function (Fig. 3A to C; also, see Fig. S5A and F in the supplemental material). We detected a substantial increase in endogenous zFGF8 transcript following exogenous zFGF8-mediated activation of FGF signaling (Fig. 4B). In parallel, injection of the human FGF8 protein into the one-cell-stage embryos resulted in increased expression of endogenous zFGF8 and broader expression of this ligand in the somites (Fig. 4C). Inhibition of FGFR activity by SU5402 markedly reduced the levels of the zFGF8 transcript and consequently, expression of erm and pea3 (Fig. 4D; also, see Fig. S6B). These results demonstrate that the expression of the FGF8 gene is triggered via the FGF signaling pathway and that, in turn, FGF8 positively modulates the FGF signaling cascade (Fig. 4E). In studies of zebrafish embryology and genetics, activation of FGF signaling has been acquired by means of gain of FGF signaling molecules, such as FGF ligands (15–17), or by loss of negative regulators of the FGF signaling pathway (15, 20, 28). In agreement with these studies, loss of zHtrA1 function exhibited an increase of zFGF8, resulting in activation of FGF signaling. Like the known negative regulators of FGF signaling Sef, Spry, and Mkp3, which are involved as regulators of the common FGF signaling pathway (15, 20, 28), zHtrA1 and zFGF8 are coordinately regulated in the FGF signaling pathway (Fig. 4; also, see Fig. S6), suggesting that HtrA1 and FGF8 may function coordinately in the common FGF signaling pathway. Moreover, these experiments provide additional molecular evidence for activation of the FGF signaling pathway via increased zFGF8 levels resulting from loss of zHtrA1 function.
Fig 4.
FGF8 positively regulates FGF8 expression through an FGF signaling feedback loop. (A) A schematic diagram of the zFGF8 mRNA. All of the indicated PCR products were amplified by the corresponding primer sets (see Table S2 in the supplemental material). (B) In vitro-transcribed zFGF8 (nucleotides 418 to 1710, 0.1 fmol/embryo) was injected into one-cell stage embryos, and transcriptional activation of endogenous zFGF8 was analyzed by qRT-PCR (4 experiments were performed; more than 13 embryos per group were used for each experiment) with the indicated primer sets. Data are means ± SEM (*, P < 0.02, and **, P < 0.01 compared with control group). (C) Recombinant human FGF8 protein (1.6 fmol/embryo) was injected into one-cell-stage embryos, and endogenous zFGF8 RNA was detected by WISH (8 embryos per group) with a zFGF8 antisense probe at 28 hpf (arrowheads, zFGF8-expressed somite borders). (D) Abrogation of the increase in FGF8 transcript levels by blockade of FGF signaling with SU5402. Embryos were treated with SU5402 at 2.5 hpf, and levels of indicated target transcripts were analyzed by qRT-PCR (3 experiments were performed; more than 15 embryos per group were used for each experiment) with the indicated primer sets. Data are means ± SEM (***, P < 0.001 compared with control group). (E) Working model of the FGF8-mediated positive feedback loop of the FGF signaling cascade. FGF8 binds to FGFR in the extracellular region and thereby activates the mitogen-activated protein kinase (MAPK) cascade (3, 42, 43). Notably, the FGF8 gene is transcriptionally activated via FGF signaling. The newly synthesized FGF8 protein is secreted into the extracellular region.
HtrA1 directly cleaves FGF8.
To explore the molecular mechanism of HtrA1 in regulation of FGF signaling, we first assessed the effects of proteolytically active zHtrA1 on regulation of this signaling pathway (Fig. 5A and B). Contrary to the effects of loss of zHtrA1 function (Fig. 3), overexpression of the active zHtrA1 protease in embryos resulted in a dramatic reduction of the FGF8 transcript (Fig. 5A) and a decrease in p-ERK (Fig. 5B). Moreover, these events did not appear in embryos that overexpressed the inactive mutant zHtrA1-S327A. Therefore, zHtrA1 negatively regulates FGF signaling in a serine protease-dependent manner.
Fig 5.
HtrA1 negatively regulates the FGF signaling pathway by cleaving FGF8. (A and B) Inhibition of zFGF8 expression and FGF signaling in the embryos overexpressing the active serine protease zHtrA1. zHtrA1 mRNA (0.3 fmol/embryo) was injected into one-cell-stage embryos. (A) The levels of zFGF8 transcript were assessed by WISH (3 embryos per group). (B) ERK activation was determined by the extent of p-ERK. (C and D) Direct cleavage of FGF8 by HtrA1. The indicated GST fusion proteins were incubated with zebrafish (C) or human (D) [35S]Met-labeled FGF8 for 3 h at 37°C. (E) Schematic diagram of the CM assay designed to assess the effect of extracellular HtrA1 on the regulation of the FGF signaling pathway. All CM were collected from HEK293 cells transfected with plasmids encoding HtrA1 at 24 h posttransfection or with siRNAs at 48 h posttransfection. HEK293 cells, which were cultured in serum-free medium for 6 h, were treated with the CM plus FGF8 for 30 min. The effect of the enzyme-substrate pair HtrA1 and FGF8 on the FGF signaling pathway in the extracellular region was assessed by the extent of p-ERK. (F and G) Inhibition of FGF signaling by HtrA1 in the extracellular region. Serum-starved HEK293 cells were treated with the indicated CM supplemented with FGF8 for 30 min. Cell lysates were resolved by 15% SDS-PAGE, followed by IB with the indicated antibodies.
Next, we examined why the levels of zFGF8 transcript are inversely related to the presence of active zHtrA1 (Fig. 3A to C and 5A). Previous studies demonstrated that both HtrA1 and FGF8 are secreted into the extracellular region (18, 26, 47). We confirmed that full-length zHtrA1 and HtrA1 are secreted into the culture medium (see Fig. S7A and C in the supplemental material). Moreover, several studies have shown that HtrA1 plays a role in the degradation and inhibition of secreted growth factors in the extracellular region and endoplasmic reticulum (24, 31, 41). Based on the FGF8 positive-feedback loop (Fig. 4E), we propose that the levels of zFGF8 expression may be negatively regulated by zHtrA1 at the posttranslational level, and thus zFGF8 may serve as a cleavage target for zHtrA1. To examine this possibility, we incubated [35S]Met-labeled zFGF8 with GST-zHtrA1 (Fig. 5C). Interestingly, zFGF8 was directly cleaved by zHtrA1 but not by the zHtrA1-S327A mutant, which lacks protease activity. The action of the human enzyme-substrate pair is comparable to that of the zebrafish pair with complete degradation of FGF8 in the presence of HtrA1 (Fig. 5D). These results indicate that FGF8 levels in vivo may be tightly associated with HtrA1-mediated FGF8 cleavage.
A novel antagonist HtrA1 in the FGF signaling pathway.
Given that secreted zFGF8 and zHtrA1 could encounter each other in the extracellular spaces between cells in close proximity in the dorsal mesoderm region, we designed a conditioned-medium (CM) system that more closely mimics the in vivo environment in order to assess the effects of HtrA1 on the regulation of FGF signaling (Fig. 5E). We used HEK293 cells because ERK was rapidly activated in response to FGF8 in a dose-dependent manner in these cells (see Fig. S7B in the supplemental material). A 3-fold increase in FGF8-induced ERK activation was observed in the CM containing secreted inactive mutant HtrA1-S328A (S328A-CM), whereas this activation was abolished when FGF8 was incubated with CM containing secreted active HtrA1 (HtrA1-CM) (Fig. 5F; also, see Fig. S7C in the supplemental material). Consistently, the decrease in HtrA1 secretion upon KD of HtrA1 expression by small interfering RNA (siRNA) (see Fig. S7D and E) was accompanied by an approximately 3-fold increase in FGF8-induced ERK activation (Fig. 5G). These results indicate that secreted HtrA1 cleaves FGF8 in the extracellular region, and this event subsequently restricts the FGF signaling pathway.
To verify the biological relevance of HtrA1-mediated FGF signaling in living organisms, we coinjected zFGF8 and zHtrA1 RNAs into embryos and further investigated the enzyme-substrate relationship of these molecules in regulating FGF signaling in zebrafish embryos (Fig. 6). Although FGF8 stimulated an increase in p-ERK, the FGF8-induced ERK activation was restored to nearly basal levels in the presence of active zHtrA1 serine protease but not in the presence of the zHtrA1-S327A mutant (Fig. 6A). Consistently, more than 70% of the dorsalized embryos were rescued back to a round phenotype, similar to that of the wild type, by overexpression of active zHtrA1 but not by overexpression of the zHtrA1-S327A mutant (Fig. 6B and C). Together, these results demonstrate that HtrA1 negatively regulates the FGF signaling pathway in an in vivo vertebrate model.
Fig 6.
Restoration of zFGF8-induced FGF signaling activation to a normal state in the presence of active zHtrA1. zFGF8 (0.1 fmol) and zHtrA1 (0.3 fmol) RNAs were coinjected into one-cell-stage embryos. (A) p-ERK was assessed by IB at 10 hpf. (B) Rescued phenotype frequencies (percent) were calculated by dividing the number of round embryos by the total number of embryos (n) at 10 hpf. (C) Embryo size from the anterior to the posterior pole, presented as means ± SEM (*, P < 0.001 compared with control group). (D) Working model of HtrA1 as a novel antagonist in the FGF8-mediated positive feedback loop of FGF signaling. HtrA1 cleaves FGF8 in the extracellular region, which may be required to maintain FGF8 and FGF signaling at a constant level.
DISCUSSION
We have demonstrated that HtrA1 is an essential gene responsible for dorsoventral patterning during the gastrula period via control of FGF signaling, as HtrA1 KD exhibited apparent dorsalization of zebrafish embryos at the phenotypic level and induced an increase of zFGF8 transcript and subsequent activation of FGF signaling at the molecular level.
Unlike the HtrA1 transcript patterns during early zebrafish embryogenesis (Fig. 1E to H), Xenopus HtrA1 (xHtrA1) is not detected from the maternal to the blastula periods (stage 9 in Xenopus) and is expressed at very low levels in the early gastrula period (stage 11) (25). This discrepancy in temporal expression of HtrA1 between zebrafish and Xenopus may translate to different mechanisms of FGF signaling regulation during early embryogenesis in the different species. Several lines of evidence, including the Xenopus study, raise the possibility that HtrA1 is associated with regulation of FGF signaling (12, 23, 25); however, the molecular mechanism by which HtrA1 regulates this pathway remains largely unknown.
Previous genetic and biophysical studies have demonstrated that concentration gradients of signaling molecules called morphogens, including FGF8, are controlled by a source sink mechanism that involves a combination of their free-random motion and removal via receptor-mediated endocytosis (35, 39, 47). This mechanism, however, may not be the only way to control morphogen concentrations in the extracellular space. A recent study with Drosophila found that MMP2 restricts FGF signaling in the extracellular region, although its target substrates have not been identified (44). Additionally, HtrA1 has been shown to degrade extracellular matrix components, such as proteoglycans (5, 19) and heparan sulfate glycosaminoglycan (5), which provide structural support to protect FGF ligands from thermal denaturation and proteolysis (2, 34). These studies support our results showing that the serine protease HtrA1 can specifically cross talk with the FGF signaling cascade through control of proteolysis of FGF signaling molecules in the extracellular region. Collectively, our findings have demonstrated the detailed molecular mechanism underlying HtrA1 function in embryonic development (Fig. 6D): HtrA1 serves as a key protease in the regulation of the FGF signaling pathway via cleavage of FGF8 in the extracellular region to establish dorsoventral patterning in zebrafish embryos.
A previous study showed that overexpression of zFGF8 by the zFGF8 injection coordinately inhibits the expression of bmp2 or bmp4 (16). In our study, KD of zHtrA1 increased the levels of zFGF8 expression, but no significant differences were observed in the levels of bmp2 or bmp4 expression (Fig. 3A to C; also, see Fig. S5A to C in the supplemental material). This discrepancy is supported by previous studies showing that the HtrA1 serine protease inhibits signaling mediated by BMP2 and BMP4 (21, 37). Because zHtrA1 was broadly expressed not only in the dorsal region but also in the ventral region, where bmp genes are predominantly expressed (Fig. 1F to H), we can postulate the possibility that HtrA1 may have novel characteristics that control the balance between FGF and BMP signaling. Notably, our study has revealed that HtrA1 is primarily responsible for the control of FGF signaling during zebrafish gastrulation.
To date, overexpression and loss-of-function studies have provided evidence that HtrA1 and FGF signaling are involved in regulating tumorigenesis, albeit by the opposing functions of tumor suppressor activity and oncogenic activity, respectively (8, 43). In agreement with these previous studies, we observed that FGF8 expression is controlled in a manner reciprocal to HtrA1 function (Fig. 3A to C; also, see Fig. S5A in the supplemental material). Accordingly, our results raise the possibility that HtrA1 functions as a tumor suppressor (1, 6–8, 23) or may have another distinct role in the different stages of cellular growth and differentiation. Importantly, our study also suggests that control of HtrA1 expression levels and protease activity may be an important target for maintaining physiological processes and for preventing life-threatening diseases associated with HtrA1 and FGF signaling, such as cancer (1, 6–8, 23), arthritis (19), AMD (11, 14, 27, 46, 48), and neuropathological disorders (18, 22, 41).
Supplementary Material
ACKNOWLEDGMENTS
We thank the Korea Zebrafish Organogenesis Mutant Bank for providing marker genes.
This work was supported by grants from the National R&D Program for cancer control, Ministry of Health & Welfare, Republic of Korea (0720300); the National Nuclear R&D Program through the National Research Foundation (NRF) of Republic of Korea, which is funded by the Ministry of Education, Science and Technology (2011-0018783); the Basic Science Research Program through the NRF of Republic of Korea funded by the Korean government (MEST) (2010-0029422); and the Ministry of Education, Science and Technology (20110007300).
We declare no financial conflict of interest.
Footnotes
Published ahead of print 4 September 2012
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1. Baldi A, et al. 2002. The HtrA1 serine protease is down-regulated during human melanoma progression and represses growth of metastatic melanoma cells. Oncogene 21:6684–6688 [DOI] [PubMed] [Google Scholar]
- 2. Beenken A, Mohammadi M. 2009. The FGF family: biology, pathophysiology and therapy. Nat. Rev. Drug Discov. 8:235–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bottcher RT, Niehrs C. 2005. Fibroblast growth factor signaling during early vertebrate development. Endocr Rev. 26:63–77 [DOI] [PubMed] [Google Scholar]
- 4. Bottcher RT, Pollet N, Delius H, Niehrs C. 2004. The transmembrane protein XFLRT3 forms a complex with FGF receptors and promotes FGF signalling. Nat. Cell Biol. 6:38–44 [DOI] [PubMed] [Google Scholar]
- 5. Chamberland A, et al. 2009. Identification of a novel HtrA1-susceptible cleavage site in human aggrecan: evidence for the involvement of HtrA1 in aggrecan proteolysis in vivo. J. Biol. Chem. 284:27352–27359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chien J, et al. 2006. Serine protease HtrA1 modulates chemotherapy-induced cytotoxicity. J. Clin. Invest. 116:1994–2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chien J, et al. 2009. Serine protease HtrA1 associates with microtubules and inhibits cell migration. Mol. Cell. Biol. 29:4177–4187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Clausen T, Kaiser M, Huber R, Ehrmann M. 2011. HTRA proteases: regulated proteolysis in protein quality control. Nat. Rev. Mol. Cell Biol. 12:152–162 [DOI] [PubMed] [Google Scholar]
- 9. Clausen T, Southan C, Ehrmann M. 2002. The HtrA family of proteases: implications for protein composition and cell fate. Mol. Cell 10:443–455 [DOI] [PubMed] [Google Scholar]
- 10. Crossley PH, Minowada G, MacArthur CA, Martin GR. 1996. Roles for FGF8 in the induction, initiation, and maintenance of chick limb development. Cell 84:127–136 [DOI] [PubMed] [Google Scholar]
- 11. Dewan A, et al. 2006. HTRA1 promoter polymorphism in wet age-related macular degeneration. Science 314:989–992 [DOI] [PubMed] [Google Scholar]
- 12. Ferrer-Vaquer A, et al. 2008. Expression and regulation of HTRA1 during chick and early mouse development. Dev. Dyn. 237:1893–1900 [DOI] [PubMed] [Google Scholar]
- 13. Fox JC, Swain JL. 1993. Auto and transactivation of FGF expression: potential mechanism for regulation of myogenic differentiation. In Vitro Cell Dev. Biol. 29A:228–230 [DOI] [PubMed] [Google Scholar]
- 14. Friedrich U, et al. 2011. Risk- and non-risk-associated variants at the 10q26 AMD locus influence ARMS2 mRNA expression but exclude pathogenic effects due to protein deficiency. Hum. Mol. Genet. 20:1387–1399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Furthauer M, Lin W, Ang SL, Thisse B, Thisse C. 2002. Sef is a feedback-induced antagonist of Ras/MAPK-mediated FGF signalling. Nat. Cell Biol. 4:170–174 [DOI] [PubMed] [Google Scholar]
- 16. Furthauer M, Thisse C, Thisse B. 1997. A role for FGF-8 in the dorsoventral patterning of the zebrafish gastrula. Development 124:4253–4264 [DOI] [PubMed] [Google Scholar]
- 17. Furthauer M, Van Celst J, Thisse C, Thisse B. 2004. Fgf signalling controls the dorsoventral patterning of the zebrafish embryo. Development 131:2853–2864 [DOI] [PubMed] [Google Scholar]
- 18. Grau S, et al. 2005. Implications of the serine protease HtrA1 in amyloid precursor protein processing. Proc. Natl. Acad. Sci. U. S. A. 102:6021–6026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Grau S, et al. 2006. The role of human HtrA1 in arthritic disease. J. Biol. Chem. 281:6124–6129 [DOI] [PubMed] [Google Scholar]
- 20. Hacohen N, Kramer S, Sutherland D, Hiromi Y, Krasnow MA. 1998. sprouty encodes a novel antagonist of FGF signaling that patterns apical branching of the Drosophila airways. Cell 92:253–263 [DOI] [PubMed] [Google Scholar]
- 21. Hadfield KD, et al. 2008. HtrA1 inhibits mineral deposition by osteoblasts: requirement for the protease and PDZ domains. J. Biol. Chem. 283:5928–5938 [DOI] [PubMed] [Google Scholar]
- 22. Hara K, et al. 2009. Association of HTRA1 mutations and familial ischemic cerebral small-vessel disease. N. Engl. J. Med. 360:1729–1739 [DOI] [PubMed] [Google Scholar]
- 23. He X, et al. 2010. Downregulation of HtrA1 promotes resistance to anoikis and peritoneal dissemination of ovarian cancer cells. Cancer Res. 70:3109–3118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hou J, Clemmons DR, Smeekens S. 2005. Expression and characterization of a serine protease that preferentially cleaves insulin-like growth factor binding protein-5. J. Cell Biochem. 94:470–484 [DOI] [PubMed] [Google Scholar]
- 25. Hou S, Maccarana M, Min TH, Strate I, Pera EM. 2007. The secreted serine protease xHtrA1 stimulates long-range FGF signaling in the early Xenopus embryo. Dev. Cell 13:226–241 [DOI] [PubMed] [Google Scholar]
- 26. Hu SI, et al. 1998. Human HtrA, an evolutionarily conserved serine protease identified as a differentially expressed gene product in osteoarthritic cartilage. J. Biol. Chem. 273:34406–34412 [DOI] [PubMed] [Google Scholar]
- 27. Jones A, et al. 2011. Increased expression of multifunctional serine protease, HTRA1, in retinal pigment epithelium induces polypoidal choroidal vasculopathy in mice. Proc. Natl. Acad. Sci. U. S. A. 108:14578–14583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kawakami Y, et al. 2003. MKP3 mediates the cellular response to FGF8 signalling in the vertebrate limb. Nat. Cell Biol. 5:513–519 [DOI] [PubMed] [Google Scholar]
- 29. Kim GY, Moon JM, Han JH, Kim KH, Rhim H. 2011. The sCMV IE enhancer/promoter system for high-level expression and efficient functional studies of target genes in mammalian cells and zebrafish. Biotechnol. Lett. 33:1319–1326 [DOI] [PubMed] [Google Scholar]
- 30. Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. 1995. Stages of embryonic development of the zebrafish. Dev. Dyn. 203:253–310 [DOI] [PubMed] [Google Scholar]
- 31. Launay S, et al. 2008. HtrA1-dependent proteolysis of TGF-beta controls both neuronal maturation and developmental survival. Cell Death Differ. 15:1408–1416 [DOI] [PubMed] [Google Scholar]
- 32. Mohammadi M, et al. 1997. Structures of the tyrosine kinase domain of fibroblast growth factor receptor in complex with inhibitors. Science 276:955–960 [DOI] [PubMed] [Google Scholar]
- 33. Mullins MC, et al. 1996. Genes establishing dorsoventral pattern formation in the zebrafish embryo: the ventral specifying genes. Development 123:81–93 [DOI] [PubMed] [Google Scholar]
- 34. Nissen NN, Shankar R, Gamelli RL, Singh A, DiPietro LA. 1999. Heparin and heparan sulphate protect basic fibroblast growth factor from non-enzymic glycosylation. Biochem. J. 338:637–642 [PMC free article] [PubMed] [Google Scholar]
- 35. Nowak M, Machate A, Yu SR, Gupta M, Brand M. 2011. Interpretation of the FGF8 morphogen gradient is regulated by endocytic trafficking. Nat. Cell Biol. 13:153–158 [DOI] [PubMed] [Google Scholar]
- 36. Nüsslein-Volhard C, Dahm R. 2002. Zebrafish: a practical approach. Oxford University Press, Oxford, United Kingdom [Google Scholar]
- 37. Oka C, et al. 2004. HtrA1 serine protease inhibits signaling mediated by Tgfbeta family proteins. Development 131:1041–1053 [DOI] [PubMed] [Google Scholar]
- 38. Schier AF. 2007. The maternal-zygotic transition: death and birth of RNAs. Science 316:406–407 [DOI] [PubMed] [Google Scholar]
- 39. Scholpp S, Brand M. 2004. Endocytosis controls spreading and effective signaling range of Fgf8 protein. Curr. Biol. 14:1834–1841 [DOI] [PubMed] [Google Scholar]
- 40. Seong YM, et al. 2004. Autocatalytic processing of HtrA2/Omi is essential for induction of caspase-dependent cell death through antagonizing XIAP. J. Biol. Chem. 279:37588–37596 [DOI] [PubMed] [Google Scholar]
- 41. Shiga A, et al. 2011. Cerebral small-vessel disease protein HTRA1 controls the amount of TGF-β1 via cleavage of proTGF-β1. Hum. Mol. Genet. 20:1800–1810 [DOI] [PubMed] [Google Scholar]
- 42. Tsang M, Dawid IB. 2004. Promotion and attenuation of FGF signaling through the Ras-MAPK pathway. Sci. STKE 2004:pe17 doi:10.1126/stke.2282004pe17 [DOI] [PubMed] [Google Scholar]
- 43. Turner N, Grose R. 2010. Fibroblast growth factor signalling: from development to cancer. Nat. Rev. Cancer 10:116–129 [DOI] [PubMed] [Google Scholar]
- 44. Wang Q, Uhlirova M, Bohmann D. 2010. Spatial restriction of FGF signaling by a matrix metalloprotease controls branching morphogenesis. Dev. Cell 18:157–164 [DOI] [PubMed] [Google Scholar]
- 45. Wittkopp N, et al. 2009. Nonsense-mediated mRNA decay effectors are essential for zebrafish embryonic development and survival. Mol. Cell. Biol. 29:3517–3528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Yang Z, et al. 2006. A variant of the HTRA1 gene increases susceptibility to age-related macular degeneration. Science 314:992–993 [DOI] [PubMed] [Google Scholar]
- 47. Yu SR, et al. 2009. Fgf8 morphogen gradient forms by a source-sink mechanism with freely diffusing molecules. Nature 461:533–536 [DOI] [PubMed] [Google Scholar]
- 48. Zhang L, et al. 2012. High temperature requirement factor A1 (HTRA1) gene regulates angiogenesis through transforming growth factor-beta family member growth differentiation factor 6. J. Biol. Chem. 287:1520–1526 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.