Abstract
The control of the cell cycle in eukaryotes is exerted in part by the coordinated action of a series of transcription factor complexes. This is exemplified by the Mcm1p-Fkh2p-Ndd1p complex in Saccharomyces cerevisiae, which controls the cyclical expression of the CLB2 cluster of genes at the G2/M phase transition. The activity of this complex is positively controlled by cyclin-dependent kinase (CDK) and polo kinases. Here, we demonstrate that the protein kinase Pkc1p works in the opposite manner to inhibit the activity of the Mcm1p-Fkh2p-Ndd1p complex and the expression of its target genes. In particular, Pkc1p causes phosphorylation of the coactivator protein Ndd1p. Reductions in Pkc1p activity and the presence of Pkc1p-insensitive Ndd1p mutant proteins lead to changes in the timing of CLB2 cluster expression and result in associated late cell cycle defects. This study therefore identifies an important role for Pkc1p in controlling the correct temporal expression of genes in the cell cycle.
INTRODUCTION
One of the underlying mechanisms for controlling the eukaryotic cell cycle is the transcriptional control of the expression of key regulatory proteins. In Saccharomyces cerevisiae, transcriptional control is exerted throughout the cell cycle, with prominent regulatory activity occurring at the G1-S and G2-M phase transitions (reviewed in references 2, 22, and 37). One important group of genes which is activated in late G2 phase is the CLB2 cluster, which contains genes such as CLB2, encoding a B-type cyclin, CDC5, encoding polo kinase, and SWI5, encoding a transcription factor. This gene cluster is coordinately regulated by a transcription factor complex containing the MADS-box transcription factor Mcm1p (1), the forkhead transcription factor Fkh2p (14, 16, 25, 38; reviewed in reference 4), and the coactivator protein Ndd1p (14, 20). A further coregulator, Sin3p, has been shown to be associated with Mcm1p and Fkh2p and contributes to the repression of genes in the CLB2 cluster at early time points in the cell cycle (36). The homeodomain repressor proteins Yox1p and Yhp1p also contribute to the repression of a subset of CLB2 cluster genes and help to shape the specific timing of their expression late in the cell cycle (7, 27). The Mcm1p-Fkh2p-Ndd1p complex is positively regulated by the cell cycle-dependent kinase complexes Clb5p-Cdc28p and Clb2p-Cdc28p and by the polo kinase Cdc5p, which together combine their activities to give maximal activation through the sequential phosphorylation of Fkh2p and Ndd1p (6, 8, 26, 28). However, to date, no regulatory phosphorylation events have been identified which restrict the activity of this transcription factor complex.
In higher eukaryotes, there are multiple protein kinase C (PKC) isoforms which have been implicated in numerous physiological processes, including cancer. This has led to PKC being identified as an attractive anticancer drug target (29). However, in S. cerevisiae, there is only one protein kinase C isoform, Pkc1p, thereby simplifying the investigation of its mode of action. Pkc1p plays a major role in cell wall integrity signaling, and genetic evidence has previously shown it to also be associated with cell cycle events (reviewed in reference 17). In particular, the observation that Pkc1p-depleted cells arrest with small- to medium-sized buds, duplicated DNA, and short nuclear spindles is strongly suggestive of a defect in passage from G2 to M phase (19). A potential link to gene expression in this context was provided by the observation that there are genetic links between PKC1 and components of the chromatin remodelling complex (RSC), where overexpression of Pkc1p suppresses RSC mutants (5, 11; reviewed in reference 17). Collectively, these results are suggestive of a role of Pkc1p in controlling the expression of genes involved in cell cycle control and hence contributing to G2-M progression. A further hint that Pkc1p is linked to regulation of cell cycle-dependent gene expression was provided by a global two-hybrid study of interacting proteins in yeast, where Ndd1p was found to associate with Ack1p (35), which was recently shown to be a component of the Pkc1p pathway (15). As Ndd1p activates CLB2 cluster gene expression and this same cluster is activated at the G2-M phase transition, these observations suggest a direct link for Pkc1p in controlling CLB2 cluster gene expression.
Here, we have investigated whether Pkc1p contributes directly to the control of CLB2 cluster gene expression. Pkc1p was found to specifically regulate the expression of CLB2 cluster genes. Furthermore, Pkc1p directly targets the coactivator Ndd1p and affects its recruitment to CLB2 gene cluster promoters. Mutant forms of Ndd1p which cannot be phosphorylated by Pkc1p trigger an earlier activation of CLB2 cluster gene expression and lead to alterations in the timing of cell cycle progression. Thus, Pkc1p plays a role in restraining the Mcm1p-Fkh2p-Ndd1p complex, thereby determining the timing of activation of this complex during the cell cycle.
MATERIALS AND METHODS
Plasmid construction and mutagenesis.
For bacterial expression, pAS1769 [encoding GST-Ndd1p(1-554)] and pAS1753 [encoding GST-Fkh2p(458-862)] were described previously (8). pAS2003 [encoding GST-Ndd1p(1-420)], pAS1994 [encoding GST-Ndd1p(1-360)], and pAS2002 [encoding GST-Ndd1p(1-150)] were obtained by inserting NcoI/XhoI-cleaved PCR products generated with primers ADS998/1658, ADS998/1657, and ADS998/1656, respectively, into the same sites of pGEX-KG. pAS2004 [encoding GST-Ndd1p(S409A)], pAS2005 [encoding GST-Ndd1p(S520A)], pAS2006 [encoding GST-Ndd1p(S527A)], and pAS2007 [encoding GST-Ndd1p(S520A/S527A)] were created by QuikChange mutagenesis (Stratagene) using the primer pairs ADS3986/3987, ADS3990/3991, and ADS3988/3989 with the pAS1769 template and ADS3988/3989 with the pAS2005 template, respectively.
For yeast expression, pVD67 (pAS1995; encoding wild-type [WT] GFP-Pkc1p) and pVD123 [pAS1997; encoding the nuclear export mutant GFP-Pkc1p(L61A,L63A)] were kindly provided by Martha Cyert (9). pUS454 (encoding HA epitope-tagged full-length Ndd1p controlled by the GAL1 promoter pGAL1-HA3-NDD1) was kindly provided by U. Surana. pAS2008 encoding pGAL1-HA3-Ndd1p(S520A) and pAS2010 encoding pGAL1-HA3-Ndd1p(S520A/S527A) were created by QuikChange mutagenesis using the primer-template combinations ADS3990/3991 with the pUS454 template and ADS3988/3989 with the pAS2008 template, respectively.
Protein production and Western blotting.
Glutathione S-transferase fusion proteins were prepared essentially as described previously (31). The source of hemagglutinin (HA) epitope-tagged Pkc1p for these assays was total yeast cell lysates isolated from the DZ2 strain.
To detect epitope-tagged derivatives by Western analysis, anti-HA (Roche) and antitubulin TAT-1 (CRUK) antibodies and Supersignal West Dura substrate (Pierce) were used.
Protein kinase assays.
Protein kinase assays were performed as described previously (8) using HA epitope-tagged Pkc1p immunoprecipitated from DZ2 cells and GST-Ndd1p derivatives as substrates. Where indicated, the Pkc1p inhibitor GF109203X (Calbiochem) was added (100 nM).
MS.
Bands of interest were excised from the gel and dehydrated using acetonitrile followed by vacuum centrifugation. Dried gel pieces were reduced with 10 mM dithiothreitol and alkylated with 55 mM iodoacetamide. Gel pieces were then washed alternately with 25 mM ammonium bicarbonate followed by acetonitrile. This was repeated, and the gel pieces were dried by vacuum centrifugation. Samples were digested with trypsin overnight at 37°C. Digested samples were analyzed by liquid chromatography-tandem mass spectrometry (LC-MS/MS) using a NanoAcquity LC (Waters, Manchester, United Kingdom) coupled to an LTQ Velos (Thermo Fisher Scientific, Waltham, MA). Peptides were concentrated on a precolumn (20 mm tall with a 180-μm inner diameter; Waters). The peptides were then separated using a gradient from 99% solution A (0.1% formic acid in water) and 1% solution B (0.1% FA in acetonitrile) to 25% solution B, in 45 min at 200 nl min−1, using a 75-mm-tall, 250-μm-inner-diameter, 1.7 μM ethylene bridged hybrid C18 analytical column (Waters). Peptides were selected for fragmentation automatically by data-dependent analysis.
Data produced were searched using Mascot (Matrix Science United Kingdom) against the Yeast_ORF database with phosphorylation at serine, threonine, and tyrosine selected as a variable modification. Data were validated using Scaffold (Proteome Software, Portland, OR).
Yeast growth, RNA analysis, and chromatin immunoprecipitation (ChIP).
The yeast strains used are listed in Table 1. KD592, expressing 13Myc epitope-tagged Pkc1p from the normal genomic locus, was constructed by introducing an integration cassette, amplified using the primers PKC1TagF/PKC1TagR with pFA6a-13Myc-kanMX6 as the template into W303-1a. The DZ2 strain expressing GAL1 promoter-regulated HA epitope-tagged Pkc1p was constructed by introducing an integration cassette, amplified using the primers ADS3984/3985 with pFA6a-kanMX6-PGAL1-3HA as the template, into W303-1a. Integrations were confirmed by PCR, DNA sequencing, and Western blot analysis.
Table 1.
Yeast strains
| Name | Genotype | Reference or source |
|---|---|---|
| W303-1a | MATa ade2-1 trp1-1 can1-100 leu2-3,112 his3-11 ura3 | |
| DL100 | MATa leu20 trp1 ura30 his4 CAN1R | 18 |
| DL523 | MATa ura30 trp1 his4 CAN1R pkc1::LEU2 Ycp50 pkc1ts | 18 |
| KD592 | MATa ade2-1 trp1-1 leu2-3,112 his3-11 ura3 PKC1-13myc KanR | This study |
| DZ2 | MATa ade2-1 trp1-1 leu2-3,112 his3-11 ura3 PGAL1-3HA-PKC1 KanR | This study |
| AP179 | MATa ade2-1 trp1-1 leu2-3,112 his3-11 ura3 ndd1::HIS3 pMW20-Ndd1 | 6 |
| DZ3 | MATa ade2-1 trp1-1 leu2-3,112 his3-11 ura3 ndd1::HIS3 pUS454 | 6 |
| DZ4 | MATa ade2-1 trp1-1 leu2-3,112 his3-11 ura3 ndd1::HIS3 pAS2010 | This study |
| BY4741 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 | EUROSCARF |
| Y01328 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 YJL095w::kanMX4 | EUROSCARF |
The in vivo function of the Ndd1p mutants was tested using a plasmid shuffle technique (3). The Ndd1p plasmid shuffle strain AP179 (containing a wild-type URA3 plasmid, pMW20Ndd1-URA, for Ndd1p maintenance [6]) was transformed with plasmids encoding wild-type or mutant variants of Ndd1p (pUS454 or pAS2010). Single transformants were plated on 5-fluoroorotic acid medium in the presence of galactose to select viable cells that had lost the wild-type pMW20Ndd1-URA plasmid.
Yeast cells were grown, GAL1-promoter-driven constructs induced, and transformations performed as described previously (8). Yeast cultures were synchronized in G1 phase by treatment with α-factor (5 μg/ml) for 3 h and in S phase by hydroxyurea (HU; 200 mM) treatment for 2.5 h prior to release into fresh medium to reenter the cell cycle. Where indicated, PKC activity was inhibited by adding 5 μg/ml cercosporamide to the medium upon release from the blocks. DNA content analyses were performed using propidium iodide-stained cells as described previously (6). Sensitivity tests were performed by plating 10-fold serial dilutions of yeast cultures onto synthetic defined (SD) plates in the presence or absence of sorbitol (1 M). The sensitivity of cells to hydroxyurea (200 mM) and 37°C was tested. Plates were incubated at either 30 or 37°C for 3 days.
RNA extraction and conventional and real-time reverse transcription-PCR (RT-PCR) analysis were carried out as described previously (6) using the following primer pairs: ADS1435-ADS1436 (CLB2), ADS1437-ADS1438 (SWI5), ADS1526-ADS1527 (CDC5), ADS3980-ADS3981 (TUB1), and ADS3978-ADS3979 (18S rRNA). All data were normalized to 18S rRNA levels in the same cells. Details of primers are shown in Table 2.
Table 2.
PCR primers
| Primer name | Locus | Sequence |
|---|---|---|
| RT-PCR | ||
| ADS1435 | CLB2-fw | CAGATGACTACGATATACAGTCTCG |
| ADS1436 | CLB2-rev | CAATTGGACTCACTAGATAATCC |
| ADS1437 | SWI5-fw | CACCTTCTAGGAGAAACAATAG |
| ADS1438 | SWI5-rev | GCT TACCTCTTGGTGTATATTG |
| ADS1526 | CDC5-fw | GAGACATTCTATCCTTAGATC |
| ADS1527 | CDC5-rev | GTGCTCTACGTATCCTTGCTTC |
| ADS3980 | TUB1-fw | TTACCCATTCTCTTGGTGGTG |
| ADS3981 | TUB1-rev | TTTGCACATGTCGTAAATAGCC |
| ADS1444 | CLN2-fw | GCAAGA ATACCACCAAGAAATCTC |
| ADS1445 | CLN2-rev | GAACTAGTTCAGAGAGTCGAGGTATAC |
| ADS3978 | 18S-fw | ACA ATTGGAGGGCAAGTCTG |
| ADS3979 | 18S-rev | ACGCCTGCTTTGAACACTCT |
| ChIP-qPCR | ||
| ADS1195 | SWI5-fw | CTGAAAGCCGATAGGAGTGGTACAC |
| ADS1196 | SWI5rev | TCCAACGATGCCTTTTTGTCTTCC |
| ADS1197 | CLB2-fw | CCAAATGCGGGTAACTATTTGTAT |
| ADS1198 | CLB2rev | ATGCCCATGCTATGAGATGCTAG |
| ADS2943 | CDC20fw | TCATGAAAGTGAATGTGAAGTG |
| ADS2944 | CDC20rev | GTCCTAAGCTTGATGACTGATCTCT |
| ADS1562 | CDC5fw | CGGGTAGGGCATTATGTAGC |
| ADS1563 | CDC5rev | GCCGCTTGAATTGGATTTAC |
| ADS2946 | CLN2fw | CTAACCTGCGAAATGTTGATCTGAC |
| ADS2945 | CLN2rev | CAGCTATAGAGTCTAGCAATGCTG |
| ADS2947 | TUB1fw | TGTTTCAGATCCTGCAATAGAAA |
| ADS2948 | TUB1rev | GGGTGGCGAGAACTGTTGTA |
| ADS3976 | RPL15aFw | CGCACCCTACATGACGTTTA |
| ADS3977 | RPL15aRev | AATTTTGCGTCAGCTCCAAG |
ChIP assays were performed as described previously (7) using anti-HA (Roche) or anti-myc (Santa Cruz) antibody and the following primer pairs: ADS1195-ADS1196 (SWI5), ADS1197-ADS1198 (CLB2), ADS2943-ADS2944 (CDC20), ADS2945-ADS2946 (CLN2), ADS3976-ADS3977 (RPL15a), ADS2947-ADS2948 (TUB1), and ADS1562-ADS1563 (CDC5) (Table 2).
Microscopy.
Nuclei and mitotic spindles were visualized by 4′,6-diamidino-2-phenylindole (DAPI) staining and immunofluorescence with Cy3-conjugated antibodies, respectively. Immunofluorescence analysis of microtubules was carried out essentially as described previously (6).
For localization of GFP-Pkc1p, overnight cultures of DZ2 cells transformed with pVD67 or pVD123 were resuspended at a final optical density at 600 nm (OD600) of 0.2 and grown at 30°C in yeast extract-peptone-dextrose (YPD) medium until mid-log phase prior to treatment and image acquisition. Cells were briefly centrifuged (800 × g for 3 min), fixed with 2% paraformaldehyde, washed and resuspended in a minimal volume of 1× phosphate-buffered saline (PBS), and spotted onto glass slides with coverslips prior to imaging. Images were acquired using an automated upright microscopic system 5500 B (Leica Microsystems GmbH) equipped with Chroma band pass emission filters (Ludl Electronic Products Ltd.) and a DFC360FX camera (Leica Microsystems GmbH).
Transcription factor activity and microarray analysis.
Duplicate RNA samples from wild-type PKC1 and temperature-sensitive pkc1 allele (pkc1ts) mutant strains were labeled and hybridized after a single round of amplification to Affymetrix Yeast Genome 2.0 array chips. The mRNA expression data were collected at two time points (blocked in HU and released from HU block for 60 min) from cells grown at 37°C in HU and in the presence of sorbitol for 2.5 h prior to release. The measurements were then normalized by using robust multichip analysis (RMA) to produce the microarray data set used in this study (see Table S1 in the supplemental material).
To determine transcription factor activation strength signals (TFAS), a recently developed bioinformatics approach (23) for transcription factor activation informatics that investigates transcription factor occupancies and mRNA expression levels was employed. The approach applies a mathematical model (30) to infer transcription factor activation strength from our microarray data in combination with the information of transcription factor binding sites obtained from the publicly available yeast genome regulatory map (10).
Microarray data accession number.
The primary data are deposited in ArrayExpress under accession number E-MEXP-3667.
RESULTS
Pkc1p controls the expression of CLB2 cluster genes.
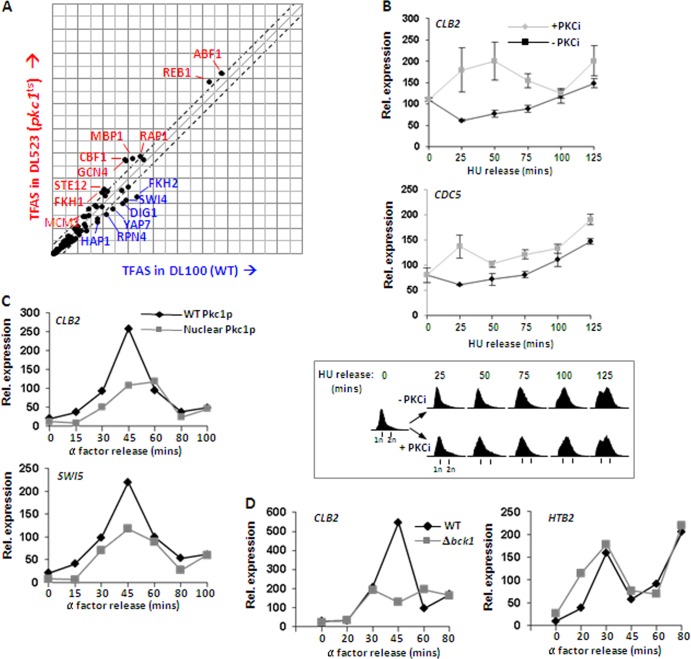
To determine the potential role of Pkc1p in controlling gene expression in S. cerevisiae, we performed microarray analysis using wild-type PKC1 (DL100) and pkc1ts (DL523) cells, grown at 37°C to inactivate Pkc1p in the pkc1ts mutant. These experiments were conducted in the presence of sorbitol to circumvent the lethality caused by disrupting Pkc1p function. To identify any effects on genes that are expressed late in the cell cycle, we performed the analysis on cells which had been synchronized in S phase by hydroxyurea (HU) treatment and released for 60 min to accumulate at G2/M. Reductions in the expression of genes such as SUN4 and PIR1 were observed upon pkc1 inactivation, and both of these genes encode cell wall components (see Table S1 in the supplemental material). Given the known association of Pkc1p with cell wall integrity (17), such changes are consistent with its predicted function.
We next associated the gene expression changes caused by Pkc1p inactivation with the prevalence of transcription factor binding events by using a discrete time-state-space model and expectation-maximization (EM) algorithm (30). The model determines transcription factor activation strength relative to target mRNA expression levels and transcription factor binding site occurrence. This transcription factor analysis identified that Fkh2, Swi4, Dig1, Yap7, Rpn4, and HapI all exhibit reductions in their activity profiles upon shifting the pkc1ts strain to the nonpermissive temperature (Fig. 1A). These transcription factors are considered potential coregulators of Pkc1p, since the factors are predicted to be highly active when Pkc1p is active, but they do not have regulatory activities when Pkc1p is inactive. Strikingly, the transcription factor Fkh2p was identified among this group of regulators with strong association with genes that were differentially expressed at higher levels in wild-type PKC1 cells (Fig. 1A). Fkh2p forms part of the Mcm1p-Fkh2p-Ndd1p complex which controls the CLB2 gene cluster. Furthermore, previous genetic studies in S. cerevisiae cells lacking Pkc1p suggested an important role for Pkc1p in G2 and/or M phase (19). We therefore focused in more detail on the role of Pkc1p in controlling expression changes of genes in the CLB2 cluster. To examine the direct consequences of Pkc1p inhibition, rather than using the temperature-sensitive pkc1 allele, we adopted an approach using the Pkc1p inhibitor cercosporamide (32), which avoids the complications associated with the heat shock response. DZ3 cells expressing wild-type Ndd1p were synchronized in early S phase by treatment with HU. Cells were either released directly from this block to reenter the cell cycle or were treated with cercosporamide upon release. The expression of two CLB2 cluster genes, CLB2 and CDC5, was found to be induced at earlier time points in the presence of the Pkc1p inhibitor, resulting in an earlier peak of expression (Fig. 1B). Significantly, this was not due to accelerated progress through the cell cycle in the presence of the inhibitor (Fig. 1B, bottom). These observations suggest that Pkc1p normally exerts a repressive effect on the expression of CLB2 cluster genes. To test this hypothesis further, we next examined the expression of genes in the CLB2 cluster in cells ectopically expressing either wild-type Pkc1p or a form of Pkc1p that accumulates in the nucleus due to the presence of a defective nuclear export signal (9). Compared to cells expressing wild-type Pkc1p, both CLB2 and SWI5 expression were substantially reduced upon expression of the constitutively nuclear mutant form of Pkc1p (Fig. 1C). A similar effect was observed in cells where the endogenous PKC1 gene had been replaced with this constitutively nuclear form of Pkc1p (data not shown).
Fig 1.
Pkc1p regulates CLB2 cluster gene expression. (A) The association of transcription factors with differential mRNA expression levels in wild-type (WT) and pkc1ts cells. Gene expression changes in WT and pkc1ts cells treated with hydroxyurea (HU) and released for 60 min were compared by microarray analysis, and changes in gene expression levels associated with transcription factor binding activities to produce a transcription factor activation strength signal (TFAS) were analyzed. (B and C) Real-time RT-PCR analysis of the indicated genes in DZ3 cells in the presence or absence of the PKC inhibitor (PKCi) cercosporamide following release from HU block at the indicated times (B) or W303-1a cells expressing either WT or a nuclear export mutant form of Pkc1p following release from α-factor block at the indicated times (C). Data are normalized to 18S rRNA levels. Cell cycle profiles were obtained by fluorescence-activated cell sorter (FACS) analysis at equivalent time points and are shown beneath panel B. (D) Real-time RT-PCR analysis of CLB2 and HTB2 expression in wild-type or Δbck1 BY4741 cells grown in sorbitol for the indicated times following release from α-factor block.
Pkc1p is known to act upstream from the Mpk1p mitogen-activated protein (MAP) kinase pathway (reviewed in reference 17). To probe whether the Mpk1p pathway also has a role in dampening down the expression of CLB2 cluster genes, we compared the expression of CLB2 throughout the cell cycle in a wild-type strain to that of a strain containing a deletion of the gene encoding the kinase Bck1p, which lies between Pkc1p and Mpk1p. In contrast to the increases in gene expression seen upon disrupting Pkc1p activity, loss of Bck1p causes reduced CLB2 expression. The expression of an early cell cycle marker, HTB2, remains largely unaltered. This suggests that Pkc1p functions to downregulate late cell cycle gene expression independently from the Mpk1p pathway.
These results are therefore consistent with a model in which Pkc1p acts in the nucleus to negatively regulate the expression of genes in the CLB2 cluster and thereby contribute to their correct temporal expression pattern in the cell division cycle.
Pkc1p is recruited to CLB2 cluster gene promoters.
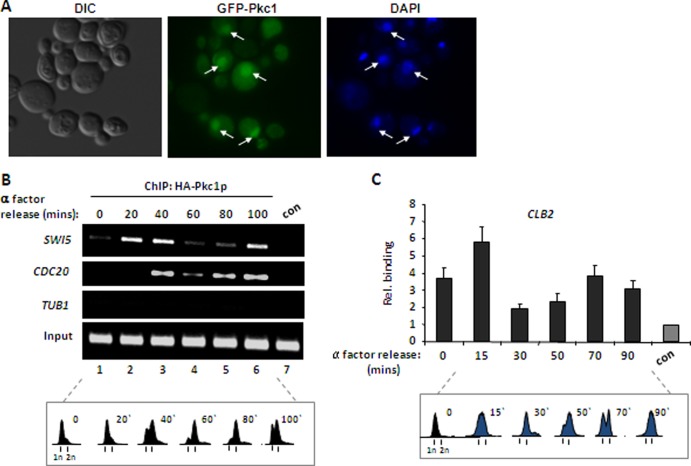
Our results suggest that Pkc1p normally has a role in the nucleus. To provide further evidence for this, we first examined whether Pkc1p could be detected in the nucleus in a strain expressing green fluorescent protein (GFP)-tagged Pkc1p (9). GFP-Pkc1p was detected in the nucleus of a subset of asynchronously growing cells, consistent with a previous study (Fig. 2A) (9). Interestingly, nuclear localization was often associated with budding cells, suggesting a potential link to late cell cycle events (e.g., the bottom two cells in Fig. 2A). Given this nuclear localization, we used ChIP analysis to examine whether Pkc1p is associated with CLB2 gene cluster promoters and hence directly implicates the protein in the responses that occur at these sites. First, we examined the occupancy of overexpressed HA-tagged Pkc1p on CLB2 gene cluster promoters at different points in the cell cycle. Little HA-Pkc1p was associated with the SWI5 and CDC20 promoters in G1 phase cells maintained in an α-factor-induced block (Fig. 2B, lane 1). However, after release from this block, Pkc1p associated with both of these promoters as cells progressed through S phase and, moreover, binding was reduced as cells progressed through G2 and M phases (Fig. 2B). To study this phenomenon further, we constructed a strain expressing a Myc epitope-tagged Pkc1p protein from its normal chromosomal locus and analyzed the association of Myc-Pkc1p with the CLB2 promoter throughout the cell cycle. In agreement with the analysis using HA-Pkc1p, enhanced promoter binding of Myc-Pkc1p was detected as cells accumulated in S phase, and this binding subsequently declined as cells progressed into the G2 and M phases (Fig. 2C). Relatively more binding of Pkc1p is detected in α-factor-blocked cells in the latter experiment. However, in addition to the use of different tags and promoters for expression of epitope-tagged Pkc1p and the different promoters analyzed, the growth conditions are not identical between the two experiments as galactose is included in the meduim when HA-Pkc1p is overexpressed.
Fig 2.
Pkc1p is dynamically recruited to CLB2 cluster gene promoters. (A) GFP fluorescence was used to detect GFP-tagged Pkc1p (middle) and DAPI staining to detect DNA (right). Nuclear Pkc1p is indicated by the arrows. A differential inference contrast (DIC) image of the cells is shown (left). (B and C) ChIP analysis of Pkc1p binding to the indicated gene promoters using either anti-HA antibody to detect HA epitope-tagged Pkc1p in DZ2 cells and analyzed by semiquantitative PCR (B) or anti-Myc antibody in cells expressing Myc epitope-tagged Pkc1p from its natural locus (KD592) measured by real-time PCR (C). Binding was analyzed following release from α-factor block in G1 phase at the indicated times. The control (con) is ChIP with either anti-HA antibody from logarithmically growing WT (W303-1a) cells (B) or with nonspecific IgG from cells expressing Myc epitope-tagged Pkc1p (KD592) (C). Data in panel C are shown relative to the control background binding to the CLB2 promoter (con) (taken as 1). Cell cycle profiles were obtained by FACS analysis at equivalent time points and are shown beneath each figure.
Collectively, these data revealed that Pkc1p associates with CLB2 gene cluster promoters in a cell cycle-dependent manner, and, importantly, the timing of binding is consistent with a role for Pkc1p as a negative regulator of the expression of CLB2 cluster genes prior to the G2 and M phases.
Pkc1p phosphorylates Ndd1p.
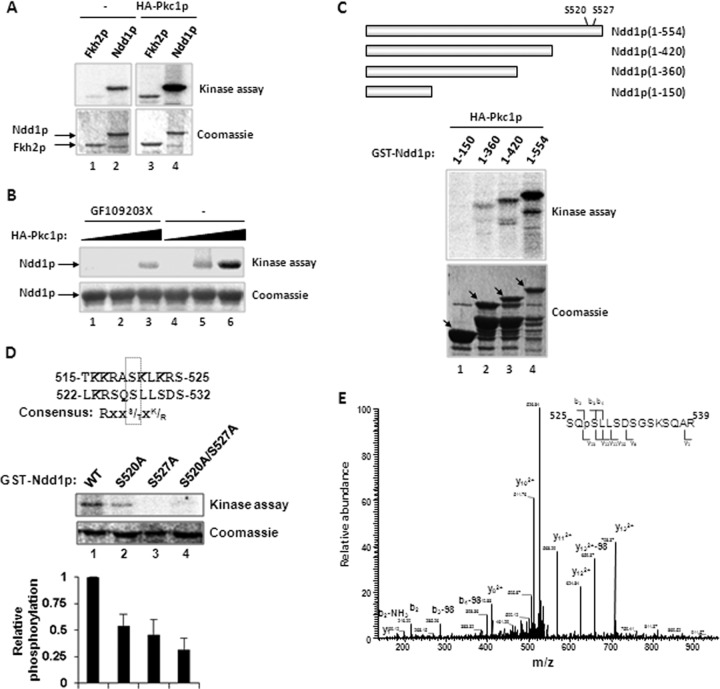
As Pkc1p associates with and regulates the expression of genes of the CLB2 cluster, this raised the possibility that the Mcm1p-Fkh2p-Ndd1p transcription factor complex which controls these promoters is a direct target. Previous work had shown that Ndd1p and the C-terminal region of Fkh2p are targeted by protein kinases to regulate their activity during the cell cycle (6, 8, 26, 28). Hence, we next tested the ability of Pkc1p to phosphorylate either Ndd1p and/or the C-terminal region of Fkh2p. HA-tagged Pkc1p precipitated from cells (DZ2) was found to strongly phosphorylate Ndd1p but only weakly phosphorylate Fkh2p (Fig. 3A). We therefore subsequently focused on Ndd1p. To establish that Pkc1p, rather than a contaminating coprecipitating kinase, was involved, the assays were repeated in the presence of increasing concentrations of the specific PKC inhibitor GF109203X. This inhibitor strongly reduced the levels of Ndd1p phosphorylation, thereby validating the role of Pkc1p in this phosphorylation event (Fig. 3B). Finally, we mapped the Pkc1p-dependent phosphorylation sites in Ndd1p. First, we created a series of C-terminal Ndd1p truncations (Fig. 3C). Analysis of the ability of Pkc1p to phosphorylate these truncations revealed a substantial drop in phosphorylation upon deletion of the C-terminal 134 amino acids of Ndd1p (Fig. 3C). Subsequent inspection of this region revealed two closely positioned sites (Ser520 and Ser527) which showed partial matches to the Pkc1p consensus sequence (Fig. 3D) (24). Importantly, mutation of either of these sites individually or in combination caused a large reduction in Pkc1p-mediated phosphorylation of Ndd1p, whereas mutation of another serine residue (Ser409) in Ndd1p had little effect (Fig. 3D; data not shown).
Fig 3.
Pkc1p phosphorylates Ndd1p. In vitro kinase assays using HA epitope-tagged Pkc1p immunoprecipitated from DZ2 cells and the following substrates: GST-Fkh2p(458-862) and GST-Ndd1p(1-554) (A), GST-Ndd1p(1-554) (B), the indicated truncation series of GST-Ndd1p constructs (C), or the indicated wild-type and mutant forms of GST-Ndd1p(1-554) (D). Mock HA-immunoprecipitated material from W303-1a cells was used as a control (A; lanes 1 and 2). (B) Increasing concentrations of HA-Pkc1p (relative molar amounts, 1, 2, and 4) were added in the presence and absence of GF109203X. The top panels in each case are phosphorimages, and the bottom panels are Coomassie-stained gels of the input GST fusion proteins. The data from four experiments were quantified and are shown graphically below panel D. The schematics in C and D show the structures of the truncation mutant versions of Ndd1p and the location and local sequence contexts of the Pkc1p target sites. (E) Product ion spectra for the triply charged precursor ion, [544.23]3+. The spectra include full annotation of product ions detected, which include both b- and y-ions on either side of the site of modification, demonstrating that the phosphate is located on Ser527.
To examine whether either of these two sites is phosphorylated in vivo, we immunoprecipitated HA-tagged Ndd1p from yeast cell lysates and subjected it to mass spectrometry analysis. A phosphopeptide match correlating to phosphorylation at Ser527 was identified with high confidence, as it was found at greater than 90.0% probability, as specified by the Peptide Prophet algorithm (12). The matched spectrum was further validated by manual inspection and was confirmed to include enough diagnostic ions to support the identity of the peptide, in addition to both b- and y-fragment ions on either side of the phosphorylated residue, Ser527 (Fig. 3E). No fragment ions were observed, supporting phosphorylation at either of the other serine residues present in the peptide. We were unable to detect phosphorylation at Ser520 due to the lack of suitable peptides caused by the large number of surrounding basic residues.
Collectively, these data indicate that Ndd1p is phosphorylated at Ser527 in vivo and, furthermore, that it is a direct target for Pkc1p in vitro, with Ser520 and Ser527 representing the two major sites of Pkc1p-dependent phosphorylation.
Mutation of the Pkc1p-dependent phosphorylation sites of Ndd1p causes temporal changes in CLB2 cluster gene expression and growth defects.
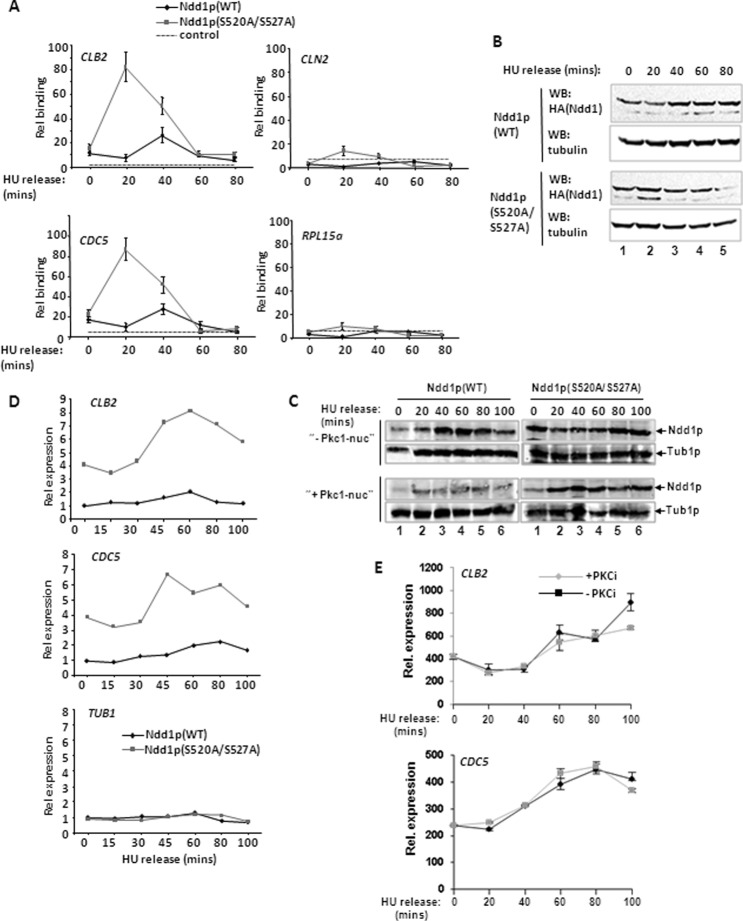
To establish the significance of Pkc1p-dependent phosphorylation of Ndd1p, we determined the effect of mutating its two target sites in Ndd1p. Endogenous Ndd1p was replaced with either HA epitope-tagged wild-type Ndd1p or the mutant Ndd1p(S520A/S527A) using the plasmid shuffle technique. First, we tested the binding of Ndd1p to the promoters of CLB2 cluster genes. Following release from an HU block, Ndd1p(S520A/S527A) binding to the CLB2 and CDC5 promoters occurred earlier in the cell cycle and in a more robust manner than the binding of wild-type Ndd1p (Fig. 4A). This enhanced binding cannot simply be attributed to increased levels of Ndd1p expression (Fig. 4B). However, in contrast to wild-type Ndd1p, Ndd1p(S520A/S527A) does not appear to be stabilized further as cells proceed through the cell cycle, suggesting a link between Pkc1p-dependent phosphorylation and protein stability (Fig. 4B). To further substantiate this link, we compared the levels of wild-type Ndd1p and the Ndd1p(S520A/S527A) mutant in the presence of the constitutively nuclear form of Pkc1p. As expected from a direct action of Pkc1p on Ndd1p, overexpression of nuclear Pkc1p caused reductions in Ndd1p levels (Fig. 4C, left). In contrast, the Ndd1p(S520A/S527A) mutant was largely refractory to such decreases in the presence of nuclear Pkc1p (Fig. 4C, right).
Fig 4.
Ndd1p(S520A/S527A) causes aberrant expression of CLB2 cluster genes. (A) ChIP analysis of Ndd1p binding to the indicated gene promoters using anti-HA antibody to detect HA epitope-tagged Ndd1p in AP179 cells expressing either wild-type (WT) Ndd1p or Ndd1p(S520A/527A). Cells were analyzed following release from the HU block at the indicated times. The dotted line represents a control ChIP with anti-HA antibody from logarithmically growing WT W303-1a cells. Data are shown relative to this control level (taken as 1). (B and C) Western blot (WB) analysis of Ndd1p expression from DZ3 cells expressing wild-type (WT) Ndd1p or DZ4 cells expressing Ndd1p(S520A/S527A) at the indicated times following release from the HU block. Tubulin levels are shown as a control. In panel C, cells also ectopically expressed WT Pkc1p (top) or a nuclear export mutant form of Pkc1p (Pkc1-nuc) (bottom). (D) Real-time RT-PCR analysis of the expression of the indicated genes in AP179 cells expressing either wild-type (WT) Ndd1p or Ndd1p(S520A/S527A) at the indicated times following release from the HU block. Data are normalized to 18S rRNA levels and are shown relative to the levels of expression in the strain expressing Ndd1p(WT) in HU-treated cells at time zero (taken as 1). (E) Real-time RT-PCR analysis of the indicated genes in DZ4 cells [expressing Ndd1p(S520A/S527A)] in the presence or absence of the PKC inhibitor (PKCi) cercosporamide following release from the HU block at the indicated times.
We next examined whether the enhanced promoter occupancy by Ndd1p(S520A/S527A) was also accompanied by increased expression of CLB2 cluster genes. Compared to cells expressing wild-type Ndd1p, cells expressing Ndd1p(S520A/S527A) exhibited much higher levels of expression of both the CLB2 and CDC5 genes (Fig. 4D). These results therefore demonstrate that the loss of the Pkc1p phosphorylation sites in Ndd1p results in deregulated CLB2 cluster gene expression and is consistent with the model that Pkc1p phosphorylation directly inhibits the expression of the CLB2 gene cluster. To further substantiate this point, we examined the consequences of Pkc1p inhibition on the expression of CLB2 cluster genes in cells expressing Ndd1p(S520A/S527A). In contrast to the early activation and increases in gene expression seen in cells expressing wild-type Ndd1p (Fig. 1B), Pkc1p inhibition had little discernible effect on CLB2 and CDC5 expression in cells expressing Ndd1p(S520A/S527A) (Fig. 4E).
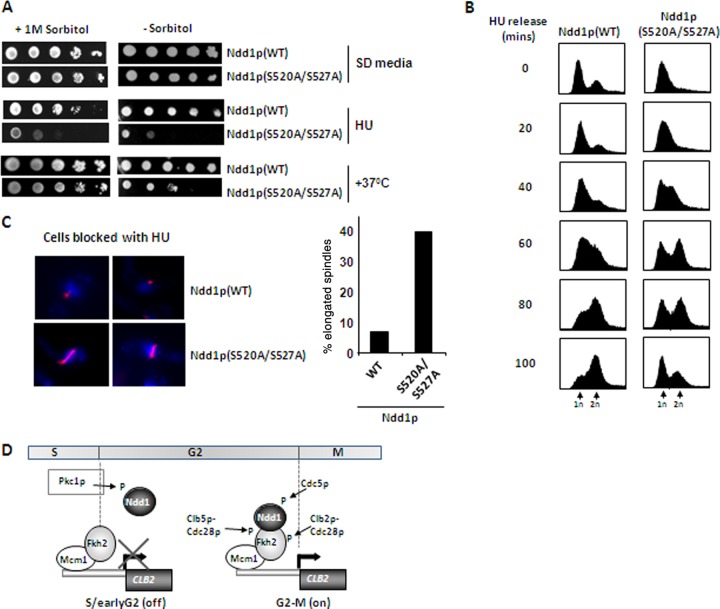
To establish the phenotypic consequences associated with loss of Pkc1p-dependent phosphorylation of Ndd1p, we first examined the growth of cells expressing either wild-type or mutant Ndd1p(S520A/S527A) under different conditions in the presence and absence of sorbitol. Sorbitol was used, as this can provide osmotic support to rescue defects caused in cell wall damage signaling via the Pkc1p pathway (18). Under normal growth conditions on SD medium, cells expressing Ndd1p(S520A/S527A) grew slightly more poorly than cells expressing wild-type Ndd1p (Fig. 5A, top). However, upon treatment with HU, which slows progression through the cell cycle, or incubation of cells at 37°C, which causes pleiotropic effects, including cell wall damage, cells expressing Ndd1p(S520A/S527A) grew much more poorly in the absence of sorbitol. Both of these treatments can affect cellular events via the Pkc1p pathway (reviewed in reference 17). Interestingly, sorbitol rescued the defects associated with growth at 37°C but not the defects associated with growth in HU, which has no known link to the cell wall integrity pathway. Hence, these results are consistent with a role for Pkc1p signaling via Ndd1p phosphorylation and, moreover, suggest that regulation of Ndd1p by Pkc1p is linked, at least partially, to the response of cells to defects in cell wall integrity. We next examined cell cycle processes more closely, as these are one of the potential causative factors for the reduced viability of Ndd1p(S520A/S527A)-expressing cells treated with HU. Compared to cells expressing wild-type Ndd1p, cells expressing Ndd1p(S520A/S527A) progressed through the cell cycle at a much higher rate upon release from HU block despite the fact that more of the cells of the mutant strain were initially in early S phase (Fig. 5B). It is not clear why Ndd1p(S520A/S527A) mutant cells consistently show better HU-induced synchrony than cells overexpressing wild-type Ndd1p, but this again emphasizes important phenotypic differences in this mutant. Furthermore, analyses of cells expressing Ndd1p(S520A/S527A) arrested by HU treatment showed that a large proportion of these cells contained elongated nuclear spindles, and a proportion of these (12%) were wrongly oriented (Fig. 5C). Thus, cells expressing Ndd1p(S520A/S527A) appear to have lost the usual inhibitory signals which impede subsequent transition to mitotic events.
Fig 5.
Mutation of the Pkc1p-dependent phosphorylation sites of Ndd1p causes cell cycle defects and different stress-sensitive phenotypes. (A) Tenfold serial dilutions of DZ3 cells expressing WT Ndd1p or DZ4 cells expressing Ndd1p(S520A/S527A) were plated onto SD medium in the presence or absence of 1 M sorbitol. Where indicated, HU was added to the medium or cells were grown at 37°C. (B) DNA content analysis of AP179 cells expressing either WT Ndd1p or Ndd1p(S520A/S527A) at the indicated times after release from an HU block. (C) Nuclear spindles were stained with antitubulin antibody (red), and nuclei were detected with DAPI (blue) in AP179 cells expressing either WT Ndd1p or Ndd1p(S520A/S527A) maintained in HU for 3 h. The percentage of cells containing elongated spindles is shown on the right. (D) Model depicting the kinases acting on the Mcm1p-Fkh2p-Ndd1p complex during the cell cycle. Pkc1p phosphorylates Ndd1p to inhibit activation during S and early G2 phases. Subsequent phosphorylation by cyclin-Cdk and Cdc5p kinases results in activation of the complex and hence CLB2 cluster gene transcription during late G2 phase.
Taken together, these results demonstrate that Ndd1p(S520A/S527A) expression causes transcriptional and phenotypic defects which are consistent with the linkage of Pkc1p to Ndd1p phosphorylation and, more generally, to the role of Pkc1p in the inhibition of G2 and M phase-specific regulatory events.
DISCUSSION
In S. cerevisiae, the Mcm1p-Fkh2p-Ndd1p transcriptional regulatory complex controls late cell cycle-dependent processes (14, 16, 20, 38; reviewed in reference 22). Furthermore, several aspects of the regulatory circuitry affecting the activity of this complex have been unraveled. In particular, it has been shown that the cell cycle-regulated protein kinases Clb5p-Cdc28p, Clb2p-Cdc28p, and Cdc5p positively control the activity of this transcription factor complex by direct phosphorylation (6, 8, 26, 28; reviewed in reference 22). Previous studies revealed that protein kinase C, Pkc1p, is linked to the regulation of late cell cycle events in yeast, although the underlying basis for this connection was unclear (reviewed in reference 17). Excitingly, here we have found that Pkc1p acts to negatively regulate the Mcm1p-Fkh2p-Ndd1p complex by direct phosphorylation of Ndd1p and, furthermore, that this regulation is essential for normal cell cycle progression in G2 and M phases.
Previous studies have hinted at a role for the Pkc1p pathway in controlling the transition between the G2 and M phases of the cell cycle (19; reviewed in reference 17), and genetic interactions with RSC components (5, 11; reviewed in reference 17) further suggested that the Pkc1p pathway functions, at least partially, at the level of gene regulation in this context. Here, we provide evidence that one direct way in which Pkc1p participates in G2 and M phase control is through phosphorylation of the transcriptional coactivator Ndd1p. We identified two Pkc1p phosphorylation sites in Ndd1p in vitro, and Ndd1p alleles containing mutations in both of these sites were refractory to Pkc1p activity in vivo. However, it is currently unclear whether one of these sites is more important than the other for the activity of Pkc1p toward Ndd1p. Furthermore, while we demonstrate an important correlation between Pkc1p-mediated phosphorylation of Ndd1p in vitro (Fig. 3) and the effects of Pkc1p on Ndd1p function in vivo (Fig. 4 and 5), it remains formally possible that there is an intermediary kinase involved which is activated by Pkc1p and phosphorylates the sites we have identified in Ndd1p. However, it appears unlikely that the Mpk1p pathway is involved, as inactivation of the gene encoding the Mpk1p regulator, BCK1, does not have the same effects on CLB2 cluster gene expression as the inhibition of Pkc1p activity.
Mechanistically, Pkc1p-mediated phosphorylation of Ndd1p delays its recruitment to CLB2 cluster gene promoters until late G2 phase. Part of the reason for this appears to be due to increased Ndd1p instability and is consistent with the recently described role for Pkc1p in promoting the degradation of Bni1 and Sec3 upon damaging the cell wall/membrane (13). Accordingly, disruption of Pkc1p-dependent phosphorylation of Ndd1p leads to its accumulation and early recruitment and causes premature activation of expression of CLB2 cluster genes. Importantly, this acceleration likely leads to inappropriate temporal production of cell cycle regulatory components, which leads to subsequent cell cycle defects. The acquisition of elongated and misaligned spindles in HU-blocked cells where the Pkc1p-dependent phosphorylation of Ndd1p has been prevented reveals some of the defects that arise. Overall, this leads to reductions in cell viability and helps explain the lethality associated with Pkc1p loss (reviewed in reference 17). It is not clear which signals contribute to the activation of Pkc1p in the context of the cell cycle, although signaling from the cell wall is an attractive possibility, as this undergoes significant changes during the cell cycle through, for example, the formation of a bud that is destined to become the new daughter cell. Indeed, the cell wall integrity pathway has previously been shown to signal through Fkh2p to control CLB2 cluster gene expression (34). In keeping with such a role, we show that sorbitol rescues defects caused by cell wall disruption by growing cells containing a Pkc1p phosphorylation-insensitive mutant version of Ndd1p at 37°C (Fig. 5A). However, in contrast, sorbitol was unable to rescue the defects associated with these mutant cells when cultured in the presence of HU. This suggests that Pkc1p is also activated by intrinsic S phase signals and/or the incomplete DNA replication caused by HU treatment. In this respect, Pkc1p could be regarded as a checkpoint kinase which acts early in the cell cycle to suppress CLB2 cluster gene expression until the correct conditions are established for transition to later cell cycle phases. Further studies are needed to dissect these upstream pathways.
Little is known about the potential nuclear function(s) of Pkc1p in S. cerevisiae, although there is growing evidence of a role for PKC isoforms in the nucleus in higher eukaryotes (reviewed in reference 21). This is exemplified by the recent demonstration that PKC-θ was found to be associated with chromatin in the regulatory regions of inducibly activated genes in human T lymphocytes (33). Previous evidence showed that Pkc1p could be found in the nucleus in S. cerevisiae (9), and indeed we confirmed this observation. Importantly, we also demonstrate temporally regulated recruitment of Pkc1p to the promoters of genes in the CLB2 cluster, providing further direct evidence that Pkc1p is present in the nucleus. It is not clear what controls the temporal recruitment of Pkc1p to chromatin, but the kinetics of recruitment are consistent with its role in suppressing premature activation of the Mcm1p-Fkh2p-Ndd1p transcription factor complex in the cell cycle. Indeed, binding of Pkc1p is lost as cells pass from G2 into M phase, which coincides with the activation of its target Ndd1p and the subsequent increased expression of the CLB2 cluster genes. It is important to also emphasize that our findings likely explain only a subset of the phenotypes caused by the loss of Pkc1p activity. Indeed, strains harboring the Pkc1p phosphorylation-insensitive Ndd1p mutant protein only partially phenocopy pkc1Δ mutant cells, with the effects largely limited to gene expression events that normally occur in late G2 phase/M phase. Thus, it is likely that Pkc1p has other nuclear roles beyond what we have described. Indeed, Pkc1p has a range of other documented functions and also signals via the MAP kinase Mpk1p through to other cell cycle-regulated transcription factor complexes, such as SBF and MBF. However, Pkc1p is not thought to act in the nucleus when driving this pathway (reviewed in reference 17). In the context of CLB2 cluster gene expression, it is possible that in addition to inhibiting Ndd1p function, it also promotes the activities of repressive proteins, such as Sin3p, Yox1p, and Yhp1p, which act on these genes (27, 36). Further studies are required to establish such putative roles.
In summary, this study has revealed a new, important, additional regulatory component to the circuitry that determines the timing of late cell cycle-dependent gene expression (Fig. 5D). Pkc1p acts to inhibit the activation of CLB2 gene cluster promoters by targeting the Ndd1p coactivator and delaying its recruitment to these promoters. The subsequent activation of Clb5p-Cdc28p begins the activation process by phosphorylation of Fkh2p, and then Clb2p-Cdc28p and Cdc5p potentiate this effect through a positive feedback loop. Thus, both negatively and positively acting kinases converge to affect the activity of the Mcm1p-Fkh2p-Ndd1p transcription factor complex. It will be very interesting to explore whether any of the PKC isoforms have an analogous regulatory role in the cell cycle in higher eukaryotes.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Karren Palmer for technical support and to staff in the core bioinformatics, flow cytometry, and microscopy facilities. We also thank members of our laboratories for comments on the manuscript and helpful discussions and Martha Cyert and David Levin for reagents.
This work was supported by the BBSRC, the Wellcome Trust, and a Royal Society-Wolfson award to A.D.S.
Footnotes
Published ahead of print 10 September 2012
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1. Althoefer H, Schleiffer A, Wassmann K, Nordheim A, Ammerer G. 1995. Mcm1 is required to coordinate G2-specific transcription in Saccharomyces cerevisiae. Mol. Cell. Biol. 15:5917–5928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bähler J. 2005. Cell-cycle control of gene expression in budding and fission yeast. Annu. Rev. Genet. 39:69–94 [DOI] [PubMed] [Google Scholar]
- 3. Boeke JD, Trueheart J, Natsoulis G, Fink GR. 1987. 5-Fluoroorotic acid as a selective agent in yeast molecular genetics. Methods Enzymol. 154:164–175 [DOI] [PubMed] [Google Scholar]
- 4. Breeden LL. 2000. Cyclin transcription: timing is everything. Curr. Biol. 10:R586–R588 [DOI] [PubMed] [Google Scholar]
- 5. Chai B, Huang J, Cairns BR, Laurent BC. 2005. Distinct roles for the RSC and Swi/Snf ATP-dependent chromatin remodelers in DNA double-strand break repair. Genes Dev. 19:1656–1661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Darieva Z, et al. 2006. Polo kinase controls cell-cycle-dependent transcription by targeting a coactivator protein. Nature 444:494–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Darieva Z, et al. 2010. A competitive transcription factor binding mechanism determines the timing of late cell cycle-dependent gene expression. Mol. Cell 38:29–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Darieva Z, et al. 2003. Cell cycle regulated transcription through the FHA domain of Fkh2p and the coactivator Ndd1p. Curr. Biology. 13:1740–1745 [DOI] [PubMed] [Google Scholar]
- 9. Denis V, Cyert MS. 2005. Molecular analysis reveals localization of Saccharomyces cerevisiae protein kinase C to sites of polarized growth and Pkc1p targeting to the nucleus and mitotic spindle. Eukaryot. Cell 4:36–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Harbison CT, et al. 2004. Transcriptional regulatory code of a eukaryotic genome. Nature 431:99–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hosotani T, Koyama H, Uchino M, Miyakawa T, Tsuchiya E. 2001. PKC1, a protein kinase C homologue of Saccharomyces cerevisiae, participates in microtubule function through the yeast EB1 homologue, BIM1. Genes Cells 6:775–788 [DOI] [PubMed] [Google Scholar]
- 12. Keller A, Nesvizhskii AI, Kolker E, Aebersold R. 2002. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal. Chem. 74:5383–5392 [DOI] [PubMed] [Google Scholar]
- 13. Kono K, Saeki Y, Yoshida S, Tanaka K, Pellman D. 2012. Proteasomal degradation resolves competition between cell polarization and cellular wound healing. Cell 150:151–164 [DOI] [PubMed] [Google Scholar]
- 14. Koranda M, Schleiffer A, Endler L, Ammerer G. 2000. Forkhead-like transcription factors recruit Ndd1 to the chromatin of G2/M-specific promoters. Nature 406:94–98 [DOI] [PubMed] [Google Scholar]
- 15. Krause SA, Xu H, Gray JV. 2008. The synthetic genetic network around PKC1 identifies novel modulators and components of protein kinase C signaling in Saccharomyces cerevisiae. Eukaryot. Cell 7:1880–1887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kumar R, et al. 2000. Forkhead transcription factors, Fkh1p and Fkh2p, collaborate with Mcm1p to control transcription required for M-phase. Curr. Biol. 10:896–906 [DOI] [PubMed] [Google Scholar]
- 17. Levin DE. 2011. Regulation of cell wall biogenesis in Saccharomyces cerevisiae: the cell wall integrity signaling pathway. Genetics 189:1145–1175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Levin DE, Bartlett-Heubusch E. 1992. Mutants in the S. cerevisiae PKC1 gene display a cell cycle-specific osmotic stability defect. J. Cell Biol. 116:1221–1229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Levin DE, Fields FO, Kunisawa R, Bishop JM, Thorner J. 1990. A candidate protein kinase C gene, PKC1, is required for the S. cerevisiae cell cycle. Cell 62:213–224 [DOI] [PubMed] [Google Scholar]
- 20. Loy CJ, Lydall D, Surana U. 1999. NDD1, a high-dosage suppressor of cdc28-1N, is essential for expression of a subset of late-S-phase-specific genes in Saccharomyces cerevisiae. Mol. Cell. Biol. 19:3312–3327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Martelli AM, Evangelisti C, Nyakern M, Manzoli FA. 2006. Nuclear protein kinase C. Biochim. Biophys. Acta 1761:542–551 [DOI] [PubMed] [Google Scholar]
- 22. McInerny CJ. 2011. Cell cycle regulated gene expression in yeasts. Adv. Genet. 73:51–85 [DOI] [PubMed] [Google Scholar]
- 23. McMaster A, et al. 2011. Ultradian cortisol pulsatility encodes a distinct, biologically important signal. PLoS One 6:e15766 doi:10.1371/journal.pone.0015766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Nishikawa K, Toker A, Johannes FJ, Songyang Z, Cantley LC. 1997. Determination of the specific substrate sequence motifs of protein kinase C isozymes. J. Biol. Chem. 272:952–960 [DOI] [PubMed] [Google Scholar]
- 25. Pic A, et al. 2000. The forkhead protein Fkh2 is a component of the yeast cell cycle transcription factor SFF. EMBO J. 19:3750–3761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Pic-Taylor A, Darieva Z, Morgan BA, Sharrocks AD. 2004. Regulation of cell cycle-specific gene expression through cyclin-dependent kinase-mediated phosphorylation of the forkhead transcription factor Fkh2p. Mol. Cell. Biol. 24:10036–10046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Pramila T, Miles S, GuhaThakurta D, Jemiolo D, Breeden LL. 2002. Conserved homeodomain proteins interact with MADS box protein Mcm1 to restrict ECB-dependent transcription to the M/G1 phase of the cell cycle. Genes Dev. 16:3034–3045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Reynolds D, Shi BJ, McLean C, Katsis F, Kemp Dalton B S. 2003. Recruitment of Thr 319-phosphorylated Ndd1p to the FHA domain of Fkh2p requires Clb kinase activity: a mechanism for CLB cluster gene activation. Genes Dev. 17:1789–1802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Roffey J, et al. 2009. Protein kinase C intervention: the state of play. Curr. Opin. Cell Biol. 21:268–279 [DOI] [PubMed] [Google Scholar]
- 30. Sanguinetti G, Lawrence ND, Rattray M. 2006. Probabilistic inference of transcription factor concentrations and gene-specific regulatory activities. Bioinformatics 22:2755–2781 [DOI] [PubMed] [Google Scholar]
- 31. Shore P, Sharrocks AD. 1994. The transcription factors Elk-1 and serum response factor interact by direct protein-protein contacts mediated by a short region of Elk-1. Mol. Cell. Biol. 14:3283–3291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sussman A, et al. 2004. Discovery of cercosporamide, a known antifungal natural product, as a selective Pkc1 kinase inhibitor through high-throughput screening. Eukaryot. Cell 3:932–943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sutcliffe EL, et al. 2011. Chromatin-associated protein kinase C-θ regulates an inducible gene expression program and microRNAs in human T lymphocytes. Mol. Cell 41:704–719 [DOI] [PubMed] [Google Scholar]
- 34. Suzuki M, et al. 2004. Dynactin is involved in a checkpoint to monitor cell wall synthesis in Saccharomyces cerevisiae. Nat. Cell Biol. 6:861–871 [DOI] [PubMed] [Google Scholar]
- 35. Uetz P, et al. 2000. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature 403:623–627 [DOI] [PubMed] [Google Scholar]
- 36. Veis J, Klug H, Koranda M, Ammerer G. 2007. Activation of the G2/M-specific gene CLB2 requires multiple cell cycle signals. Mol. Cell. Biol. 27:8364–8373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wittenberg C, Reed SI. 2005. Cell cycle-dependent transcription in yeast: promoters, transcription factors, and transcriptomes. Oncogene 24:2746–2755 [DOI] [PubMed] [Google Scholar]
- 38. Zhu G, et al. 2000. Two yeast forkhead genes regulate the cell cycle and pseudohyphal growth. Nature 406:90–94 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.