Abstract
Initiation of transcription of RNA polymerase II (RNAPII)-dependent genes requires the participation of a host of basal transcription factors. Among genes requiring RNAPII for transcription, small nuclear RNAs (snRNAs) display a further requirement for a factor known as snRNA-activating protein complex (SNAPc). The scope of the biological function of SNAPc and its requirement for transcription of protein-coding genes has not been elucidated. To determine the genome-wide occupancy of SNAPc, we performed chromatin immunoprecipitation followed by high-throughput sequencing using antibodies against SNAPC4 and SNAPC1 subunits. Interestingly, while SNAPC4 occupancy was limited to snRNA genes, SNAPC1 chromatin residence extended beyond snRNA genes to include a large number of transcriptionally active protein-coding genes. Notably, SNAPC1 occupancy on highly active genes mirrored that of elongating RNAPII extending through the bodies and 3′ ends of protein-coding genes. Inhibition of transcriptional elongation resulted in the loss of SNAPC1 from the 3′ ends of genes, reflecting a functional association between SNAPC1 and elongating RNAPII. Importantly, while depletion of SNAPC1 had a small effect on basal transcription, it diminished the transcriptional responsiveness of a large number of genes to two distinct extracellular stimuli, epidermal growth factor (EGF) and retinoic acid (RA). These results highlight a role for SNAPC1 as a general transcriptional coactivator that functions through elongating RNAPII.
INTRODUCTION
Small nuclear RNAs (snRNAs) specify a class of small noncoding RNAs that are assembled into ribonucleoprotein complexes to regulate various nuclear processes such as transcriptional elongation (7SK) and mRNA splicing (UsnRNAs). The snRNA-activating protein complex (SNAPc) (also called PTF) is a five-subunit complex (SNAPC1 to -5) that acts as a basal transcription factor to mediate transcription of snRNAs (2, 9–11, 22, 24, 26, 27, 29). Small nuclear RNAs are transcribed by both RNA polymerase II (RNAPII) and RNAPIII. SNAPc was first described as a TATA binding protein (TBP)-containing complex required for the activation of transcription of the UsnRNAs in vitro (25, 26, 28). SNAPc recognizes a conserved DNA sequence, known as the proximal sequence element (PSE), located approximately 50 bases upstream from the transcription start site of the UsnRNAs to drive the assembly of the preinitiation complex (17). Two subunits, SNAPC3 and SNAPC4, were shown to directly bind DNA in vitro through a zinc finger and Myb DNA binding domain, respectively (14, 27). Additionally, recent in vitro experiments, using the Drosophila homolog of SNAPc, suggested that SNAPC1 might also bind DNA (16). The confirmation of SNAPc binding to the UsnRNA promoters in vivo was recently provided by the chromatin immunoprecipitation of SNAPC2 (4).
While the precise role of SNAPC1 in the complex is only partially understood, it was shown to serve as a bridge to connect SNAPC3 and SNAPC4 proteins (19). This interaction is required to mediate the formation of a “minimal” SNAPc (comprising SNAPC1, SNAPC3, and the N-terminal portion of SNAPC4) that can recapitulate DNA binding, TBP recruitment, and transcription activation (21). Interestingly, all three subunits were reported to directly bind TBP (12, 24, 29). Moreover, SNAPC1 is also able to interact with Rb and p53 (8, 13) and might therefore play a role in the regulation of UsnRNA expression during the cell cycle.
Here we present the analysis of the genome-wide occupancy of SNAPC1 and SNAPC4 in nontumorigenic mammary epithelial MCF10A cells. We show that SNAPC4 predominantly occupies UsnRNA genes, consistent with its role in UsnRNA transcription, whereas SNAPC1 localization extends beyond UsnRNA genes to include a large number of protein-coding genes. We show that SNAPC1 is functionally associated with the elongating form of RNAPII, suggesting a role for this protein in transcriptional elongation. Functional analysis of SNAPC1 revealed a role for this protein in both basal and activator-induced transcription.
MATERIALS AND METHODS
Cell culture.
Breast epithelial MCF10A cells were cultivated in serum-free Dulbecco modified Eagle medium (DMEM)–F-12 (1:1) (Invitrogen) medium supplemented with 2 mM l-glutamine, 50 ng/ml cholera toxin, 10 μg/ml bovine insulin, 500 ng/ml hydrocortisone, 10 ng/ml epidermal growth factor (EGF), and 50 μg/ml bovine pituitary extract. HeLa cells were grown in high-glucose DMEM supplemented with 2 mM l-glutamine and 10% fetal bovine serum (FBS).
Antibodies.
Rabbit anti-SNAPC1 antibodies were obtained from Sigma. Antibodies against RNAPII (N-20; rabbit polyclonal antibodies recognizing all forms of RNAPII) and SNAPC4 (SNAAD17A; mouse monoclonal) were obtained from Santa Cruz. Phospho-Ser2 CTD antibodies were purchased from Bethyl Laboratories.
ChIP-seq.
See the supplemental material for the complete protocol for chromatin immunoprecipitation followed by high-throughput sequencing (ChIP-seq). A total of 25 × 106 to 30 × 106 asynchronously growing MCF10A cells were cross-linked with 1% formaldehyde for 10 min at room temperature. Single immunoprecipitations (IPs) of 2.5 × 106 cells were set up using a specific antibody or a total rabbit IgG control along with protein A magnetic beads. Protein G beads were used for SNAPC4 ChIP-seq. The immunoprecipitated DNA was purified and measured with a Quantit PicoGreen double-stranded DNA (dsDNA) kit (Invitrogen), and 5 to 10 ng was used to generate the sequencing libraries. DNA fragments of ∼150 to 400 bp were isolated by agarose gel purification, ligated to primers, and then subjected to Solexa sequencing using the manufacturer's recommendations (Illumina, Inc.).
ChIP-seq data analysis.
ChIP-seq data were obtained using an Illumina Genome Analyzer II. The 36-bp reads were filtered for duplicated reads and aligned to the human genome hg18 using BOWTIE (v = 0, n = 0, m = 1) (18) without allowing any mismatch; reads with more than one reported alignment were also discarded. Snapshots of raw ChIP-seq data presented throughout the figures were obtained as follows: BigWiggle files for every ChIP-seq were generated using Bed Tools and the utility bedGraphToBigWig (http://hgdownload.cse.ucsc.edu/admin/exe/linux.x86_64/), and these tracks were then uploaded into the UCSC Genome Browser hg18. Peak analysis of SNAPC1, SNAPC4, and RNAPII was performed using MACS (30). Rabbit IgG ChIP-seq was used as a control for all the samples. The analysis was run with a P value threshold of 1 × 10−7, and the bandwidth was set at 300 bp (the other parameters were set to default). We reported peaks containing at least 30 reads, provided that the fold change over the result from the IgG experiment was higher than 10. In the RNAPII experiment, the threshold was set to 20 reads (fold change of >10). Annotation of peaks was obtained using CARPET (5) and the RefSeq Genes table or RNA Genes table downloaded from UCSC Genome Browser (hg18).
Motif analysis.
Motif analysis was performed on the 250 bp upstream from the transcription start sites (TSS) of the 29 UsnRNA genes. We used MEME (3) with default parameters and a maximum width of 25 bases.
Clustering and heat map analysis.
ChIP-seq data were subjected to unbiased clustering, with respect to a list of unique RefSeq genes or UsnRNA genes (extracted from the RNA genes list on hg18), using the seqMINER 1.3.2 platform 50. We used Kmeans linear as the method of clustering, with the following parameters for UsnRNA gene analysis: left and right extension = 1 kb, internal bins = 10, flanking region bins = 80, and number of clusters = 5. For the analysis of RefSeq genes we employed the following: left and right extension = 1.5 kb, internal bins = 160, flanking region bins = 20, and number of cluster = 5. seqMINER was also used to generate all the heat maps and the average profiles of read density.
Quantitative ChIP (qChIP).
ChIP was performed in HeLa and MCF10A cells as previously described (7). ChIP eluates from the specific antibodies, control IgG, and input were assayed by real-time quantitative PCR (qPCR) in a 20-μl reaction mixture with 0.4 μM each primer, 10 μl of iQ SYBR green Supermix (Bio-Rad), and 5 μl of template DNA (corresponding to 1/40 of the elution material). Thermal cycling parameters were as follows: 3 min at 95°C, followed by 40 cycles of 10 s at 95°C, and 30 s at 60°C. The strength of the ChIP signal was calculated as the amount of immunoprecipitated DNA relative to that present in the input chromatin.
Short hairpin RNA (shRNA).
pSUPER.retro.puro constructs against SNAPC1 and a nontargeting control (see the supplemental material for sequences) were transfected in HeLa cells using MetafectenePro (Biontex) as a carrier. After 24 h, cells were selected with 2.5 μg/ml puromycin for 72 h. RNA was extracted before and after EGF stimulation.
EGF and flavopiridol treatments.
Wild-type or shRNA-transfected HeLa cells were plated at 50 to 60% confluence and starved in DMEM supplemented with 0.5% FBS for 24 h. EGF (Invitrogen) was then added to the medium (100 ng/ml). Samples were collected at different time points and subjected to qChIP or RNA isolation. Exponentially growing HeLa cells (70 to 80% confluence) were treated with flavopiridol (2 μM). Treated and untreated cells were collected after 6 h and subjected to ChIP analysis.
Gene expression arrays.
HeLa cells were transfected with different shRNA constructs, and 400 ng of total RNA was amplified according to Illumina protocols and hybridized to an Illumina HumanHT-12 v4 Expression BeadChip. Three biological replicates were analyzed for each condition. Data were processed using the bead array library in R. Raw data were log2 transformed and normalized by quantile normalization, and fold changes and statistics were calculated using the LIMMA (Linear Models for Microarray Data) library in R. Heat maps were created using GPLOT library with default parameters.
qRT-PCR.
See the supplemental material for the complete quantitative reverse transcription-PCR (qRT-PCR) protocol. cDNAs were synthesized from 2 μg of total RNA using random primers. qPCR was performed as described above for ChIP samples, using 15 ng of cDNA. Each sample was run in triplicate. The GUSB gene was used as a normalizer.
Microarray data accession number.
ChIP-seq data have been deposited under GEO accession number GSE37403.
RESULTS AND DISCUSSION
Genome-wide analyses of SNAPC1 and SNAPC4 identify active UsnRNA genes in MCF10A cells.
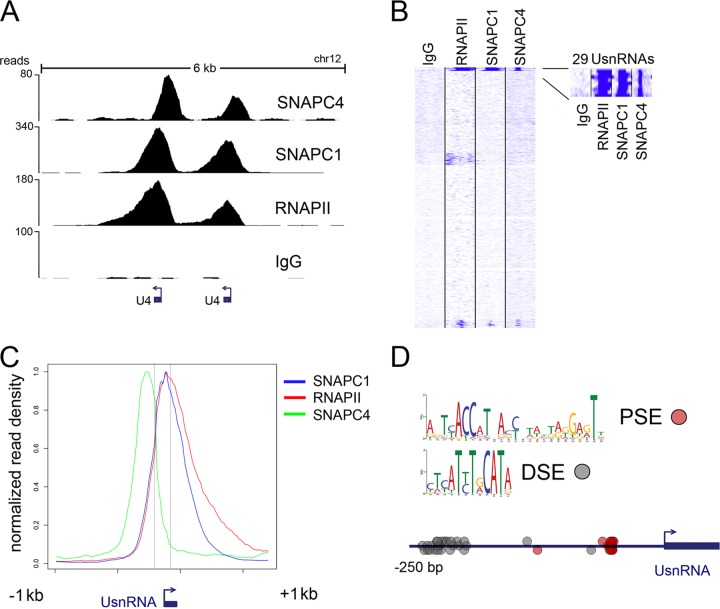
SNAPc regulates the transcription of a small set of noncoding RNAs known as small nuclear RNAs (snRNAs). While the majority of UsnRNA genes are transcribed by RNAPII, U6 and U6ATAC are known to require RNAPIII for their transcription. To assess SNAPc genome-wide localization, we performed chromatin immunoprecipitation followed by high-throughput sequencing (ChIP-seq) using antibodies against the subunits SNAPC1 and SNAPC4. Furthermore, to gain a comprehensive picture of the transcriptional landscape, we performed ChIP-seq using RNAPII antibodies (N-20, which recognizes RPB1 independent of its phosphorylation status). We performed all ChIP-seq experiments with MCF10A cells, a nontumorigenic mammary epithelial cell line. Unbiased clustering of SNAPC1 and SNAPC4 across the genomic coordinates of 1,721 predicted UsnRNA loci (RNA genes hg18) identified 29 UsnRNA genes. Analysis of RNAPII occupancy confirmed the 29 genes to be the actively transcribed UsnRNAs in MCF10A cells (Fig. 1A and B; see Table S1 in the supplemental material).
Fig 1.
Genome-wide analyses of SNAPC1 and SNAPC4 identify active UsnRNA genes in MCF10A cells. (A) Occupancy of SNAPC1 and SNAPC4 at a tandem of U4 snRNA genes in MCF10A cells. These loci are actively transcribed as suggested by concomitant binding of RNAPII. ChIP-seq tracks are aligned to release hg18 of the UCSC genome browser; values on the y axis indicate the actual number of reads. (B) Unbiased clustering of SNAPC1, SNAPC4, and RNAPII with respect to all human predicted UsnRNAs (1,721 predicted loci, hg18 annotation tables) reveals a set of 29 RNAs where both SNAPC1 and SNAPC4 colocalize. These appear to be the only fraction of UsnRNAs that are actively transcribed in MCF10A cells, according to RNAPII occupancy. (C) Average profiles of SNAPC1, SNAPC4, and RNAPII at active UsnRNAs. Normalized average read density is shown for the group of 29 UsnRNA genes described for panel B. The x axis depicts an average UsnRNA gene (100 to 200 bp) and the surrounding 2 kb. (D) Probability matrices for the PSE and DSE motifs as identified from the 29 UsnRNA promoters occupied by SNAPC1, SNAPC4, and RNAPII. The distribution of PSE and DSE motifs along the promoter is also shown; the PSE clusters around 50 bp upstream from the UsnRNA TSS, while the DSE is distributed mostly from bp −200 to −250.
Due to the repetitive nature of these loci, we employed highly stringent criteria for the mapping and alignment of ChIP-seq reads to the human genome. Therefore, we cannot exclude that a limited additional number of active UsnRNAs may have been filtered out by this analysis (see Materials and Methods for details).
The average binding profiles across all 29 UsnRNAs showed that SNAPC1 and SNAPC4 peaks do not coincide (Fig. 1C). Interestingly, SNAPC1 occupancy resembles that of RNAPII, peaking at the core of UsnRNA genes and trailing into the 3′ untranslated region (UTR). On the other hand, SNAPC4 was localized further upstream, peaking 150 to 200 bp prior to the SNAPC1 and RNAPII peaks at all loci (Fig. 1C). In agreement with its role in the recognition and binding to the PSE (27), the average SNAPC4 profile peaks between nucleotides −80 and −70 from the transcription start site (TSS). We searched the DNA sequences of all 29 UsnRNA promoters for the presence of the octamer binding motifs and the proximal sequence elements (PSEs) that determine SNAPc recruitment. We performed ab initio motif analysis on the promoter regions (250 bp upstream of the TSS) of all active UsnRNA genes. The analysis identified an octamer-like matrix in all 29 promoters that is likely to represent the distal sequence elements (DSEs) previously described (14) and is located on average at bp −210 from the TSS (Fig. 1D; see Table S2 in the supplemental material).
We next focused our attention on the proximal promoter in search for the PSE. The base composition of the PSE was previously determined through the manual annotation of a limited number of UsnRNA promoter sequences known for their ability to drive transcription in vitro; therefore, a computationally annotated matrix has never been reported. We retrieved a novel probability matrix in 18/29 UsnRNAs that embodies the PSE and is located at an average 56 bp upstream from the TSS. We further looked into the group of UsnRNAs that did not contribute to define this matrix and found an additional set of 8 U1 and U1-related snRNAs, sharing a higher sequence similarity, that delivers a slightly different PSE-like matrix (see Table S2 in the supplemental material). This analysis confirms the previously reported in vitro analyses of the SNAPc complex, supporting the requirement of the PSE and DSE elements for UsnRNA transcription in vivo.
SNAPC1 occupancy parallels that of RNAPII at transcriptionally active genes, suggesting functions beyond UsnRNA transcription.
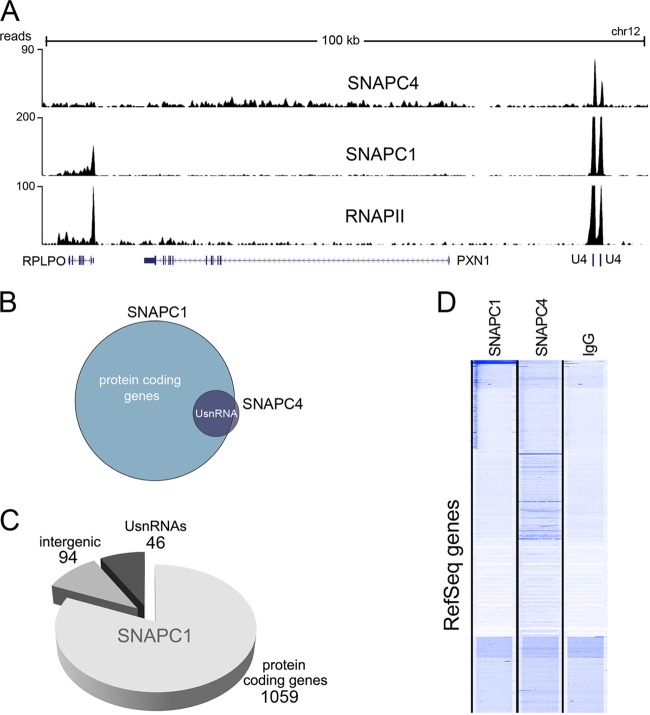
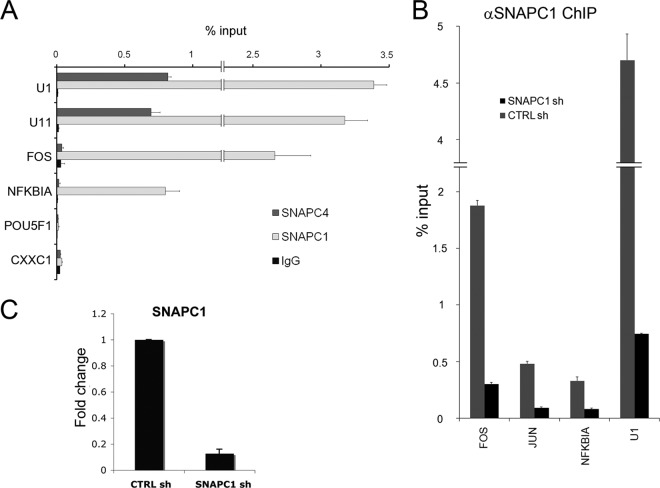
While we found an overlap between SNAPC4 and SNAPC1 occupancy at UsnRNA genes, SNAPC1 displayed a more complex pattern of genome-wide localization. In contrast to SNAPC4 localization, SNAPC1 occupied a large number of highly active RNAPII genes (Fig. 2A to C; see Table S3 in the supplemental material). Indeed, in addition to snRNA genes, SNAPC1 occupied nearly 1,000 protein-coding genes and a smaller number of intergenic sites (Fig. 2B and C). Importantly, while unbiased clustering of ChIP-seq data showed an association of SNAPC1 and SNAPC4 at UsnRNA genes (Fig. 1B), only SNAPC1 could be shown to cluster at RefSeq genes (Fig. 2D). The differences in SNAPC1 and SNAPC4 occupancy were validated at several protein-coding genes using conventional ChIP followed by real-time PCR in MCF10A cells (Fig. 3A). Moreover, SNAPC1 occupancy was verified in HeLa cells at a number of loci by conventional ChIP following SNAPC1 depletion by small interfering RNAs (Fig. 3B and C), confirming the specificity of SNAPC1 binding and extending our observations to a different cell line.
Fig 2.
SNAPC1 genome-wide occupancy extends beyond UsnRNA genes. (A) SNAPC1, SNAPC4, and RNAPII binding at a 100-kb region on chr12 encompassing the U4 loci displayed in Fig. 1A. ChIP-seq reads of SNAPC1 also occupy the TSS and gene body of the ribosomal gene RPLP0, an actively transcribed gene that is also displaying widespread RNAPII binding. (B) SNAPC1 targets an additional set of protein-coding genes. Peak calling analysis of SNAPC4 reveals a very small number of bona fide, high-stringency, peaks (n = 43), compared to a much larger set of SNAPC1 peaks (n = 1,176). SNAPC4 peaks intersect (31/43) the subgroup of SNAPC1 peaks at UsnRNAs, while additional SNAPC1 peaks are found at protein-coding genes. (C) Annotation plot of SNAPC1 binding regions. SNAPC1 peaks are annotated based on RefSeq and RNA Gene tables from the hg18 assembly of the human genome. “Intergenic” refers to binding regions without any GenBank ID or small RNA annotation within a 1-kb radius. Other minor groups of noncoding RNAs are omitted from the pie chart (see Table S3 in the supplemental material). (D) Unbiased clustering of SNAPC1, SNAPC4, and IgG suggests occupancy of a subset of RefSeq genes by SNAPC1 but not SNAPC4.
Fig 3.
SNAPC1 occupies a large number of highly active RNAPII genes. (A) SNAPC1 targets protein-coding genes. ChIP analysis of UsnRNA and coding gene loci shows comparable amounts of SNAPC1 but not SNAPC4, which is enriched only at UsnRNAs. POU5F1 and CXXC1 are negative-control loci: POU5F1 (OCT4) is not expressed in MCF10A, while CXXC1 is expressed but not a target of SNAPC1 according to ChIP-seq data. qChIP was performed in MCF10A cells. (B) Anti-SNAPC1 antibody specifically recognizes SNAPC1 at both UsnRNAs and coding genes. HeLa cells were transfected with a specific SNAPC1 or a nontarget shRNA vector (CTRL sh), and cells were selected with puromycin and harvested 96 h after transfection. The SNAPC1 ChIP signal dramatically decreases after shRNA-mediated depletion, at both UsnRNAs and coding genes. (C) Verification of knockdown efficiency for SNAPC1. HeLa cells were transfected with the corresponding shRNA vectors for the different genes and selected with puromycin for 72 h. Total RNA was extracted, reverse transcribed, and analyzed by qPCR. GUSB was used as a reference gene.
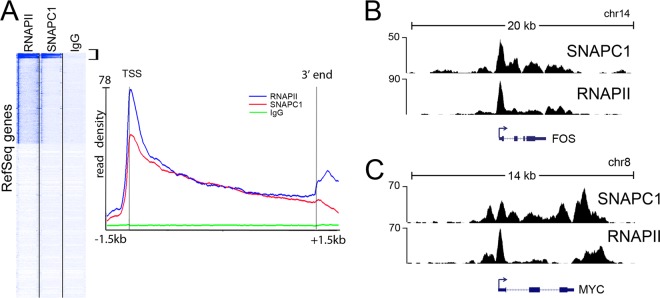
Unbiased clustering of ChIP-seq data for SNAPC1, RNAPII, and IgG showed that SNAPC1 occupied highly active protein-coding genes as evidenced by RNAPII localization (Fig. 4A). Indeed, genes with the highest peaks of RNAPII displayed the largest SNAPC1 occupancy. These included 267 active genes in which RNAPII and SNAPC1 average profiles are similarly distributed: they both peak at the transcription start site and extend into the reading frame beyond the annotated 3′ end of the gene (Fig. 4A to C; see Table S4 in the supplemental material). These genes comprise highly expressed housekeeping genes such as those for histones and ribosomal proteins, key regulators of cellular growth such as FOS, MYC, and JUN, some highly abundant noncoding RNAs (MALAT1 and NEAT1), and genes involved in the oxidative stress response (SOD2, DUSP1, and TXNIP).
Fig 4.
SNAPC1 parallels RNAPII occupancy at protein-coding genes. (A) Average profile of occupancy of SNAPC1. Unbiased clustering of SNAPC1 and RNAPII with respect to Refseq genes (23,316 unique genes) identifies a class of 267 genes with the highest number of reads in both ChIP-seq experiments (bracketed in the left panel). The average read densities of SNAPC1, RNAPII, and rabbit IgG at these genes are shown in the right panel. Profiles extend from 1.5 kb before the transcription start site up to 1.5 kb beyond the 3′ ends of the genes. (B) Binding profile of SNAPC1 and RNAPII at the FOS locus. A snapshot from UCSC Genome Browser on chr14 displays SNAPC1 and RNAPII peaking at the TSS of the gene, with additional occupancy into the gene body and the 3′ end. (C) Binding profile of SNAPC1 and RNAPII at the MYC locus on chr8.
SNAPC1 functionally associates with elongating RNAPII.
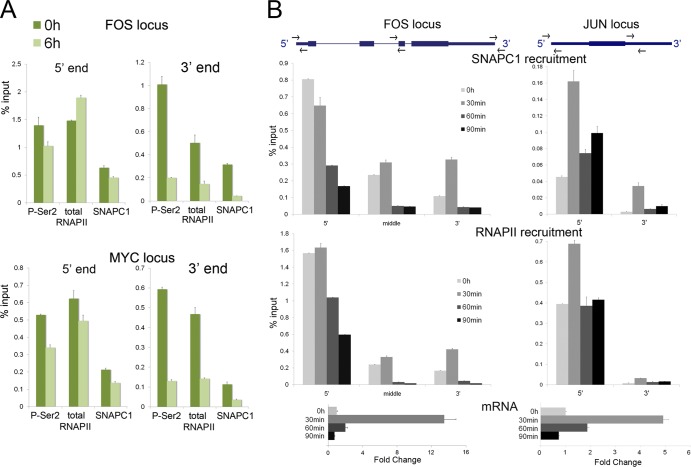
To determine whether the pattern of occupancy that we observed with SNAPC1 across the body of protein-coding genes depends on elongating RNAPII, we treated cells with the positive transcription elongation factor b (P-TEFb) inhibitor flavopiridol (Fig. 5A). Previous experiments indicated that treatment of cells with flavopiridol prevented the release of promoter-proximal RNAPII and therefore resulted in diminished occupancy of RNAPII at the 3′ ends of protein-coding genes (6, 23). We treated HeLa cells with 2 μM flavopiridol for 6 h and monitored the occupancy of RNAPII and SNAPC1 (Fig. 5A). We also used antibodies against the phosphorylated serine 2 (a mark of transcriptional elongation) of the C-terminal domain of the largest subunit of RNAPII to assess the occupancy of the elongating form of RNAPII (Fig. 5A). While treatment of cells with flavopiridol led to a small decrease in the occupancy of RNAPII and SNAPC1 on the 5′ ends of FOS and MYC, we observed a large decrease in SNAPC1 and RNAPII localization at the 3′ ends of both genes, suggesting an intimate connection of SNAPC1 with the elongating RNAPII (Fig. 5A).
Fig 5.
SNAPC1 functionally associates with elongating RNAPII. (A) SNAPC1 binding dramatically decreases upon inhibition of elongating RNAPII. qChIP analyses were performed before and after flavopiridol treatment in HeLa cells, revealing major changes at the 3′ ends of FOS and MYC gene loci and a moderate effect at the TSS (5′ end). ChIP values are represented as the average percentage of input from three IPs. (B) SNAPC1 is recruited at the FOS locus during EGF-induced transcriptional activation. qChIP analysis was performed before EGF treatment and 30, 60, and 90 min after induction. The TSS, middle, and 3′ end of FOS were analyzed. ChIP assays were performed in triplicate for each antibody, and average values are shown.
To further assess the functional link between elongating forms of RNAPII and SNAPC1, we used conventional ChIP to analyze SNAPC1 occupancy at the FOS locus in HeLa cells following induction by epidermal growth factor (EGF) (Fig. 5B). We reasoned that the concomitant increase in RNAPII and SNAPC1 occupancy on the body and the 3′ end of the FOS gene following its activation by EGF will further support a role for this protein in transcriptional elongation. While addition of EGF resulted in stimulation of FOS expression (∼12-fold) at 30 min, by 90 min FOS transcript levels had returned to basal levels. Analysis of FOS occupancy by SNAPC1 and RNAPII revealed that EGF stimulation of HeLa cells resulted in a simultaneous increased occupancy by RNAPII and SNAPC1 on the body and the 3′ end of FOS after 30 minutes. Moreover, at 90 min after stimulation, when transcription had returned to basal levels, there was a decrease in both RNAPII and SNAPC1 occupancy across the entire gene. We observed a similar effect at the JUN locus, where RNAPII and SNAPC1 are recruited at the TSS as well as across the body of the gene after 30 minutes of EGF induction, while they return at prestimulation levels after 1 h. These results indicate that SNAPC1 occupancy of FOS and JUN loci during transcriptional activation parallels that of RNAPII, confirming its functional association with polymerase and reflecting a potential role for SNAPC1 in transcriptional elongation.
SNAPC1 regulates the responsiveness to transcriptional activation.
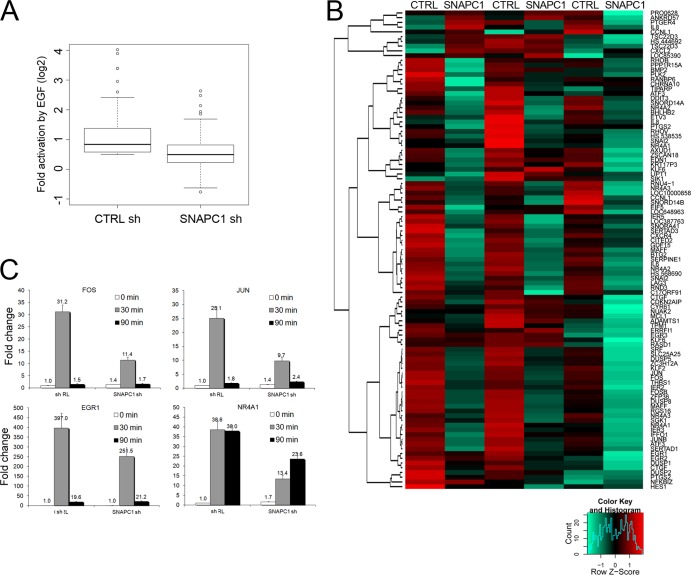
To assess the function of SNAPC1 in transcription of RNA polymerase II-dependent genes, we examined the transcriptional responsiveness to epidermal growth factor (EGF) using gene expression arrays. We focused on the response to EGF stimulation since SNAPC1 was present on the bodies of many canonical immediate-early genes, such as FOS and JUN. We conducted these experiments with HeLa cells since these cells are more permissive to the effect of short hairpin RNAs (shRNAs) and display a greater responsiveness to EGF stimulation (1). We confirmed the occupancy of SNAPC1 in HeLa cells on many of the target genes identified in MCF10A cells, with comparable or at times higher levels (Fig. 3B and data not shown). Importantly, depletion of SNAPC1 resulted in a pronounced attenuation of EGF responsiveness of nearly all EGF-responsive genes in HeLa cells (Fig. 6A and B). We validated these results by examining four canonical immediate-early genes, FOS, JUN, EGR1, and NR4A1 (Fig. 6C). Interestingly, with the exception of EGR1, depletion of SNAPC1 also resulted in a small increase of the basal level of expression of these genes (Fig. 6C).
Fig 6.
SNAPC1 regulates the responsiveness to EGF-mediated transcriptional activation. (A) SNAPC1 knockdown globally reduces EGF activation of immediate-early response genes. The box plot represents fold activation (expressed as log2) of the 100 most responsive microarray probes after 30 min of induction of HeLa cells with 100 ng/ml EGF. (B) Depletion of SNAPC1 impairs EGF-activated transcription. The heat map representation covers the top 100 microarray probes upregulated by EGF under normal conditions (CTRL shRNA, t = 30 min over t = 0). The color scale represents the modified log2 ratio (“sweep” function R, scaled by row) between the induced and the basal states. The color variation accounts for the difference of induction across the 2 conditions (red, augmented induction; green, decreased induction). Results from three independent experiments are shown. (C) Detailed expression analysis of the effect of SNAPC1 knockdown on the induction by EGF of four immediate-early genes. Cells were harvested at 0, 30, and 90 min after induction. GUSB was used as a control gene. The control shRNA at time zero was used as a reference. Data are the averages from three independent experiments.
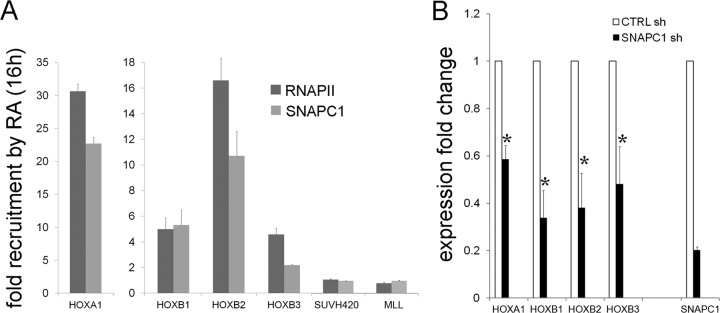
To address the importance of SNAPC1 in transcriptional activation for other activating stimuli, we assessed its requirement for responsiveness to retinoic acid (RA). NT2/D1 cells respond to RA by terminally differentiating into neurons (20). We measured the recruitment of RNAPII and SNAPC1 to the set of HOX cluster genes known to be targets of RA in NT2/D1 cells. Interestingly, we saw a concomitant recruitment of RNAPII and SNAPC1 to HOXA1, HOXB1, HOXB2, and HOXB3 (Fig. 7A). Importantly, depletion of SNAPC1 diminished transcriptional activation of these genes by RA treatment, supporting a role for SNAPC1 in RA-induced transcriptional activation (Fig. 7B).
Fig 7.
SNAPC1 regulates the responsiveness to retinoic acid in NT2/D1 cells. (A) SNAPC1 and RNAPII are recruited to the transcriptional start site of HOX genes. ChIP analysis was performed on human teratocarcinoma-derived NT2/D1 cells upon 16 h of induction with retinoic acid (ATRA). Quantitative ChIP data were obtained for the TSS of HOXA1, HOXB1, HOXB2, and HOXB3. Data are plotted as relative enrichment compared to the noninduced state. (B) SNAPC1 depletion impairs transcriptional activation at HOXA and HOXB clusters. NT2/D1 cells were interfered for SNAPC1 and stimulated with 10 μM ATRA. RNA was collected at t = 16 h, and the relative induction over the control shRNA (CTRL sh) was calculated. Data were normalized to GUSB expression. Results from three independent experiments are plotted (*, P < 0.05).
In this work, we dissected the role of two components of the SNAPc complex using a functional genomics approach. Our data defined the genomic landscape of RNAPII-dependent UsnRNAs and revealed an unexpected association of SNAPC1 with a set of protein-coding genes. We found that while SNAPC1 and SNAPC4 colocalized at a similar set of active UsnRNA genes in human cells, they displayed a difference in their promoter occupancy. SNAPC4 occupancy peaked prior to the transcription start site, where it could contribute to the recruitment of the general transcription machinery. In contrast, SNAPC1 occupancy overlapped that of RNAPII at the body of the UsnRNAs and extended into the 3′ UTR, reflective of a prolonged association with RNAPII.
While the association of SNAPC1 with snRNA genes was expected, we were surprised to find SNAPC1 occupancy of a large number of highly active protein-coding genes. Interestingly, the SNAPC1 occupancy at a large number of these genes mirrored that of RNAPII chromatin residency (Fig. 4). We often observed a peak of SNAPC1 at the 5′ ends of the protein-coding genes followed by SNAPC1 occupancy extending into the body of the genes. This pattern of genomic occupancy was not only suggestive of an association of SNAPC1 with elongating RNAPII but also reflective of a role for SNAPC1 in transcriptional elongation. This contention was confirmed following experiments where SNAPC1 occupancy of the 3′ ends of the FOS and MYC genes was abrogated following treatment of cells with the transcriptional elongation inhibitor flavopiridol. Moreover, we observed a dynamic interaction of SNAPC1 with elongating RNAPII during activation of the FOS gene by EGF (Fig. 5B).
Finally, we showed that SNAPC1 is a critical component of the transcriptional responsiveness to EGF and RA stimulation. Depletion of SNAPC1 potently reduced the transcriptional responsiveness to both stimuli (Fig. 6 and 7). These results implicate SNAPC1 as a general cofactor signaling through elongating RNAPII to confer transcriptional activation. Considering the broad occupancy of SNAPC1 on protein-coding genes, it is likely that it may regulate the responsiveness to a number of other transcriptional activators involved in regulating cellular growth and tissue homeostasis.
Supplementary Material
ACKNOWLEDGMENTS
We thank Nitya Krishnan and the other members of the Wistar Institute Genomics facility for processing of ChIP-seq samples and expression arrays, and we also thank the bioinformatics core unit for help with Solexa data analysis.
This work was supported by grant R01-GM 078455 (R.S.) from the National Institutes of Health. A.G. is supported by an American-Italian Cancer Foundation postdoctoral research fellowship.
Footnotes
Published ahead of print 10 September 2012
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1. Amit I, et al. 2007. A module of negative feedback regulators defines growth factor signaling. Nat. Genet. 39:503–512 [DOI] [PubMed] [Google Scholar]
- 2. Bai L, Wang Z, Yoon JB, Roeder RG. 1996. Cloning and characterization of the beta subunit of human proximal sequence element-binding transcription factor and its involvement in transcription of small nuclear RNA genes by RNA polymerases II and III. Mol. Cell. Biol. 16:5419–5426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bailey TL, et al. 2009. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 37:W202–W208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Canella D, Praz V, Reina JH, Cousin P, Hernandez N. 2010. Defining the RNA polymerase III transcriptome: genome-wide localization of the RNA polymerase III transcription machinery in human cells. Genome Res. 20:710–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cesaroni M, Cittaro D, Brozzi A, Pelicci PG, Luzi L. 2008. CARPET: a web-based package for the analysis of ChIP-chip and expression tiling data. Bioinformatics 24:2918–2920 [DOI] [PubMed] [Google Scholar]
- 6. Chao SH, Price DH. 2001. Flavopiridol inactivates P-TEFb and blocks most RNA polymerase II transcription in vivo. J. Biol. Chem. 276:31793–31799 [DOI] [PubMed] [Google Scholar]
- 7. Frank SR, Schroeder M, Fernandez P, Taubert S, Amati B. 2001. Binding of c-Myc to chromatin mediates mitogen-induced acetylation of histone H4 and gene activation. Genes Dev. 15:2069–2082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gridasova AA, Henry RW. 2005. The p53 tumor suppressor protein represses human snRNA gene transcription by RNA polymerases II and III independently of sequence-specific DNA binding. Mol. Cell. Biol. 25:3247–3260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Henry RW, Ma B, Sadowski CL, Kobayashi R, Hernandez N. 1996. Cloning and characterization of SNAP50, a subunit of the snRNA-activating protein complex SNAPc. EMBO J. 15:7129–7136 [PMC free article] [PubMed] [Google Scholar]
- 10. Henry RW, Mittal V, Ma B, Kobayashi R, Hernandez N. 1998. SNAP19 mediates the assembly of a functional core promoter complex (SNAPc) shared by RNA polymerases II and III. Genes Dev. 12:2664–2672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Henry RW, Sadowski CL, Kobayashi R, Hernandez N. 1995. A TBP-TAF complex required for transcription of human snRNA genes by RNA polymerase II and III. Nature 374:653–656 [DOI] [PubMed] [Google Scholar]
- 12. Hinkley CS, Hirsch HA, Gu L, LaMere B, Henry RW. 2003. The small nuclear RNA-activating protein 190 Myb DNA binding domain stimulates TATA box-binding protein-TATA box recognition. J. Biol. Chem. 278:18649–18657 [DOI] [PubMed] [Google Scholar]
- 13. Hirsch HA, Gu L, Henry RW. 2000. The retinoblastoma tumor suppressor protein targets distinct general transcription factors to regulate RNA polymerase III gene expression. Mol. Cell. Biol. 20:9182–9191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Jawdekar GW, et al. 2006. The unorthodox SNAP50 zinc finger domain contributes to cooperative promoter recognition by human SNAPC. J. Biol. Chem. 281:31050–31060 [DOI] [PubMed] [Google Scholar]
- 15. Reference deleted.
- 16. Kim MK, et al. 2010. Identification of SNAPc subunit domains that interact with specific nucleotide positions in the U1 and U6 gene promoters. Mol. Cell. Biol. 30:2411–2423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kuhlman TC, Cho H, Reinberg D, Hernandez N. 1999. The general transcription factors IIA, IIB, IIF, and IIE are required for RNA polymerase II transcription from the human U1 small nuclear RNA promoter. Mol. Cell. Biol. 19:2130–2141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Langmead B, Trapnell C, Pop M, Salzberg SL. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10:R25 doi:10.1186/gb-2009-10-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ma B, Hernandez N. 2001. A map of protein-protein contacts within the small nuclear RNA-activating protein complex SNAPc. J. Biol. Chem. 276:5027–5035 [DOI] [PubMed] [Google Scholar]
- 20. Mavilio F, Simeone A, Boncinelli E, Andrews PW. 1988. Activation of four homeobox gene clusters in human embryonal carcinoma cells induced to differentiate by retinoic acid. Differentiation 37:73–79 [DOI] [PubMed] [Google Scholar]
- 21. Mittal V, Ma B, Hernandez N. 1999. SNAP(c): a core promoter factor with a built-in DNA-binding damper that is deactivated by the Oct-1 POU domain. Genes Dev. 13:1807–1821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Murphy S, Yoon JB, Gerster T, Roeder RG. 1992. Oct-1 and Oct-2 potentiate functional interactions of a transcription factor with the proximal sequence element of small nuclear RNA genes. Mol. Cell. Biol. 12:3247–3261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Rahl PB, et al. 2010. c-Myc regulates transcriptional pause release. Cell 141:432–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sadowski CL, Henry RW, Kobayashi R, Hernandez N. 1996. The SNAP45 subunit of the small nuclear RNA (snRNA) activating protein complex is required for RNA polymerase II and III snRNA gene transcription and interacts with the TATA box binding protein. Proc. Natl. Acad. Sci. U. S. A. 93:4289–4293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sadowski CL, Henry RW, Lobo SM, Hernandez N. 1993. Targeting TBP to a non-TATA box cis-regulatory element: a TBP-containing complex activates transcription from snRNA promoters through the PSE. Genes Dev. 7:1535–1548 [DOI] [PubMed] [Google Scholar]
- 26. Waldschmidt R, Wanandi I, Seifart KH. 1991. Identification of transcription factors required for the expression of mammalian U6 genes in vitro. EMBO J. 10:2595–2603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wong MW, et al. 1998. The large subunit of basal transcription factor SNAPc is a Myb domain protein that interacts with Oct-1. Mol. Cell. Biol. 18:368–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yoon JB, Murphy S, Bai L, Wang Z, Roeder RG. 1995. Proximal sequence element-binding transcription factor (PTF) is a multisubunit complex required for transcription of both RNA polymerase II- and RNA polymerase III-dependent small nuclear RNA genes. Mol. Cell. Biol. 15:2019–2027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yoon JB, Roeder RG. 1996. Cloning of two proximal sequence element-binding transcription factor subunits (gamma and delta) that are required for transcription of small nuclear RNA genes by RNA polymerases II and III and interact with the TATA-binding protein. Mol. Cell. Biol. 16:1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zhang Y, et al. 2008. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9:R137 doi:10.1186/gb-2008-9-9-r137 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.