Abstract
Factors interacting with core circadian clock components are essential to achieve transcriptional feedback necessary for metazoan clocks. Here, we show that all three members of the Drosophila behavior human splicing (DBHS) family of RNA-binding proteins play a role in the mammalian circadian oscillator, abrogating or altering clock function when overexpressed or depleted in cells. Although these proteins are members of so-called nuclear paraspeckles, depletion of paraspeckles themselves via silencing of the structural noncoding RNA (ncRNA) Neat1 did not affect overall clock function, suggesting that paraspeckles are not required for DBHS-mediated circadian effects. Instead, we show that the proteins bound to circadian promoter DNA in a fashion that required the PERIOD (PER) proteins and potently repressed E-box-mediated transcription but not cytomegalovirus (CMV) promoter-mediated transcription when they were exogenously recruited. Nevertheless, mice with one or both copies of these genes deleted show only small changes in period length or clock gene expression in vivo. Data from transient transfections show that each of these proteins can either repress or activate, depending on the context. Taken together, our data suggest that all of the DBHS family members serve overlapping or redundant roles as transcriptional cofactors at circadian clock-regulated genes.
INTRODUCTION
The circadian oscillator governs diurnal timing for most aspects of mammalian physiology (8). Its mechanism is cell autonomous and consists of interlocked feedback loops of circadian transcription, translation, and protein modification. In one loop, the CLOCK/NPAS2 and BMAL1/ARNTL transcriptional activators drive expression of the period (Per1 and Per2) and cryptochrome (Cry1 and Cry2) gene families, whose products subsequently multimerize and repress their own transcription. In a second loop, the transcriptional repressor REV-ERBα, whose transcription is also driven by CLOCK and BMAL1, represses the expression of Bmal1 itself (8). Beyond these “dedicated” clock genes, a large number of other factors are necessary to the circadian clock or for its regulation of physiology, including kinases and phosphatases, chromatin modifying factors, and other proteins (26, 34). We have shown previously that the NONO protein in mammalian cells (or its ortholog NON-A in flies) plays such a role by modulating PERIOD (PER)-mediated transcriptional repression via unknown mechanisms (5).
NONO (also known as p54nrb in humans) has two RNA-binding (RNA recognition motif [RRM]) domains and has been shown to regulate a variety of processes outside the circadian clock (39). These include transcriptional activation and repression (17, 27), pre-mRNA processing (20), and RNA transport in neurons (19). For example, NONO has been shown to regulate the transcriptional activation of the TORC family of growth and metabolic factors via recruitment of the RNA polymerase II (1). In an apparently unrelated nuclear function, it also mediates the nuclear retention of edited RNAs in nuclear paraspeckles, which are thought to be RNA holding structures (31). These structures contain the NONO, SFPQ, and PSPC1 proteins, as well as the scaffolding noncoding RNA (ncRNA) Neat1 (4). Both SFPQ and PSPC1 share significant structural and functional similarities with NONO, and for this reason all three proteins have been grouped into the DBHS (Drosophila behavior human splicing) family of nuclear factors. Nevertheless, to date only NONO (5) and SFPQ (11) have been implicated in the circadian clock mechanism.
Herein, we show that all three DBHS factors play important roles in the circadian clock by binding directly to the promoter of the Rev-Erbα clock gene in a circadian- and PER protein-dependent fashion. In addition, although overexpression or silencing of any one of them influences clock period and amplitude in cells, depletion of paraspeckles themselves has no effect on the circadian oscillator. Mice deficient for two of these proteins show circadian phenotypes, albeit less prominently than in vitro. We therefore suggest that all three proteins play redundant roles in circadian transcriptional modulation.
MATERIALS AND METHODS
Animal husbandry.
Chimeric mice were obtained from Nono gene-trapped (Nonogt) embryonic stem (ES) cells (C57BL/6J genotype) via standard blastocyst injection of the ES cell clone YHA266 into SV129 mice by the University of California, Davis. Individual chimeric mice were backcrossed 4 to 10 generations against the C57BL/6J background. The same procedure was chosen to obtain Pspc1gt/gt and Sfpqgt/+ mutant mice, using ES cell clones RRS358 and BC0256, respectively. Individual chimeric mice were backcrossed two to four generations against the C57BL/6J background. All experiments were performed by comparing wild-type and mutant littermates. Animal housing and experimental procedures are in agreement with veterinary law of the canton of Zurich.
Animal activity measurements.
For period measurements of Nonogt mice, 24 mice of each genotype were habituated to a controlled 12/12 light-dark (LD) cycle in the presence of running wheels for 2 weeks and then kept in constant dim red light for an additional 2 weeks. Data recording and period analysis were performed using the Clocklab software package (Actimetrics). Period measurements of Pspc1gt/gt and Sfpqgt/+ mice were performed identically except that 12 mice of each genotype were used, and measurements were performed twice on each mouse. For skeleton photoperiod measurements, the same mice were given 1 h of normal room light at each LD transition of a normal day and otherwise kept in constant dim red light. Running-wheel activity was measured as in period experiments but plotted as the sum of activities of all the mice over a 24-h day using the Clocklab software.
Plasmids.
The bioluminescence reporter construct pBmal1-luc has been described previously (28). Overexpression of NONO, SFPQ, and PSPC1 (tagged with the myc epitope) were achieved using the plasmids described in Kuwahara et al. (22). Plasmids expressing PER1 and PER2 proteins tagged with the FLAG epitope were a gift of T. Wallach (Kramer lab, Charite Universitätsmedizin, Berlin, Germany). To create GAL4 fusion constructs, the same constructs were obtained as entry vectors from NITE (the Japanese Bioresource Information Center) and recombined into a destination vector (Invitrogen) containing the GAL4 DNA-binding domain (DBD) (amino acids [aa] 1 to 93). This vector was made by cloning PCRed recombination sites from pEF-DEST51 (Invitrogen) into pSCT-GALVP80 (gift of W. Schaffner, University of Zurich). The Neat1 overexpression vector is described in Clemson et al. (7). RNAi vectors against NONO have been described previously in Brown et al. (5). Vectors targeting SFPQ and PSPC1 were purchased from Open Biosystems (clone numbers RRM3981-98064499 TRCN0000102241 and RMM3981-98064691 TRCN0000102470, respectively). p4xEbox-luc is described in Brown et al. (5). pGAL4-E box-luc was made by inserting five copies of a multimerized GAL4 (5×GAL4) site (cut from pFR-luc; Invitrogen) upstream of the E boxes in p4xEbox-luc. pGAL4-CMV-luc (where CMV is cytomegalovirus) was made by inserting the same fragment the same distance upstream relative to the transcription start site of the CMV promoter.
Primary cell isolation and culture.
Primary adult dermal fibroblasts (ADFs) were taken from a 0.5-cm piece of mouse tail that was cut into several small pieces by using a razor blade. Digestion occurred in 1.8 ml of Dulbecco's modified Eagle's medium (DMEM) containing 20% fetal bovine serum (FBS), 1% penicillin-streptomycin, and 1% amphotericin B supplemented with 0.7 unit of Liberase Blendzyme (Roche) at 37°C and 5% CO2 for 8 h. After centrifugation in 1× phosphate-buffered saline (PBS), the pellet was resuspended in DMEM containing 20% FBS, 100 U/ml penicillin, 100 μg/ml streptomycin, and 2.5 μg/ml amphotericin B and kept at 37°C and 5% CO2. The day after, medium was exchanged, and remaining tail pieces were removed. Another medium exchange was done 3 days later. After a week the medium was exchanged for medium without amphotericin B. ADFs were cultured at 37°C and 5% CO2 in DMEM supplemented with 20% FBS and 1% penicillin-streptomycin.
Transient transfections.
For p4xEbox-luc reporter transfection studies in NIH 3T3 cells, Lipofectamine LTX with Plus reagent (Invitrogen) was used according to the manufacturer's instructions; cells were cultivated in 24-well plates and transfected with a total of 850 ng of DNA, of which 50 ng was the promoter luciferase reporter construct. Various amounts of plasmid were “balanced” by the addition of pcDNA3.1 to a total of 800 ng. Cells were harvested after 60 h by one wash with 1× PBS, and luciferase was extracted with a luciferase assay kit (Promega) and normalized against the amount of total protein in each extract (measured by Coomassie staining compared to a bovine serum albumin [BSA] standard curve). Transfections in primary cells were performed identically, except that twice the amount of cells was used for each reaction.
Lentiviral infections.
Measurements were conducted in U2OS cells stably transfected with a circadian Bmal1-luc reporter and then infected with Open Biosystems RNA interference (RNAi) lentivectors (pGIPZ), as described previously (25).
Measurement of circadian bioluminescence in cultured cells.
After transfection or infection as described above, circadian rhythms in cell populations were synchronized with dexamethasone and then measured for 3 to 5 days via real-time luminometry in normal culture medium lacking phenol red but supplemented with 0.2 mM luciferin and 25 mM HEPES, as described previously (28). Data were analyzed using the LumiCyle analysis program (Actimetrics).
cDNA production and quantitative real-time PCR.
RNA was extracted as described by Xie and Rothblum (43). Five hundred nanograms of total RNA was transcribed to cDNA with SuperScript II (Invitrogen) using random hexamer primers according to the manufacturer's instructions. For quantitative real-time PCR, 20 ng of cDNA was used, and single transcript levels of genes were detected by TaqMan probes used with the TaqMan PCR mix protocol (Roche) and an AB7900 thermocycler. Primers used for detection of NOPS transcripts were as follows: Nono, TGC GCT TCG CCT GTC A (sense), GCA GTT CGT TCG ACA GTA CTG (antisense), and FAM-AGT GCA CCC TTA CAG TCC GCA ACC TT-TAMRA (probe; FAM is 6-carboxyfluorescein, and TAMRA is 6-carboxytetramethylrhodamine); Pspc1, GAA CTA TAC CTG GCC CAC CAA T (sense), ACT GCG CC ATTA TCT GGT ATC A (antisense), and FAM-ATA TTT GCA GCT CCT TCT GGT CCC ATG-TAMRA (probe); Sfpq, TTT GAA AGA TGC AGT GAA GGT GTT (sense), CCT GCT TCA CCA CCT TCT TGA (antisense), and FAM-TCC TAC TGA CAA CGA CTC CTC GCC CA-TAMRA (probe). Primers for detection of circadian genes and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) can be found in Preitner et al. (32).
Protein extraction and Western blotting.
For in vitro immunoprecipitation, a 10-cm culture dish of HEK 293T cells was cotransfected with 5 μg each of NONO-myc, SPFQ-myc, or PSCP-myc together with 5 μg of PER1-FLAG or PER2-FLAG, via polyethyleneimine transfection (JetPEI; Polyplus) following the manufacturer's instructions. The cells were harvested 24 h later by rinsing with PBS and resuspending the sample in a total of 100 μl of lysis buffer, as described previously for liver nuclei by Lopez-Molina et al. (24). Extracts were stored in 500-μl aliquots at −80°C until used. Liver nuclei were prepared by sucrose cushion centrifugation as described by Lopez-Molina et al. (24) and then extracted exactly as for cells. Western blotting was performed using standard procedures (2). Equal loading and size detection using a protein ladder were verified by Ponceau-S staining of membranes prior to probing.
Immunohistochemistry.
Immunohistochemistry was performed according to the protocols described at http://www.pharma.uzh.ch/research/neuromorphology/researchareas/neuromorphology/Protocols/protocol_immuno.pdf. Substrates were either brains collected in isopentane at −20°C and cryostatically sliced or cells grown on glass coverslips, rinsed with PBS, and fixed for 5 min at room temperature in PBS–4% paraformaldehyde.
Antibodies.
Polyclonal antibodies against NONO, SFPQ, PSPC1, and PER2 were produced from rabbits by Charles River Laboratories using bacterially overexpressed proteins. Antibody from each serum was immunopurified over a column whose resin consisted of the relevant antigen covalently coupled to Affygel 10 (Bio-Rad). Anti-PSPC1 is described in Fox et al. (13). For detection in coimmunoprecipitation (co-IP) experiments, primary anti-MYC antibody (catalog number 11667149001; Roche) was diluted at 1:2,000, primary anti-FLAG antibody (F3167; Sigma) was used at 1:2,000, primary anti-NONO antibody was used at 1:2,000, primary anti-PSPC1 was used at 1:1,000, primary anti-SPFQ antibody was used at 1:2,000, and primary anti-PER2 antibody was used at 1:1,000. The probing of the secondary antibody was done at 1:10,000 for IRDye 680–goat anti-mouse IgG (926-32220; Licor) and 1:10,000 for IRDye 800–goat anti-rabbit IgG (926-33210; Licor). For immunoprecipitations, primary anti-cMYC antibody was diluted at 1:500, primary anti-FLAG antibody was used at 1:500, primary anti-NONO antibody for IP was used at 1:100, primary anti-SFPQ antibody for IP was used at 1:100, primary anti-PSCP1 antibody for IP was used at 1:100, and primary anti-PER2 antibody for IP was used at 1:100.
Immunoprecipitation.
Immunoprecipitation was performed using standard procedures with the below-mentioned adjustments (2). Extracts were precleared by incubation of the crude extracts with protein A beads (catalog number IP06; Calbiochem) and 0.1% BSA for 1 h at 4°C. Precleared extract (500 μg) was bound for 2 h to antibody with co-IP buffer. The antibody-protein complex was then incubated for 1 h with protein A beads. The beads were washed gently with co-IP buffer (without protease inhibitor mix) and denatured for 15 min at 65°C with 2× SDS sample buffer containing β-mercaptoethanol. Equal amounts of IP reaction mixtures were loaded on a 7% (for IP of overexpressed proteins in cells) or 9% (for IP of liver nuclear extracts) SDS-PAGE gel together with 1/10 of the IP amounts of precleared extract as input. The protein gel electrophoresis and blotting were performed as described in the Western blotting and immunohistochemistry sections above.
Chromatin immunoprecipitation.
Chromatin from mouse liver and tissue culture cells was obtained as described previously (35). Equal amounts of precleared chromatin were incubated overnight at 4°C with 1 μl of anti-NONO antibody or anti-PER2 antibody. The capture of the DNA-protein complexes, the washing conditions, and the purification of the DNA fragments prior to quantitative PCR (qPCR) as well the control antibodies have been described previously (37). The region-specific primer/probe pairs are listed in references 35 and 37.
Paraspeckle quantification.
For paraspeckle detection, after immunodetection of PSPC1 as described above, cells were analyzed with an LSM710 Zeiss confocal microscope. Pictures taken were with a 40× (numerical aperture [NA], 1.3) objective, and the pinhole was kept at 1 arbitrary unit (AU) or 0.8 to 0.9 μm. Nuclei were manually detected using ImageJ software routines (http://rsbweb.nih.gov/ij/index.html). Speckles were determined by subtracting background nucleoplasmic PSPC1 protein staining and thereafter counting remaining pixel clusters in nuclei. The total amount of paraspeckles per cell was estimated by counting all pixels brighter than 140 (arbitrary units) with spot sizes between 0.25 to 10 μm2. Nuclei smaller than 200 pixels or 100 μm2 as well as dividing cells were excluded. The average number of speckles was normalized to the mean area and compared to the control transfected cells (hairpin NEAT-S).
RESULTS
NONO-deficient mice show significant changes in circadian period.
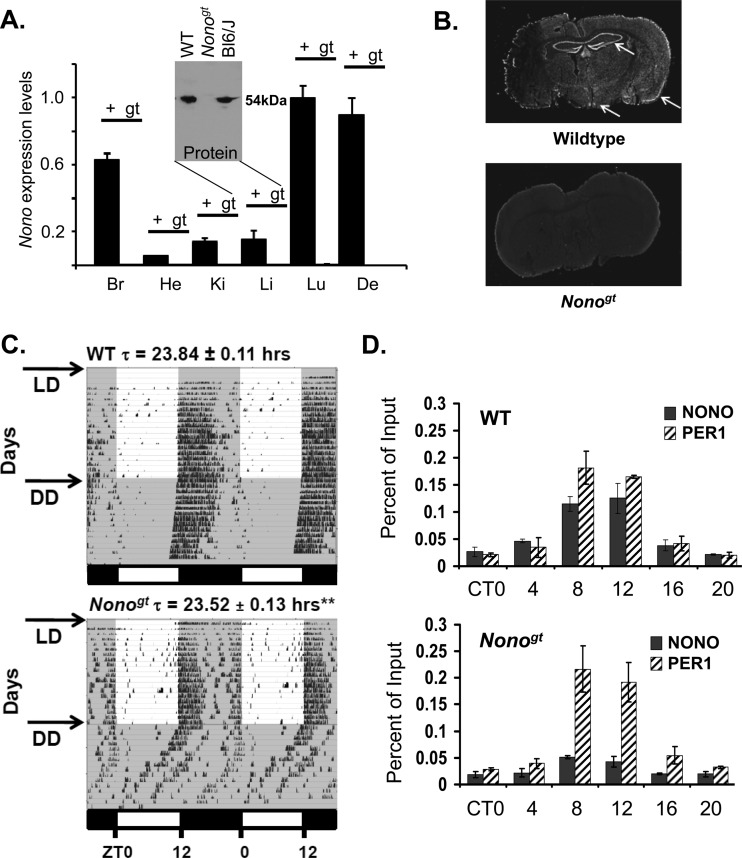
To better understand the function of NONO in the circadian clock and in mammalian physiology, we obtained NONO-deficient mice from ES cells bearing a gene trap in the intron preceding the Nono translational start site (see Fig. S1A and B in the supplemental material). In wild-type mice, NONO is expressed in most tissues including the suprachiasmatic nuclei in the brain. Nonogt mice showed no expression of Nono mRNA or protein in any of the tissues examined (Fig. 1A and B). These mice showed a 20-min reduction in circadian behavioral period when under constant dark conditions (Fig. 1C). This reduction in period length was highly significant, but it was nevertheless far less dramatic than that in a Drosophila hypomorphic nonA strain that we observed previously to become arrhythmic (5). Hence, we suspected that in mammals the lack of Nono may be compensated by other factors.
Fig 1.
(A) NONO RNA expression measured by qPCR in various tissues taken from wild-type (WT; black) and Nonogt animals (gray, not detectable). Inset, NONO protein measured in liver nuclear extract from the same animals, as well as in unrelated C57BL/6J mice (BL6/J). (B) NONO protein expression in brain coronal sections from wild-type and Nonogt animals, visualized by immunohistochemistry using a polyclonal anti-NONO antibody. Arrows from left to right show principal areas of NONO expression in wild-type mouse brain: suprachiasmatic nuclei, hippocampus, and neocortex. (C) Wheel-running activity of wild-type and Nonogt mice in 12/12 light-dark cycles (LD) and in constant darkness (DD). Darkness is indicated by gray shading. n = 23. (D) Chromatin immunoprecipitation of NONO and PER1 at the Rev-Erbα promoter in liver nuclei harvested at different circadian times (CT) of day in constant darkness. CT0, beginning of subjective day. (n = 3 experiments; values represent means ± standard deviations; **, P ≤ 0.01).
NONO binds to the circadian promoter of the Rev-Erbα gene.
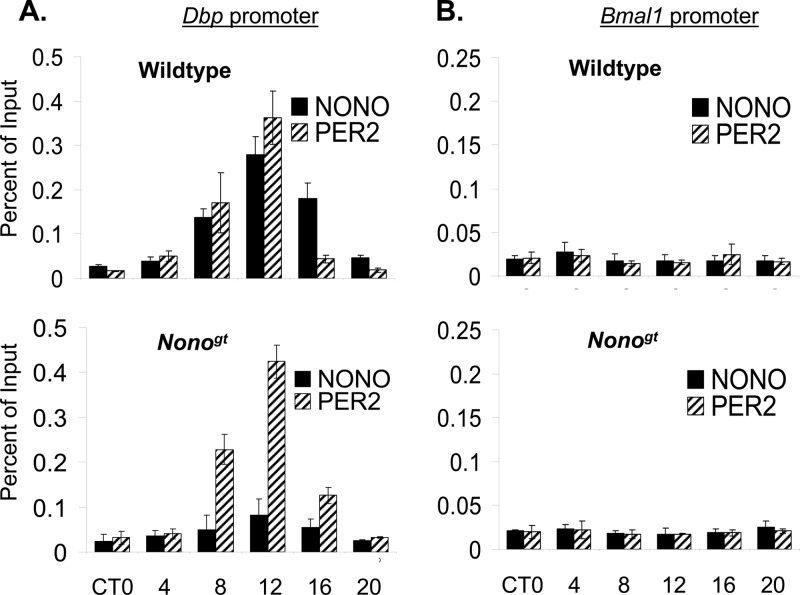
Next, we verified the relevance of NONO in vivo by looking for its presence at the promoters of clock genes. Since we showed previously that NONO interacted with PER proteins (5), we guessed that it ought to be found at PER-regulated clock genes. Chromatin immunoprecipitation experiments confirmed that this was indeed the case: NONO interacted with the promoter of the Rev-Erbα gene in circadian fashion, sharing the same kinetics as the PER1 protein (Fig. 1D, top). This interaction was considerably reduced but surprisingly not absent in Nonogt mice, which completely lack NONO transcript and protein (Fig. 1D, bottom). Equivalent results were seen for the Dbp promoter (Fig. 2A), and no binding was observed at the promoter of the antiphasic Bmal1 gene (Fig. 2B). Based upon the residual binding observed at the Rev-Erbα and Dbp promoters, we considered the possibility that NONO is redundant with homologous DBHS family factors with which our antibody might weakly cross-react. Conserved domain analysis (with the conserved domain architecture tool [CDART]) (15) showed that the other two known DBHS proteins, PSPC1 and SFPQ, shared both high homology with NONO and similar domain architectures (see Fig. S1C and D in the supplemental material). In addition, SFPQ was recently shown to play a role in circadian transcriptional repression (11). Therefore, we speculated that all three proteins might have similar functions in the circadian oscillator.
Fig 2.
(A) Chromatin immunoprecipitation of NONO and PER2 at the Dbp promoter in liver nuclei harvested from wild-type mice and Nonogt mice at different times of day in constant darkness (n = 3 experiments; values are ±SD). (B) Identical experiments for the Bmal1 promoter.
Overexpression or silencing of DBHS proteins interferes with circadian function.
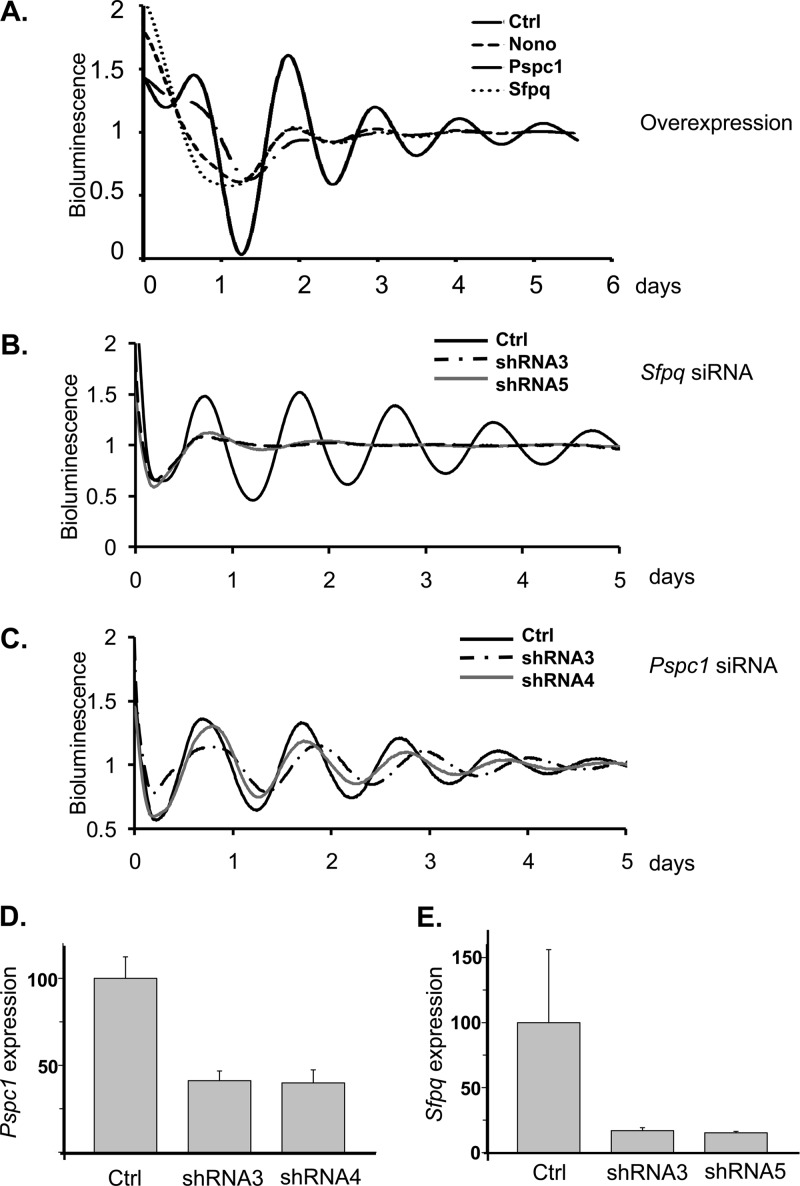
To test this idea, we transfected vectors expressing each of the three proteins into cultured cells together with a luciferase reporter under the control of the circadian Rev-Erbα gene promoter. After synchronizing circadian clocks in these transfected cells with dexamethasone (3), we monitored reporter bioluminescence in real time. Overexpression of any of the three proteins in human U2OS fibroblasts perturbed circadian rhythmicity (Fig. 3A).
Fig 3.
(A) Bioluminescence from U2OS cells transiently transfected with the Rev-Erbα-luc circadian reporter and constructs expressing either NONO, SFPQ, or PSPC1. Data shown are detrended and expressed in arbitrary units relative to mean expression. Control, cells transfected with the empty vector; Values for cells overexpressing NONO, PSPC1, and SFPQ are shown. (B) Bioluminescence from U2OS cells containing an integrated Bmal1-luc circadian reporter, infected with viruses expressing two different RNAi hairpins targeting the Sfpq gene and then clock synchronized with dexamethasone. Data shown are detrended and expressed in arbitrary units relative to mean expression. Control, scrambled-sequence shRNA. (C) Similar experiment with RNAi constructs targeting Pspc1. (D and E) Transcript levels of Pspc1 (D) and Sfpq (E) in U2OS cells infected with lentiviruses expressing the indicated RNAi targeting vectors used in the experiment shown in Fig. 2 (n = 3; values are ±standard errors).
We next undertook loss-of-function experiments based upon RNA interference (RNAi), in which U2OS human osteosarcoma cells containing an integrated Bmal1-luc reporter were infected with lentiviruses expressing short hairpin RNAs (shRNAs) targeting Pspc1 or Sfpq. RNAi hairpins against SFPQ dampened circadian oscillations dramatically (Fig. 3B) similar to what was observed previously for NONO (5), but those against PSPC1 lengthened it and somewhat dampened amplitude (Fig. 3C). Measurement of Sfpq and Pspc1 RNA levels in these cells showed that these hairpins reduced expression of Sfpq by 7-fold and of Pspc1 by 2.5-fold (Fig. 3D and E).
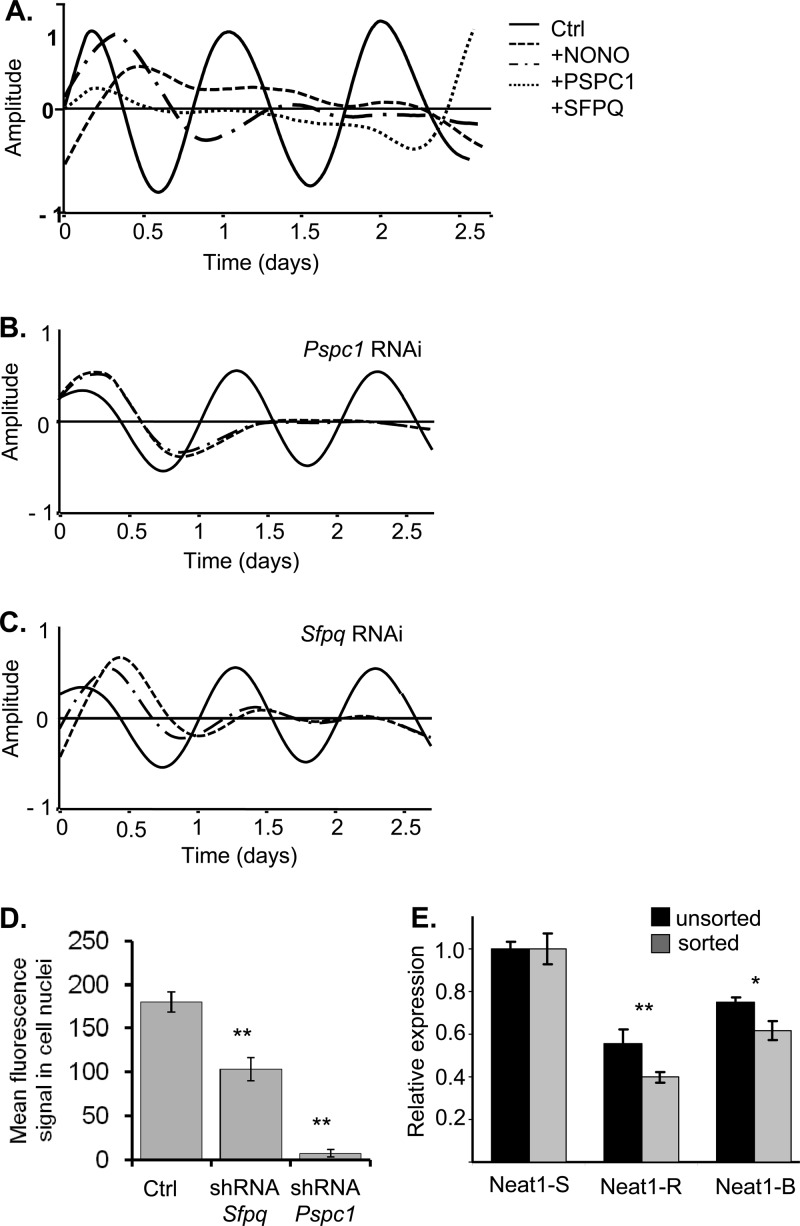
To ensure that the effects that we observed were not cell type specific, identical experiments were conducted using NIH 3T3 mouse fibroblasts. Again, overexpression of any of the three proteins perturbed circadian rhythmicity (Fig. 4A). Suppression of circadian rhythmicity was also seen in NIH 3T3 cells transiently transfected with the circadian Rev-Erbα promoter reporter together with RNAi hairpins targeting Pspc1 or Sfpq (Fig. 4B and C). In this case, immunofluorescence experiments showed that these hairpins reduced expression of SFPQ 2-fold and of PSPC1 10-fold (Fig. 4D).
Fig 4.
(A) Bioluminescence from 3T3 cells transiently transfected with the Rev-Erbα-luc circadian reporter and constructs expressing either NONO, SFPQ, or PSPC1. Data shown are detrended and expressed in arbitrary units relative to mean expression. Ctrl, wild-type cells. Values for cells overexpressing NONO, PSPC1, and SFPQ are shown. (B and C) Bioluminescence from 3T3 cells transiently transfected with the Rev-Erbα-luc circadian reporter and RNAi constructs targeting either Pspc1 (B) or Sfpq (C). After synchronization with dexamethasone, cultures were measured 3 days. Data shown are detrended and expressed in arbitrary units relative to mean expression. Solid line, wild-type cells. Dashed lines, duplicate plates of cells expressing an Sfpq- or Pspc1-targeting vector. (D) Quantification of depletion of SFPQ and PSPC1 proteins from experiments above. Relative repression from 3T3 cells cotransfected with a green fluorescent protein-expressing plasmid and a plasmid expressing an RNAi interference construct targeting Sfpq or Pspc1. Averages shown are from 10 cells each (±standard errors). Mean fluorescence is expressed in arbitrary units. (E) Quantification (values are ±standard errors; n = 2 independent experiments, performed in triplicate) of Neat1 levels for two different RNAi constructs (R and B), as well as a scrambled hairpin (S) used in the experiment shown in Fig. 4, quantified from RNA of bulk-transfected cells (unsorted) or from cells cotransfected with a green fluorescent protein-expressing plasmid and then subjected to fluorescence-activated cell sorting to isolate green fluorescent protein-expressing cells (sorted).
Depletion of paraspeckles does not perturb overall circadian clock function.
Since the three NONO-related proteins are also the three known members of nuclear paraspeckles, we speculated that the paraspeckle itself might serve a circadian role. This subnuclear domain requires the nuclear noncoding RNA Neat1, probably as a scaffold, and depletion of Neat1 has been shown to eliminate paraspeckles themselves (6, 7). By transiently transfecting shRNAs complementary to Neat1 into U2OS cells, we were able to reduce Neat1 levels (Fig. 4E) and thereby deplete paraspeckles, measured by counting the number of punctate PSPC1 foci (Fig. 5A and B). However, cotransfection of the circadian Bmal1-luc reporter showed that the circadian clock retained normal period length in these paraspeckle-depleted cells (Fig. 5C), making it unlikely that paraspeckles per se play a significant role in the circadian oscillator. Therefore, it is probable that nucleoplasmic, non-paraspeckle-associated pools of NONO, SFPQ, and PSPC1 proteins were responsible for the circadian effects that we have documented.
Fig 5.
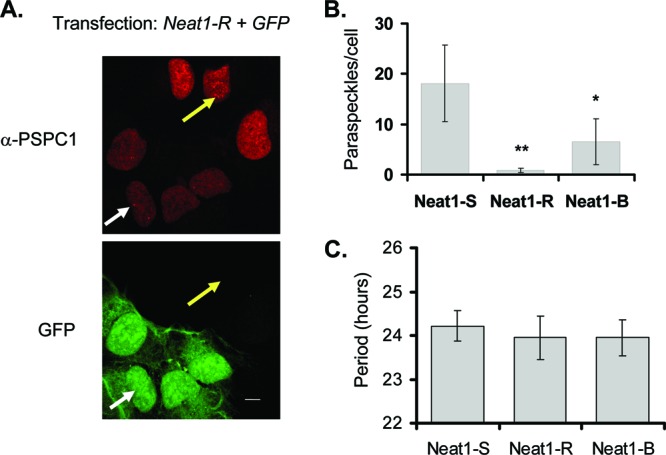
(A) Immunofluorescence from cells transfected with a plasmid expressing green fluorescent protein (GFP) and an RNAi construct targeting Neat1 (Neat-R). White arrow, paraspeckle in transfected cell; yellow arrow, paraspeckle in untransfected cell; α, anti. Scale bar, 10 μm. (B) Quantification (±standard deviation) of paraspeckles per cell for two different RNAi constructs (R and B), as well as a scrambled hairpin (S), quantified by immunostaining as described for panel A (n = 12 cells for Neat1-R, 24 for Neat1-B, and 18 for Neat1-S). *, P < 0.05; **, P <0.01 (Student t test.) (C) Period length of circadian reporter expression for U2OS cells cotransfected with the hairpins described for panel A and the Bmal1-luc circadian reporter. (n = 6 per sample; no significant differences, as determined by a Student t test).
DBHS proteins bind to clock promoter DNA and repress clock gene transcription.
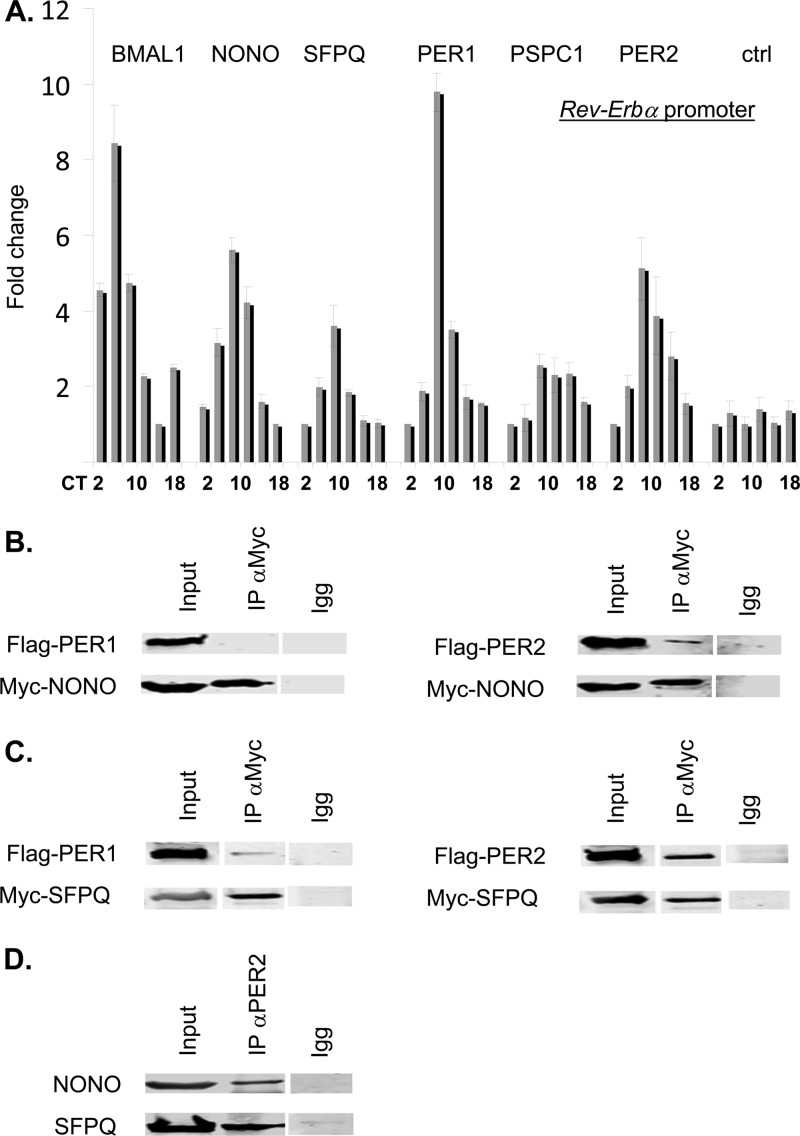
Since NONO can bind to circadian clock gene promoters in vivo (Fig. 1 and 2), it was logical to imagine that the other DBHS factors might do the same. Indeed, similarly to NONO, SFPQ and PSPC1 could also be immunoprecipitated at the Rev-Erbα promoter in a circadian fashion in liver nuclear extracts (Fig. 6A).
Fig 6.
(A) Chromatin immunoprecipitation of the indicated proteins at the Rev-Erbα promoter in liver nuclei harvested at different circadian times (CT) of day in constant darkness (n = 4; values are ±standard deviations, expressed relative to time point of minimum binding). Control reactions used an unrelated antibody raised at the same time in the same species (anti-PAR-BZIP). (B) Immunoprecipitations from whole-cell extracts from 293T cells cotransfected with myc-tagged NONO and Flag-tagged PER1 or PER2. For each panel, the left lane is 1/10 input. Subsequent to immunoprecipitation, all blots were probed with both anti-myc and anti-FLAG antibodies. (C) Identical experiments performed with whole-cell extracts from 293T cells cotransfected with myc-tagged SFPQ and Flag-tagged PER1 or PER2. (D) Mouse liver nuclear extracts from CT16 were immunoprecipitated with anti-PER2 and probed with anti-NONO or anti-SFPQ. Left lane, 1/10 input.
All three proteins are present at clock gene promoters with the same temporal profile as PER proteins, suggesting a corepressor function. Moreover, both NONO and SFPQ were identified as PER-interacting proteins and show interactions with PER1 and PER2 in various contexts (5, 11) (Fig. 6B to D). However, actual functions of these proteins are less clear. For example, we have shown previously that NONO can antagonize PER-mediated transcriptional repression when transfected into immortalized cells (5). In fact, the same is seen with SFPQ and to a lesser extent with PSPC1 (see Fig S2A and B in the supplemental material), whereas overexpression of NEAT does not influence BMAL1/CLOCK-mediated transcriptional activation (see Fig. S2C). In other reports, NONO and SFPQ have been reported by different investigators as either transcriptional coactivators or corepressors (17, 27), and SFPQ has been shown to act as a transcriptional repressor in the circadian clock (11).
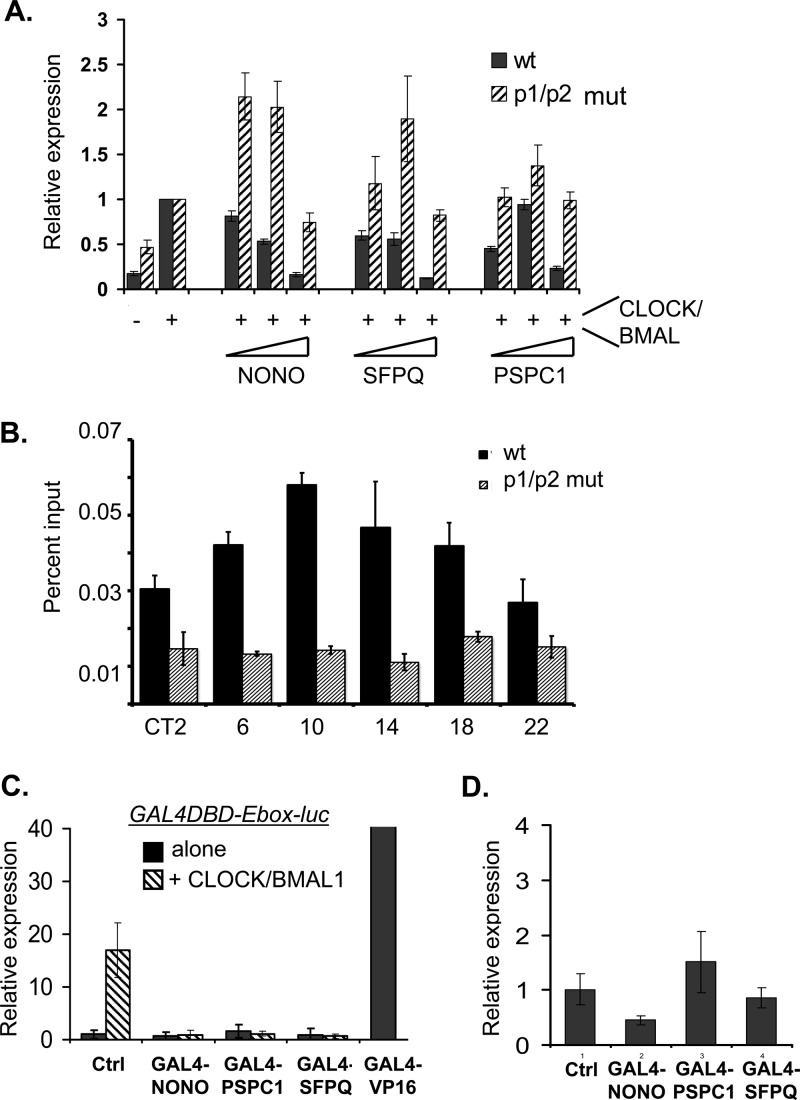
Within the circadian clock, we favor a repressive role of these factors because of the following experiments. First, when we transfected primary mouse fibroblasts with an E-box-driven luciferase reporter, together with the transcriptional activators CLOCK and BMAL1 and either NONO, PSPC1, or SFPQ, both NONO and SFPQ repressed CLOCK-BMAL-mediated transcription from the reporter and, to a lesser extent, PSPC1, which was initially activating and then repressing at higher concentrations (Fig. 7A). When equivalent transfections were performed using fibroblasts from Per1brdm/brdm/Per2brdm/brdm mice that lack functional PER proteins and circadian clocks (45), repression was no longer observed, but instead weak activation was shown (Fig. 7A). Similarly, in Per1brdm/brdm/Per2brdm/brdm mice, no circadian immunoprecipitation of NONO was observed at the Rev-Erbα promoter (Fig. 7B). Therefore, PER recruits NONO and, presumably, the other family members too.
Fig 7.
(A) Bioluminescence measured after transient transfection of mouse primary fibroblasts from wild-type (wt) or per1brdm/brdm/per2brdm/brdm (p1/p2) double mutant animals transfected with an E-box-luc reporter and vectors expressing CLOCK and BMAL proteins and NONO, SFPQ, or PSPC1 as indicated (n = 3 experiments in duplicate; values are ±standard errors). (B) Chromatin immunoprecipitation of NONO in wild-type (wt) or PER-deficient (p1/p2 mut) mice at the Rev-Erbα promoter in liver nuclei harvested at different circadian times (CT) of day in constant darkness (±standard deviations). CT0, beginning of subjective day. n = 3. (C) Transient transfection of fibroblasts with a GAL4 DNA-binding domain-E box-luciferase reporter and vectors expressing GAL4-NONO, -PSPC1, -SFPQ, or -VP16. “alone,” no exogenous activator added. (D) Identical experiments showing no statistically significant effects using a GAL4-CMV-luciferase reporter without exogenous activator.
Second, to confirm that DBHS factors are repressors at circadian promoters, we designed a hybrid GAL4 DNA-binding domain (GAL4DBD)-E box-luciferase reporter and fusions of NONO, PSPC1, and SFPQ with the GAL4 DNA-binding domain in order to enable their direct recruitment to DNA independently of PER proteins. When the GAL4DBD-E box-luciferase construct was transfected into primary mouse fibroblasts together with the GAL4-VP16 transcriptional activator, strong activation was observed, demonstrating the functionality of the construct. When GAL4-NONO, -PSPC1, and -SFPQ were cotransfected into primary mouse fibroblasts together with the GAL4 DNA-binding domain-E box-luciferase reporter, all three proteins strongly repressed CLOCK-BMAL-mediated transcription (Fig. 7C) though they had no statistically significant effect when similarly recruited to the constitutively active CMV promoter (Fig. 7D).
Importance of DBHS proteins to circadian behavior.
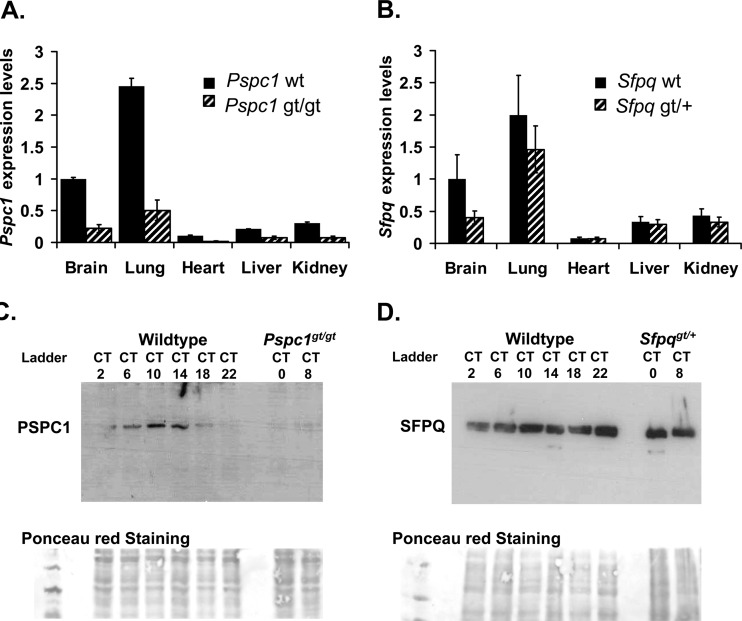
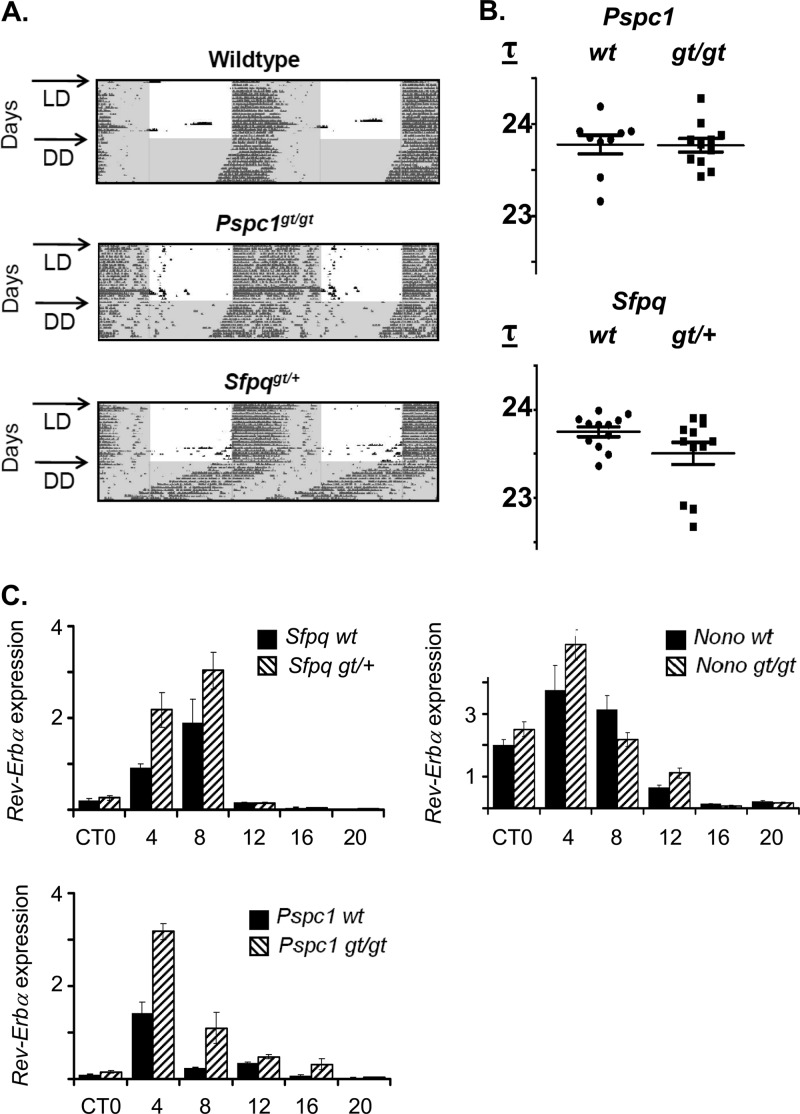
Finally, in order to verify the relevance of these factors to the circadian clock in vivo, we obtained mice with gene trap-based inactivations of Pspc1 and Sfpq to match the Nonogt mouse described earlier in this paper. Homozygous Pspc1-gene-trapped mice showed a 5-fold reduction in Pspc1 transcript levels in multiple tissues (Fig. 8A) and no detectable levels of PSPC1 protein in liver nuclear extracts (Fig. 8C). Although the Sfpq gene trap was homozygous lethal, heterozygous mice showed up to a 2-fold reduction in both RNA and protein (Fig. 8B and D). When tested for circadian wheel-running behavior, these Sfpqgt/+ mice also showed a trend toward shortening of period similar to that of Nonogt in some animals (Fig. 9A and B), as well as altered entrainment in a minimal-light skeleton photoperiod in all animals (see Fig. S3A to C in the supplemental material). Pspc1gt/gt mice showed no abnormalities (Fig. 9A and B; see also Fig. S3).
Fig 8.
(A) Pspc1 RNA levels measured by qPCR from different tissues of gene-trapped mice and wild-type littermates. For panels A and B, n = 2 mice per measurement, measured four times in duplicate, ±standard errors. (B) Sfpq RNA levels measured by qPCR from different tissues of gene-trapped mice and wild-type littermates. (C) PSPC1 protein levels in liver nuclear extracts harvested at different times of day from wild-type and gene-trapped animals kept in darkness. Top panel, Western blot probed with anti-PSPC1; bottom panel, Ponceau-S staining of filter to show equal loading. (D) SFPQ protein levels in liver nuclear extracts harvested at different times of day from wild-type and gene-trapped animals kept in darkness. Top panel, Western blot probed with anti-SFPQ; bottom panel, Ponceau-S staining of filter to show relative loading.
Fig 9.
(A) Left, wheel-running activity of wild-type, Pspc1gt/gt, and Sfpqgt/+ mice in 12/12 LD (arrow) and in constant darkness (DD). Darkness is indicated by gray shading. (B) Period lengths of 12 mice of each genotype, together with wild-type littermates. Values for Pspc1 or Sfpq are not significantly different from wild-type values (Student t test). (C) Rev-Erbα RNA expression from Nono, Sfpq, and Pspc1 gene-trapped mice (gt) and wild-type littermates (wt), measured by quantitative reverse transcription-PCR from liver extracts harvested at different circadian times (CT) of day from mice in constant darkness. RNA was measured four times in technical duplicates. Data shown are ±standard errors; n = 2 mice per time point.
Consistent with the proposed repressive role of these factors, at the gene expression level, Rev-Erbα RNA showed modestly increased expression in liver extracts from all three knockouts at the time (circadian time 8 to 12 [CT8-12]) that coincides with binding of NONO and PER2 (Fig. 9C). Interestingly, its timing coincides with the peak of Rev-Erbα expression levels and the beginning of their decline but not with maximum repression. Hence, it is possible that these factors are associated with the establishment of repression but not its maintenance. Similar but smaller gene expression effects were seen upon Per2 transcript levels, but the expression levels of other clock genes remained mostly unchanged (see Fig. S4 in the supplemental material).
DISCUSSION
Because of their homologies, shared functions, and abilities to interact with one another, the three factors NONO, PSPC1, and SFPQ have recently been classified by multiple authors as a family of proteins: the NOPS family (for NONO and PSPC1 [41]) or DBHS family (for Drosophila behavior human splicing [4]). Our data and that of others point to another important role of these proteins within the circadian oscillator. We initially isolated NONO as a PER-interacting protein (5), and Duong et al. recently isolated SFPQ in the same way (11). Here, we present data that all three DBHS proteins likely play overlapping roles within the circadian clock.
Nuclear paraspeckles and the circadian clock.
All three DBHS proteins are part of nuclear paraspeckles (14), subnuclear bodies probably involved in splicing and RNA storage. Nevertheless, our data suggest that the paraspeckle per se is not important for circadian function: depletion of these nuclear bodies by targeting the structural ncRNA Neat1 (7) has no effect upon the circadian clock, nor does transfection of this ncRNA into cells alter E-box-mediated transcription. Instead, our results imply that DBHS proteins likely exist in at least two nuclear pools. One of these pools is present in paraspeckles and appears to play no role so far in the circadian clock though it may be important for nuclear retention of edited RNAs as reported by others (6, 31, 44). A second pool is nucleoplasmic and could be in part responsible for the transcriptional roles reported for DBHS proteins.
DBHS proteins as transcription factors.
Besides their roles in nuclear paraspeckles, the previously reported functions of DBHS proteins have ranged widely. They have been implicated in splicing (18, 30) and axonal transport of RNA (19). They are players in the regulation of pre-mRNA processing and transcription termination (20) and in the DNA damage response (33, 36). NONO has also been characterized as a nonclassical carbonic anhydrase (21). In addition, however, all have been implicated in transcription. In some cases they have been implicated as activators (1, 17, 22) and in other cases as repressors (9, 27, 40), even for the same gene (38). Interestingly, a mechanism has been proposed in both cases: whereas NONO and SFPQ can interact directly with the RNA polymerase II C-terminal domain (CTD) in a way that might explain transcriptional activation (12), SFPQ has been proposed to recruit the mSIN3A histone deacetylase to promote repression (11, 27). For the circadian clock we originally identified NONO as an antagonist of PER-mediated repression (5), and other investigators demonstrated PER-mediated repression by SFPQ (11).
Apparently, the roles of DBHS proteins depend on the cellular context. In this paper alone, we show that transient transfection of SFPQ into U2OS cells can activate transcription from E-box reporters, but its transfection into mouse primary fibroblasts represses it. To try to resolve the role of these factors within the circadian clock, we therefore created GAL4 fusion proteins to unambiguously recruit these factors to promoters. All three had no effect on the CMV promoter but strongly repressed transcription mediated by the circadian transcription factors CLOCK and BMAL1 at a circadian E-box promoter. With this experiment, we show (i) that the effects of these factors upon transcription are context specific and (ii) that they are likely to be repressors in the circadian context.
Overlapping functions of DBHS proteins in the circadian clock.
One possible explanation for our results and those of others suggesting activation or repression is the overlapping function of related genes; if two repressors have different repressive potentials, for example, then titrating increasing exogenous amounts of the weaker one results in an increase in transcription as the endogenous stronger one is displaced. In this paper, we present considerable evidence for overlapping functions of the three DBHS proteins within the circadian clock. Depletion or overexpression of all three unambiguously affects circadian function in cells and in cellular transcription assays, but depletion of any one in mice results in only small circadian phenotypes, and generation of double DBHS mutants—or even complete knockouts of Sfpq—is complicated by embryonic lethality. Moreover, we along with others have shown by chromatin immunoprecipitation that all three DBHS proteins can bind directly to clock promoters or clock-controlled promoters in a circadian fashion in vivo and in cells (11, 16, 27). At least for circadian function, it is likely that this binding requires PER proteins. SFPQ and NONO were identified as PER-interacting proteins and clearly immunoprecipitate with them, as shown here and elsewhere (5, 11). Moreover, we show here that in PER-deficient mice, binding of NONO to circadian promoters is no longer observed. Thus, we think it is likely that PER proteins recruit DBHS proteins to clock-controlled genes to control and orchestrate PER-mediated transcriptional repression. The degree of this repression could be precisely controlled by the mix of the factors recruited.
Functional redundancy of this family of proteins is also highlighted by the recently published crystal structure of a NONO-PSPC1 complex (29). Not only do these proteins probably form obligate heterodimers, which would suggest a role for multiple DBHS family members within the circadian clock, but their structure also allows for possible higher-order oligomers, which might provide an ideal platform for the recruitment of other factors that have been found associated with these factors in various contexts.
DBHS proteins as orchestrators of circadian physiology.
Although we have shown clear roles of DBHS proteins in a cellular context, the circadian behavioral phenotypes of DBHS protein-deficient mice were relatively minor. As discussed above, functional redundancy could account for this lack of phenotype. In addition, however, the unique coupling of SCN cells into a network renders them more resilient to the effects of mutation (23). Therefore, it is also possible that more severe circadian effects of DBHS proteins occur in peripheral tissues.
Indeed, it is likely that considerable further circadian physiology directed by DBHS proteins remains to be elucidated. Mice deficient in these factors show a spectrum of unique phenotypes, ranging from embryonic lethality (Sfpq) to neurological phenotypes (Nono). Pspc1 protein is strongly regulated in circadian fashion although the other two factors are not (Fig. 8 and data not shown). Moreover, the E box is a standard motif for orchestrating clock-controlled physiology (35) and directs circadian transcription at thousands of promoters (42). DBHS factor binding has been observed at multiple clock-regulated promoters containing this motif, including prolactin (16), progesterone (10), Rev-Erbα (Fig. 1B), and androgen receptor (9). Through their interaction with PER proteins, we show here that DBHS factors play an important role directly in the circadian oscillator. Binding to clock gene promoters and modulating transcriptional repression, they regulate a portion of the transcriptional feedback which is the hallmark of metazoan circadian clocks.
Supplementary Material
ACKNOWLEDGMENTS
We thank Robert Dallmann for a critical reading of the manuscript and W. Schaffner (University of Zurich) for donation of Gal4 vectors.
This work was supported by the Swiss National Science Foundation and the University of Zurich Fonds der Akademischen Nachwuchses. Further support to S.A.B. was provided by the Neurosciences Center Zurich and the Molecular Life Sciences programs.
Footnotes
Published ahead of print 10 September 2012
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1. Amelio AL, et al. 2007. A coactivator trap identifies NONO (p54nrb) as a component of the cAMP-signaling pathway. Proc. Natl. Acad. Sci. U. S. A. 104:20314–20319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ausubel FM, et al. 2003. Current protocols in molecular biology. J. Wiley & Sons, Inc., New York, NY [Google Scholar]
- 3. Balsalobre A, et al. 2000. Resetting of circadian time in peripheral tissues by glucocorticoid signaling. Science 289:2344–2347 [DOI] [PubMed] [Google Scholar]
- 4. Bond CS, Fox AH. 2009. Paraspeckles: nuclear bodies built on long noncoding RNA. J. Cell Biol. 186:637–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Brown SA, et al. 2005. PERIOD1-associated proteins modulate the negative limb of the mammalian circadian oscillator. Science 308:693–696 [DOI] [PubMed] [Google Scholar]
- 6. Chen LL, Carmichael GG. 2009. Altered nuclear retention of mRNAs containing inverted repeats in human embryonic stem cells: functional role of a nuclear noncoding RNA. Mol. Cell 35:467–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Clemson CM, et al. 2009. An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol. Cell 33:717–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dibner C, Schibler U, Albrecht U. 2010. The mammalian circadian timing system: organization and coordination of central and peripheral clocks. Annu. Rev. Physiol. 72:517–549 [DOI] [PubMed] [Google Scholar]
- 9. Dong X, Sweet J, Challis JR, Brown T, Lye SJ. 2007. Transcriptional activity of androgen receptor is modulated by two RNA splicing factors, PSF and p54nrb. Mol. Cell. Biol. 27:4863–4875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dong X, et al. 2009. p54nrb is a transcriptional corepressor of the progesterone receptor that modulates transcription of the labor-associated gene, connexin 43 (Gja1). Mol. Endocrinol. 23:1147–1160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Duong HA, Robles MS, Knutti D, Weitz CJ. 2011. A molecular mechanism for circadian clock negative feedback. Science 332:1436–1439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Emili A, et al. 2002. Splicing and transcription-associated proteins PSF and p54nrb/nonO bind to the RNA polymerase II CTD. RNA 8:1102–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Fox AH, Bond CS, Lamond AI. 2005. P54nrb forms a heterodimer with PSP1 that localizes to paraspeckles in an RNA-dependent manner. Mol. Biol. Cell 16:5304–5315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Fox AH, Lamond AI. 2010. Paraspeckles. Cold Spring Harb Perspect Biol. 2:a000687 doi:10.1101/cshperspect.a000687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Geer LY, Domrachev M, Lipman DJ, Bryant SH. 2002. CDART: protein homology by domain architecture. Genome Res. 12:1619–1623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Guillaumond F, et al. 2011. Chromatin remodeling as a mechanism for circadian prolactin transcription: rhythmic NONO and SFPQ recruitment to HLTF. FASEB J. 25:2740–2756 [DOI] [PubMed] [Google Scholar]
- 17. Ishitani K, et al. 2003. p54nrb acts as a transcriptional coactivator for activation function 1 of the human androgen receptor. Biochem. Biophys. Res. Commun. 306:660–665 [DOI] [PubMed] [Google Scholar]
- 18. Kameoka S, Duque P, Konarska MM. 2004. p54(nrb) associates with the 5′ splice site within large transcription/splicing complexes. EMBO J. 23:1782–1791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kanai Y, Dohmae N, Hirokawa N. 2004. Kinesin transports RNA: isolation and characterization of an RNA-transporting granule. Neuron 43:513–525 [DOI] [PubMed] [Google Scholar]
- 20. Kaneko S, Rozenblatt-Rosen O, Meyerson M, Manley JL. 2007. The multifunctional protein p54nrb/PSF recruits the exonuclease XRN2 to facilitate pre-mRNA 3′ processing and transcription termination. Genes Dev. 21:1779–1789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Karhumaa P, et al. 2000. Nuclear NonO/p54(nrb) protein is a nonclassical carbonic anhydrase. J. Biol. Chem. 275:16044–16049 [DOI] [PubMed] [Google Scholar]
- 22. Kuwahara S, et al. 2006. PSPC1, NONO, and SFPQ are expressed in mouse Sertoli cells and may function as coregulators of androgen receptor-mediated transcription. Biol. Reprod. 75:352–359 [DOI] [PubMed] [Google Scholar]
- 23. Liu AC, et al. 2007. Intercellular coupling confers robustness against mutations in the SCN circadian clock network. Cell 129:605–616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lopez-Molina L, Conquet F, Dubois-Dauphin M, Schibler U. 1997. The DBP gene is expressed according to a circadian rhythm in the suprachiasmatic nucleus and influences circadian behavior. EMBO J. 16:6762–6771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Maier B, et al. 2009. A large-scale functional RNAi screen reveals a role for CK2 in the mammalian circadian clock. Genes Dev. 23:708–718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Masri S, Sassone-Corsi P. 2010. Plasticity and specificity of the circadian epigenome. Nat. Neurosci. 13:1324–1329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mathur M, Tucker PW, Samuels HH. 2001. PSF is a novel corepressor that mediates its effect through Sin3A and the DNA binding domain of nuclear hormone receptors. Mol. Cell. Biol. 21:2298–2311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Nagoshi E, et al. 2004. Circadian gene expression in individual fibroblasts: cell-autonomous and self-sustained oscillators pass time to daughter cells. Cell 119:693–705 [DOI] [PubMed] [Google Scholar]
- 29. Passon DM, et al. 2012. Structure of the heterodimer of human NONO and paraspeckle protein component 1 and analysis of its role in subnuclear body formation. Proc. Natl. Acad. Sci. U. S. A. 109:4846–4850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Patton JG, Porro EB, Galceran J, Tempst P, Nadal-Ginard B. 1993. Cloning and characterization of PSF, a novel pre-mRNA splicing factor. Genes Dev. 7:393–406 [DOI] [PubMed] [Google Scholar]
- 31. Prasanth KV, et al. 2005. Regulating gene expression through RNA nuclear retention. Cell 123:249–263 [DOI] [PubMed] [Google Scholar]
- 32. Preitner N, et al. 2002. The orphan nuclear receptor REV-ERBα controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell 110:251–260 [DOI] [PubMed] [Google Scholar]
- 33. Proteau A, et al. 2005. The multifunctional nuclear protein p54nrb is multiphosphorylated in mitosis and interacts with the mitotic regulator Pin1. J. Mol. Biol. 346:1163–1172 [DOI] [PubMed] [Google Scholar]
- 34. Reischl S, Kramer A. 2011. Kinases and phosphatases in the mammalian circadian clock. FEBS Lett. 585:1393–1399 [DOI] [PubMed] [Google Scholar]
- 35. Ripperger JA, Schibler U. 2006. Rhythmic CLOCK-BMAL1 binding to multiple E-box motifs drives circadian Dbp transcription and chromatin transitions. Nat. Genet. 38:369–374 [DOI] [PubMed] [Google Scholar]
- 36. Salton M, Lerenthal Y, Wang SY, Chen DJ, Shiloh Y. 2010. Involvement of matrin 3 and SFPQ/NONO in the DNA damage response. Cell Cycle 9:1568–1576 [DOI] [PubMed] [Google Scholar]
- 37. Schmutz I, Ripperger JA, Baeriswyl-Aebischer S, Albrecht U. 2010. The mammalian clock component PERIOD2 coordinates circadian output by interaction with nuclear receptors. Genes Dev. 24:345–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sewer MB, et al. 2002. Transcriptional activation of human CYP17 in H295R adrenocortical cells depends on complex formation among p54(nrb)/NonO, protein-associated splicing factor, and SF-1, a complex that also participates in repression of transcription. Endocrinology 143:1280–1290 [DOI] [PubMed] [Google Scholar]
- 39. Shav-Tal Y, Zipori D. 2002. PSF and p54(nrb)/NonO—multi-functional nuclear proteins. FEBS Lett. 531:109–114 [DOI] [PubMed] [Google Scholar]
- 40. Song KS, Kim K, Chung KC, Seol JH, Yoon JH. 2008. Interaction of SOCS3 with NonO attenuates IL-1beta-dependent MUC8 gene expression. Biochem. Biophys. Res. Commun. 377:946–951 [DOI] [PubMed] [Google Scholar]
- 41. Staub E, Fiziev P, Rosenthal A, Hinzmann B. 2004. Insights into the evolution of the nucleolus by an analysis of its protein domain repertoire. Bioessays 26:567–581 [DOI] [PubMed] [Google Scholar]
- 42. Ueda HR, et al. 2005. System-level identification of transcriptional circuits underlying mammalian circadian clocks. Nat. Genet. 37:187–192 [DOI] [PubMed] [Google Scholar]
- 43. Xie WQ, Rothblum LI. 1991. Rapid, small-scale RNA isolation from tissue culture cells. Biotechniques 11:324, 326–327 [PubMed] [Google Scholar]
- 44. Zhang Z, Carmichael GG. 2001. The fate of dsRNA in the nucleus: a p54(nrb)-containing complex mediates the nuclear retention of promiscuously A-to-I edited RNAs. Cell 106:465–475 [DOI] [PubMed] [Google Scholar]
- 45. Zheng B, et al. 2001. Nonredundant roles of the mPer1 and mPer2 genes in the mammalian circadian clock. Cell 105:683–694 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.