Abstract
While the functions of hypoxia-inducible factor 1α (HIF1α)/aryl hydrocarbon receptor nuclear translocator (ARNT) and HIF2α/ARNT (HIF2) proteins in activating hypoxia-inducible genes are well established, the role of other transcription factors in the hypoxic transcriptional response is less clear. We report here for the first time that the basic helix-loop-helix-leucine-zip transcription factor upstream stimulatory factor 2 (USF2) is required for the hypoxic transcriptional response, specifically, for hypoxic activation of HIF2 target genes. We show that inhibiting USF2 activity greatly reduces hypoxic induction of HIF2 target genes in cell lines that have USF2 activity, while inducing USF2 activity in cells lacking USF2 activity restores hypoxic induction of HIF2 target genes. Mechanistically, USF2 activates HIF2 target genes by binding to HIF2 target gene promoters, interacting with HIF2α protein, and recruiting coactivators CBP and p300 to form enhanceosome complexes that contain HIF2α, USF2, CBP, p300, and RNA polymerase II on HIF2 target gene promoters. Functionally, the effect of USF2 knockdown on proliferation, motility, and clonogenic survival of HIF2-dependent tumor cells in vitro is phenocopied by HIF2α knockdown, indicating that USF2 works with HIF2 to activate HIF2 target genes and to drive HIF2-depedent tumorigenesis.
INTRODUCTION
A hypoxic microenvironment is frequently found in solid tumors. The transcriptional response mediated by hypoxia-inducible factor 1α (HIF1α)/aryl hydrocarbon receptor nuclear translocator (ARNT) (HIF1) and HIF2α/ARNT (HIF2) plays a critical role in malignant progression by increasing expression of genes involved in angiogenesis, anaerobic metabolism, and other processes that enable tumor cells to survive and/or escape their O2-deficient microenvironment (25, 53, 56, 93).
It is well established that multiple transcription factors (TFs) are required to achieve maximal activation of target genes in response to a specific stimulus. This multifactorial transcription complex has been termed the “enhanceosome” (100). Individual factors in the enhanceosome complex may promote transcription initiation by recruiting RNA polymerase II (Pol II)/general transcription factors and/or recruiting chromatin-modifying enzymes, such as histone acetylases and chromatin remodeling complexes. In addition, TFs such as Myc increase gene expression by recruiting elongation factors to regulate Pol II pause release (77). Thus, reduced levels of transcription could occur in the absence of factors that have redundant functions within the enhanceosome, while other transcription factors having unique functions are absolutely required for gene activation.
The role of HIF1 and HIF2 in activating hypoxia-inducible genes is well established (21, 37, 48, 79, 103). However, the other transcription factors required for hypoxic activation of HIF target genes have been much less studied. Based on the enhanceosome concept, we hypothesized that another transcription factor(s) is required to activate HIF target genes during hypoxia. We found that many HIF target genes, including HMOX1 (41, 42, 63, 64, 84, 88, 89, 112), SFTPA1 (13, 32, 33), CXCR4 (73, 90, 96), PAI1 (2, 18, 27, 29, 47, 57, 58, 66), BDNF (48, 97), hTERT (3, 9, 35, 44, 52, 61, 67, 72, 75, 110, 113), CTSB (111) (107), and P4Hα(I) (10, 43, 98), are also reported to be activated by the transcription factor upstream stimulatory factor 1 (USF1) or USF2, suggesting a possible role of USF1/USF2 in the hypoxic response.
USFs (USF1 and USF2) are basic helix-loop-helix–leucine zipper (bHLH-LZ) transcription factors that are expressed ubiquitously, albeit at different levels depending on the tissue type (14, 36, 94, 95, 101). They exert their transcriptional function by binding to E boxes (the consensus sequence is CANNTG, where the NN nucleotides are in most cases either GC or CG) (36, 95), noncanonical E boxes (16, 40, 83, 102), or pyridine (Py)-rich initiator (Inr) sites (Py−2Py−1A+1N+2T+3 or A+3Py+4Py+5) (7, 8, 19) as either USF1/USF1 or USF2/USF2 homodimers or USF1/USF2 heterodimers. The major functional USF complexes in most cell types are USF1/USF2 heterodimers (94, 101). USF activates gene expression by recruiting chromatin-modifying enzymes, including histone acetylases PCAF, CBP, p300, and histone methylase SET7/92 (6, 51, 106). In addition, USF can interact with the TATA box binding protein of TFIID and TATA box binding protein-associated factors (TAFs) to directly promote preinitiation complex formation (12, 59, 70, 80).
Here we characterize the role of USF in the hypoxic transcriptional response. We find that USF2 but not USF1 function is required for HIF target gene activation during hypoxia. Interestingly, USF2 activity is required for activation of HIF2 but not HIF1 target genes. Additionally, we show that USF2 but not HIF2 is primarily responsible for recruiting the CBP and p300 coactivator(s) to HIF2 target gene promoters during hypoxia. Importantly, USF2 not only is required for HIF2 target gene activation but also is required for HIF2-dependent tumorigenicity in vitro. Identification of USF2 as a HIF2-specific cotranscriptional factor lays the foundation for the use of inhibitors of USF2 activity or the HIF2-USF2 interaction to specifically inhibit HIF2 function for tumors whose progression is dependent upon HIF2 activity.
MATERIALS AND METHODS
Cell culture.
Hep3B cells were cultured in minimal essential medium-Earle's balanced salt solution (HyClone) containing 10% fetal bovine serum (FBS), 2 mM l-glutamine, 1 mM sodium pyruvate, 100,000 units/liter penicillin-streptomycin, 1.5 g/liter sodium bicarbonate, and 1× nonessential amino acids (NEAA). HEK293T, PRC3, PRC3/pVHL, RCC4, and RCC4/pVHL cells were grown in high-glucose Dulbecco modified Eagle medium (DMEM; HyClone) with 10% FBS, 2 mM l-glutamine, 100,000 units/liter penicillin-streptomycin, and 1× NEAA. Mouse embryonic stem (mES) cells were cultured in high-glucose DMEM (HyClone) with 15% ES cell-approved FBS, 100,000 units/liter penicillin-streptomycin, 1× NEAA, 4 mM l-glutamine, 8 μl/liter 12.5 M beta-mercaptoethanol (BME), and 530 μl/L leukemia inhibitory factor (LIF). ES cells were plated on culture dishes pretreated with 0.2% bovine gelatin. For hypoxia treatment, 25 mM HEPES was added to the growth medium and cells were incubated in 21% or 1.5% O2 for 12 to 16 h before collection.
Knockdown of endogenous mRNAs using siRNAs or shRNA.
Control small interfering RNAs (siRNAs; 1027281) or siRNAs specific for human HIF1α (SI02664053), HIF2α (SI00380212), ARNT (an equal mix of SI00304220, SI00304227, SI00304234, SI03020913), USF1 (SI02780778), and USF2 (SI02780785) mRNAs were purchased from Qiagen. Hep3B or RCC4 cells were transfected with 10 nM siRNAs at 60% confluence using HiPerFect transfection reagent (Qiagen) according to the manufacturer's protocol. At 24 h posttransfection, cells were cultured in 21% or 1.5% O2 for 12 h and then collected to analyze HIF RNA and protein and their target gene mRNAs. To stably knock down USF2 or HIF2α mRNA levels, RCC4 and PRC3 cells were transduced with pLKO.1 lentiviruses expressing short hairpin RNA (shRNA)-degrading mRNA of USF2 (TRCN0000020736; Open Biosystems) or HIF2α (TRCN0000003803; Open Biosystems) and selected by hygromycin for transduced cells.
Generation of the mouse Hif1α−/−/USF2 shRNA/hUSF2-Flag ES cell line.
Hif1α−/− mES cells have been described previously (46). To knock down endogenous USF2 expression, Hif1α−/− mouse ES cells were infected with lentivirus expressing shRNA against mouse USF2 mRNA at a region of HLH that is shared by full-length USF2 and the exon 4 deletion isoform (RMM3981-97059962; Open Biosystems). The Hif1α−/−/USF2 shRNA ES cells were then electroporated with a human, full-length, Flag-tagged USF2 expression plasmid. Human USF2 (hUSF2)-Flag expression was under the control of the elongation factor 1 (EF1) promoter, which maintains gene expression well in primary cells such as mouse ES cells (46). In addition, the human USF2 cDNA had 6 nucleotides (but not amino acids) changed, allowing it to be resistant to shRNA against mouse USF2 mRNA. Hif1α−/−/USF2 shRNA/hUSF2-Flag ES cell populations were selected using hygromycin. The Hif1α−/−/USF2 shRNA/hUSF2-Flag ES cells were transfected with HIF2α siRNA (SI02712192; Qiagen) to knock down endogenous mouse HIF2α mRNA.
Plasmid construction.
Human USF1 and USF2 full-length cDNAs were cloned from Hep3B cDNA using Advantage GC cDNA polymerase (Clontech) and primers containing NheI/BamHI restriction sites for USF1 and NheI/HindIII restriction sites for USF2. The cDNAs were inserted into the pcDNA3.1hygro vector (Invitrogen) containing a 2× Flag tag between the BstXI and EcoRV sites. USF2ΔE4 was made by deleting exon 4 of USF2 from the full-length USF2 plasmid. The promoter for the HIF target gene PAI1 was cloned from human genomic DNA using GC-Melt genomic DNA polymerase (Clontech) and inserted into the pGL3basic luciferase vector (Promega). Deletion and single- and multiple-USF/HIF-binding-site mutants of the PAI1/Luc reporter were generated by Pfu Ultra II polymerase (Invitrogen)-mediated PCR. All DNA constructs were sequenced to ensure the fidelity of the PCR. pcDNA3HIF1αTM (referred to as HIF1αTM; a mutant with triple mutations [TMs] of P402A/P577A/N813A to make the HIF1α protein normoxia active) and pcDNA3HIF2αTM (referred to as HIF2αTM) have been previously described (47).
Transient and stable transfection. (i) Reporter assay.
All promoter reporter assays were conducted using the Fugene 6 transfection reagent (Roche) to transfect DNA into HEK293T cells. Typically, ∼50% confluent HEK293T cells in 6-well plates were transfected with 200 ng reporter DNA, 200 ng β-galactosidase (β-Gal), and 400 ng HIFαTM or USF expression vector (or 200 ng each of HIFαTM and USF). At 36 h after transfection, cells were collected into 400 μl 1× reporter lysis buffer (Promega) and assayed for β-Gal activity and luciferase activity using a luminometer. Promoter activation by HIF and USF was corrected for β-Gal transfection efficiency and presented as the fold induction relative to the promoter activities from an empty control vector. Results are the averages of at least three experiments.
(ii) Transfection of Hep3B cells with HIFαTM/USF to assess endogenous target gene activation.
Sixty percent confluent Hep3B cells in 6-cm dishes were transfected with 3 μg of HIFαTM or USF DNA or 1.5 μg each of HIFαTM and USF using Lipofectamine and Lipofectamine Plus reagent (Invitrogen). At 48 h posttransfection, RNA was collected from cells for the HIFαTM/USF target gene study. Results are the averages of at least three experiments.
RNA preparation, microarray, and qPCR to analyze mRNA levels.
RNA was isolated from cells using a Qiagen RNeasy-Plus kit that has a step for DNase digestion of genomic DNA. RNA was reverse transcribed using random hexamers and SuperScript III reverse transcriptase (Invitrogen). Levels of mRNA were quantified by Sybr green quantitative reverse transcription-PCR (qPCR) using a CFX384 real-time system (Bio-Rad). All primer sets designed for detection of target gene mRNA or used in chromatin immunoprecipitation (ChIP) by Sybr green qPCR were validated for their product specificity and amplification efficiency using melt curve analysis, qPCR product sequencing, and standard dilution analysis. The efficiencies of the primer sets used were between 90 and 110%. Primer sequences for qPCR are presented in Table S1 in the supplemental material. qPCR results were normalized using 18S rRNA and beta-actin. Results are the averages of a minimum of three independent experiments performed in triplicate. For microarrays, cDNAs were generated from total RNA using a Superscript Choice system (Gibco BRL Life Technologies) and T7-(dT)24 primers. Subsequently, biotin-labeled ribonucleotides were synthesized with a BioArray high-yield RNA transcript labeling kit (Enzo Diagnostic, Inc.). Fragmented cRNA was first tested for quality by a test array and then subjected to hybridization with Affymetrix Human Genome U133 Plus (version 2.0) arrays. The Gene Expression Core of the University of Colorado Cancer Center performed the hybridization and scans. Gene expression levels were normalized with expression levels for data sets generated from normoxic cells and then compared with those for the data set generated from cells of the same genotype treated with hypoxia. Genes upregulated under hypoxia are hypoxia-responsive genes. Data for cells targeted with HIF1α, HIF2α, or HIF1α and HIF2α siRNA were then compared with those for cells targeted with green fluorescent protein (GFP) siRNA and with each other to determine genes activated by HIF1, HIF2, or HIF1/HIF2.
Protein analysis.
Western blot analysis was performed using standard protocols with the following primary antibodies: anti-HIF1α monoclonal antibody (MAb) (NB100-105; Novus Biologicals, Littleton, CO), anti-HIF2α polyclonal antibodies (pAb) (NB100-122; Novus Biologicals), anti-HIF2α pAb (DE-93 for Fig. 4D; Cell Signaling), anti-ARNT MAb (NB100-124; Novus Biologicals), anti-Flag M2 MAb (F-3165; Sigma-Aldrich), anti-USF2 pAb (N-18, SC-861; Santa Cruz Biotechnology), anti-hemagglutinin (MMS-101P; Covance), and anti-p300 (SC-584; Santa Cruz Biotechnology).
Fig 4.
Increased FL USF2 expression in Hif1α−/− mouse ES cells restores hypoxic induction of HIF2 target genes. (A) Western blot analysis of USF2 protein in HIF2 functional (PRC3, PRC3/pVHL, RCC4, RCC4/pVHL, and Hep3B) and not functional (mouse ES) cells. Full-length USF2 and the splicing variant of USF2 are indicated. (B) Western blot of USF2 in mouse ES, Hep3B, 293T, or 293T cells transfected with USF2ΔE4 expression vectors. The Western blot gel was run longer to separate a nonspecific band from USF2ΔE4. (C) Western blot of endogenous mouse USF2 and Flag-tagged human USF2 proteins in Hif1α−/− or Hif1α−/−/USF2 shRNA mES cells or Hif1α−/−/USF2 shRNA/hUSF2-Flag ES cells, using anti-USF2 or anti-Flag antibodies. NS, nonspecific band. (D) Western blot of endogenous HIF2α and HIF1α proteins in normoxic and hypoxic (Hx) ES cells. (E and F) qPCR analysis of endogenous known HIF1/HIF2 target genes (E) and HIF1-specific target genes (F) in normoxic and hypoxic WT, Hif1α−/−, Hif1α−/−/USF2 shRNA/hUSF2-Flag, or Hif1α−/−/USF2 shRNA/hUSF2-Flag/HIF2α siRNA cells.
Electrophoretic mobility shift assay.
Nuclear extracts (NEs) were prepared from HEK293T cells transfected with USF2 expression plasmid DNA using an nuclear and cytoplasmic extraction reagent (NE-PER) kit (Pierce). USF binding to the biotin-labeled double-stranded oligonucleotides was performed in 20-μl binding reaction mixtures by mixing the following components: 2 μl 10× binding buffer (100 mM Tris, 500 mM KCl, 10 mM dithiothreitol [DTT], pH 7.5), 0.5 μg of poly(dI-dC), 1 μl of 50% glycerol, 6 μg NE, and 10 fmol of biotin-labeled, double-stranded oligonucleotides and H2O to 20 μl. For competition experiments, cold wild-type (WT) or USF2-binding-site-mutated competitor (200-fold molar excess) was simultaneously added with biotin-labeled oligonucleotide. The binding reaction mixture was loaded on a native, 6% polyacrylamide-TBE (Tris-borate-EDTA) gel to separate DNA-protein complexes from free probe and was then transferred to a nylon membrane. After cross-linking of DNA-bound complexes to the membrane, the position of biotin-labeled oligonucleotide was detected by streptavidin-horseradish peroxidase using a chemoluminescent nucleic acid detection module (Pierce).
ChIP and re-ChIP.
ChIP assays were performed as described previously. Anti-HIF1α (NB100-134B3; Novus Biologicals), anti-HIF2α (NB100-122; Novus Biologicals), anti-USF1 (SC-229; Santa Cruz), anti-USF2 (C-20, SC-862; Santa Cruz), anti-Pol II (H-224, SC-9001; Santa Cruz), anti-p300 (SC-584; Santa Cruz), and anti-CBP (SC-369; Santa Cruz) were used for protein-DNA complex precipitation, whereas rabbit preimmune serum served as a control. DNA from input or immunoprecipitated samples was assayed using Sybr green-based quantitative PCR methods (real-time detection system; Bio-Rad) with specific primers designed to amplify a specific region of the PAI1 promoter or EPO enhancer or promoter (see the primer sequences in Table S1 in the supplemental material). For ChIP and re-ChIP, the first immunoprecipitation (IP) was performed as described above, with the binding complexes from the first IP eluted from the Sepharose beads using re-ChIP buffer (0.5 mM DTT, 1% Triton X-100, 2 mM EDTA, 150 mM NaCl, 20 mM Tris-HCl, pH 8.1) at 37°C for 30 min. The eluted protein-DNA complexes were then diluted in radioimmunoprecipitation assay buffer and resubmitted to ChIP using a different antibody.
Coimmunoprecipitation.
HEK293T cells were transfected with HIF2αTM-Flag or USF2-Flag constructs for analyzing USF2-p300 and HIF2α-p300 interactions or hypoxic Hep3B cells for endogenous HIF1α-USF2 or HIF2α-USF2 interactions. Cells were washed in phosphate-buffered saline (PBS) and collected in nondenaturing lysis buffer (50 mM Tris, pH 7.4, 150 mM NaCl, 5 mM sucrose, 0.5% NP-40, 10% glycerol, 1× protease inhibitor cocktail [Thermo], 1 mM phenylmethylsulfonyl fluoride). Lysates were rotated at 4°C for 20 min and then homogenized by passage through a 21-gauge syringe needle 5 times, followed by another 20-min rotation at 4°C. Insoluble material was cleared from the lysates by centrifugation at 12,000 × g for 15 min. A sample of the lysate was set aside for analysis by Western blotting (lysate) to detect expression of transfected DNA or endogenous proteins. The remainder of the cleared lysates was incubated with M2–anti-Flag agarose beads (Sigma) or HIF1α, HIF2α, or USF2 antibody and protein A/protein G beads for 3 h. After binding, the beads were washed 5 times with washing buffer (50 mM Tris, pH 7.4, 150 mM NaCl, 0.5% NP-40). Immunocomplexes were eluted from the beads in 50 μl 2× SDS sample buffer by heating for 5 min at 80°C. The eluate and lysate samples were then assayed for protein expression and precipitation by Western blot protocols as described above.
In vitro tumorigenic assays. (i) Proliferation assays.
Five hundred cells per well were plated in growth medium in 24-well plates. Medium was changed every 2 days, and every 24 h, 3 wells of cells were collected by trypsinization and counted with a hemacytometer to yield an average number of cells per day.
(ii) Scratch assays.
Cells were plated at 80% confluence in normal growth medium in duplicate in 6-well plates. On the next day, when cells had formed a monolayer, the monolayers were scratched with a pipette tip. The cells were then washed with PBS twice to remove the scratched cells and serum and then incubated for the remainder of the experiment in serum-free medium. The width of the scratch at the same location in the well was measured daily. Duplicate measurements were averaged and graphed as percent closure of the scratch.
(iii) Clonogenic survival assays.
A total of 500, 1,000, or 1,500 cells/well were plated in duplicate in 6-well plates in normal growth medium. Medium was changed every 2 days, and then the cells were washed and the colonies were stained with crystal violet dye 7 to 10 days after plating. The colonies were counted, and knockdown cell colony numbers were graphed as the percent survival of the wild-type cells for each number of cells originally plated. Percent survival values were averaged and graphed relative to the clonogenic survival of wild-type cells.
RESULTS
USF2 knockdown significantly reduced the hypoxic induction of several known HIF2 but not HIF1 target genes in Hep3B and RCC4 cells.
We started studying the role of USF1/USF2 in the hypoxic response by testing whether reported USF and HIF common target genes are indeed activated by USF and HIF. To do this, Hep3B cells in 6-cm dishes were transiently transfected with 3 μg of HIF1αTM (a mutant with a triple mutation to make the HIF1α protein stable and active under normoxia), HIF2αTM, USF2, or 1.5 μg USF2 plus 1.5 μg of HIF1αTM (or HIF2αTM) plasmid. qPCR analysis indicated that HIF1 or HIF2, but not USF2, weakly activated CTSB and P4HA1 expression in Hep3B cells (Fig. 1). In addition, SFTPA1 and BDNF levels were too low to be detected by qPCR in Hep3B cells (data not shown). However, CXCR4, hTERT, HMOX, and PAI1 were indeed activated by USF2, HIF1α, or HIF2α (Fig. 1). Interestingly, HIF2α plus USF2, but not HIF1α plus USF2, exhibited cooperative activation of HMOX and PAI1 but not CXCR4 and hTERT (Fig. 1), suggesting a unique relationship between the HIF2α (but not HIF1α) and USF2 proteins and a role of HIF2/USF2 in select HIF target gene activation. In addition, USF1 was unable to activate any of the above-described genes alone or cooperatively with HIF1α or HIF2α (data not shown).
Fig 1.

Some reported HIF/USF common target genes are indeed activated by USF2 or HIF2αTM in Hep3B cells. qPCR detection of reported HIF/USF common target gene expression levels in normoxic Hep3B cells or Hep3B cells transfected with 3 μg of HIF1αTM, HIF2αTM, or USF2 plasmid or with 1.5 μg USF2 plus 1.5 μg of HIF1αTM (or HIF2αTM).
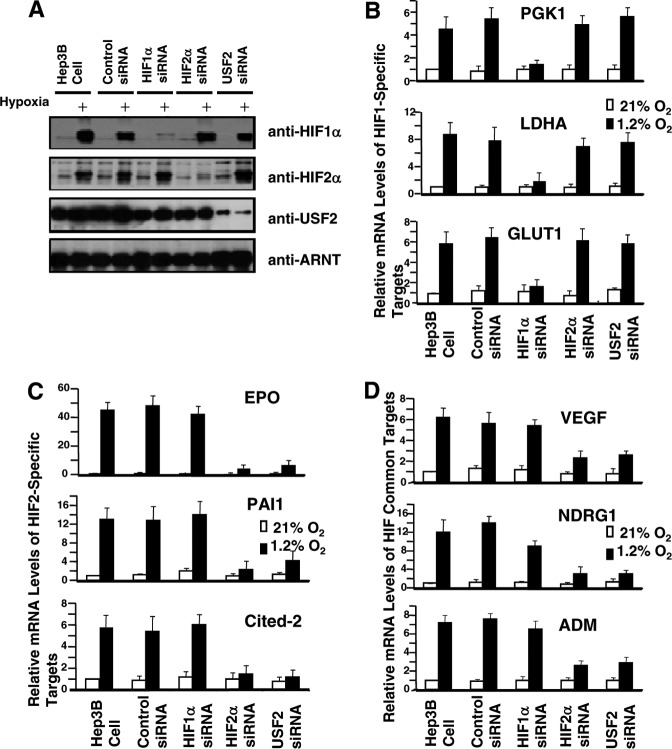
To formally test the role of USF2 in the hypoxic response, HIF target genes were analyzed in normoxic and hypoxic Hep3B cells targeted with siRNAs against human USF2, HIF1α, HIF2α, or GFP (for nonspecific control) mRNA. As shown in Fig. 2A, hypoxia stabilized HIF1α and HIF2α protein levels but had no effect on USF2 protein levels (Fig. 2A, Hep3B cell and control siRNA). As expected, hypoxic induction of HIF1α and HIF2α protein was significantly reduced by HIF siRNAs, while USF2 siRNA strongly decreased USF2 protein levels under both normoxia and hypoxia (Fig. 2A). qPCR detection of mRNA levels supported the Western blot data that Hep3B cells exhibited 80% knockdown of HIF1α, HIF2α, or USF2 mRNA, and there was no cross regulation among these genes (data not shown). Next, we used Sybr green qPCR to assess several known HIF target genes in these Hep3B cells (26, 47, 104). As expected, the previously identified HIF target genes exhibited hypoxic induction in Hep3B cells or cells targeted with control siRNA (Fig. 2B to D, Hep3B cell or control siRNA). In addition, HIF1α siRNA but not HIF2α siRNA reduced the hypoxic induction of several known HIF1-specific target genes (PGK1, LDHA, and GLUT1) (Fig. 2B); HIF2α siRNA but not HIF1α siRNA reduced hypoxic induction of HIF2-specific target genes CITED2, EPO, and PAI1 (Fig. 2C); and HIF1α siRNA and, in particular, HIF2α siRNA reduced hypoxic activation of HIF1/HIF2 common target genes VEGF, NDRG1, and ADM (Fig. 2D). These data confirmed the HIF target gene specificity in Hep3B cells (47, 104). Interestingly, USF2 knockdown had no effect on hypoxic induction of HIF1-specific target genes (Fig. 2B) but greatly reduced the hypoxic activation of HIF2-specific target genes (Fig. 2C). In addition, the hypoxic induction of HIF1/HIF2 common target genes was also reduced by USF2 siRNA at levels similar to those for cells targeted with HIF2α siRNA (Fig. 2D). To further test the role of USF2 in regulating HIF target genes in another cell type, VHL-deficient RCC4 cells were targeted with HIF1α, HIF2α, USF2, or control siRNA (Fig. 3A). Interestingly, USF2 siRNA specifically lowered the levels of HIF2-specific targets (CST and PAI1) (Fig. 3C) and HIF1/HIF2 common targets (VEGF and NDRG1) (Fig. 3D) but not HIF1-specific targets (PGK and LDHA) (Fig. 3B) in normoxic RCC4 cells, confirming the importance of USF2 for HIF2 target gene expression. Furthermore, we found that siRNA knockdown of USF1 in Hep3B or RCC4 cells had no effect on the expression of any HIF target gene assessed (data not shown). Collectively, these data support a possible role of USF2 (but not USF1) in mediating the hypoxic activation of HIF2 but not HIF1 target genes.
Fig 2.
USF2 siRNA reduces hypoxic induction of several known HIF2 but not HIF1 target genes in Hep3B cells. (A) Western blot analysis of HIF1α, HIF2α, USF2, and ARNT proteins in normoxic and hypoxic Hep3B cells or Hep3B cells transfected with control siRNA or siRNA against HIF1α, HIF2α, or USF2. (B to D) qPCR analysis of known HIF1-specific target genes (B), HIF2-specific target genes (C), and HIF1/HIF2 common targets (D) in normoxic and hypoxic Hep3B cells or Hep3B cells targeted with the indicated siRNAs. Results are presented relative to those for WT Hep3B cells cultured under normoxia. qPCR results were normalized to 18S rRNA expression; error bars are ±1 SD from three independent experiments in this and the other figures.
Fig 3.
USF2 siRNA reduces HIF2 but not HIF1 target gene expression in normoxic RCC4 cells. (A) qPCR analysis of HIF1α, HIF2α, and USF2 mRNA in normoxic RCC4 cells or RCC4 cells transfected with control, HIF1α, HIF2α, or USF2 siRNA. (B to D) qPCR analysis of known HIF1-specific (B), HIF2-specific (C), and HIF1/HIF2 common (D) target genes in normoxic RCC4 cells or RCC4 cells transfected with control, HIF1α, HIF2α, or USF2 siRNA.
USF2 is required for hypoxic induction of global HIF2 target genes in Hep3B cells.
The siRNA results presented above suggested that USF2 is required for the activation of a number of HIF2 but not HIF1 target genes in Hep3B and RCC4 cells. We wondered if this trend was true for all HIF2 or HIF1 target genes. To address this question, we first identified global HIF1-specific (downregulated by HIF1α but not by HIF2α siRNA), HIF2-specific (downregulated by HIF2α but not by HIF1α siRNA), or HIF1/HIF2 common (downregulated by HIF1α and HIF2α siRNAs much more than HIF1α or HIF2α siRNA alone) target genes by conducting Affymetrix DNA expression array analysis in normoxic or hypoxic Hep3B cells or Hep3B cells transfected with HIF1α, HIF2α, or HIF1α plus HIF2α siRNA. Table 1 lists the top 10 (or more) most likely HIF1- or HIF2-specific or HIF1/HIF2 common target genes (Table 1, microarray), based on levels of reduction by HIF1α, HIF2α, or HIF1α plus HIF2α siRNA. Furthermore, using qPCR and RNA prepared from normoxic or hypoxic parental, HIF1α, HIF2α, ARNT, or USF2 siRNA-transfected Hep3B cells, we validated the microarray-identified top 10 (or more) HIF1-specific, HIF2-specific, and HIF1/HIF2 common target genes. Importantly, USF2 was required for hypoxic induction of all HIF2 (including HIF2-specific and HIF1/HIF2 common genes) but not HIF1 target genes assessed (Table 1, qPCR), demonstrating the functional requirement of USF2 for HIF2 target gene activation by hypoxia. Since USF2 is required only for HIF2 target gene activation, the following studies focus on USF2's role in HIF2 target gene activation.
Table 1.
DNA microarray to determine global HIF1- and HIF2-specific and HIF1/HIF2 target genes and qPCR to confirm HIF target gene specificity and to determine the role of USF2 in hypoxic induction of HIF target genes in Hep3B cellsa
| Target | Probe identifier | GenBank accession no. | Gene symbol | Fold hypoxic induction measured by: |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Microarray |
qPCR |
|||||||||||
| Hep3B cell | HIF1α siRNA | HIF2α siRNA | HIF1α + HIF2α siRNA | Hep3B cell | HIF1α siRNA | HIF2α siRNA | USF2 siRNA | ARNT siRNA | ||||
| HIF1 specific | 203685_at | NM_000633 | BCL2 | 2.3 | 0.47 | 2.17 | 1.07 | 5.31 | 1.44 | 5.08 | 5.18 | 0.92 |
| 202095_s_at | NM_001012270 | BIRC5 | 2.43 | 0.87 | 2.14 | 0.93 | 2.01 | 1.02 | 2.09 | 2.12 | 0.83 | |
| 200737_at | NM_000291 | PGK1 | 3.03 | 1.07 | 2.86 | 1.15 | 3.97 | 1.3 | 4.45 | 4.1 | 1.27 | |
| 200650_s_at | NM_001135239 | LDHA | 3.52 | 1.23 | 3.11 | 1.23 | 7.33 | 0.97 | 6.66 | 5.73 | 0.93 | |
| 201250_s_at | NM_006516 | GLUT1 | 4.92 | 1.23 | 4.59 | 1.23 | 5.99 | 1.02 | 6.23 | 5.67 | 0.99 | |
| 206686_at | NM_002610 | PDK1 | 4.29 | 1.32 | 3.87 | 1.52 | 8.6 | 2.34 | 7.75 | 9.06 | 1.85 | |
| 1553036_at | NM_153839 | GPR111 | 3.42 | 1.32 | 3.22 | 0.87 | 6.7 | 3.29 | 5.24 | 8.17 | 2.3 | |
| 202364_at | NM_001008541 | MXI1 | 5.57 | 1.52 | 5.36 | 1.52 | 11.97 | 3.07 | 8.91 | 7.66 | 1.86 | |
| 221478_at | NM_004331 | BNIP3L | 6.06 | 1.62 | 5.73 | 0.81 | 9.68 | 1.82 | 9.01 | 12.1 | 1.22 | |
| 206367_at | NM_001002914 | KCTD11 | 8.14 | 1.62 | 11.38 | 1.87 | 13.87 | 5.51 | 19.93 | 24.26 | 3.27 | |
| 213943_at | NM_000474 | TWIST1 | 12.13 | 3.12 | 10.56 | 2.29 | 3.53 | 1.21 | 3.27 | 3.84 | 1.29 | |
| 219888_at | NM_003116 | SPAG4 | 13.93 | 3.25 | 12.56 | 1.15 | 13.3 | 3.67 | 8.9 | 11.45 | 2.65 | |
| 201313_at | NM_001975 | ENO2 | 14.93 | 4 | 16.74 | 2.46 | 12.71 | 3.05 | 16.18 | 9.18 | 2.18 | |
| 205199_at | NM_001216 | CA9 | 39.4 | 6.5 | 36.22 | 2.64 | 45.31 | 5.7 | 34.48 | 47.29 | 2.75 | |
| HIF2 specific | 205220_at | NM_006018 | GPR109B | 4 | 3.54 | 1 | 1.15 | 3.33 | 2.71 | 0.83 | 0.94 | 1.43 |
| 207980_s_at | NM_001168388 | CITED2 | 3.03 | 3.25 | 1 | 1.15 | 5.91 | 5.52 | 1.86 | 2.13 | 1.27 | |
| 209242_at | NM_001146184 | PEG3 | 2.83 | 2.3 | 1.15 | 0.62 | 2.64 | 2.54 | 1.45 | 1.23 | 1.3 | |
| 1552846_s_at | NM_152304 | RAB42 | 6.5 | 6.06 | 1.23 | 0.76 | 4.39 | 3.53 | 1.11 | 1.58 | 1.33 | |
| 208286_x_at | NM_001173531 | POU5F1 | 2.82 | 2.21 | 1.23 | 0.81 | 2.32 | 2.2 | 1.57 | 0.81 | 1.14 | |
| 227970_at | NM_024980 | GPR157 | 4.29 | 3.48 | 1.41 | 1.07 | 6.8 | 7.57 | 3.28 | 4.19 | 0.95 | |
| 202627_s_at | NM_000602 | PAI1 | 4.92 | 4.13 | 1.52 | 1.62 | 14.98 | 12.72 | 2.5 | 4.06 | 2.25 | |
| 227353_at | NM_152468 | TMC8 | 8 | 7.18 | 1.52 | 1.23 | 5.63 | 4.47 | 2.04 | 2.12 | 1.45 | |
| 203066_at | NM_015892 | CHST15 | 4.92 | 4.01 | 1.52 | 0.93 | 3.54 | 3.11 | 1.63 | 1.87 | 1.27 | |
| 205258_at | NM_002193 | INHBB | 4 | 3.48 | 1.62 | 1.23 | 5.78 | 5.58 | 3.37 | 3.78 | 1.08 | |
| 219014_at | NM_001130715 | PLAC8 | 9.19 | 8 | 2.64 | 0.76 | 4.12 | 3.89 | 2.67 | 2.74 | 1.37 | |
| 230250_at | NM_001109754 | PTPRB | 7.46 | 6.75 | 4.29 | 1.41 | 8.71 | 7.63 | 3.87 | 3.85 | 0.97 | |
| 217254_s_at | NM_000799 | EPO | 16 | 14.93 | 4.92 | 1.07 | 31.32 | 27.41 | 8.05 | 11.94 | 1.78 | |
| HIF1/HIF2 common | 211919_s_at | NM_001008540 | CXCR4 | 2.64 | 1.52 | 0.87 | 0.58 | 3.57 | 2.32 | 2.11 | 2.24 | 1.32 |
| 217287_s_at | NM_004621 | TRPC6 | 9.19 | 6.25 | 3.25 | 0.76 | 3.94 | 1.66 | 1.4 | 1.62 | 0.97 | |
| 221748_s_at | NM_022648 | TNS1 | 9.19 | 5.66 | 2.14 | 0.81 | 7.66 | 3.29 | 4.19 | 4.95 | 0.97 | |
| 213004_at | NM_012098 | ANGPTL2 | 6.5 | 4.36 | 1.74 | 1.07 | 4.55 | 1.1 | 1.7 | 2.85 | 0.63 | |
| 228212_at | NM_182509 | ISM2 | 4.59 | 2.13 | 1.51 | 1.15 | 4.02 | 0.95 | 1.53 | 1.85 | 0.93 | |
| 210512_s_at | NM_001025366 | VEGFA | 2.46 | 2.46 | 2.14 | 1.23 | 6.25 | 4.74 | 2.57 | 2.84 | 1.33 | |
| 220041_at | NM_025163 | PIGZ | 6.96 | 3.48 | 3.25 | 1.32 | 3.06 | 2.31 | 2.52 | 2.2 | 1.38 | |
| 206424_at | NM_000783 | CYP26A1 | 5.28 | 2.92 | 1.41 | 1.41 | 3.2 | 1.63 | 1.02 | 1.93 | 1.4 | |
| 200632_s_at | NM_001135242 | NDRG1 | 13.93 | 9.64 | 4.29 | 1.41 | 10.32 | 6.64 | 3.26 | 3.9 | 1.91 | |
| 202912_at | NM_001124 | ADM | 9.85 | 7.27 | 3.25 | 1.52 | 7.13 | 5.52 | 3.15 | 4.14 | 1.27 | |
Listed is the fold hypoxic induction of HIF target genes in Hep3B cells or Hep3B cells targeted with HIF1α, HIF2α, or HIF1α and HIF2α siRNA, determined by DNA microarray expression analysis or qPCR. Hypoxia-inducible genes are grouped on the basis of their dependence upon either or both HIFα subunits for their hypoxic induction. The most likely 10 to 14 HIF1- and HIF2-specific or HIF1/HIF2 common genes are listed (microarray). HIF target gene specificity determined by microarray was validated using qPCR. In addition, the role of USF2 in activating the listed genes in Hep3B/USF2 cell siRNA is also determined (qPCR).
Increased USF2 expression in Hif1−/− mouse ES cells restores hypoxic induction of HIF2 target genes.
We and others have shown that HIF2α is functional in PRC3, PRC3/pVHL, RCC4, RCC4T/pVHL, and Hep3B cells (26, 47, 50, 79, 104). In contrast, HIF2α is not functional in mES cells, despite the fact that these cells express similar levels of HIF2α mRNA and protein under hypoxia relative to cells in which HIF2 is functional (46). The absolute requirement of USF2 for hypoxic induction of global HIF2 target genes in Hep3B cells and known HIF2 target genes in RCC4 cells prompted us to look at whether there is a correlation between a lack of USF2 expression and a lack of hypoxic induction of HIF2 target genes. Western blot analysis of USF2 indicated that all cell types expressed similar levels of full-length USF2, while HIF2α nonfunctional mES cells expressed an additional low-molecular-weight band (Fig. 4A, isoform). The low-molecular-weight band is a USF2 isoform since, like the full-length USF2 protein, the levels of the low-molecular-weight band were reduced by USF2 shRNA in Hif1−/− mES cells (Fig. 4C). This USF2 isoform is likely the USF2b isoform (also called USF2ΔExon4), since we identified one USF2b clone from six cDNAs amplified from mES cells but none from six clones generated from Hep3B cells (data not shown). Additionally, using qPCR and primers designed to specifically amplify either full-length USF2 or USF2b, we determined that USF2b mRNA was approximately 27-fold more abundant in mES cells than in Hep3B cells (threshold cycle [CT] numbers for USF2b mRNA in Hep3B and mES cells, 32.86 and 28.10, respectively), despite relatively similar quantities of full-length USF2 mRNA in both cell lines (CT numbers for full-length USF2 mRNA in Hep3B and mES cells, 22.64 and 23.54, respectively). Furthermore, the USF2 isoform in mouse ES cells is similar in size to the USF2b protein expressed in 293T cells transfected with the USF2b expression vector (Fig. 4B). USF2b is created through the exclusion of the positive regulatory region in exon 4 of USF2 during RNA splicing and functions as an endogenous dominant-negative inhibitor of full-length USF2 (45, 101). To increase USF2 activity and thus possible HIF2 activity, Hif1−/− mES cells were targeted with mouse USF2 shRNA, which significantly reduced full-length and USF2b isoform proteins (Fig. 4B), followed by stable transfection with a human USF2 cDNA vector resistant to the anti-mouse USF2 shRNA due to an introduced multinucleotide mutation. The Hif1−/−/USF2 shRNA/hUSF2-Flag cells expressed high levels of USF2 protein, as detected by anti-USF2 or anti-Flag Western blot assays (Fig. 4C). qPCR analysis indicated that our previously determined HIF target genes (46) were indeed induced by hypoxia in WT mES cells but not in Hif1−/− ES cells (Fig. 4E and F) or Hif1−/−/USF2 shRNA cells (data not shown). However, Hif1−/−/USF2 shRNA/hUSF2-Flag cells exhibited significant restoration of the hypoxic induction of known HIF1/HIF2 common target genes, including Adrp, Anxa2, Ndrg1, Stra14, Vegf, Hmox1, Aoc3, Adm, and Ccng2 (Fig. 4E and data not shown) but not known HIF1 target genes of A2m, Ca12, Glut1, P4ha1, and Pgk1 (Fig. 4F). To determine the role of HIF2 in hypoxic induction of genes observed in Hif1−/−/USF2 shRNA/hUSF2-Flag cells, the levels of endogenous HIF2α mRNA and, consequently, HIF2α protein in Hif1−/−/USF2 shRNA/hUSF2-Flag cells were reduced by HIF2α siRNA (Fig. 4D). As expected, Hif1−/−/USF2 shRNA/hUSF2-Flag/HIF2α siRNA cells exhibited significantly reduced hypoxic activation of HIF1/HIF2 common genes (Fig. 4E). Overexpression of USF2 restored hypoxic induction of HIF2 but not HIF1 target genes, providing additional evidence for a positive role of USF2 in hypoxic activation of HIF2 but not HIF1 target genes.
USF2 binding increases PAI1 basal promoter activity and HIF2-mediated activation of PAI1.
After establishing the function of USF2 in hypoxic activation of endogenous HIF2 target genes in several cell types, we wanted to study why USF2 is required for HIF2 target gene activation, starting with determining whether USF2 binds to the HIF2 target gene promoter and the function of USF2 binding for HIF2 target gene activation during hypoxia. We selected the human PAI1 promoter as a model to address this question since PAI1 is a clear HIF2 and USF2 target gene in Hep3B and RCC4 cells (Fig. 2C and 3C). We first cloned the human PAI1 promoter fragment from positions −806 to +26 into a pGL3Basic/Luc reporter (Fig. 5A). Like the endogenous PAI1 gene (Fig. 1), the PAI1/Luc reporter exhibited activation by USF2, HIF2αTM, or HIF1αTM individually and by USF2 plus HIF2αTM in a cooperative manner (Fig. 5B), indicating the usefulness of the reporter to study the role of USF2 in hypoxic activation of a HIF2 target gene.
Fig 5.

USF2 binds to the human PAI1 promoter at multiple sites. (A) Cloned human PAI1 promoter/Luc reporter, showing the locations of consensus USF/HIF common (CACGTG), HIF (ACGTG), or USF2 (CANNTG, where the NN nucleotides are CG or GC) binding sites. The previously reported HIF or USF binding sites are represented by squares with bold borders, and the additional potential HIF or USF binding sites are represented by white squares. (B) Relative luciferase activity of PAI1/Luc reporter in response to activation by indicated plasmids in nanograms. (C) Gel shift assay of USF2 binding to potential USF2 binding sites on the PAI1 promoter using 30-mer double-stranded oligonucleotides centered at the indicated number and nuclear extracts prepared from USF2-transfected HEK293T cells. A 200-fold excess of cold WT competitor oligonucleotide or cold USF2-binding-site-mutated oligonucleotide (Mut) was added in competition assays.
For PAI1/Luc, a functional HIF binding site (HBS) at positions −194 to −187 and two USF2 binding sites at positions −681 to −674 and −566 to −559 have been reported (18, 27). However, these previous studies did not address if HIF1 or HIF2 activates the PAI1 gene or the function of USF2 in hypoxic activation of PAI1. To study the function of reported USF2 binding sites, we created PAI1/Luc deletion mutants that lacked the reported USF2 binding sites at position −684 (Fig. 6A, Δ−808/−684) or both positions −684 and −565 (Fig. 6A, Δ−808/−565). However, both deletion reporters exhibited almost WT fold activation by USF2, suggesting the existence of other USF2 binding sites on our cloned PAI1 promoter (positions −806 to +26). We searched for USF consensus sequences (CANNTG, where the NN nucleotides are GC or CG) and initiator elements (YYA+1NT/AYY), both of which have been reported to be USF binding sites (14). In our cloned PAI1 promoter, we identified three additional potential USF binding sites, including one classical USF site at position +5 and two initiator elements at positions −35 and −47 (Fig. 5A). By gel shift using nuclear extracts from USF2-transfected 293T cells, we saw that, like the known USF2 binding sites at positions −684 and −565 (18), the potential USF2 site at position +5 but not initiators at positions −35 and −47 was able to serve as a USF2 binding site (Fig. 5C and data not shown). In addition, USF2 was able to bind to the consensus HIF binding site at position −383 (AACGTG) but not at either position −451 (CACGTT) or −191 (CACGTA) (Fig. 5C).
Fig 6.
USF2 binding to the PAI1 promoter is important to maintain basal activity of the PAI1 promoter and important for HIF2 to activate the PAI1 promoter. (A) Schematic representation of PAI1/Luc reporters in which USF binding sites were sequentially deleted. In addition, all the deletion mutants were based on PAI1 MUSF2 +5/Luc, in which the USF2 binding site at position +5 was mutated. (B) Activation of the PAI1 MUSF2 +5/Luc deletion reporters by 400 ng of empty vector as a control (activation of the vector is 100%), HIF2αTM, or USF2 or 200 ng each of HIF2αTM and USF2 expression plasmids in HEK293T cells. The relative basal activity of these deletion reporters to the full-length PAI1/Luc reporter is also indicated. (C) Gel shift assay of USF2 binding to additional USF2 binding sites on the PAI1 promoter using 30-mer double-stranded oligonucleotides centered at the indicated number. (D) Activation of single or combined USF2/HIF-binding-site-mutated PAI1/Luc reporters by the same activators used for panel B in HEK293T cells. The basal activity of these reporters relative to that of the WT PAI1/Luc reporter is also indicated.
To characterize the functional importance of USF2 binding for HIF2-mediated activation of the PAI1 promoter, the USF2 binding site at position −383 was also deleted from the PAI1 MUSF2 +5/Luc reporter in which the USF2 binding site at position +5 was mutated (Fig. 6A). PAI1 Δ−808/−383 MUSF2 +5/Luc exhibited significantly reduced activation by USF2, likely due to the lack of all identified USF2 binding sites at positions −684, −565, −383, and +5; interestingly, PAI1 Δ−808/−383 MUSF2 +5/Luc also exhibited significantly reduced activation by HIF2α, even though the HBS at position −191 was still intact, demonstrating the functional importance of USF2 binding sites for HIF2-mediated activation of the PAI1 promoter. As expected, the PAI1 Δ−808/−191 MUSF2 +5/Luc construct exhibited no activation by HIF2αTM, USF2, or HIF2αTM plus USF2 (Fig. 6B).
Deletion constructs removed USF2 binding sites and also other potential positive/negative binding sites for other factors, as PAI1 Δ808-684 MUSF2 +5/Luc had only 4% basal activity of the full-length PAI1/Luc promoter (Fig. 6B). To further confirm the importance of USF2 binding sites for hypoxic activation of the PAI1 promoter, we generated PAI1 promoter mutants having the binding sites for USF2 (CANNTG) mutated to CTTAAT individually or in combination. All the single-USF2-binding-site mutants, such as mutants with mutations at MUSF2 position −684, −565, −383, or +5, exhibited a similarly slightly reduced activation by HIF2αTM, USF2, or HIF2αTM plus USF2, suggesting that all the USF2 binding sites play some role in HIF2 activation of the PAI1/Luc reporter, but no particular USF2 binding site was more critical than the others for activation by HIF2αTM or USF2 (Fig. 6D). The PAI1/Luc reporter in which all four identified USF2 binding sites were mutated (MUSF2 positions −684, −565, −383, and +5) exhibited weaker activation by USF2, HIF2αTM, or HIF2αTM plus USF2, although the HBS at position −191 was intact, suggesting the functional relevance of USF2 binding for HIF2 activation of the PAI1 promoter (Fig. 6D). However, the most striking observation was the significant reduction in basal activity of the PAI1 MUSF2 −684, −565, −383, and +5/Luc reporter (1.54% of the WT PAI1/Luc reporter) (Fig. 6D), indicating that USF2 binding to the HIF2 target gene promoter of PAI1 was not only important for optimum activation by HIF2 but also critical for the basal activity of the PAI1 promoter. However, the PAI1 MUSF2 −684, −565, −383, and +5/Luc reporter still exhibited notable activation by USF2, HIF2αTM, and USF2 plus HIF2αTM, suggesting that not all USF2 binding sites in the cloned PAI1 promoter were identified. Indeed, when the USF2 consensus site is expanded to CANNTG (where the NN nucleotides were not limited to CG or GC, as before), we found that USF2 could bind to the −643 site strongly and to the −214 site weakly (Fig. 6C) but could not bind to the −151 site. When all six identified USF2 binding sites in the PAI1/Luc reporter were mutated, promoter activation by HIF2α was no longer observed (Fig. 6D, PAI1 MUSF2 −684, −643, −565, −383, −214, and +5). Mutation of HBS at −191 (M HBS −191), a known HIF binding site (27), resulted in significantly decreased activation by HIF2, confirming that position −191 is indeed critical for HIF activity. Unexpectedly, but consistent with a previous report (18), the HBS at the −191 site was also important for USF2 activation of the PAI1 promoter (Fig. 6D, MHBS-191) although HBS −191 was not bound by USF2 in our gel shift experiment (Fig. 5C, −191). In summary, these data indicate that the PAI1 promoter has multiple USF2 binding sites and USF2 binding is critical to maintain PAI1 promoter basal activity and for PAI1 promoter activation by HIF2. In contrast to full-length USF2, USF2Δbasic, which lacks the DNA binding basic region, is unable to activate PAI1/Luc reporter or PAI1 and HMOX endogenous genes, while USF2Δbasic plus HIF2αTM exhibited no cooperation in activating the PAI1/Luc reporter or PAI1 and HMOX endogenous genes (data not shown), further supporting our conclusion that USF2 activates HIF2 target genes in a DNA binding-dependent manner.
USF2 recruits CBP and p300 coactivators to HIF2 target gene (PAI1 and EPO) promoters/enhancers in vivo.
Next, we wanted to test if USF2 binds to HIF2 target genes in vivo and to determine the molecular mechanisms for why USF2 is required for HIF2 target gene activation during hypoxia in vivo. USF proteins activate gene transcription by recruiting acetylases (PCAF, CBP, and p300), the H3K4 methyltransferase SET7/92, and basal components of the RNA polymerase II transcription complex (14). Thus, we performed ChIP experiments to assess the binding of CBP, p300, the largest subunit of RNA Pol II, USF2, and HIF2α to HIF2 target gene promoter/enhancers in normoxic or hypoxic Hep3B cells. Pol II, CBP, p300, and USF2 exhibited normoxic binding to the region of the PAI1 promoter from positions −248 to −128 (a region close to the transcriptional start site and HBS at position −191), while serum controls precipitated minimal amounts of the PAI1 promoter (Fig. 7A), indicating the specificity of these antibodies to pull down the associated genomic DNA. In addition, these data also supported a previous report that HIF2 target gene promoters have an open chromatin structure (108). As expected, HIF2α exhibited increased binding to the PAI1 promoter region from positions −248 to −128 in the hypoxia-treated cells (Fig. 7A). Interestingly, the binding of the coactivators CBP and p300 was also significantly enhanced by hypoxia (Fig. 7A), consistent with a reported critical role of CBP and p300 in hypoxic transcriptional response (5, 17, 20, 22, 23, 30, 54, 82).
Fig 7.
USF2 is critical for CBP and p300 protein recruitment to HIF2 target genes (PAI1 and EPO) in vivo. ChIP analysis of Pol II, CBP, p300, USF2, and HIF2α binding to the HIF2 target gene PAI1 promoter at the region close to the transcription start site and HRE (PAI1 from positions −248 to −128) (A), the PAI promoter away from the transcription start site and HRE (PAI1 from positions −808 to −608) (B), the HIF2 target gene EPO promoter close to the EPO transcription start site (EPO from positions −128 to −23) (C), and the EPO enhancer that is close to the validated HRE (EPO from positions +2970 to +3058) (D) in normoxic (Nx) or hypoxic (Hx) parental Hep3B, Hep3B/ARNT shRNA, or Hep3B/USF2 shRNA cells. No Chro., no chromatin.
To determine if USF2 contributed to the binding of any of the assayed factors to the PAI1 promoter, ChIP analysis was performed in normoxic or hypoxic Hep3B/USF2 shRNA cells that had 80% reduction of USF2 mRNA and significantly reduced USF2 binding to the PAI1 promoter (Fig. 7A, USF2). Interestingly, HIF2α binding was significantly increased in hypoxic Hep3B/USF2 shRNA cells (Fig. 7A), which might be explained by HIF2 binding to USF2 binding sites in the absence of USF2 protein, as E boxes at positions −684 and −565 are common consensus HIF/USF2 binding sites and USF2 binding site −383 is a consensus HIF binding site. In addition, increased Pol II binding to the PAI1 promoter region from positions −248 to −128 was also observed in normoxic and hypoxic Hep3B/USF2 shRNA cells (Fig. 7A) and might be caused by increased pausing of Pol II around the transcription start site. However, USF2 inhibition resulted in a significant decrease of the binding of coactivators CBP and p300 in hypoxic cells (Fig. 7A), indicating that USF2 is responsible for hypoxia-mediated increased CBP and p300 recruitment to the PAI1 promoter.
HIF's role in activating HIF target genes was thought to be recruitment of CBP and p300 histone acetylases by C-terminal transactivation domains (C-TADs) as well as N-terminal TADs (N-TADs) of HIF1α and HIF2α (55, 81). Thus, we conducted ChIP experiments in normoxic or hypoxic Hep3B/ARNT shRNA cells that have 80% ARNT reduction. Selection of ARNT but not ARNT plus ARNT2 knockdown is due to the fact that Hep3B cells expressed high levels of ARNT (CT, 24.52) and very low levels of ARNT2 (CT, 31.13) and ARNT siRNA is sufficient to inhibit hypoxic induction of HIF target genes (Table 1). As expected, knockdown of ARNT significantly decreased HIF2α binding to the PAI1 promoter (Fig. 7A). However, reduction of ARNT did not alter binding of USF2 and Pol II to the PAI1 promoter. Consistent with a reported role of HIF in recruiting CBP and p300 (5, 81), hypoxic Hep3B/ARNT shRNA cells exhibited a noticeable reduction of CBP and p300 binding to the PAI1 promoter (Fig. 7A). However, the reduction was much less than what we observed in USF2-knockdown cells (Fig. 7A), demonstrating that USF2, but not HIF2, functions as the major recruiter of CBP and p300 to the PAI1 promoter, even at the region with a functional Change text to: hypoxia-responsive element (HRE). Analysis of the region of the PAI1 promoter from positions −808 to −608 produced similar results, although most tested proteins (except USF2) displayed a lower level of binding at this end of the promoter (Fig. 7B). In addition, the hypoxia-mediated increase in CBP and p300 binding to this region was totally USF2 dependent, as no reduction was observed in Hep3B/ARNT shRNA cells (Fig. 7B), likely due to a lack of significant HIF2α binding to the PAI1 region from positions −808 to −608 in hypoxic Hep3B cells (Fig. 7B).
EPO is a well-known HIF2 target gene (38, 47, 78, 104), and its hypoxic induction is USF2 and HIF2 dependent in Hep3B cells (Fig. 2C and Table 1), consistent with a validated HBS at position +3020 (part of HRE) and 6 consensus (CANNTG) USF2 binding sites in the region from positions −976 to +56 of the EPO promoter. The EPO gene is transcriptionally activated by hypoxia via DNA looping facilitated by interaction of the 3′ enhancer that serves as the major HIF binding element and the 5′ promoter that is the binding site for Pol II and basal transcription machinery (28, 68). To determine the role of USF2 in activation of another HIF2 target gene, we analyzed factors binding to the EPO promoter (positions −128 to −23) (Fig. 7C) and enhancer (positions +2970 to +3058, overlapping HRE) regions (Fig. 7D). ChIP analysis of these two regions in normoxic and hypoxic WT Hep3B cells demonstrated that the EPO promoter is the major region bound by USF2, Pol II, CBP, p300, and even HIF2, even though HRE is located in the EPO enhancer (Fig. 7C and D). On the EPO promoter and enhancer, binding of Pol II, CBP, p300, and HIF2α was increased by hypoxia, whereas USF2 binding was unaffected (Fig. 7C and D). Interestingly, similar to the PAI1 promoter, the hypoxia-mediated increase in p300 and CBP binding to the EPO promoter region was more heavily dependent upon USF2 than ARNT binding (Fig. 7C), while ARNT and USF2 contributed to the hypoxia-mediated increase in CBP and p300 binding to the EPO enhancer region almost equally (Fig. 7D). In summary, these ChIP experiments demonstrated that USF2 binds to HIF2 target genes in vivo and USF2 contributes significantly to the hypoxia-mediated increase in CBP and p300 binding to the HIF2 target genes PAI1 and EPO.
HIF2α and USF2 proteins exhibit similar affinities of binding to the p300 protein.
It was clear from our ChIP experiments that the amount of CBP and p300 proteins on HIF2 target genes PAI1 and EPO depends more on USF2 than the HIF2α/ARNT protein (Fig. 7). This could be explained by a stronger interaction of USF2/p300 than HIF2α/p300. To test this hypothesis, HEK293T cells were transfected with Flag-tagged HIF2αTM or Flag-tagged USF2 or were not transfected as a control. We then performed Western blot analysis to detect endogenous p300 protein coprecipitated with HIF2αTM or USF2 protein. We showed that similar amounts of HIF2α and USF2 protein pulled down similar amounts of p300 protein (Fig. 8A), confirming previously reported interactions of USF2 and p300 (6, 51) and HIF2α and p300 (54) and demonstrating the similar binding affinity of USF2 and HIF2α/ARNT for p300 protein for the first time.
Fig 8.

HIF2α, USF2, and p300 physically interact in vitro and in vivo. (A) Western blot analysis of p300 protein coprecipitated by HIF2α or USF2 protein (top), p300 protein in lysates (middle), and HIF2α/USF2 protein (bottom) in lysates of HEK293T cells or HEK293T cells transfected with HIF2αTM-Flag or USF2-Flag. (B) Western blot detection of HIF1α (top), HIF2α (middle), and USF2 protein (bottom) in hypoxic Hep3B cell lysates or coprecipitated with beads or the USF2, HIF1α, or HIF2α protein. (C) The PAI1 promoter indicating the locations of ChIP qPCR primers in relation to the positions of validated USF/HIF binding sites. (D) ChIP and re-ChIP were conducted in hypoxic Hep3B cells, and precipitated DNA was analyzed for the indicated regions of the PAI1 promoter. The antibodies listed first were used in the first precipitation, while the antibodies listed second were used to precipitate the DNA-protein complex immunoprecipitated by the first antibody. PAI intron 4 served as a negative control.
HIF2α, USF2, and p300 proteins physically interact in vitro and in vivo.
The transcription factors in the enhanceosome complex typically interact with each other directly or indirectly. Although we showed binding of HIF2α, USF2, and p300 to both the HIF2 target gene PAI1 and EPO promoters/enhancers in vivo and interactions of HIF2α or USF2 with p300 protein, it is not clear if USF2 interacts with the HIF2α protein on HIF2 target gene promoters to form the enhanceosome. To test this, we first assessed the physical interaction of the USF2 protein with the HIF2 protein in vivo. Pull down of the USF2 protein coprecipitated the HIF2α protein, while pull down of the HIF2α protein coprecipitated the USF2 protein in hypoxic Hep3B cells, demonstrating the physical interaction of the HIF2α and USF2 proteins (Fig. 8B). However, we did not detect an HIF1α-USF2 protein interaction (Fig. 8B), consistent with our functional results showing that HIF1 and USF2 did not cooperatively activate HIF1 target genes (Fig. 1 and Fig. 5B). To directly test the interaction of HIF2α, USF2, and p300 on a HIF2 target gene promoter, ChIP/re-ChIP experiments were performed in hypoxic Hep3B cells in which chromatin was subjected to ChIP analysis first with anti-HIF2α, anti-USF2, or anti-p300 antibody or serum control. Following immunoprecipitation, the DNA-protein complexes were eluted and resubjected to a second ChIP using a different (anti-USF2, anti-HIF2α, or anti-USF1) antibody. The DNA precipitated from the secondary ChIP was analyzed by qPCR at several regions of the PAI1 promoter containing HIF2 and/or USF2 binding elements or the PAI1 gene intron 4 as a control (Fig. 8C). Interestingly, HIF2α/USF2 and USF2/HIF2α ChIP/re-ChIP both precipitated similar amounts of the PAI1 promoter across the analyzed regions but not at intron 4, suggesting that the two transcription factors are bound in proximity on the same PAI1 promoter fragments (Fig. 8D). In addition, we found USF2 and p300 bound together on the PAI1 promoter (Fig. 8D, p300/USF2). However, USF1 was not coprecipitated with HIF2α (Fig. 8D, HIF2α/USF1), in agreement with our findings that USF1 is not involved in PAI1 gene activation (data not shown). In addition, we also detected interaction of HIF2α with Pol II and USF2 with Pol II across the PAI1 promoter with a strong signal around the region from positions −248 to −128 (data not shown). Taken together, the ChIP/re-ChIP results, in conjunction with the co-IP results showing that these factors can physically interact, support the hypothesis that HIF2, USF2, p300, and RNA Pol II form an interacting transcriptional complex on the PAI1 promoter.
USF2 is required for HIF2α-dependent tumorigenic properties in RCC4 cells and PRC3 cells.
Our hypoxia target gene studies indicated that USF2 is required to activate global or known HIF2 target genes in Hep3B, mES, and RCC4 cells (Fig. 1 to 4 and Table 1). RCC4 cells exhibit normoxic functional HIF2 activity due to a VHL mutation and have been shown to be dependent on HIF2α activity for their tumorigenic properties in cell culture (34). We wanted to test whether inhibition of USF2 in RCC4 cells could phenocopy the effects of inhibition of HIF2α in functional assays. To test this hypothesis, USF2 or HIF2α was stably knocked down in RCC4 cells using shRNA (Fig. 9A). As expected, RCC4/HIF2α shRNA cells exhibited slow proliferation relative to wild-type RCC4 cells (Fig. 9B). Interestingly, identically to RCC4/HIF2α shRNA cells, RCC4/USF2 shRNA cells also exhibited slower cellular proliferation (Fig. 9B). In addition, both HIF2 shRNA and USF2 shRNA likewise similarly inhibited wound healing (Fig. 9C) and clonogenic survival (Fig. 9D and E). We also tested the function of USF2 in another HIF2-dependent tumor cell line, PRC3, by stably knocking down USF2 or HIF2α (Fig. 10A). Consistent with previous reports (60), HIF2α is not critical for PRC3 cell proliferation in cell culture; interestingly, USF2 knockdown also did not reduce PRC3 cell proliferation (data not shown). However, like PRC3/HIF2α shRNA cells, PRC3/USF2 shRNA cells also exhibited reduced wound healing and clonogenic survival (Fig. 10B to D). Taken together, these results functionally demonstrate that USF2 works with HIF2 to activate HIF2 target genes and promote tumor cell proliferation, motility, and clonogenic survival.
Fig 9.
USF2 is required for HIF2α-dependent tumorigenic properties in RCC4 cells. (A) Western blot analysis of HIF2α and USF2 proteins in normoxic RCC4, RCC4/USF2 shRNA, or RCC4/HIF2α shRNA cells. (B) Cell number of normoxic RCC4, RCC4/USF2 shRNA, or RCC4/HIF2α shRNA cells on different days. Assays were performed in triplicate, and results are presented as averages from three independent experiments. (C) USF2 or HIF2α shRNA similarly reduced RCC4 cell motility in scratch assays. Assays were performed in duplicate, and the results are graphed as the average percent scratch closure at each time point from three experiments. (D) Clonogenic survival assays demonstrated that USF2 and HIF2 shRNA decreased colony formation. Assays were conducted in duplicate with plating of 500, 1,000, or 1,500 cells. Representative plates from 1,000 cells are shown. (E) The average number of CFU from 1,000 cells was graphed. The raw numbers are also shown.
Fig 10.
USF2 is required for HIF2-dependent tumorigenicity in PRC3 cells in vitro. PRC3 cells lack functional pVHL and HIF1α gene expression. HIF2α protein and HIF2 target genes are constitutively expressed. (A) The efficiency of USF2 and HIF2α protein knockdown in PRC3 cells by USF2 or HIF2 shRNA is demonstrated by Western blotting. (B) In vitro cellular motility was measured by scratch assay and found to be decreased in both PRC3/USF2 shRNA and PRC3/HIF2α shRNA cells. (C) Colony formation in a clonogenic survival assay where 1,000 cells were plated was likewise decreased by USF2 or HIF2 shRNA in comparison to that for wild-type controls. (D) Average colony counts from 1,000 cells are quantified in PRC3, PRC3/USF2 shRNA, and PRC3/USF2 shRNA cells. The raw clone number is given at the top of each bar.
DISCUSSION
While the role of HIF proteins in activating the hypoxic transcriptional response has been well characterized (25, 56, 93), the involvement of other transcription factors in mediating this process has been less well studied. With the understanding that optimal activation of most human genes requires multiple transcription factors in an enhanceosome complex, it is highly unlikely that HIF proteins function alone to activate hypoxia-inducible genes. Indeed, multiple studies, including ours, have previously reported transcriptional cooperation between HIF1/HIF2 and other transcription factors in mediating hypoxic induction of cloned HIF target gene promoters or some endogenous HIF target genes (1, 4, 20, 21, 24, 47, 50, 85, 86, 99). Here we have demonstrated that USF2 is required for hypoxic activation of all the endogenous HIF2 target genes that we analyzed in several cell lines (Hep3B, RCC4, and mES cells). This response is specific for HIF2 target genes, as USF2 is not required for hypoxic activation of HIF1 target genes in RCC4, Hep3B, and mES cells. To our knowledge, this is the first study to have identified a critical cotranscription factor that is differentially required for hypoxia-inducible gene activation by HIF1 and HIF2 on a genomewide level. Our finding explains why Hif1α−/− mES cells have no functional HIF2, even though mES cells express HIF2α mRNA and HIF2α protein under hypoxia (46). It will be interesting to see whether a specific functional requirement of USF2 and the physical interaction of USF2 with the HIF2α protein could explain HIF1/HIF2 target gene specificity.
We found that USF2 is not required for HIF1 target gene activation. Conversely, reduction of USF2 expression by siRNA increased hypoxic activation of some HIF1 target genes, such as KCTD11, BNIP3L, and GRP111, in hypoxic Hep3B cells (Table 1). This inhibitory role of USF2 for some HIF1 target genes is consistent with previous reports indicating that HREs of some HIF1 target genes, such as LDHA, BNIP3, and LPK, are bound by USF1/USF2 under normoxia or in response to glucose activation but by HIF1 in hypoxic cells (49, 62). However, while the previous reports indicated that USF1/USF2 and HIF1 compete for the same binding site on some HIF1 target promoters during hypoxia, we show that USF2 and HIF2 bind to different sites of HIF2 target genes in an uncompetitive manner in vitro (Fig. 5) and in vivo (Fig. 7). In addition, our ChIP/re-ChIP data indicate that both the USF2 and HIF2α proteins can be simultaneously detected on the HIF2 target gene PAI1 promoter in hypoxic cells (Fig. 8D). In agreement with our data, the USF2 and HIF proteins have been reported to bind to different sites on promoters of HIF1/HIF2 common target genes (CXCR4 and hTERT) and the HIF2-specific gene HMOX (35, 42, 63, 73, 96, 113). It will be important to test in the future if the number of USF binding sites on HIF target genes is important for HIF1/HIF2 target gene specificity, since HIF2 target genes have multiple USF binding sites, while some HIF1 target genes seem to have only one binding site for the USF protein. In addition, the type of USF binding site may also be important for HIF target gene specificity, since HIF2 target genes have USF2/USF2 binding sites that are not shared with HIF2, while some HIF1 target genes seem to contain USF1/USF2 binding sites that could be occupied by HIF1 during hypoxia.
CBP and p300 are coactivators required for the HIF-mediated transcriptional response (5, 17, 20, 22, 23, 30, 54, 82). Besides having a role in promoting chromatin opening through histone acetyltransferase activity, CBP and p300 have also been shown to function as a bridge between transcription factor activation domains and RNA polymerase II, as well as to act as a scaffold for additional activating factors to facilitate the assembly of enhanceosome complexes on target promoters (71). While previous reports indicate a role for C-TADs and N-TADs of both HIFα subunits in recruiting CBP and p300, our data demonstrate that USF2 is more critical than HIF2 for the recruitment of the CBP and p300 proteins to HIF2 target promoters in vivo, particularly in the promoter region away from the HRE (Fig. 7). It will be interesting to determine the major function of HIF2α/ARNT in activation of HIF2 target genes, since HIF2 is not the primary factor recruiting coactivators CBP and p300 in vivo. Our data suggest that USF2-dependent recruitment of p300 is not due to the differential binding affinities of HIF2α or USF2 for p300. While most HIF target genes thus far identified have one or two HREs (105), the HIF2 target gene PAI1 promoter has six confirmed USF2 binding sites in the region from positions +26 to −806. Multiple USF2 binding sites and very few HREs could potentially explain why USF2 functions as the major recruiter of CBP and p300 to these promoters. While we showed the functional importance of USF2 in recruiting CBP and p300, it is possible that USF2's other functions, including recruiting histone methylase SET7/92, a H3K4 methylase that typically activates gene transcription, also contribute to USF2's role in HIF2 target gene activation.
Although USF2 binding on HIF2 target promoters is not regulated by hypoxia, the hypoxia-mediated increase in CBP and p300 binding to the HIF2 target gene promoters is USF2 dependent (Fig. 7). USF1 proteins are posttranslationally modified by a number of kinases, including those activated by cellular stress and low oxygen tension (11, 15, 31, 76, 109). Modification of USF1 by phosphorylation at specific residues enhances its physical association with coactivators, including CBP and p300 (77). However, whether hypoxia modifies the USF2 protein is still unclear. Increased USF2-dependent recruitment of CBP and p300 by hypoxia might also be explained by direct modification of the CBP and p300 proteins by hypoxia-activated mitogen-activated protein kinase signaling to change their protein conformation and subsequently increase CBP and p300 binding affinities with its partners, such as USF2 (39, 65, 87). Taken together, these findings suggest potential explanations for the USF2-dependent hypoxic increase in CBP and p300 binding to HIF2 target promoters and provide an interesting basis for further study.
Because of the global requirement of USF2 for HIF2 target gene activation, it is likely that inhibition of either one of these factors could lead to functional inhibition of HIF2-driven tumorigenic properties in an HIF2-dependent tumor cell line. Indeed, we showed that many in vitro qualities associated with a tumorigenic phenotype, including proliferation, motility, and clonogenic survival, could be similarly inhibited by HIF2α shRNA or USF2 shRNA in RCC4 and PRC3 renal carcinoma cell lines (Fig. 9 and 10). The discovery of a HIF2/USF2 protein interaction suggests an alternative means of blocking HIF2 activity by interrupting this interaction, which may prove beneficial in the treatment of cancers in which HIF2 plays a significant role in tumor progression.
Our results thus far have demonstrated a critical role of USF2 in the activation of HIF2 target genes in Hep3B, RCC4, and mouse ES cell lines; however, it remains to be determined if USF2/HIF2α cooperation is involved in normal physiology. While Hif2α−/− mice exhibit deficiencies in red blood cell production and EPO expression (38, 78, 91, 92), Usf2−/− mice exhibit increased tissue iron accumulation and relatively normal red blood cell, hemoglobin, and hematocrit levels (74). However, these Usf2−/− mice additionally have deficiencies in hepcidin-1 and -2 genes due to the proximity of these genes to the USF2 gene locus (74). Interestingly, Hepc1−/− mice exhibit an initial increase in red blood cell, hemoglobin, and hematocrit levels (74). Thus, it is possible that in Usf2−/−/Hepc1−/− mice a reduction of red blood cell production by USF2 knockdown is counteracted by the increase in red blood cell from hepcidin deficiency. Furthermore, Hepc1−/− mice exhibit increased levels of the HIF2 target gene DMT1, while Hepc1−/−/Hif2α−/− mice (69) as well as Hepc1−/−/Usf2−/− mice (74) exhibited similar WT levels of DMT1 gene expression. Although these data suggested a possible USF2/HIF2 interaction in normal physiology, USF2-only-knockout mice are required to study the role of USF2 in normal physiology.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grants from the National Cancer Institute (RO1CA134687 to C.-J. Hu) and Cancer Leagues of Colorado (to C.-J. Hu). M. R. Pawlus was supported by training grant NIHT32-GM08730 (Molecular Biology Graduate Program) from 1 July 2008 to 30 June 2009.
Footnotes
Published ahead of print 10 September 2012
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1. Alfranca A, et al. 2002. c-Jun and hypoxia-inducible factor 1 functionally cooperate in hypoxia-induced gene transcription. Mol. Cell. Biol. 22:12–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Allen RR, Qi L, Higgins PJ. 2005. Upstream stimulatory factor regulates E box-dependent PAI-1 transcription in human epidermal keratinocytes. J. Cell. Physiol. 203:156–165 [DOI] [PubMed] [Google Scholar]
- 3. Anderson CJ, Hoare SF, Ashcroft M, Bilsland AE, Keith WN. 2006. Hypoxic regulation of telomerase gene expression by transcriptional and post-transcriptional mechanisms. Oncogene 25:61–69 [DOI] [PubMed] [Google Scholar]
- 4. Aprelikova O, Wood M, Tackett S, Chandramouli GV, Barrett JC. 2006. Role of ETS transcription factors in the hypoxia-inducible factor-2 target gene selection. Cancer Res. 66:5641–5647 [DOI] [PubMed] [Google Scholar]
- 5. Arany Z, et al. 1996. An essential role for p300/CBP in the cellular response to hypoxia. Proc. Natl. Acad. Sci. U. S. A. 93:12969–12973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Breen GA, Jordan EM. 1999. Transcriptional activation of the F(1)F(0) ATP synthase alpha-subunit initiator element by USF2 is mediated by p300. Biochim. Biophys. Acta 1428:169–176 [DOI] [PubMed] [Google Scholar]
- 7. Breen GA, Jordan EM. 1998. Upstream stimulatory factor 2 activates the mammalian F1F0 ATP synthase alpha-subunit gene through an initiator element. Gene Expr. 7:163–170 [PMC free article] [PubMed] [Google Scholar]
- 8. Breen GA, Jordan EM. 2000. Upstream stimulatory factor 2 stimulates transcription through an initiator element in the mouse cytochrome c oxidase subunit Vb promoter. Biochim. Biophys. Acta 1517:119–127 [DOI] [PubMed] [Google Scholar]
- 9. Chang JT, Yang HT, Wang TC, Cheng AJ. 2005. Upstream stimulatory factor (USF) as a transcriptional suppressor of human telomerase reverse transcriptase (hTERT) in oral cancer cells. Mol. Carcinog. 44:183–192 [DOI] [PubMed] [Google Scholar]
- 10. Chen L, et al. 2006. Human prolyl-4-hydroxylase alpha(I) transcription is mediated by upstream stimulatory factors. J. Biol. Chem. 281:10849–10855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cheung E, Mayr P, Coda-Zabetta F, Woodman PG, Boam DS. 1999. DNA-binding activity of the transcription factor upstream stimulatory factor 1 (USF-1) is regulated by cyclin-dependent phosphorylation. Biochem. J. 344(Pt 1):145–152 [PMC free article] [PubMed] [Google Scholar]
- 12. Chiang CM, Roeder RG. 1995. Cloning of an intrinsic human TFIID subunit that interacts with multiple transcriptional activators. Science 267:531–536 [DOI] [PubMed] [Google Scholar]
- 13. Compernolle V, et al. 2002. Loss of HIF-2alpha and inhibition of VEGF impair fetal lung maturation, whereas treatment with VEGF prevents fatal respiratory distress in premature mice. Nat. Med. 8:702–710 [DOI] [PubMed] [Google Scholar]
- 14. Corre S, Galibert MD. 2005. Upstream stimulating factors: highly versatile stress-responsive transcription factors. Pigment Cell Res. 18:337–348 [DOI] [PubMed] [Google Scholar]
- 15. Corre S, et al. 2009. Target gene specificity of USF-1 is directed via p38-mediated phosphorylation-dependent acetylation. J. Biol. Chem. 284:18851–18862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Coulson JM, et al. 2003. Upstream stimulatory factor activates the vasopressin promoter via multiple motifs, including a non-canonical E-box. Biochem. J. 369:549–561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Dames SA, Martinez-Yamout M, De Guzman RN, Dyson HJ, Wright PE. 2002. Structural basis for Hif-1 alpha/CBP recognition in the cellular hypoxic response. Proc. Natl. Acad. Sci. U. S. A. 99:5271–5276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Dimova EY, Kietzmann T. 2006. Cell type-dependent regulation of the hypoxia-responsive plasminogen activator inhibitor-1 gene by upstream stimulatory factor-2. J. Biol. Chem. 281:2999–3005 [DOI] [PubMed] [Google Scholar]
- 19. Du H, Roy AL, Roeder RG. 1993. Human transcription factor USF stimulates transcription through the initiator elements of the HIV-1 and the Ad-ML promoters. EMBO J. 12:501–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ebert BL, Bunn HF. 1998. Regulation of transcription by hypoxia requires a multiprotein complex that includes hypoxia-inducible factor 1, an adjacent transcription factor, and p300/CREB binding protein. Mol. Cell. Biol. 18:4089–4096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Elvert G, et al. 2003. Cooperative interaction of hypoxia-inducible factor-2alpha (HIF-2alpha) and Ets-1 in the transcriptional activation of vascular endothelial growth factor receptor-2 (Flk-1). J. Biol. Chem. 278:7520–7530 [DOI] [PubMed] [Google Scholar]
- 22. Ema M, et al. 1999. Molecular mechanisms of transcription activation by HLF and HIF1alpha in response to hypoxia: their stabilization and redox signal-induced interaction with CBP/p300. EMBO J. 18:1905–1914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Emerling BM, Weinberg F, Liu JL, Mak TW, Chandel NS. 2008. PTEN regulates p300-dependent hypoxia-inducible factor 1 transcriptional activity through Forkhead transcription factor 3a (FOXO3a). Proc. Natl. Acad. Sci. U. S. A. 105:2622–2627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Figueroa YG, et al. 2002. NF-kappaB plays a key role in hypoxia-inducible factor-1-regulated erythropoietin gene expression. Exp. Hematol. 30:1419–1427 [DOI] [PubMed] [Google Scholar]
- 25. Finger EC, Giaccia AJ. 2010. Hypoxia, inflammation, and the tumor microenvironment in metastatic disease. Cancer Metastasis Rev. 29:285–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Fink T, Ebbesen P, Zachar V. 2001. Quantitative gene expression profiles of human liver-derived cell lines exposed to moderate hypoxia. Cell. Physiol. Biochem. 11:105–114 [DOI] [PubMed] [Google Scholar]
- 27. Fink T, Kazlauskas A, Poellinger L, Ebbesen P, Zachar V. 2002. Identification of a tightly regulated hypoxia-response element in the promoter of human plasminogen activator inhibitor-1. Blood 99:2077–2083 [DOI] [PubMed] [Google Scholar]
- 28. Firth JD, Ebert BL, Pugh CW, Ratcliffe PJ. 1994. Oxygen-regulated control elements in the phosphoglycerate kinase 1 and lactate dehydrogenase A genes: similarities with the erythropoietin 3′ enhancer. Proc. Natl. Acad. Sci. U. S. A. 91:6496–6500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Fitzpatrick TE, Graham CH. 1998. Stimulation of plasminogen activator inhibitor-1 expression in immortalized human trophoblast cells cultured under low levels of oxygen. Exp. Cell Res. 245:155–162 [DOI] [PubMed] [Google Scholar]
- 30. Freedman SJ, et al. 2002. Structural basis for recruitment of CBP/p300 by hypoxia-inducible factor-1 alpha. Proc. Natl. Acad. Sci. U. S. A. 99:5367–5372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Galibert MD, Carreira S, Goding CR. 2001. The Usf-1 transcription factor is a novel target for the stress-responsive p38 kinase and mediates UV-induced tyrosinase expression. EMBO J. 20:5022–5031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Gao E, Wang Y, Alcorn JL, Mendelson CR. 1997. The basic helix-loop-helix-zipper transcription factor USF1 regulates expression of the surfactant protein-A gene. J. Biol. Chem. 272:23398–23406 [DOI] [PubMed] [Google Scholar]
- 33. Gao E, Wang Y, Alcorn JL, Mendelson CR. 2003. Transcription factor USF2 is developmentally regulated in fetal lung and acts together with USF1 to induce SP-A gene expression. Am. J. Physiol. 284:L1027–L1036 [DOI] [PubMed] [Google Scholar]
- 34. Gordan JD, Bertout JA, Hu CJ, Diehl JA, Simon MC. 2007. HIF-2alpha promotes hypoxic cell proliferation by enhancing c-myc transcriptional activity. Cancer Cell 11:335–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Goueli BS, Janknecht R. 2003. Regulation of telomerase reverse transcriptase gene activity by upstream stimulatory factor. Oncogene 22:8042–8047 [DOI] [PubMed] [Google Scholar]
- 36. Gregor PD, Sawadogo M, Roeder RG. 1990. The adenovirus major late transcription factor USF is a member of the helix-loop-helix group of regulatory proteins and binds to DNA as a dimer. Genes Dev. 4:1730–1740 [DOI] [PubMed] [Google Scholar]
- 37. Greijer AE, et al. 2005. Up-regulation of gene expression by hypoxia is mediated predominantly by hypoxia-inducible factor 1 (HIF-1). J. Pathol. 206:291–304 [DOI] [PubMed] [Google Scholar]
- 38. Gruber M, et al. 2007. Acute postnatal ablation of Hif-2alpha results in anemia. Proc. Natl. Acad. Sci. U. S. A. 104:2301–2306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Gusterson R, et al. 2002. The transcriptional co-activators CBP and p300 are activated via phenylephrine through the p42/p44 MAPK cascade. J. Biol. Chem. 277:2517–2524 [DOI] [PubMed] [Google Scholar]
- 40. Harris VK, Coticchia CM, List HJ, Wellstein A, Riegel AT. 2000. Mitogen-induced expression of the fibroblast growth factor-binding protein is transcriptionally repressed through a non-canonical E-box element. J. Biol. Chem. 275:28539–28548 [DOI] [PubMed] [Google Scholar]
- 41. Hartsfield CL, Alam J, Choi AM. 1999. Differential signaling pathways of HO-1 gene expression in pulmonary and systemic vascular cells. Am. J. Physiol. 277:L1133–L1141 [DOI] [PubMed] [Google Scholar]
- 42. Hock TD, Nick HS, Agarwal A. 2004. Upstream stimulatory factors, USF1 and USF2, bind to the human haem oxygenase-1 proximal promoter in vivo and regulate its transcription. Biochem. J. 383:209–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hofbauer KH, et al. 2003. Oxygen tension regulates the expression of a group of procollagen hydroxylases. Eur. J. Biochem. 270:4515–4522 [DOI] [PubMed] [Google Scholar]
- 44. Horikawa I, et al. 2002. Downstream E-box-mediated regulation of the human telomerase reverse transcriptase (hTERT) gene transcription: evidence for an endogenous mechanism of transcriptional repression. Mol. Biol. Cell 13:2585–2597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Howcroft TK, et al. 1999. Upstream stimulatory factor regulates major histocompatibility complex class I gene expression: the U2DeltaE4 splice variant abrogates E-box activity. Mol. Cell. Biol. 19:4788–4797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Hu CJ, et al. 2006. Differential regulation of the transcriptional activities of hypoxia-inducible factor 1 alpha (HIF-1alpha) and HIF-2alpha in stem cells. Mol. Cell. Biol. 26:3514–3526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Hu CJ, Sataur A, Wang L, Chen H, Simon MC. 2007. The N-terminal transactivation domain confers target gene specificity of hypoxia-inducible factors HIF-1alpha and HIF-2alpha. Mol. Biol. Cell 18:4528–4542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hu CJ, Wang LY, Chodosh LA, Keith B, Simon MC. 2003. Differential roles of hypoxia-inducible factor 1alpha (HIF-1alpha) and HIF-2alpha in hypoxic gene regulation. Mol. Cell. Biol. 23:9361–9374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Hu J, et al. 2011. Interaction of HIF and USF signaling pathways in human genes flanked by hypoxia-response elements and E-box palindromes. Mol. Cancer Res. 9:1520–1536 [DOI] [PubMed] [Google Scholar]
- 50. Huang LE, et al. 1997. Erythropoietin gene regulation depends on heme-dependent oxygen sensing and assembly of interacting transcription factors. Kidney Int. 51:548–552 [DOI] [PubMed] [Google Scholar]
- 51. Huang S, Li X, Yusufzai TM, Qiu Y, Felsenfeld G. 2007. USF1 recruits histone modification complexes and is critical for maintenance of a chromatin barrier. Mol. Cell. Biol. 27:7991–8002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Jiang S, Galindo MR, Jarrett HW. 2010. Purification and identification of a transcription factor, USF-2, binding to E-box element in the promoter of human telomerase reverse transcriptase (hTERT). Proteomics 10:203–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Kaelin WG., Jr 2011. Cancer and altered metabolism: potential importance of hypoxia-inducible factor and 2-oxoglutarate-dependent dioxygenases. Cold Spring Harbor Symp. Quant. Biol. 76:335–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Kallio PJ, et al. 1998. Signal transduction in hypoxic cells: inducible nuclear translocation and recruitment of the CBP/p300 coactivator by the hypoxia-inducible factor-1alpha. EMBO J. 17:6573–6586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Kasper LH, et al. 2005. Two transactivation mechanisms cooperate for the bulk of HIF-1-responsive gene expression. EMBO J. 24:3846–3858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Keith B, Johnson RS, Simon MC. 2012. HIF1alpha and HIF2alpha: sibling rivalry in hypoxic tumour growth and progression. Nat. Rev. Cancer 12:9–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Kietzmann T, Roth U, Jungermann K. 1999. Induction of the plasminogen activator inhibitor-1 gene expression by mild hypoxia via a hypoxia response element binding the hypoxia-inducible factor-1 in rat hepatocytes. Blood 94:4177–4185 [PubMed] [Google Scholar]
- 58. Kimura D, et al. 2002. Hypoxia enhances the expression of plasminogen activator inhibitor-1 in human lung cancer cells, EBC-1. Tohoku J. Exp. Med. 196:259–267 [DOI] [PubMed] [Google Scholar]
- 59. Kokubo T, et al. 1993. Identification of TFIID components required for transcriptional activation by upstream stimulatory factor. J. Biol. Chem. 268:17554–17558 [PubMed] [Google Scholar]
- 60. Kondo K, Kim WY, Lechpammer M, Kaelin WG., Jr 2003. Inhibition of HIF2alpha is sufficient to suppress pVHL-defective tumor growth. PLoS Biol. 1:E83 doi:10.1371/journal.pbio.0000083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Koshiji M, et al. 2004. HIF-1alpha induces cell cycle arrest by functionally counteracting Myc. EMBO J. 23:1949–1956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Krones A, Jungermann K, Kietzmann T. 2001. Cross-talk between the signals hypoxia and glucose at the glucose response element of the L-type pyruvate kinase gene. Endocrinology 142:2707–2718 [DOI] [PubMed] [Google Scholar]
- 63. Lee PJ, et al. 1997. Hypoxia-inducible factor-1 mediates transcriptional activation of the heme oxygenase-1 gene in response to hypoxia. J. Biol. Chem. 272:5375–5381 [PubMed] [Google Scholar]
- 64. Li B, Takeda K, Yokoyama S, Shibahara S. 2008. A prolyl-hydroxylase inhibitor, ethyl-3,4-dihydroxybenzoate, induces haem oxygenase-1 expression in human cells through a mechanism independent of hypoxia-inducible factor-1alpha. J. Biochem. 144:643–654 [DOI] [PubMed] [Google Scholar]
- 65. Li QJ, et al. 2003. MAP kinase phosphorylation-dependent activation of Elk-1 leads to activation of the co-activator p300. EMBO J. 22:281–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Liao H, Hyman MC, Lawrence DA, Pinsky DJ. 2007. Molecular regulation of the PAI-1 gene by hypoxia: contributions of Egr-1, HIF-1alpha, and C/EBPalpha. FASEB J. 21:935–949 [DOI] [PubMed] [Google Scholar]
- 67. Lou F, et al. 2007. The opposing effect of hypoxia-inducible factor-2alpha on expression of telomerase reverse transcriptase. Mol. Cancer Res. 5:793–800 [DOI] [PubMed] [Google Scholar]
- 68. Madan A, Curtin PT. 1993. A 24-base-pair sequence 3′ to the human erythropoietin gene contains a hypoxia-responsive transcriptional enhancer. Proc. Natl. Acad. Sci. U. S. A. 90:3928–3932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Mastrogiannaki M, et al. 2012. Deletion of HIF-2alpha in the enterocytes decreases the severity of tissue iron loading in hepcidin knockout mice. Blood 119:587–590 [DOI] [PubMed] [Google Scholar]
- 70. Meisterernst M, Horikoshi M, Roeder RG. 1990. Recombinant yeast TFIID, a general transcription factor, mediates activation by the gene-specific factor USF in a chromatin assembly assay. Proc. Natl. Acad. Sci. U. S. A. 87:9153–9157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Merika M, Williams AJ, Chen G, Collins T, Thanos D. 1998. Recruitment of CBP/p300 by the IFN beta enhanceosome is required for synergistic activation of transcription. Mol. Cell 1:277–287 [DOI] [PubMed] [Google Scholar]
- 72. Minamino T, Mitsialis SA, Kourembanas S. 2001. Hypoxia extends the life span of vascular smooth muscle cells through telomerase activation. Mol. Cell. Biol. 21:3336–3342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Moriuchi M, Moriuchi H, Margolis DM, Fauci AS. 1999. USF/c-Myc enhances, while Yin-Yang 1 suppresses, the promoter activity of CXCR4, a coreceptor for HIV-1 entry. J. Immunol. 162:5986–5992 [PubMed] [Google Scholar]
- 74. Nicolas G, et al. 2001. Lack of hepcidin gene expression and severe tissue iron overload in upstream stimulatory factor 2 (USF2) knockout mice. Proc. Natl. Acad. Sci. U. S. A. 98:8780–8785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Nishi H, et al. 2004. Hypoxia-inducible factor 1 mediates upregulation of telomerase (hTERT). Mol. Cell. Biol. 24:6076–6083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Nowak M, et al. 2005. Insulin-mediated down-regulation of apolipoprotein A5 gene expression through the phosphatidylinositol 3-kinase pathway: role of upstream stimulatory factor. Mol. Cell. Biol. 25:1537–1548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Rahl PB, et al. 2010. c-Myc regulates transcriptional pause release. Cell 141:432–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Rankin EB, et al. 2007. Hypoxia-inducible factor-2 (HIF-2) regulates hepatic erythropoietin in vivo. J. Clin. Invest. 117:1068–1077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Raval RR, et al. 2005. Contrasting properties of hypoxia-inducible factor 1 (HIF-1) and HIF-2 in von Hippel-Lindau-associated renal cell carcinoma. Mol. Cell. Biol. 25:5675–5686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Roy AL, Meisterernst M, Pognonec P, Roeder RG. 1991. Cooperative interaction of an initiator-binding transcription initiation factor and the helix-loop-helix activator USF. Nature 354:245–248 [DOI] [PubMed] [Google Scholar]
- 81. Ruas JL, et al. Complex regulation of the transactivation function of hypoxia-inducible factor-1 alpha by direct interaction with two distinct domains of the CREB-binding protein/p300. J. Biol. Chem. 285:2601–2609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Ruas JL, Poellinger L, Pereira T. 2002. Functional analysis of hypoxia-inducible factor-1 alpha-mediated transactivation. Identification of amino acid residues critical for transcriptional activation and/or interaction with CREB-binding protein. J. Biol. Chem. 277:38723–38730 [DOI] [PubMed] [Google Scholar]
- 83. Salero E, Gimenez C, Zafra F. 2003. Identification of a non-canonical E-box motif as a regulatory element in the proximal promoter region of the apolipoprotein E gene. Biochem. J. 370:979–986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Samoylenko A, et al. 2008. Opposite expression of the antioxidant heme oxygenase-1 in primary cells and tumor cells: regulation by interaction of USF-2 and Fra-1. Antioxid. Redox Signal. 10:1163–1174 [DOI] [PubMed] [Google Scholar]
- 85. Sanchez-Elsner T, et al. 2001. Synergistic cooperation between hypoxia and transforming growth factor-beta pathways on human vascular endothelial growth factor gene expression. J. Biol. Chem. 276:38527–38535 [DOI] [PubMed] [Google Scholar]
- 86. Sanchez-Elsner T, Botella LM, Velasco B, Langa C, Bernabeu C. 2002. Endoglin expression is regulated by transcriptional cooperation between the hypoxia and transforming growth factor-beta pathways. J. Biol. Chem. 277:43799–43808 [DOI] [PubMed] [Google Scholar]
- 87. Sang N, et al. 2003. MAPK signaling up-regulates the activity of hypoxia-inducible factors by its effects on p300. J. Biol. Chem. 278:14013–14019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Sato M, et al. 1989. Transcriptional control of the rat heme oxygenase gene by a nuclear protein that interacts with adenovirus 2 major late promoter. J. Biol. Chem. 264:10251–10260 [PubMed] [Google Scholar]
- 89. Sato M, Ishizawa S, Yoshida T, Shibahara S. 1990. Interaction of upstream stimulatory factor with the human heme oxygenase gene promoter. Eur. J. Biochem. 188:231–237 [DOI] [PubMed] [Google Scholar]
- 90. Schioppa T, et al. 2003. Regulation of the chemokine receptor CXCR4 by hypoxia. J. Exp. Med. 198:1391–1402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Scortegagna M, et al. 2003. Multiple organ pathology, metabolic abnormalities and impaired homeostasis of reactive oxygen species in Epas1−/− mice. Nat. Genet. 35:331–340 [DOI] [PubMed] [Google Scholar]
- 92. Scortegagna M, et al. 2005. HIF-2alpha regulates murine hematopoietic development in an erythropoietin-dependent manner. Blood 105:3133–3140 [DOI] [PubMed] [Google Scholar]
- 93. Semenza GL. 2012. Hypoxia-inducible factors in physiology and medicine. Cell 148:399–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Sirito M, Lin Q, Maity T, Sawadogo M. 1994. Ubiquitous expression of the 43- and 44-kDa forms of transcription factor USF in mammalian cells. Nucleic Acids Res. 22:427–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Sirito M, et al. 1992. Members of the USF family of helix-loop-helix proteins bind DNA as homo- as well as heterodimers. Gene Expr. 2:231–240 [PMC free article] [PubMed] [Google Scholar]
- 96. Staller P, et al. 2003. Chemokine receptor CXCR4 downregulated by von Hippel-Lindau tumour suppressor pVHL. Nature 425:307–311 [DOI] [PubMed] [Google Scholar]
- 97. Tabuchi A, Sakaya H, Kisukeda T, Fushiki H, Tsuda M. 2002. Involvement of an upstream stimulatory factor as well as cAMP-responsive element-binding protein in the activation of brain-derived neurotrophic factor gene promoter I. J. Biol. Chem. 277:35920–35931 [DOI] [PubMed] [Google Scholar]
- 98. Takahashi Y, Takahashi S, Shiga Y, Yoshimi T, Miura T. 2000. Hypoxic induction of prolyl 4-hydroxylase alpha (I) in cultured cells. J. Biol. Chem. 275:14139–14146 [DOI] [PubMed] [Google Scholar]
- 99. Tendler DS, et al. 2001. Intersection of interferon and hypoxia signal transduction pathways in nitric oxide-induced tumor apoptosis. Cancer Res. 61:3682–3688 [PubMed] [Google Scholar]
- 100. Thanos D, Maniatis T. 1995. Virus induction of human IFN beta gene expression requires the assembly of an enhanceosome. Cell 83:1091–1100 [DOI] [PubMed] [Google Scholar]
- 101. Viollet B, et al. 1996. Immunochemical characterization and transacting properties of upstream stimulatory factor isoforms. J. Biol. Chem. 271:1405–1415 [DOI] [PubMed] [Google Scholar]
- 102. Virolle T, et al. 2002. Binding of USF to a non-canonical E-box following stress results in a cell-specific derepression of the lama3 gene. Nucleic Acids Res. 30:1789–1798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Wang V, Davis DA, Haque M, Huang LE, Yarchoan R. 2005. Differential gene up-regulation by hypoxia-inducible factor-1alpha and hypoxia-inducible factor-2alpha in HEK293T cells. Cancer Res. 65:3299–3306 [DOI] [PubMed] [Google Scholar]
- 104. Warnecke C, et al. 2004. Differentiating the functional role of hypoxia-inducible factor (HIF)-1alpha and HIF-2alpha (EPAS-1) by the use of RNA interference: erythropoietin is a HIF-2alpha target gene in Hep3B and Kelly cells. FASEB J. 18:1462–1464 [DOI] [PubMed] [Google Scholar]
- 105. Wenger RH, Stiehl DP, Camenisch G. 2005. Integration of oxygen signaling at the consensus HRE. Sci. STKE 2005:re12 doi:10.1126/stke.3062005re12 [DOI] [PubMed] [Google Scholar]
- 106. West AG, Huang S, Gaszner M, Litt MD, Felsenfeld G. 2004. Recruitment of histone modifications by USF proteins at a vertebrate barrier element. Mol. Cell 16:453–463 [DOI] [PubMed] [Google Scholar]
- 107. Wickramasinghe NS, Banerjee K, Nagaraj NS, Vigneswaran N, Zacharias W. 2005. Hypoxia alters cathepsin B/inhibitor profiles in oral carcinoma cell lines. Anticancer Res. 25:2841–2849 [PubMed] [Google Scholar]
- 108. Xia X, et al. 2009. Integrative analysis of HIF binding and transactivation reveals its role in maintaining histone methylation homeostasis. Proc. Natl. Acad. Sci. U. S. A. 106:4260–4265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Xiao Q, Kenessey A, Ojamaa K. 2002. Role of USF1 phosphorylation on cardiac alpha-myosin heavy chain promoter activity. Am. J. Physiol. Heart Circ. Physiol. 283:H213–H219 [DOI] [PubMed] [Google Scholar]
- 110. Yago M, Ohki R, Hatakeyama S, Fujita T, Ishikawa F. 2002. Variant forms of upstream stimulatory factors (USFs) control the promoter activity of hTERT, the human gene encoding the catalytic subunit of telomerase. FEBS Lett. 520:40–46 [DOI] [PubMed] [Google Scholar]
- 111. Yan S, Jane DT, Dufresne MJ, Sloane BF. 2003. Transcription of cathepsin B in glioma cells: regulation by an E-box adjacent to the transcription initiation site. Biol. Chem. 384:1421–1427 [DOI] [PubMed] [Google Scholar]
- 112. Yang ZZ, Zou AP. 2001. Transcriptional regulation of heme oxygenases by HIF-1alpha in renal medullary interstitial cells. Am. J. Physiol. Renal Physiol. 281:F900–F908 [DOI] [PubMed] [Google Scholar]
- 113. Yatabe N, et al. 2004. HIF-1-mediated activation of telomerase in cervical cancer cells. Oncogene 23:3708–3715 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.