Abstract
NAD-isocitrate dehydrogenase (NAD-IDH) from the eukaryotic microalga Chlamydomonas reinhardtii was purified to electrophoretic homogeneity by successive chromatography steps on Phenyl-Sepharose, Blue-Sepharose, diethylaminoethyl-Sephacel, and Sephacryl S-300 (all Pharmacia Biotech). The 320-kD enzyme was found to be an octamer composed of 45-kD subunits. The presence of isocitrate plus Mn2+ protected the enzyme against thermal inactivation or inhibition by specific reagents for arginine or lysine. NADH was a competitive inhibitor (Ki, 0.14 mm) and NADPH was a noncompetitive inhibitor (Ki, 0.42 mm) with respect to NAD+. Citrate and adenine nucleotides at concentrations less than 1 mm had no effect on the activity, but 10 mm citrate, ATP, or ADP had an inhibitory effect. In addition, NAD-IDH was inhibited by inorganic monovalent anions, but l-amino acids and intermediates of glycolysis and the tricarboxylic acid cycle had no significant effect. These data support the idea that NAD-IDH from photosynthetic organisms may be a key regulatory enzyme within the tricarboxylic acid cycle.
IDH catalyzes the oxidative decarboxylation of isocitrate to produce 2-oxoglutarate. According to the specificity for the electron acceptor, two enzymes with IDH activity are known, NAD-IDH (EC 1.1.1.41) and NADP-IDH (EC 1.1.1.42) (Chen and Gadal, 1990a).
In photosynthetic organisms NADP-IDH has been detected in the cytosol, chloroplasts, mitochondria, and peroxisomes. Cytosolic NADP-IDH has been purified from higher plants (Chen et al., 1988) and eukaryotic algae (Martínez-Rivas et al., 1996), and its cDNA has been cloned from alfalfa (Shorrosh and Dixon, 1992), soybean (Udvardi et al., 1993), potato (Fieuw et al., 1995), and tobacco (Gálvez et al., 1996). This 80-kD isoenzyme is a dimer, and it is likely to be involved in the synthesis of NADPH for biosynthetic purposes in the cytosol (Chen et al., 1988), in the synthesis of 2-oxoglutarate for ammonium assimilation (Chen and Gadal, 1990b), and in the cycling, redistribution, and export of amino acids (Fieuw et al., 1995). Chloroplastic NADP-IDH has been studied in higher plants (Gálvez et al., 1994) and eukaryotic algae (Martínez-Rivas and Vega, 1994). It is a 154-kD dimer that has been proposed to be involved in the supply of NADPH for biosynthetic reactions in the chloroplast when photosynthetic NADPH production is low (Gálvez et al., 1994). The mitochondrial NADP-IDH of higher plants may have a physiological role in the production of NADPH, which can be converted to NADH by a transhydrogenase or used to reduce glutathione in the mitochondrial matrix (Rasmusson and Møller, 1990). NADP-IDH activity has also been detected in peroxisomes from spinach leaves (Yamazaki and Tolbert, 1970).
NAD-IDH is localized exclusively in the mitochondria in association with the TCA cycle. This enzyme has been purified from several nonphotosynthetic eukaryotes such as fungi (Keys and McAlister-Henn, 1990; Alvarez-Villafañe et al., 1996) and animals (Giorgio et al., 1970), in which it appears to be a 300-kD octamer. Its key regulatory role in the TCA cycle is well documented. The NAD-IDH from yeast is activated by AMP and citrate (Hathaway and Atkinson, 1963), whereas the animal enzyme is activated by ADP and citrate (Cohen and Colman, 1972). In addition, the NAD-IDH cDNAs have been cloned from yeast (Cupp and McAlister-Henn, 1991, 1992) and animals (Nichols et al., 1995; Zeng et al., 1995). In these organisms, the enzyme is composed of two (yeast) or more (animals) different subunits encoded by different genes.
To our knowledge, no NAD-IDH from photosynthetic organisms has yet been purified to homogeneity, mainly because of the low stability of the enzyme (Oliver and McIntosh, 1995). However, partial purifications have been reported from pea (Cox and Davies, 1967; Cox, 1969; McIntosh and Oliver, 1992), potato (Laties, 1983), spruce (Cornu et al., 1996), and the eukaryotic microalga Chlamydomonas reinhardtii (Martínez-Rivas and Vega, 1994). Matrix and membrane forms of the enzyme have been detected in potato (Tezuka and Laties, 1983) and pea (McIntosh, 1997). Although it is an allosteric enzyme that exhibits sigmoidal kinetics with respect to isocitrate (Cox and Davies, 1967; McIntosh and Oliver, 1992) and is activated in vitro by ABA (Tezuka et al., 1990), the regulatory importance of NAD-IDH in photosynthetic organisms is still under debate.
To elucidate the regulatory significance of NAD-IDH in photosynthetic organisms and its apparent contribution to the 2-oxoglutarate supply for ammonium assimilation, we have purified and characterized the NAD-IDH from C. reinhardtii.
MATERIALS AND METHODS
Chemicals
Metabolites, standard proteins, PDP, and PGL were purchased from Sigma. NAS and BTD were supplied by ICN Biomedicals. Phenyl-Sepharose, Blue-Sepharose, DEAE-Sephacel, and Sephacryl S-300 were from Pharmacia Biotech. Chemicals for electrophoresis were purchased from Bio-Rad. All other chemicals were supplied by Merck (Darmstadt, Germany).
Organism and Culture Conditions
The eukaryotic microalga Chlamydomonas reinhardtii, wild-type strain 21gr, was grown at 25°C in liquid medium (Martínez-Rivas et al., 1991) with 10 mm KNO3 as the nitrogen source. The cultures were flushed with air supplemented with 5% (v/v) CO2 and continuously illuminated with white light from fluorescent lamps (50 W m−2).
Enzyme Assay and Protein Determination
The NAD-IDH activity was determined in a 1-mL reaction containing 50 mm potassium phosphate buffer, pH 7.5, 0.5 mm MnCl2, 1.5 mm NAD+, 4 mm d,l-isocitrate, and the appropriate amount of enzyme. The reaction was started by the addition of isocitrate and followed by the increase in A340. One unit of activity was defined as the amount of enzyme that catalyzed the production of 1 μmol NADH min−1.
Protein was estimated by the method of Bradford (1976) using protein reagent dye (Bio-Rad) with BSA as a standard.
Enzyme Purification
Cells were harvested during the logarithmic phase of growth, and were then broken by freezing in liquid nitrogen for 2 min and thawing in buffer A (10 mm potassium phosphate buffer, pH 7.5, and 14 mm 2-mercaptoethanol) including 1 mm PMSF. The homogenate was centrifuged at 16,000g for 30 min at 4°C, and the supernatant was used as a crude extract. All subsequent steps were carried out at 4°C.
A solution of 2% (w/v) protamine sulfate, pH 7.5, was slowly added to the crude extract up to 0.16% (w/v) final concentration. After 10 min of incubation with gentle stirring, the suspension was centrifuged at 16,000g for 30 min and the pellet was discarded.
A Phenyl-Sepharose column (1.6 × 40 cm) was equilibrated at a flow rate of 30 mL h−1 with buffer A adjusted to 15% saturation with solid (NH4)2SO4. The column was loaded with the supernatant previously adjusted to similar ionic strength, and then washed with 300 mL of the same buffer. The NADP-IDH activity was eluted with 300 mL of buffer A containing (NH4)2SO4 to a saturation of 5%. After another washing with 300 mL of buffer A, the NAD-IDH activity was eluted with 300 mL of this buffer, including 50% (v/v) ethylene glycol. Fractions (3 mL) containing enzyme activity were pooled and diluted with buffer A to 10% (v/v) ethylene glycol (buffer B).
The resulting NAD-IDH preparation was applied onto a Blue-Sepharose column (1.6 × 12 cm) preequilibrated with buffer B at a flow rate of 15 mL h−1. The column was washed with 60 mL of this buffer, and the enzyme was eluted with 60 mL of buffer B containing 50 mm KCl. Active fractions (fraction size, 1 mL) were pooled and dialyzed against 10 mm Tris-HCl, pH 8.5, containing 14 mm 2-mercaptoethanol and 10% (v/v) ethylene glycol (buffer C) overnight.
The dialysate was loaded onto a DEAE-Sephacel column (0.7 × 6 cm), which had been previously equilibrated with buffer C at a flow rate of 15 mL h−1. The column was washed with 20 mL of buffer C. Bound protein was eluted with a 20-mL linear gradient from 0 to 100 mm KCl in buffer C. Fractions (0.5 mL) containing NAD-IDH activity were pooled and concentrated to 2 mL by ultrafiltration.
The DEAE-Sephacel-purified NAD-IDH was applied onto a Sephacryl S-300 column (1.6 × 100 cm) equilibrated with 10 mm potassium phosphate buffer, pH 7.5, at a flow rate of 15 mL h−1 and calibrated with blue dextran, ferritin (364 kD), catalase (240 kD), alcohol dehydrogenase (150 kD), BSA (66 kD), ovalbumin (45 kD), and Cyt c (12 kD). The enzyme was eluted with the same buffer and 1-mL fractions were collected. The pooled peak-activity fractions were concentrated by ultrafiltration and kept at 4°C for immediate use.
Gel Electrophoresis
Nondenaturing PAGE and SDS-PAGE were performed as described previously (Laemmli, 1970), using 5% and 12% polyacrylamide gels, respectively. Proteins were located in the gel by staining with 0.1% (w/v) Coomassie brilliant blue R-250 in 25% (v/v) ethanol and 10% (v/v) acetic acid. NAD-IDH activity was located by submerging the gel in a reaction mixture containing 100 mm Tris-HCl, pH 7.5, 10 mm MnCl2, 7 mm NAD+, 40 mm d,l-isocitrate, 0.05% (w/v) nitroblue tetrazolium, and 0.005% (w/v) phenazine methosulfate. After 30 min at room temperature in the dark, the activity was shown by a blue band.
RESULTS AND DISCUSSION
Purification of NAD-IDH from C. reinhardtii
Cells of C. reinhardtii contain two IDH enzymes specific for either NAD+ or NADP+, which can be separated by hydrophobic chromatography on Phenyl-Sepharose. In addition, the NADP-IDH preparation can be further resolved in two different isoenzymes by affinity chromatography on Blue-Sepharose (Martínez-Rivas and Vega, 1994). The main problem in achieving complete purification of the NAD-IDH from photosynthetic organisms has been its low stability (McIntosh and Oliver, 1992). To stabilize the NAD-IDH activity from C. reinhardtii, we found that it was critical to keep ethylene glycol in the buffer after elution from the Phenyl-Sepharose chromatography step. The high concentration of ethylene glycol (50%) required for the elution of the enzyme showed its high hydrophobicity.
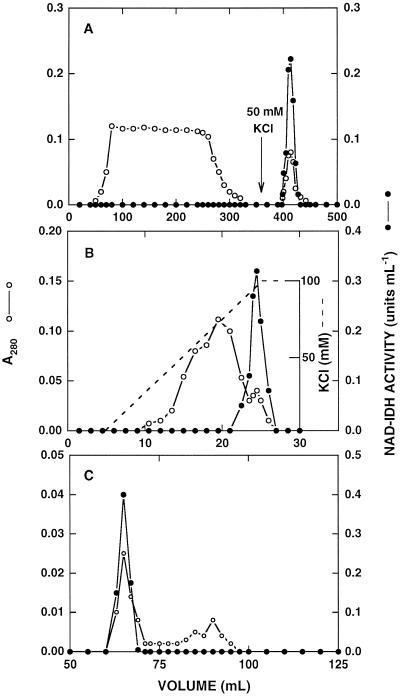
Before it was applied to the Blue-Sepharose column, we reduced the concentration of ethylene glycol in the preparation from 50% to 10% to avoid its interaction with the gel matrix. A typical profile corresponding to this chromatography step is shown in Figure 1A. The eluate obtained in this step was dialyzed against 10 mm Tris-HCl, pH 8.5, containing 14 mm 2-mercaptoethanol and 10% (v/v) ethylene glycol to change the pH of the buffer. This was required to bind the NAD-IDH activity to the DEAE-Sephacel matrix, in contrast to previous studies in which the enzyme was partially purified using a similar approach but keeping the pH of the buffer at 7.5. The enzyme was eluted with a salt concentration of 75 mm KCl in a linear gradient from 0 to 100 mm, and separated from other proteins that eluted at different ionic strengths (Fig. 1B). The combination of the affinity chromatography with the anion-exchange matrix allowed us to obtain a preparation of high purity. Finally, we used gel-filtration chromatography on Sephacryl S-300 to remove some minor contaminant proteins of low Mr (Fig. 1C).
Figure 1.
Purification of NAD-IDH from C. reinhardtii by column chromatography. NAD-IDH activity (•) was measured using the standard assay conditions and protein (○) was monitored by A280. A, Affinity chromatography on Blue-Sepharose. B, Anion-exchange chromatography on DEAE-Sephacel. C, Gel-filtration chromatography on Sephacryl S-300.
The purification procedure summarized in Table I produced 0.12 mg of NAD-IDH from 148 g of cells, with a total purification of about 922-fold, a yield of 6%, and a specific activity of 16.6 units mg−1 protein. Comparable specific activities have been reported for the purified preparations of NAD-IDH from Phycomyces blakesleeanus (Alvarez-Villafañe et al., 1996) and Saccharomyces cerevisiae (Keys and McAlister-Henn, 1990).
Table I.
Purification of NAD-IDH from 148 g (fresh weight) of C. reinhardtii cells
| Step | Volume | Activity | Protein | Specific Activity | Yield | Purification |
|---|---|---|---|---|---|---|
| mL | units | mg | units mg−1 | % | -fold | |
| Crude extract | 755 | 32 | 1796 | 0.02 | 100 | 1 |
| Protamine sulfate | 775 | 32 | 883 | 0.04 | 100 | 2 |
| Phenyl-Sepharose | 42 | 31 | 32 | 0.97 | 97 | 54 |
| Blue-Sepharose | 20 | 7 | 1.75 | 4.0 | 22 | 222 |
| DEAE-Sephacel | 4 | 3 | 0.20 | 15.0 | 9 | 833 |
| Sephacryl S-300 | 10 | 2 | 0.12 | 16.6 | 6 | 922 |
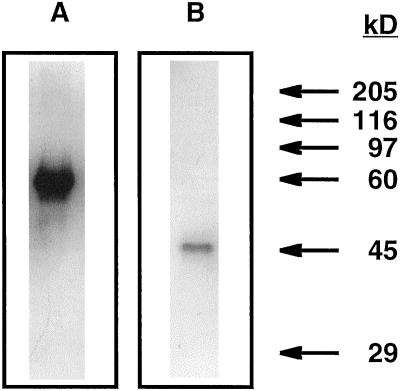
The purified enzyme migrated as a single band in a nondenaturing gel stained for NAD-IDH activity (Fig. 2A). The homogeneity of the preparation was also confirmed by SDS-PAGE (Fig. 2B). According to the purification method described here, isolation of the mitochondrial fraction was not required to purify the NAD-IDH from C. reinhardtii, as was reported previously for the plant enzyme (Tezuka and Laties, 1983; McIntosh and Oliver, 1992). In contrast to the NAD-IDH from pea, where 35% to 50% of the activity was found tightly associated with the membrane (McIntosh, 1997), no NAD-IDH activity was detected in the membrane pellet obtained in the crude extract step after solubilization with 0.5% Triton X-100. However, we cannot discount that a membrane form was also present but was denatured during the treatment with the detergent.
Figure 2.
Nondenaturing PAGE and SDS-PAGE of purified NAD-IDH from C. reinhardtii. A, Five percent polyacrylamide gel stained for NAD-IDH activity. B, SDS-12% polyacrylamide gel stained with Coomassie blue. Five-microgram samples of protein were loaded in each well.
Structural Characterization of NAD-IDH from C. reinhardtii
The native molecular mass of NAD-IDH from C. reinhardtii, as estimated by gel-filtration chromatography on Sephacryl S-300, was 320 kD. A subunit molecular mass of 45 kD was deduced from SDS-PAGE (Fig. 2B). Therefore, the enzyme in its native form appears to be an octamer, but from our data we cannot determine if it is composed of identical or different subunits. The octameric structure and the native molecular mass value are consistent with those reported for the NAD-IDH from bovine heart (333 kD; Giorgio et al., 1970) and from P. blakesleeanus (338 kD; Alvarez-Villafañe et al., 1996), and for the matrix (300 kD; McIntosh and Oliver, 1992) and membrane forms (320 kD; McIntosh, 1997) from pea.
McIntosh and Oliver (1992) reported the correlation between the loss of activity during subsequent purification steps and a change in the native molecular mass of the protein from pea. However, we did not detect any dissociation of the C. reinhardtii NAD-IDH protein during the purification. To check this, preparations from each purification step were analyzed by gel-filtration chromatography, in which a single activity peak corresponding to 320 kD was always observed, and by nondenaturing PAGE followed by activity staining, in which a single band with the same electrophoretic mobility was always observed (data not shown).
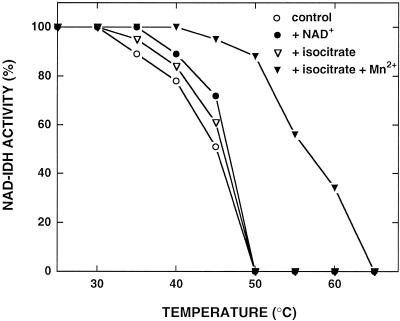
The heat-inactivation profile of NAD-IDH from C. reinhardtii is shown in Figure 3. The temperature at which 50% of the enzyme activity was recovered after 10 min of incubation was 42°C. These data are similar to those from the cytosolic NADP-IDH isoenzyme (Martínez-Rivas et al., 1996) and the NAD-IDH from spruce (Cornu et al., 1996). However, in this plant, mitochondrial NADP-IDH was more resistant than NAD-IDH to heat inactivation. As reported earlier for the NADP-IDH isoenzymes (González-Villaseñor and Powers, 1985; Martínez-Rivas et al., 1996), the presence of the pyridin nucleotide, isocitrate, and, especially, isocitrate plus divalent cation, protects NAD-IDH activity against thermal inactivation. The binding of the substrate to the enzyme may produce a conformational modification that increases the enzyme's resistance to heat inactivation.
Figure 3.
Heat-inactivation profiles of NAD-IDH from C. reinhardtii. Enzyme samples were incubated at the indicated temperature in 10 mm potassium phosphate buffer, pH 7.5, and the indicated substrates at a 5 mm final concentration in a final volume of 0.3 mL. After 10 min the samples were cooled rapidly on ice and assayed for NAD-IDH activity (100% activity = 0.1 unit mL−1).
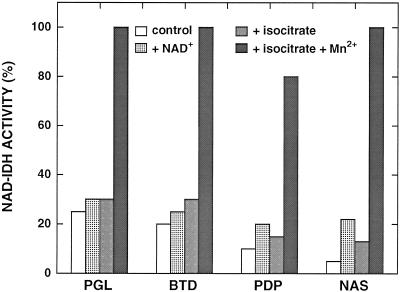
The essential nature of amino acids with sulfhydryl, amino, and carboxyl side groups was demonstrated previously for the NAD-IDH activity from C. reinhardtii (Martínez-Rivas and Vega, 1994). To better characterize those observations, we tested the effects of specific reagents for Arg (PGL or BTD) and Lys (PDP or NAS) on the NAD-IDH activity. The enzyme was almost completely inactivated in the presence of any of these compounds at a concentration of 1 mm (Fig. 4). The simultaneous presence of isocitrate and Mn2+, but not isocitrate alone, prevented this inactivation. These results indicate that the true substrate for the enzyme is the complex formed by isocitrate and the divalent cation, as has been demonstrated for the NAD-IDH from pea leaves (Duggleby and Dennis, 1970a), pig heart (Cohen and Colman, 1974), and P. blakesleeanus (Alvarez-Villafañe et al., 1996).
Figure 4.
Effect of specific reagents on the NAD-IDH activity from C. reinhardtii. The enzyme was incubated at 4°C in 10 mm potassium phosphate buffer, pH 7.5, containing 1 mm specific reagents for Arg (PGL or BTD) or Lys (PDP or NAS), and the indicated substrate at 5 mm final concentration in a final volume of 0.3 mL. After 2 h, NAD-IDH activity was measured (100% activity = 0.13 unit mL−1).
In addition, these data show the involvement of Arg and Lys residues in the active site for the isocitrate-divalent cation substrate, as has been reported previously for the NAD-IDH from bovine heart (Fan and Plaut, 1974). However, these amino acids are not involved in the active site for NAD+, because this substrate did not protect against inactivation. Arg and Lys residues are conserved in the deduced amino acid sequences of the corresponding NAD-IDH cDNA clones from yeast (Cupp and McAllister-Henn, 1991, 1992) and animals (Nichols et al., 1995; Zeng et al., 1995). The presence of these residues in the active site for the isocitrate-divalent cation substrate, but not for NADP+, has been well established for the NADP-IDH from Escherichia coli (Hurley et al., 1989).
Kinetic and Regulatory Properties of NAD-IDH from C. reinhardtii
In a previous study (Martínez-Rivas and Vega, 1994), we found that the NAD-IDH from C. reinhardtii showed sigmoidal kinetics for isocitrate (S0.5 = 0.37 mm, n = 1.82, k = 0.16), but standard Michaelis-Menten kinetics for NAD+ (Km = 0.15 mm) and Mn2+ (Km = 0.03 mm). Other kinetic parameters were also determined: optimum pH (8.0), optimum temperature (40°C), and activation energy (78.1 kJ mol−1). In the present study we investigated the kinetic properties of this enzyme to search for possible regulatory mechanisms.
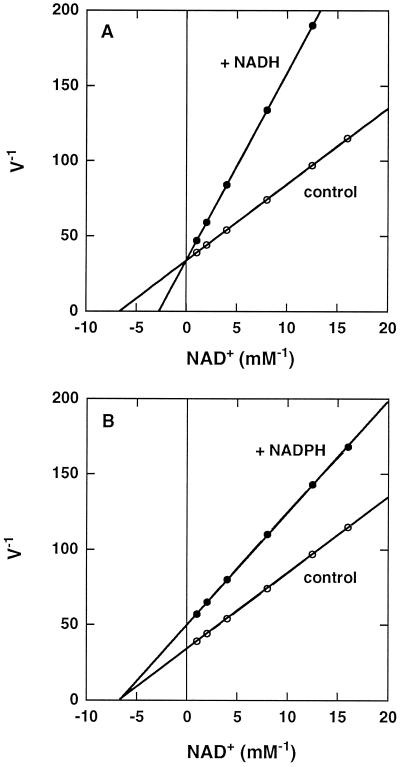
NAD-IDH from C. reinhardtii was not inhibited by an excess of isocitrate, NAD+, or NADP+, but 10 mm 2-oxoglutarate, the product of the reaction, inhibited it slightly (Martínez-Rivas and Vega, 1994). This metabolite had no effect on the enzyme from swede (Coultate and Dennis, 1969) and spruce (Cornu et al., 1996). In contrast, NADH was a competitive inhibitor (Ki = 0.14 mm) with respect to NAD+ (Fig. 5A), as has been reported for the NAD-IDH from pea (Duggleby and Dennis, 1970b; McIntosh and Oliver, 1992; McIntosh, 1997) and swede (Coultate and Dennis, 1969). The inhibition by NADH may be an important regulatory mechanism of the TCA cycle. NADPH was also an inhibitor of NAD-IDH activity, but it was noncompetitive (Ki = 0.42 mm) with respect to NAD+ (Fig. 5B). The same effect was reported for the matrix form (McIntosh and Oliver, 1992) and the membrane form (McIntosh, 1997) of the enzyme from pea, and these authors suggested that there is a separate binding site for NADPH on the NAD-IDH enzyme that is not the catalytic site for NAD+ and NADH binding. A physiological role for the NADPH inhibition has been suggested in the control and balance of NADH and NADPH production (McIntosh and Oliver, 1992).
Figure 5.
Effect of NADH (A) and NADPH (B) on the NAD-IDH activity from C. reinhardtii. Reaction mixtures were as described for the standard assay except that the concentration of NAD+ was varied as indicated in the figure. NADH or NADPH was added to a 0.2 mm final concentration. Initial rates (V−1) are expressed as micromoles of NADH produced per minute.
Unlike previous studies using the NAD-IDH from pea (Cox and Davies, 1967) and swede (Coultate and Dennis, 1969), the enzyme from C. reinhardtii was not activated by citrate at concentrations less than 1 mm (Table II), and 1 mm citrate did not produce a change in the kinetics with respect to isocitrate from sigmoidal to Michaelis-Menten (data not shown). These differences could be explained by the fact that we always used freshly purified enzyme, thus avoiding problems of subunit dissociation caused by freezing and thawing of the enzyme preparation (McIntosh and Oliver, 1992). At concentrations greater than 1 mm, citrate inhibited C. reinhardtii NAD-IDH activity. This effect was caused by the chelation of the divalent cation (Coultate and Dennis, 1969) and the competitive inhibition with respect to isocitrate (Cox and Davies, 1969). However, the high concentration required for it to act as an inhibitor indicates that the regulatory role of citrate may be very limited.
Table II.
Effect of citrate and nucleotides on the NAD-IDH from C. reinhardtii
| Metabolite | Concentration | Enzyme Activity |
|---|---|---|
| mm | % | |
| Citrate | 0.01 | 100 |
| 0.1 | 82 | |
| 1.0 | 64 | |
| 10.0 | 36 | |
| ATP | 0.01 | 109 |
| 0.1 | 91 | |
| 1.0 | 54 | |
| 10.0 | 9 | |
| ADP | 0.01 | 109 |
| 0.1 | 100 | |
| 1.0 | 91 | |
| 10.0 | 9 | |
| AMP | 0.01 | 109 |
| 0.1 | 100 | |
| 1.0 | 91 | |
| 10.0 | 82 |
Reaction mixtures were as described for the standard assay except that the corresponding metabolite was included. One hundred percent NAD-IDH activity was 0.58 unit mL−1.
In contrast to the NAD-IDH from yeast or animals, no activation was observed when the enzyme was incubated with AMP or ADP (Table II). In contrast, ATP or ADP at concentrations greater than 1 mm showed an inhibitory effect at least partially attributable to the formation of a complex with the divalent cation (Omran and Dennis, 1971). Similar results were reported for this enzyme from swede (Coultate and Dennis, 1969), potato (Tezuka and Laties, 1983), pea (McIntosh and Oliver, 1992), and spruce (Cornu et al., 1996). However, this inhibition is considered to be of little regulatory significance (Meixner-Monori et al., 1986). Incubation of the enzyme with 10 mm GTP or GDP brought about a 30% inactivation of the enzyme (data not shown).
Glycolysis intermediates were checked for inhibitory effects. Pyruvate, PEP, dihydroxyacetone phosphate, Fru-1,6-bisP, Fru-6-P, Glc-6-P, and Glc-1-P at a concentration of 10 mm had a slight or no inhibitory effect on the NAD-IDH activity from C. reinhardtii (data not shown). TCA intermediates such as acetyl-CoA, cis-aconitate, succinate, fumarate, l-malate, and oxaloacetate also had no significant effect. In addition, no significant inhibition was observed when any of the proteinogenic amino acids were tested at a final concentration of 10 mm. On the other hand, NAD-IDH activity from C. reinhardtii was strongly inhibited by monovalent inorganic anions such as chloride, iodide, and nitrate at a concentration of 100 mm. No significant effect was observed when divalent anions such as sulfate or arsenate were tested at the same concentration (data not shown). Similar results were obtained by Cox and Davies (1967) with the pea enzyme. It has been proposed that the monovalent inorganic anions may cause a conformational change in NAD-IDH (Coultate and Dennis, 1969), but such an effect would have no physiological regulatory significance because of the high concentrations required.
CONCLUSIONS
The molecular properties of NAD-IDH from C. reinhardtii (molecular mass, number of subunits, and configuration of the active site) are similar to those reported from nonphotosynthetic eukaryotes such as S. cerevisiae (Keys and McAlister-Henn, 1990) or pig heart (Giorgio et al., 1970). However, the NAD-IDH from C. reinhardtii did not exhibit the same regulatory properties, since citrate, ADP, and AMP did not act as effectors. The main differences between the NAD-IDH enzyme from unicellular and multicellular photosynthetic organisms were: (a) the requirement for a pH change to retain the C. reinhardtii NAD-IDH on an anion-exchange column, thereby showing different electrostatic properties, and (b) the high hydrophobicity of the algal enzyme.
The most important regulatory effect was the strong inhibition by NADH and NADPH, which, together with the concentration of the divalent cations (Duggleby and Dennis, 1970a), seem to be the main regulators. Therefore, NAD-IDH from photosynthetic organisms can be considered a key regulatory point in the carbon flow through the TCA cycle. Furthermore, because of its low in vitro activity, it has been suggested that NAD-IDH catalyzes the rate-limiting step in the TCA cycle in these organisms (Møller and Palmer, 1984). On the other hand, the participation of NAD-IDH in the 2-oxoglutarate supply for ammonium assimilation cannot be discounted. The TCA cycle is stimulated under conditions in which there is a high demand for this metabolite (Weger et al., 1988), and NAD-IDH activity increases under nitrogen starvation (Martínez-Rivas and Vega, 1993). Future studies, including the cloning of the gene, are required to understand the molecular basis of the structure and the regulatory mechanisms of NAD-IDH.
ACKNOWLEDGMENT
We thank Dr. Jodi Scheffler for revising the English version of the manuscript.
Abbreviations:
- BTD
2,3-butanedione
- IDH
isocitrate dehydrogenase
- NAS
N-acetyl-succinimide
- PDP
pyridoxal 5′-phosphate
- PGL
phenylglyoxal
- TCA
tricarboxylic acid
Footnotes
This work was supported by research grant no. PB96-1367 from Dirección General de Investigación Científica y Técnica, Spain. J.M.M.-R. was the recipient of a postdoctoral contract from the Ministerio de Educación y Ciencia, Spain.
LITERATURE CITED
- Alvarez-Villafañe E, Soler J, del Valle P, Busto F, de Arriaga D. Two NAD+-isocitrate dehydrogenase forms in Phycomyces blakesleeanus: induction in response to acetate growth and characterization, kinetics, and regulation of both enzyme forms. Biochemistry. 1996;35:4741–4752. doi: 10.1021/bi951268j. [DOI] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Chen RD, Gadal P. Structure, functions and regulation of NAD and NADP dependent isocitrate dehydrogenases in higher plants and in other organisms. Plant Physiol Biochem. 1990a;28:411–427. [Google Scholar]
- Chen RD, Gadal P. Do the mitochondria provide the 2-oxoglutarate needed for glutamate synthesis in higher plant chloroplasts? Plant Physiol Biochem. 1990b;28:141–145. [Google Scholar]
- Chen RD, LeMaréchal P, Vidal J, Jacquot JP, Gadal P. Purification and comparative properties of the cytosolic isocitrate dehydrogenases (NADP) from pea (Pisum sativum) roots and green leaves. Eur J Biochem. 1988;175:565–572. doi: 10.1111/j.1432-1033.1988.tb14229.x. [DOI] [PubMed] [Google Scholar]
- Cohen PF, Colman RF. Diphosphopyridine nucleotide dependent isocitrate dehydrogenase from pig heart: characterization of the active substrate and modes of regulation. Biochemistry. 1972;11:1501–1508. doi: 10.1021/bi00758a027. [DOI] [PubMed] [Google Scholar]
- Cohen PF, Colman RF. Role of manganous ion in the kinetics of pig-heart NAD-specific isocitrate dehydrogenase. Eur J Biochem. 1974;47:35–45. doi: 10.1111/j.1432-1033.1974.tb03665.x. [DOI] [PubMed] [Google Scholar]
- Cornu S, Pireaux JC, Gerard J, Dizengremel P. NAD(P)+-dependent isocitrate dehydrogenases in mitochondria purified from Picea abies seedlings. Physiol Plant. 1996;96:312–318. [Google Scholar]
- Coultate TP, Dennis DT. Regulatory properties of a plant NAD:isocitrate dehydrogenase: the effect of inorganic ions. Eur J Biochem. 1969;7:153–158. doi: 10.1111/j.1432-1033.1969.tb19586.x. [DOI] [PubMed] [Google Scholar]
- Cox GF. Isocitrate dehydrogenase (NAD-specific) from pea mitochondria. Methods Enzymol. 1969;13:47–51. [Google Scholar]
- Cox GF, Davies DD. Nicotinamide-adenine dinucleotide-specific isocitrate dehydrogenase from pea mitochondria: purification and properties. Biochem J. 1967;105:729–734. doi: 10.1042/bj1050729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox GF, Davies DD. The effects of pH and citrate on the activity of nicotinamide-adenine dinucleotide-specific isocitrate dehydrogenase from pea mitochondria. Biochem J. 1969;113:813–820. doi: 10.1042/bj1130813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cupp JR, McAlister-Henn L. NAD+-dependent isocitrate dehydrogenase: cloning, nucleotide sequence, and disruption of the IDH2 gene from Saccharomyces cerevisiae. J Biol Chem. 1991;266:22199–22205. [PubMed] [Google Scholar]
- Cupp JR, McAlister-Henn L. Cloning and characterization of the gene encoding the IDH1 subunit of NAD+-dependent isocitrate dehydrogenase from Saccharomyces cerevisiae. J Biol Chem. 1992;267:16417–16423. [PubMed] [Google Scholar]
- Duggleby RG, Dennis DT. Nicotinamide adenine dinucleotide-specific isocitrate dehydrogenase from a higher plant: the requirement for free and metal-complexed isocitrate. J Biol Chem. 1970a;245:3745–3750. [PubMed] [Google Scholar]
- Duggleby RG, Dennis DT. Regulation of the nicotinamide adenine dinucleotide-specific isocitrate dehydrogenase from a higher plant: the effect of reduced nicotinamide adenine dinucleotide and mixtures of citrate and isocitrate. J Biol Chem. 1970b;245:3751–3754. [PubMed] [Google Scholar]
- Fan CC, Plaut GWE. Functional groups of diphosphopyridine nucleotide-linked isocitrate dehydrogenase from bovine heart. J Biol Chem. 1974;249:4839–4845. [PubMed] [Google Scholar]
- Fieuw S, Müller-Röber B, Gálvez S, Willmitzer L. Cloning and expression analysis of the cytosolic NADP+-dependent isocitrate dehydrogenase from potato. Implications for nitrogen metabolism. Plant Physiol. 1995;107:905–913. doi: 10.1104/pp.107.3.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gálvez S, Bismuth E, Sarda C, Gadal P. Purification and characterization of chloroplastic NADP-isocitrate dehydrogenase from mixotrophic tobacco cells. Comparison with the cytosolic enzyme. Plant Physiol. 1994;105:593–600. doi: 10.1104/pp.105.2.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gálvez S, Hodges M, Decottignies P, Bismuth E, Lancien M, Sangwan RS, Dubois F, LeMaréchal P, Crétin C, Gadal P. Identification of a tobacco cDNA encoding a cytosolic NADP-isocitrate dehydrogenase. Plant Mol Biol. 1996;30:307–320. doi: 10.1007/BF00020116. [DOI] [PubMed] [Google Scholar]
- Giorgio NA, Yip JAT, Fleming J, Plaut GWE. Diphosphopyridine nucleotide-linked isocitrate dehydrogenase from bovine heart. J Biol Chem. 1970;245:5469–5477. [PubMed] [Google Scholar]
- González-Villaseñor LI, Powers DA. A multilocus system for studying tissue and subcellular specialization: the three NADP-dependent isocitrate dehydrogenase isoenzymes of the fish Fundulus heteroclitus. J Biol Chem. 1985;260:9106–9113. [PubMed] [Google Scholar]
- Hathaway JA, Atkinson DE. The effect by adenylic acid on yeast nicotinamide adenine dinucleotide isocitrate dehydrogenase: a possible metabolic control mechanism. J Biol Chem. 1963;238:2875–2881. [PubMed] [Google Scholar]
- Hurley JH, Thorsness PE, Ramalingam V, Helmers NH, Koshland DE, Jr, Stroud RM. Structure of a bacterial enzyme regulated by phosphorylation, isocitrate dehydrogenase. Proc Natl Acad Sci USA. 1989;86:8635–8639. doi: 10.1073/pnas.86.22.8635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keys DA, McAlister-Henn L. Bacteriol. 1990;172:4280–4287. doi: 10.1128/jb.172.8.4280-4287.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Laties GG. Membrane-associated NAD-dependent isocitrate dehydrogenase in potato mitochondria. Plant Physiol. 1983;72:953–958. doi: 10.1104/pp.72.4.953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Rivas JM, García-Díaz MT, Vega JM. Purification and properties of cytosolic NADP-isocitrate dehydrogenase from Chlamydomonas reinhardtii. Plant Physiol Biochem. 1996;34:673–676. [Google Scholar]
- Martínez-Rivas JM, Vega JM. Plant. 1993;88:599–603. doi: 10.1111/j.1399-3054.1993.tb01377.x. [DOI] [PubMed] [Google Scholar]
- Martínez-Rivas JM, Vega JM. Studies on the isoforms of isocitrate dehydrogenase from Chlamydomonas reinhardtii. J Plant Physiol. 1994;143:129–134. doi: 10.1104/pp.118.1.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Rivas JM, Vega JM, Márquez AJ. Differential regulation of the nitrate-reducing and ammonium-assimilatory systems in synchronous cultures of Chlamydomonas reinhardtii. FEMS Microbiol Lett. 1991;78:85–88. [Google Scholar]
- McIntosh CA. Partial purification and characteristics of membrane-associated NAD+-dependent isocitrate dehydrogenase activity from etiolated pea mitochondria. Plant Sci. 1997;129:9–20. [Google Scholar]
- McIntosh CA, Oliver DJ. NAD+-linked isocitrate dehydrogenase: isolation, purification, and characterization of the protein from pea mitochondria. Plant Physiol. 1992;100:69–75. doi: 10.1104/pp.100.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meixner-Monori B, Kubicek CP, Harrer W, Schreferl G, Rohr M. NADP-specific isocitrate dehydrogenase from the citric acid-accumulating fungus Aspergillus niger. Biochem J. 1986;236:549–557. doi: 10.1042/bj2360549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Møller IM, Palmer JM. Regulation of the tricarboxylic acid cycle and organic acid metabolism. Soc Exp Biol Semin Ser. 1984;20:105–122. [Google Scholar]
- Nichols BJ, Perry ACF, Hall L, Denton RM. Molecular cloning and deduced amino acid sequences of the α- and β-subunits of mammalian NAD+-isocitrate dehydrogenase. Biochem J. 1995;310:917–922. doi: 10.1042/bj3100917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver DJ, McIntosh CA (1995) The biochemistry of the mitochondrial matrix. In CS Levings III, IK Vasil, eds, The Molecular Biology of Plant Mitochondria, Vol 3: Advances in Cellular and Molecular Biology of Plants. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 237–280
- Omran RG, Dennis DT. Nicotinamide adenine dinucleotide phosphate-specific isocitrate dehydrogenase from a higher plant. Isolation and characterization. Plant Physiol. 1971;47:43–47. doi: 10.1104/pp.47.1.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmusson AG, Møller IM. NADP-utilizing enzymes in the matrix of plant mitochondria. Plant Physiol. 1990;94:1012–1018. doi: 10.1104/pp.94.3.1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shorrosh BS, Dixon RA. Molecular characterization and expression of an isocitrate dehydrogenase from alfalfa (Medicago sativa L.) Plant Mol Biol. 1992;20:801–807. doi: 10.1007/BF00027151. [DOI] [PubMed] [Google Scholar]
- Tezuka T, Laties GG. Isolation and characterization of inner membrane- associated and matrix NAD-specific isocitrate dehydrogenase in potato mitochondria. Plant Physiol. 1983;72:959–963. doi: 10.1104/pp.72.4.959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tezuka T, Yamamoto Y, Kondo N. Activation of O2 uptake and NAD-specific isocitrate dehydrogenase in mitochondria isolated from cotyledons of castor bean by cis,trans-abscisic acid. Plant Physiol. 1990;92:147–150. doi: 10.1104/pp.92.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udvardi MK, McDermott TR, Kahn ML. Isolation and characterization of a cDNA encoding NADP+-specific isocitrate dehydrogenase from soybean (Glycine max) Plant Mol Biol. 1993;21:739–752. doi: 10.1007/BF00027108. [DOI] [PubMed] [Google Scholar]
- Weger HG, Birch DG, Elrifi IR, Turpin DH. Ammonium assimilation requires mitochondrial respiration in the light. Plant Physiol. 1988;86:688–692. doi: 10.1104/pp.86.3.688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki RK, Tolbert NE. Enzymic characterization of leaf peroxisomes. J Biol Chem. 1970;245:5137–5144. [PubMed] [Google Scholar]
- Zeng Y, Weiss C, Yao TT, Huang J, Siconolfi-Baez L, Hsu P, Rushbrook JI. Isocitrate dehydrogenase from bovine heart: primary structure of subunit 3/4. Biochem J. 1995;310:507–516. doi: 10.1042/bj3100507. [DOI] [PMC free article] [PubMed] [Google Scholar]