Abstract
This study aimed to compare the clonal distribution of common pneumococcal strains not included in the 7-valent pneumococcal conjugate vaccine (PCV7) that were isolated from cases of acute otitis media (AOM) and invasive pneumococcal disease (IPD) in two distinct ethnic populations in southern Israel during the decade (1999 to 2008) preceding PCV7 implementation. Isolates recovered from Jewish and Bedouin children <5 years old were characterized by antibiotic resistance and molecular epidemiology using pulsed-field gel electrophoresis and multilocus sequence typing. Of 5,236 AOM and 425 IPD isolates, 43% and 57% were from Jewish and Bedouin children, respectively. PCV7 accounted for 54% and 45% of the AOM and IPD episodes, respectively. Eleven major non-PCV7 serotypes (1, 3, 5, 6A, 7F, 12F, 15B/C, 19A, 21, 33F, and 35B) constituted 31% and 42% of the AOM and IPD episodes, respectively. The clonal distributions of the 11 non-PCV7 serotypes and their antibiotic susceptibilities were significantly different among the two ethnic populations in both the AOM and IPD groups. About half of the AOM and IPD cases resulted from non-PCV7 pneumococci, even before PCV7 implementation. The significant differences between the two ethnic populations suggest that lifestyle and microenvironment are major determinants in the clonal distribution of disease-causing pneumococci. Post-PCV7 surveillance is important in understanding non-PCV7 clonal expansion in the two distinct populations.
INTRODUCTION
The 7-valent pneumococcal conjugate vaccine (PCV7) has had a substantial impact on the prevention of pneumococcal disease in children, with indirect benefits extending to individuals not in the targeted age group (6). Other benefits include reductions in disease caused by antibiotic-resistant strains and reduction in antibiotic use. However, the potential impact of conjugate vaccines can be negatively influenced by the issue of serotype replacement, wherein serotypes not included in the vaccine are being increasingly implicated in pneumococcal disease (6). While the evidence for serotype replacement in the nasopharynx after PCV7 introduction is universal and beyond doubt, what remains to be understood is its relationship to replacement disease (7).
While it is widely accepted that the capsular type is the key determinant of the ability of pneumococci to cause disease (3, 4, 12, 29–31), the extent of the additional role of other determinants has not yet been fully clarified. A single pneumococcal serotype typically includes a number of genetically divergent clones (based on a comparison of sequence types deposited at http://www.mlst.net), presumably as a consequence of the occasional horizontal transfer of the capsular biosynthetic genes into new lineages (3). The relative importance of serotype and genotype in determining the potential of the organism to cause infection is still debated. Hence, a detailed characterization of the genetic background of isolates will provide invaluable insights concerning the clonal composition of non-PCV7 strains involved in replacement disease.
Sequential trends of non-PCV7 serotypes/clones causing infections before PCV7 introduction have rarely been documented. PCV7 was introduced to the national immunization program in Israel in July 2009. We have been collecting pneumococcal strains from cases of acute otitis media (AOM) and invasive pneumococcal disease (IPD) in children of <5 years of age since 1999 in a prospective way. Since the population in southern Israel includes two distinct ethnic groups, differing in their lifestyles, we aimed at comparing the dynamics of the various non-PCV7 strains in the two study populations. The main goal of this study was to compare the clonal distributions of the major non-PCV7 serotypes, isolated from AOM and IPD cases, between children in these two populations during a decade preceding PCV7 implementation. These data will help to reveal which trends occur even without vaccination and to what extent.
MATERIALS AND METHODS
Setting.
This prospective study was conducted in the Negev area in southern Israel from January 1999 to December 2008. The study population in this area consists of Jews with a lifestyle resembling that of developed populations and Moslem Bedouins with standards of living resembling those of developing populations (8). Contact between children of the two populations is rare. Over 95% of the children in the area are born at the Soroka University Medical Center (SUMC), where they also receive all emergency and inpatient services. Over 60% of the children in the Negev region are medically insured in the largest health plan in Israel, the General Health Insurance Plan. During the study period, PCV7 had not yet been introduced in Israel, and <3% of the population had participated in clinical trials with one of the experimental pneumococcal conjugate vaccines.
Two groups of disease were studied based on the isolation site: (i) the IPD group consisting of all children with positive blood or cerebrospinal fluid (CSF) S. pneumoniae culture; (ii) the AOM group consisting of all children diagnosed with AOM with a positive pneumococcal middle ear fluid (MEF) culture. Blood and CSF cultures were obtained in the emergency department or the pediatric wards (no routine blood or CSF cultures are collected in primary care clinics in Israel) and inoculated into Bactec 9240 (Becton, Dickinson, Cockeysville, MD). Middle ear fluid cultures were obtained either in the primary care clinics or at the SUMC, either after tympanocentesis or following spontaneous drainage, and applied onto transport swabs (Transwab; Medical Wire and Equipment, England). In the case of multiple IPD or AOM isolates from an individual, a new episode was defined if >30 days had elapsed since the beginning of a previous episode with the same serotype. If a different serotype was determined, there was no limitation on the period of time between isolations. Parents or legal guardians signed informed consent statements. The study was approved by the SUMC ethics committee.
Bacteriology.
Blood and CSF cultures were inoculated into Bactec 9240 and incubated at 35°C. Cultures were monitored with the Bactec system twice daily on the first 2 days and then once daily on days 3, 5, and 7. Swabs obtained from AOM were inoculated onto Columbia agar with 5% sheep blood and 5.0 μg/ml gentamicin and incubated aerobically at 35°C for 48 h in a 5% enriched CO2 atmosphere. Identification, serotyping, and antimicrobial susceptibility testing to penicillin, erythromycin, clindamycin, tetracycline, chloramphenicol, and sulfamethoxazole/trimethoprim (SXT) were performed as previously described (27). Isolates with penicillin MICs of <0.1 μg/ml were defined as susceptible to penicillin, and those with MICs of ≥0.1 μg/ml were considered nonsusceptible. Isolates with resistance to three or more antibiotic classes were defined as multidrug resistant.
Pulsed-field gel electrophoresis (PFGE).
Chromosomal DNA fragments generated by SmaI or ApaI digestion were prepared and analyzed as described previously (27). For strains belonging to serotypes 6A and 12F, ApaI digestion was done, too, to obtain better distinction among the various clones.
MLST.
Pneumococcal isolates were unambiguously characterized by multilocus sequence typing (MLST) as described by Enright and Spratt (9). The sequences (alleles) at each locus were compared to those at the MLST website (www.mlst.net) and were assigned allele numbers if they corresponded to sequences already submitted to the pneumococcal MLST database. The allelic profiles of isolates (the allele numbers at the seven loci) were compared to those at the MLST website, and sequence types (STs) were assigned.
Data analysis.
Each isolate was counted only once per episode (episodes were separated by ≥30 days for the same serotype or by any interval for different serotypes). Statistical analysis was done using SPSS, version 14.0, software for Windows (Chicago, IL). A chi-square test was used to compare the distribution of serotypes and clones between the two ethnic populations. A P value of <0.05 was considered significant.
RESULTS
A total of 5,718 S. pneumoniae isolates from the MEF and blood/CSF of children of <5 years old during a 10-year period before the introduction of PCV7 were available. The AOM group included 5,281 isolates (2,288 from Jewish children and 2,993 from Bedouin children), of which 5,236 (99.1%) were available for serotyping. Non-PCV7 strains accounted for 2,427/5,236 (46.4%). The proportion of these strains was higher among the Bedouin than among the Jewish children, with 1,513/2,969 (50.1%) versus 914/2,267 (40.3%) isolates, respectively (P < 0.001). The IPD group included 437 strains consisting of 142 and 295 isolates from Jewish and Bedouin children, respectively, of which 425 (97.3%) were available for serotyping. Non-PCV7 strains accounted for 234/425 (55.1%), with 67/140 (47.9%) and 167/285 isolates (58.6%) from Jewish and Bedouin children, respectively (P < 0.05).
In the present study, we analyzed 11 common non-PCV7 serotypes, namely, 1, 3, 5, 6A, 7F, 12F, 15B/C, 19A, 21, 33F, and 35B, of which six are included in PCV13. PCV13 was introduced to the national immunization program in Israel in November 2010. Theoretically, PCV13 could have prevented 78.2% (4,096/5,236 episodes) and 81.4% (346/425 episodes) of the total AOM and IPD episodes, respectively, during the 10-year period (1999 to 2008) preceding PCV7 implementation in Israel.
Serotype and clonal distribution among AOM isolates.
The 11 studied serotypes accounted for 31.0% (1,623/5,236) of the AOM isolates available for serotyping and for 66.9% (1,623/2,427) of the non-PCV7 isolates in the AOM group. Of the 1,623 AOM isolates, 94.9% (1,540/1,623) originated from single subjects, and only 5.1% (83/1,623) were from multiple episodes. Moreover, only 1.3% (21/1,623) of the 11 non-PCV7 strains represented multiple episodes per subject with the same serotype. Hence, 98.7% of the strains analyzed in the AOM group originated from either a single episode per subject (94.9%) or multiple episodes with different serotypes (3.7%). Due to the small proportion of multiple episodes with the same serotypes, data analysis was based on the number of isolates by each serotype.
The most common non-PCV7 serotype was 19A (514/5,236 isolates, 9.8%), followed by 6A (285/5,236, 5.4%), 3 (197/5,236, 3.8%), 5 (145/5,236, 2.8%), 15B/C (101/5,236, 1.9%), 1 (92/5,236, 1.8%), 35B (82/5,236, 1.6%), 21 (71/5,236, 1.4%), 7F (54/5,236, 1.0%), 33F (49/5,236, 0.9%), and 12F (33/5,236, 0.6%) (Table 1). The distribution of these serotypes between the two study populations was significantly different (P < 0.05), except for serotypes 7F, 15B/C, and 19A. Additional analysis which took into account only the first episode of each subject and thus excluded all multiple episodes gave the same results except for serotype 3, where the P value changed from 0.03 to 0.19.
Table 1.
Serotype distribution of AOM isolates among Jewish and Bedouin children
| Serotype by group and/or ethnicity | No. of positive isolates (%) by groupb |
Pc | ||
|---|---|---|---|---|
| Jewish children (n = 2,267) | Bedouin children (n = 2,969) | Total (n = 5,236) | ||
| Studied serotypesa | ||||
| 1 | 29 (1.3) | 63 (2.1) | 92 (1.8) | 0.022 |
| 3 | 100 (4.4) | 97 (3.3) | 197 (3.8) | 0.031 |
| 5 | 43 (1.9) | 102 (3.4) | 145 (2.8) | <0.001 |
| 6A | 154 (6.8) | 131 (4.4) | 285 (5.4) | <0.001 |
| 7F | 24 (1.1) | 30 (1.0) | 54 (1.0) | NS |
| 12F | 5 (0.2) | 28 (0.9) | 33 (0.6) | <0.001 |
| 15B/C | 35 (1.5) | 66 (2.2) | 101 (1.9) | NS |
| 19A | 225 (9.9) | 289 (9.7) | 514 (9.8) | NS |
| 21 | 14 (0.6) | 57 (1.9) | 71 (1.4) | <0.001 |
| 33F | 9 (0.4) | 40 (1.3) | 49 (0.9) | <0.001 |
| 35B | 23 (1.0) | 59 (2.0) | 82 (1.6) | 0.005 |
| Total for group | 661 (29.2) | 962 (32.4) | 1,623 (31.0) | 0.012 |
| Total for all non-PCV7 serotypes | 914 (40.3) | 1,513 (50.1) | 2,427 (46.4) | <0.001 |
| Total for all PCV7 serotypes | 1,353 (59.7) | 1,456 (49.9) | 2,809 (53.6) | <0.001 |
| Overall total | 2,267 | 2,969 | 5,236 | |
The group consists of 11 non-PCV7 serotypes.
Percentages are calculated with respect to the total number (n) in the group.
The P value was calculated for each serotype relative to the total number of isolates for each ethnic group. NS, not significant.
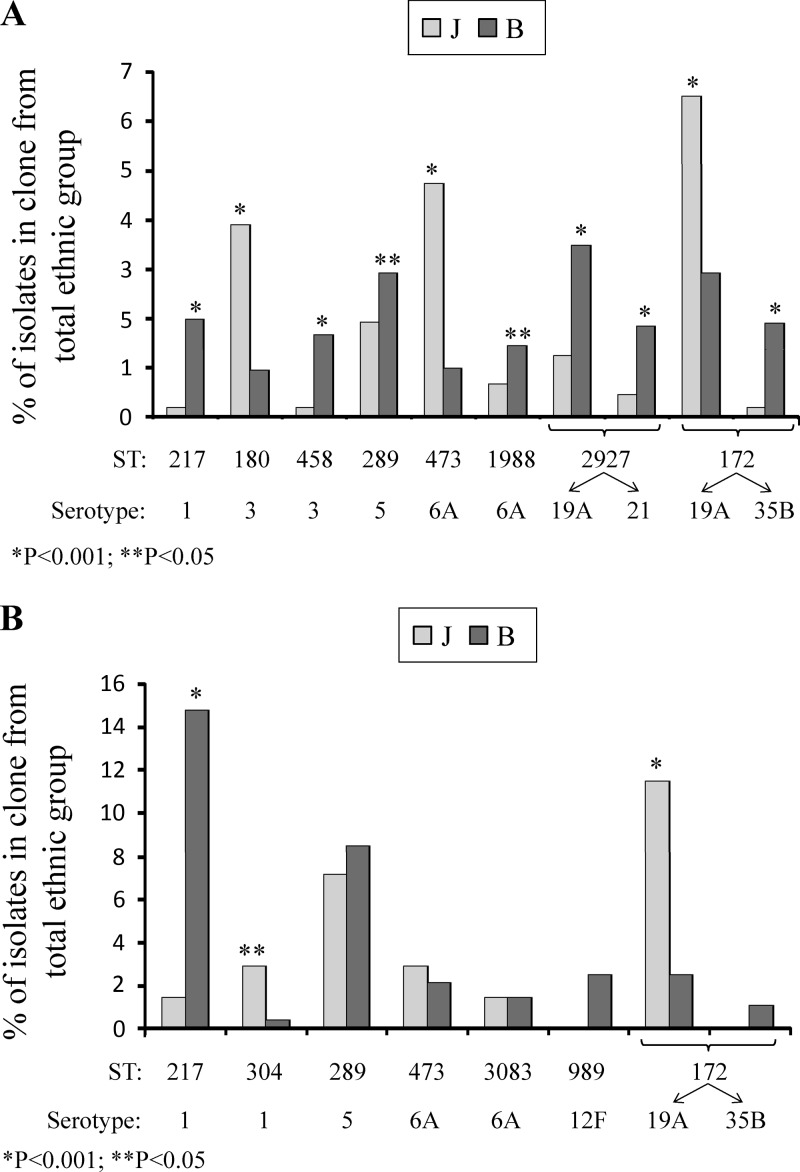
A total of 1,534/1,623 (94.5%) isolates of the 11 non-PCV7 serotypes were available for PFGE; 2 to 12 representative strains from each PFGE cluster and all the strains with unique PFGE patterns were further characterized by MLST (193/1,534 isolates, 12.6%). MLST yielded 81 STs (see Table S1 in the supplemental material), comprising 55 known and 26 novel STs. The eight major AOM STs (STs which constituted ≥1.0% of the total AOM isolates) covered 67.9% (1,041/1,534 isolates) of the 11 non-PCV7 serotypes and 19.9% (1,041/5,236 isolates) of the total number of AOM isolates in this study. Of the major clones, two were nonsusceptible to penicillin (ST2927 [serotype 21] and ST1988), two were nonsusceptible to penicillin and SXT (ST172 and ST2927 [serotype 19A]), one was nonsusceptible to penicillin and erythromycin (ST473), and two were nonsusceptible to SXT (ST217 and ST289) (see Table S1). The distributions of the major STs were significantly different among the two ethnic groups (P < 0.05) (Fig. 1A): five STs were more frequently isolated from Bedouin children, and three STs were more frequently isolated from Jewish children. Noteworthy is the fact that ST2927 and ST172 were associated with two different serotypes each (see Table S1): ST2927 was associated with serotypes 19A and 21, and ST172 was associated with serotypes 19A and 35B. However, while ST2927 was significantly more prevalent among Bedouin than among Jewish children in both serotypes (P < 0.001), ST172 was more frequent among Jewish children as a 19A strain and more frequent among Bedouin children as a 35B strain (P < 0.001 for both serotypes), suggesting that the distribution of ST172 among the two ethnic groups is determined also by the capsular type.
Fig 1.
(A) Distribution of the major STs among Jewish (J) and Bedouin (B) children in the AOM group. P values were calculated for each ST relative to the total number of isolates in each ethnic group (Jewish children, 2,267; Bedouin children, 2,969). (B) Distribution of the major STs among Jewish (J) and Bedouin (B) children in the IPD group. P values were calculated from total number of isolates in each ethnic group (Jewish children, 140; Bedouin children, 285).
The total number of clones per serotype varied from 3 to 20 but was not proportional to the total number of isolates in each individual serotype (see Table S1 in the supplemental material). For example, serotype 5, with 141 isolates available for genotyping, was represented by 1 major clone, ST289, covering 91% of serotype 5 isolates, and 3 minor clones, whereas serotype 15B/C, with 99 isolates available for genotyping, was represented by 12 minor clones. This suggests that different serotypes vary in their abilities to undergo genetic transformation, which may allow their adaptation to various ecological niches of the human host.
Serotype and clonal distribution among IPD isolates.
The 11 studied serotypes accounted for 41.9% (178/425 isolates) of the IPD isolates available for serotyping and 76.1% (178/234) of the non-PCV7 isolates in the IPD group. Of the 178 IPD isolates, 99.4% (177/178) originated from single subjects, and only one strain (1/178, 0.6%) came from a child who had two episodes with different serotypes. Hence, analysis excluding the multiple-serotype episode had no effect on the distribution of the 11 common non-PCV7 serotypes between the two study populations.
The most common non-PCV7 serotype in the IPD group was 1 (55/425 isolates, 12.9%), followed by 5 (40/425, 9.4%), 19A (31/425, 7.3%), 6A (25/425, 5.9%), 12F (9/425, 2.1%), 15B/C (6/425, 1.4%), 35B (4/425, 0.9%), 33F (3/425, 0.7%), 3 and 7F (2/425, 0.5% each), and 21 (1/425, 0.2%) (Table 2). The distributions of the various non-PCV7 serotypes among the two study populations were significantly different for four serotypes: serotypes 1 and 12F were isolated significantly more often from Bedouin children, whereas serotypes 3 and 19A were isolated more often from Jewish children (P < 0.05).
Table 2.
Serotype distribution of IPD isolates among Jewish and Bedouin children
| Serotype by group and/or ethnicity | No. of positive isolates (%) by groupb |
Pc | ||
|---|---|---|---|---|
| Jewish children (n = 140) | Bedouin children (n = 285) | Total (n = 425) | ||
| Studied serotypesa | ||||
| 1 | 7 (5.0) | 48 (16.8) | 55 (12.9) | <0.001 |
| 3 | 2 (1.4) | 0 (0.0) | 2 (0.5) | 0.043 |
| 5 | 11 (7.9) | 29 (10.2) | 40 (9.4) | NS |
| 6A | 8 (5.7) | 17 (6.0) | 25 (5.9) | NS |
| 7F | 1 (0.7) | 1 (0.4) | 2 (0.5) | NS |
| 12F | 0 (0.0) | 9 (3.2) | 9 (2.1) | 0.034 |
| 15B/C | 3 (2.1) | 3 (1.1) | 6 (1.4) | NS |
| 19A | 19 (13.6) | 12 (4.2) | 31 (7.3) | <0.001 |
| 21 | 0 (0.0) | 1 (0.4) | 1 (0.2) | NS |
| 33F | 1 (0.7) | 2 (0.7) | 3 (0.7) | NS |
| 35B | 0 (0.0) | 4 (1.4) | 4 (0.9) | NS |
| Total for group | 52 (37.1) | 126 (44.2) | 178 (41.9) | NS |
| Total for all non-PCV7 serotypes | 67 (47.9) | 167 (58.6) | 234 (55.1) | 0.036 |
| Total for PCV7 serotypes | 73 (52.1) | 118 (41.4) | 191 (44.9) | 0.036 |
| Overall total | 140 | 285 | 425 | |
The group consists of 11 non-PCV7 serotypes.
Percentages are calculated with respect to the total number (n) in the group.
The P value was calculated for each serotype relative to the total number of isolates for each ethnic group. NS, not significant.
A total of 167/178 (93.8%) IPD isolates of the 11 non-PCV7 serotypes were available for PFGE, of which 42/167 (25.1%) were further characterized by MLST. MLST yielded 30 STs (see Table S2 in the supplemental material): 25 clones shared identical STs with the AOM clones in our collection, and only 5 STs were unique. Noteworthy is the fact that 61.1% of the 11 non-PCV7 IPD isolates were represented by only 3/30 STs carrying the three most prevalent serotypes, namely, ST217 with serotype 1, ST289 with serotype 5, and ST172 with serotype 19A. ST172 was also found in three out of four isolates of serotype 35B.
Of the 30 STs in the IPD group, the seven major STs (STs represented by isolates numbering≥1.0% of the total IPD isolates) covered 79.0% (132/167) of the 11 non-PCV7 serotypes and 31.1% (132/425) of the total number of IPD isolates in this study. The antibiotic resistance pattern of the clones in the IPD group resembled that found in the AOM group. Three out of the seven major STs in the IPD group showed resistance to more than one antimicrobial agent: ST172, ST473, and ST989 (see Table S2). Figure 1B shows the distribution of the seven major STs in the IPD group among the two ethnic groups. The prevalence of three out of these seven clones was significantly different among the two study populations (P < 0.05) (Fig. 1B): two STs were more frequently isolated from Jewish children, and one ST was more frequently isolated from Bedouin children. The distribution of the seven major STs in the IPD group among Jewish and Bedouin children resembled that found in the AOM group, except for ST289 of serotype 5 and ST473 of serotype 6A (see Tables S1 and S2 in the supplemental material). ST289 was more frequently isolated from the Bedouin population in both the IPD and AOM groups but reached significance only in the AOM group, whereas ST473 was more frequently isolated from the Jewish children in both groups but reached significance only in the AOM group.
DISCUSSION
Despite the success of PCV7 in reducing mucosal and invasive disease, critical questions remain about the changing pneumococcal reservoir. Efficacy studies have already demonstrated that the use of the vaccine was associated with increased rates of AOM and IPD caused by pneumococcal serotypes immunologically unrelated to the vaccine serotypes, which would suggest the involvement of the replacement phenomenon (7, 10, 14, 16, 19, 21, 32). Recent data from the United States, Europe, and Australia have shown an increase in IPD caused by non-PCV7 serotypes in both infants and adults (13, 14, 17, 21, 22, 25, 33). This increase was observed mainly due to serotype 19A, which is characterized by its antibiotic resistance profile (13, 21, 32). In addition, increased rates of complicated pneumonia due to serotypes 1, 3, 5, 7F, and 19A in Europe and North America have been reported (5, 23). However, these data are deficient since they derive only from calculations of proportions of non-PCV7 serotypes after the introduction of the vaccine, which caused a significant reduction in the prevalence of vaccine-type strains. In order to determine the extent of replacement, data regarding secular trends of PCV7 and non-PCV7 strains before PCV7 introduction have to be taken into consideration. This study provides a baseline of a 10-year comprehensive database, which includes demographic and clinical information of patients from two distinct pediatric populations and the phenotypic and genotypic characteristics of their S. pneumoniae strains isolated from AOM and IPD cases before the introduction of PCV7.
Of the 93 S. pneumoniae serotypes, only 20 to 30 types are associated with the great majority of human diseases. Hence, there is an association between serotype and the potential of pneumococci to cause mucosal and invasive disease. Our data show that non-PCV7 strains accounted for 46% and 55% of the AOM and IPD episodes, respectively, even before the introduction of PCV7 to our region, and non-PCV13 strains accounted for 21.8% of the AOM episodes and 18.6% of the IPD episodes. Of the non-PCV7 strains, we studied 11 serotypes which constituted 67% and 76% of the AOM and IPD episodes, respectively. The incidence of these serotypes differed in the AOM and IPD groups. Serotype 19A was the most prevalent one in the AOM group, followed by 6A, 3, and 5, and these four serotypes accounted for 47% of the non-PCV7 isolates. However, serotypes 1 and 5 were the most prevalent ones in the IPD group, followed by 19A and 6A, and these four serotypes accounted for 65% of the non-PCV7 isolates. Previous surveillance from the same study population during 2000 to 2004 (31) suggested that serotypes 3, 5, 1, 12F, and 19A have the highest disease potential to cause AOM in patients being colonized. Similarly, it was suggested that serotypes 1, 5, and 12F had the highest disease potential for IPD. The increased proportions of serotypes 19A and 6A with high antibiotic resistance in the last years of the study (data not shown) in both AOM and IPD cases, even before the introduction of PCV7, suggests that this phenomenon can be related to factors other than vaccination, such as excessive use of antimicrobial agents (8, 26).
Significantly different distributions of non-PCV7 serotypes among the two study populations were found in 8/11 and 4/11 serotypes from AOM and IPD isolates, respectively. The fact that the different distributions between the two ethnic groups reached significance in only four serotypes in the IPD group might be due to the small number of isolates. Genotyping methods showed the same trend. Previous studies from our group have already shown the differential circulation of non-PCV7 clones among Jewish and Bedouin children in AOM cases (8, 26, 28). The current study expands this observation also to IPD cases. We suggest that this phenomenon is due to the rare contacts of the two pediatric populations.
Our data show that all seven major STs in the IPD group were represented in the AOM group as well. Moreover, four of these STs, namely, ST217 (serotype 1), ST289 (serotype 5), ST473 (serotype 6A), and ST172 (serotypes 19A and 35B), which carry the most prevalent invasive serotypes, were also highly abundant in the AOM group. These four clones were associated with invasive diseases in other studies. For example, ST217 and ST289 were previously isolated from outbreaks of meningitis and bacteremia in northern Ghana (18) and in Israel (1). In the current study, we found that these two clones were susceptible to all antimicrobial agents tested. However, ST5056, a single locus variant of ST289 isolated from cases of AOM and IPD during 2001 and 2002, was multidrug resistant. This clone disappeared after 2002. The two other prevalent clones among the IPD isolates, ST473 and ST172, were penicillin nonsusceptible; ST172 was also resistant to SXT, and ST473 was also resistant to erythromycin. These clones were also common in bacteremic patients from the United States, where each clonal type shared more than one capsular type, i.e., ST473 carrying serotypes 6A, 6B, and 6C (2, 11) and ST172 carrying serotypes 19A and 23F (2, 24).
The exact mechanisms that explain how some serotypes can go beyond colonization to cause disease remain unclear. The capsule is the major determinant of disease potential but not the only one (12, 15, 20). The genetic background of the organism also plays a critical role in dictating virulence. In the present study, two clonal types were associated with two serotypes each: ST172 was the most prevalent clone among serotype 19A and 35B AOM strains, and ST2927 was the most prevalent clone among serotype 21 AOM strains and the second most prevalent clone among serotype 19A AOM isolates. These data suggest that the clonal type is an important determinant in the disease potential of S. pneumoniae. Understanding the underlying genetic mechanisms of virulent genotypes is a priority in the era of pneumococcal conjugate vaccines (15).
Since the introduction of PCV7, the epidemiology of S. pneumoniae has been changing in many countries (33). It was postulated that the disappearance of vaccine serotypes has promoted the emergence of nonvaccine strains, reducing the benefits of vaccination. However, our data show that non-PCV7 strains were frequently isolated in AOM and IPD cases even before the implementation of the vaccine. Hence, long-term surveillance of post-PCV7 disease is crucial to determine the true effect of the vaccine on the expansion of non-PCV7 strains.
Supplementary Material
ACKNOWLEDGMENT
This work was partially supported by grant 0887X1-4463 from Wyeth (now Pfizer).
Footnotes
Published ahead of print 8 August 2012
Supplemental material for this article may be found at http://jcm.asm.org/.
REFERENCES
- 1. Balicer RD, et al. 2010. Control of Streptococcus pneumoniae serotype 5 epidemic of severe pneumonia among young army recruits by mass antibiotic treatment and vaccination. Vaccine 28:5591–5596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Beall B, et al. 2006. Pre- and postvaccination clonal compositions of invasive pneumococcal serotypes for isolates collected in the United States in 1999, 2001, and 2002. J. Clin. Microbiol. 44:999–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Brueggemann AB, et al. 2003. Clonal relationships between invasive and carriage Streptococcus pneumoniae and serotype- and clone-specific differences in invasive disease potential. J. Infect. Dis. 187:1424–1432 [DOI] [PubMed] [Google Scholar]
- 4. Brueggemann AB, et al. 2004. Temporal and geographic stability of the serogroup-specific invasive disease potential of Streptococcus pneumoniae in children. J. Infect. Dis. 190:1203–1211 [DOI] [PubMed] [Google Scholar]
- 5. Byington CL, et al. 2006. Impact of the pneumococcal conjugate vaccine on pneumococcal parapneumonic empyema. Pediatr. Infect. Dis. J. 25:250–254 [DOI] [PubMed] [Google Scholar]
- 6. Dagan R. 2009. New insights on pneumococcal disease: what we have learned over the past decade. Vaccine 27:C3–C5 [DOI] [PubMed] [Google Scholar]
- 7. Dagan R. 2009. Serotype replacement in perspective. Vaccine 27:C22–C24 [DOI] [PubMed] [Google Scholar]
- 8. Dagan R, Givon-Lavi N, Leibovitz E, Greenberg D, Porat N. 2009. Introduction and proliferation of multidrug-resistant Streptococcus pneumoniae serotype 19A clones that cause acute otitis media in an unvaccinated population. J. Infect. Dis. 199:776–785 [DOI] [PubMed] [Google Scholar]
- 9. Enright MC, Spratt BG. 1998. A multilocus sequence typing scheme for Streptococcus pneumoniae: identification of clones associated with serious invasive disease. Microbiology 144:3049–3060 [DOI] [PubMed] [Google Scholar]
- 10. Eskola J, et al. 2001. Efficacy of a pneumococcal conjugate vaccine against acute otitis media. N. Engl. J. Med. 344:403–409 [DOI] [PubMed] [Google Scholar]
- 11. Green MC, et al. 2011. Increase in prevalence of Streptococcus pneumoniae serotype 6C at eight children's hospitals in the United States from 1993 to 2009. J. Clin. Microbiol. 49:2097–2101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hanage WP, et al. 2005. Invasiveness of serotypes and clones of Streptococcus pneumoniae among children in Finland. Infect. Immun. 73:431–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hanna JN, Humphreys JL, Murphy DM. 2008. Invasive pneumococcal disease in Indigenous people in north Queensland: an update, 2005–2007. Med. J. Aust. 189:43–46 [DOI] [PubMed] [Google Scholar]
- 14. Hicks LA, et al. 2007. Incidence of pneumococcal disease due to non-pneumococcal conjugate vaccine (PCV7) serotypes in the United States during the era of widespread PCV7 vaccination, 1998–2004. J. Infect. Dis. 196:1346–1354 [DOI] [PubMed] [Google Scholar]
- 15. Hsieh Y, Lee W, Shao P, Chang L, Huang L. 2008. The transforming Streptococcus pneumoniae in the 21st century Chang. Gung. Med. J. 31:117–124 [PubMed] [Google Scholar]
- 16. Håhman Kilpi T, et al. 2003. Protective efficacy of a second pneumococcal conjugate vaccine against pneumococcal acute otitis media in infants and children: randomized, controlled trial of a 7-valent pneumococcal polysaccharide-meningococcal outer membrane protein complex conjugate vaccine in 1666 children. Clin. Infect. Dis. 37:1155–1164 [DOI] [PubMed] [Google Scholar]
- 17. Lehmann D, et al. 2010. The changing epidemiology of invasive pneumococcal disease in aboriginal and non-aboriginal western Australians from 1997 through 2007 and emergence of nonvaccine serotypes. Clin. Infect. Dis. 50:1477–1486 [DOI] [PubMed] [Google Scholar]
- 18. Leimkugel J, et al. 2005. An outbreak of serotype 1 Streptococcus pneumoniae meningitis in northern Ghana with features that are characteristic of Neisseria meningitidis meningitis epidemics. J. Infect. Dis. 192:192–199 [DOI] [PubMed] [Google Scholar]
- 19. McEllistrem MC, et al. 2005. Acute otitis media due to penicillin-nonsusceptible Streptococcus pneumoniae before and after the introduction of the pneumococcal conjugate vaccine. Clin. Infect. Dis. 40:1738–1744 [DOI] [PubMed] [Google Scholar]
- 20. Melin M, Trzciński K, Meri S, Käyhty H, Väkeväinen M. 2010. The capsular serotype of Streptococcus pneumoniae is more important than the genetic background for resistance to complement. Infect. Immun. 78:5262–5270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Moore MR, et al. 2008. Population snapshot of emergent Streptococcus pneumoniae serotype 19A in the United States, 2005. J. Infect. Dis. 197:1016–1027 [DOI] [PubMed] [Google Scholar]
- 22. Muñoz-Almagro C, et al. 2008. Emergence of invasive pneumococcal disease caused by nonvaccine serotypes in the era of 7-valent conjugate vaccine. Clin. Infect. Dis. 46:174–182 [DOI] [PubMed] [Google Scholar]
- 23. Obando I, et al. 2008. Pediatric parapneumonic empyema, Spain. Emerg. Infect. Dis. 14:1390–1397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Pai R, Gertz RE, Whitney CG, Beall B, Active Bacterial Core Surveillance Team 2005. Clonal association between Streptococcus pneumoniae serotype 23A, circulating within the United States, and an internationally dispersed clone of serotype 23F. J. Clin. Microbiol. 43:5440–5444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Pilishvili T, et al. 2010. Sustained reductions in invasive pneumococcal disease in the era of conjugate vaccine. J. Infect. Dis. 201:32–41 [DOI] [PubMed] [Google Scholar]
- 26. Porat N, Amit U, Givon-Lavi N, Leibovitz E, Dagan R. 2010. Increasing importance of multidrug-resistant serotype 6A Streptococcus pneumoniae clones in acute otitis media in southern Israel. Pediatr. Infect. Dis. J. 29:126–130 [DOI] [PubMed] [Google Scholar]
- 27. Porat N, Barkai G, Jacobs MR, Trefler R, Dagan R. 2004. Four antibiotic-resistant Streptococcus pneumoniae clones unrelated to the pneumococcal conjugate vaccine serotypes, including 2 new serotypes, causing acute otitis media in southern Israel. J. Infect. Dis. 189:385–392 [DOI] [PubMed] [Google Scholar]
- 28. Porat N, Park IH, Nahm MH, Dagan R. 2010. Differential circulation of Streptococcus pneumoniae serotype 6C clones in two Israeli pediatric populations. J. Clin. Microbiol. 48:4649–4651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sá-Leão R, et al. 2011. Analysis of invasiveness of pneumococcal serotypes and clones circulating in Portugal before widespread use of conjugate vaccines reveals heterogeneous behavior of clones expressing the same serotype. J. Clin. Microbiol. 49:1369–1375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sandgren A, et al. 2004. Effect of clonal and serotype-specific properties on the invasive capacity of Streptococcus pneumoniae. J. Infect. Dis. 189:785–796 [DOI] [PubMed] [Google Scholar]
- 31. Shouval DS, Greenberg D, Givon-Lavi N, Porat N, Dagan R. 2006. Site-specific disease potential of individual Streptococcus pneumoniae serotypes in pediatric invasive disease, acute otitis media and acute conjunctivitis. Pediatr. Infect. Dis. J. 25:602–607 [DOI] [PubMed] [Google Scholar]
- 32. Singleton RJ, et al. 2007. Invasive pneumococcal disease caused by nonvaccine serotypes among Alaska native children with high levels of 7-valent pneumococcal conjugate vaccine coverage. JAMA 297:1784–1792 [DOI] [PubMed] [Google Scholar]
- 33. Weinberger DM, Malley R, Lipsitch M. 2011. Serotype replacement in disease after pneumococcal vaccination. Lancet 378:1962–1973 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.