Abstract
Novel lineages of human astrovirus (HAstV) types 2, 2c, and 2d have been identified. Upon sequencing of the 3′ end of the genome, the type 2c and 2d HAstVs were found to be open reading frame 1b (ORF1b)-ORF2 recombinant, with ORF1b being derived from type 3 and type 1 HAstVs, respectively. An ORF2 interlineage recombinant strain, 2c/2b, was also identified.
TEXT
Human astroviruses (HAstVs) are an important cause of gastroenteritis in young children (17). HAstVs have a single-stranded positive-sense RNA genome containing three open reading frames (ORFs). The capsid protein precursor (ORF2) can be divided into a highly conserved N-terminal domain (amino acids [aa] 1 to 424), a hypervariable (HVR) domain (aa 425 to 688), and a highly acidic C-terminal domain (25). The HVR of HAstV is believed to form the capsid spike and to control binding to cell receptors as neutralizing epitopes have been mapped inside this capsid portion (7). On the basis of the capsid protein precursor, HAstVs are classified genetically and antigenically into eight types (HAstV-1 to -8) (18). The predominant types appear to vary seasonally and geographically (3, 5, 10, 12, 15, 23, 24), although the relevance of this genetic and antigenic heterogeneity and these temporal and geographical fluctuations is unclear. Epidemiological investigations of HAstVs worldwide have shown that HAstV-1 is predominant, while the prevalence of HAstV-2 can vary markedly (1, 5, 13, 15, 21), reaching epidemiological relevance in some geographical settings and seasons (4, 9, 11, 16). Discrete sequence variation has been observed within some HAstV types, allowing for distinction of different genetic lineages. Surveillance studies for HAstV in Italy identified the appearance of novel HAstV-2 lineages in 2002 (3, 5) and again in 2009 (15). Based on a small sequence generated by the diagnostic primers Mon269 and Mon270 (18) in the 5′ region of ORF2 the (D5′ region, nt 4571 to 4918 of accession no. L13745), the 2002 Italian HAstV-2 strains were clearly distinguishable phylogenetically from type 2a and type 2b strains, and they were classified as lineage 2c, along with a number of other strains detected worldwide since the mid-1990s (5). In a similar fashion, in 2009 a novel lineage, 2d, was identified (15). In order to investigate better the extent of the genetic heterogeneity observed in HAstV-2 strains, a 3.2-kb portion at the 3′ end of the genome, encompassing an ∼740-nt-long portion at the 3′ end of ORF1b, the full-length ORF2 (2,400 nt), and the 3′ untranslated region (3′ UTR) (82 nt), was sequenced for two representative strains, ITA/2002/PA65R/type2c (accession no. JX087963) and ITA/2009/PR5142/type2d (accession no. JX087964). The sequences were compared with complete and partial ORF2 sequences of HAstV-2 strains retrieved from the databases (Table 1 and Fig. 1). Sequence editing, multiple alignments, and phylogenetic analysis were performed with Geneious software v5.6 and MEGA 5.0 software (22).
Table 1.
Nucleotide and amino acid similarity comparisons among type 2 lineagesa
| Strain (accession no.) | Region | % of similarity to strain (accession no.) shown |
Region | ||||||
|---|---|---|---|---|---|---|---|---|---|
| GBR/1993/Oxford-H2/2b (L13745) | GBR/Oxford-S2/2b (AB000288) | NOR/1993/NORAS1128/r(AB000290) | ITA/2002/PA65R/2c (JX087963) | USA/1999/1299-CA/2c (EF138827) | ITA/2009/PR5142/2d (JX087964) | GBR/1993/Oxford-AS8811/2a (AB000289) | |||
| GBR/1993/Oxford-H2/2b (L13745) | 96.4 | 95.9 | 98.6 | 97.8 | 97.6 | 95.7 | N | ||
| 90.0 | 95.2 | 90.9 | 94.4 | 92.6 | 70.2 | HVR | |||
| 94.6 | 96.4 | 94.6 | 95.9 | 93.8 | 85.3 | Cap | |||
| GBR/Oxford-S2/2b (AB000288) | D5′ | 99.5 | 94.2 | 95.4 | 94.9 | 94.4 | 93.2 | N | |
| D3′ | 100 | 88.7 | 85.7 | 88.3 | 87.4 | 66.0 | HVR | ||
| Cap | 97.5 | 93.0 | 91.4 | 92.5 | 90.5 | 82.7 | Cap | ||
| NOR/1993/NORAS1128/r (AB000290) | D5′ | 94.3 | 94.3 | 97.1 | 96.6 | 94.9 | 94.2 | N | |
| D3′ | 100 | 100 | 88.7 | 91.3 | 88.7 | 67.7 | HVR | ||
| Cap | 95.0 | 93.8 | 93.1 | 94.3 | 91.1 | 83.7 | Cap | ||
| ITA/2002/PA65R/2c (JX087963) | D5′ | 94.9 | 94.8 | 96.8 | 99.3 | 97.8 | 95.9 | N | |
| D3′ | 94.0 | 94.0 | 94.0 | 95.7 | 93.1 | 68.9 | HVR | ||
| Cap | 92.7 | 91.1 | 93.7 | 97.3 | 94.4 | 84.5 | Cap | ||
| USA/1999/1299-CA/2c (EF138827) | D5′ | 95.0 | 94.5 | 97.5 | 98.0 | 97.6 | 95.9 | N | |
| D3′ | 94.5 | 94.5 | 94.5 | 97.5 | 95.7 | 71.5 | HVR | ||
| Cap | 93.1 | 91.4 | 94.6 | 96.0 | 95.6 | 85.7 | Cap | ||
| ITA/2009/PR5142/2d (JX087964) | D5′ | 93.8 | 93.3 | 92.5 | 93.3 | 92.8 | 95.4 | N | |
| D3′ | 93.5 | 93.5 | 93.5 | 93.5 | 94.0 | 70.6 | HVR | ||
| Cap | 92.0 | 90.6 | 90.1 | 90.8 | 91.3 | 84.3 | Cap | ||
| GBR/1993/Oxford-AS8811/2a (AB000289) | D5′ | 88.3 | 87.8 | 87.1 | 87.1 | 86.3 | 87.8 | ||
| D3′ | 91.0 | 91.0 | 91.0 | 88.5 | 89.5 | 88.5 | |||
| Cap | 83.5 | 82.0 | 81.9 | 81.6 | 81.9 | 81.8 | |||
The values in the bottom left portion of the table represent the nucleotide comparisons in the 449-bp region (D5′) targeted by diagnostic primers Mon269/Mon270 at the 5′ end of ORF2 (nt 4526 to 4974 of accession no. L13745), in the 239-bp region (D3′) targeted by primers prBEG/Mon2 at the 3′ end of ORF2 (nt 6477 to 6715 of accession no. L13745), and in the full-length (2.4-kb) ORF2 protein (Cap). The values in the top right portion of the table represent the amino acid comparisons among the type 2 lineages in the conserved N terminus (N) (aa 1 to 415), in the hypervariable region (HVR) (aa 416 to 650), and in the full-length capsid protein (Cap). The recombinant (r) strain 2c/2b is also included. The sequence of strain GBR/1993/Oxford-ref/2b (L06802) showed 99.9% nucleotide and amino acid similarity to that of strain GBR/1993/Oxford-H2 and was not included in the table.
Fig 1.
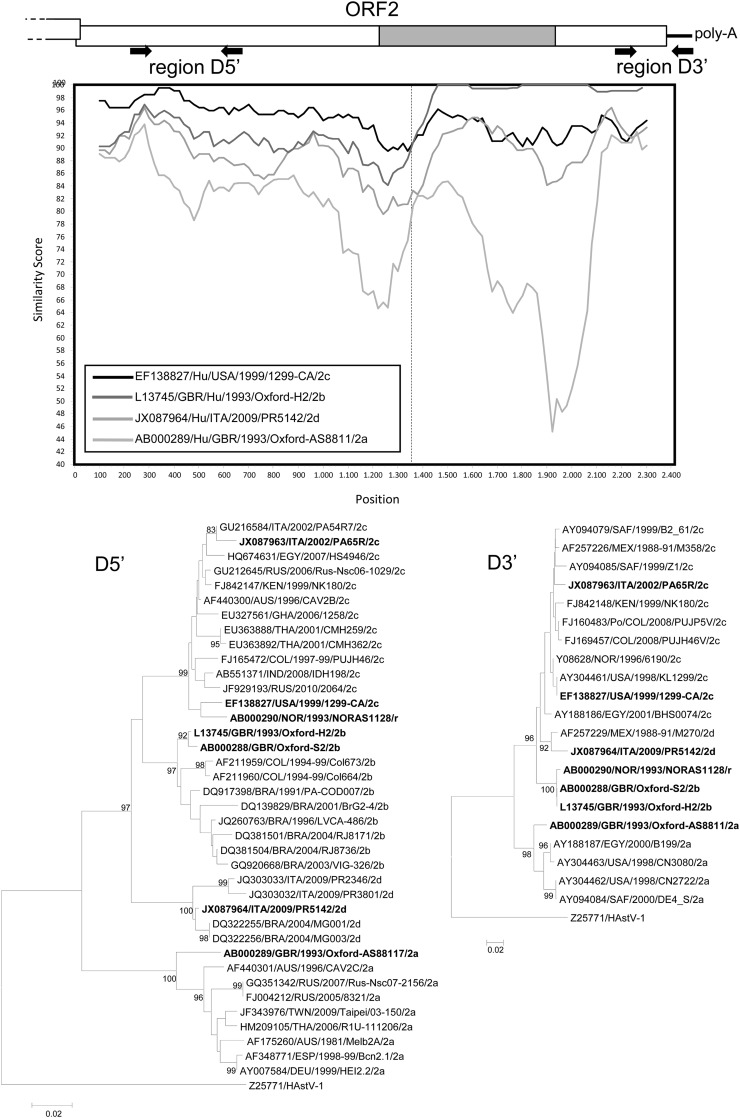
Interlineage recombination in type-2 HAstVs. Lineage 2a (accession no. AB000289), 2b (L13745), 2c (EF138827), and 2d (JX087964) HAstVs were plotted against the recombinant strain NOR/1993/NORAS1128 (AB000290). Sequences were analyzed with Simplot (14) using a window size of 200 and step size of 20 with gap strip off and Hamming correction on. The recombination breakpoint is shown as a vertical dashed line. The genome organization of HAstV from the 3′ end of ORF1b to the poly(A) tail is also shown. The HVR region is in gray. The locations of the diagnostic regions D5′ and D3′ in the ORF2 are shown. Phylogenetic trees were constructed using the D5′ and D3′ regions. The D5′ tree was constructed using an ∼350-nt ORF2 fragment of a selection of 38 sequences of type 2 HAstV strains retrieved from the databases. The D3′ tree was constructed using an ∼200-nt ORF2 fragment of 21 sequences. For strains in boldface, the full-length ORF2 is available. The trees were generated by the neighbor-joining method and the Kimura 2-parameter distance correction, with 1,000 bootstrap replicates, using the software Mega 5 (22). Bootstrap values lower than 95% are not shown.
In the ORF1b strains, ITA/2002/PA65R/type2c and ITA/2009/PR5142/type2d differed markedly from each other (nucleotide identity of 86.0%) and from the type 2b strain GBR/1993/Oxford-H2 (83.4% and 92.5% nucleotide identity, respectively). Strain ITA/2002/PA65R/type2c displayed the highest nucleotide identity (97.8%) to the African HAstV-2c strain KEN/1999/NK180, regarded as a recombinant virus (26), while strain ITA/2009/PR5142/type2d showed the highest nucleotide identity (96.4 to 96.9%) to HAstV-1 detected in India and China.
In the full-length ORF2, strain ITA/2002/PA65R/type2c resembled a strain detected in the United States in 1999 (96.0% nucleotide identity and 97.3% amino acid identity). Lineage 2c HAstVs differed by 6.9 to 8.9% nucleotides and 4.1 to 8.6% amino acids from HAstV-2b strains and by 18.1 18.3% nucleotides and 14.3 to 15.5% amino acids from the 2a strain prototype GBR/1993/Oxford-AS881. Strain ITA/2009/PR5142/type2d differed by 8.0 to 9.4% nucleotides and 6.2 to 9.5% amino acids from lineage 2b HAstVs, by 8.7 to 9.2% nucleotides and 4.4 to 5.6% amino acids from lineage 2c HAstVs, and by 18.2% nucleotides and 15.7% amino acids from the type 2a strain GBR/1993/Oxford-AS881 (Table 1). The four lineages (2a to 2d) were clearly resolved by phylogenetic analysis both in the full-length ORF2 and in the D5′ region (Fig. 1), and they were strongly supported statistically, with high bootstrap values (>90%). Cutoff values of 6% nucleotides in the full-length ORF2 and of 5% nucleotides in the D5′ region could be defined among the four lineages. These values are similar to values calculated in the D5′ region in other studies (9, 12).
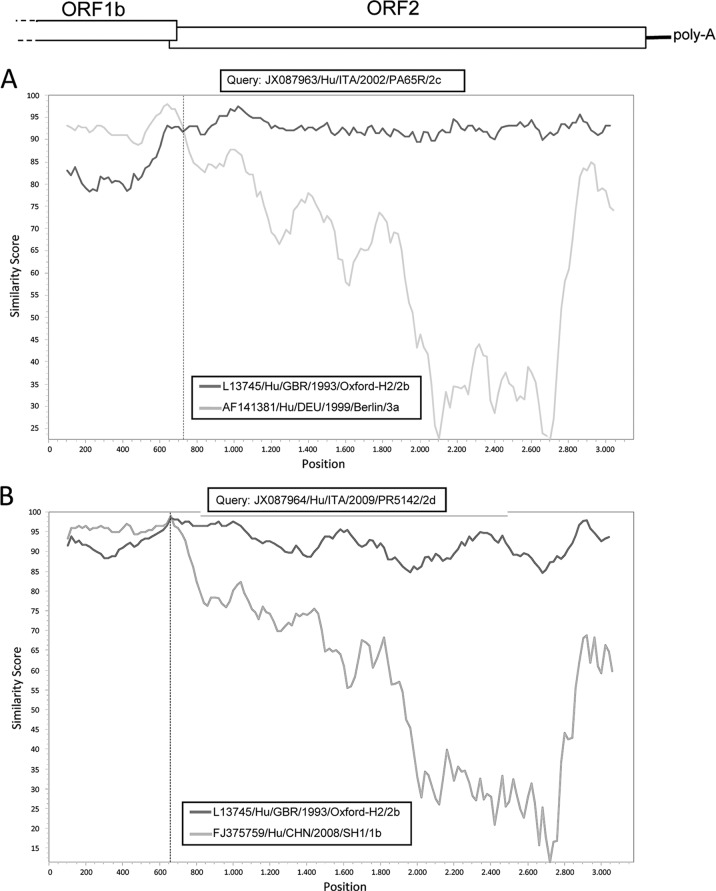
By analyzing region D5′ in parallel with the diagnostic region D3′, located at the 3′ end of ORF2 (nt 6477 to 6715 of accession no. L13745), inconsistencies were observed in the clustering patterns for a unique strain, NOR/1993/NORAS1128. The virus was grouped with HAstV-2c in the D5′ region and with HAstV-2b in the D3′ region, suggesting a recombination event. By using SimPlot software (version 3.2) (14), the crossover site was mapped at the beginning of the HVR between nt 1350 and 1400 (Fig. 1). Accordingly, the chimeric capsid protein of strain NOR/1993/NORAS1128 possessed the N-terminal domain from a HAstV-2c strain and the HVR from a HAstV-2b strain. In addition, by Simplot analysis both the type 2c and 2d strains also appeared to have a recombinant nature, with the crossover sites mapped within or close to the ORF1b-ORF2 junction region (Fig. 2). The type 2c strain ITA/2002/PA65R retained the same ORF1b-ORF2 combination as the African HAstV-2c strain KEN/1999/NK180 (26), with an ORF1b gene likely derived from HAstV-3 strains, suggesting that this genomic asset is stable in HAstV-2c viruses. In the opposite case, the type 2d strain ITA/2009/PR5142 displayed a novel ORF1b-ORF2 combination, with the ORF1b likely acquired from type 1 HAstVs. The exchange of genome fragments via recombination is common in single-stranded RNA viruses and appears to occur at higher frequency in highly conserved genomic regions and between genetically related strains (2). Virus recombination can affect phylogenetic groupings, increase the virulence/fitness of the virus, confuse molecular epidemiological studies, and have major implications in vaccine design (2). Evidence of recombination has been documented in both human and animal astroviruses (19, 20, 26). Examples of recombination within the ORF2 of astroviruses are uncommon, as this region undergoes a strong genetic diversification that likely decreases the frequency of recombination, and to our knowledge, the interlineage 2c/2b ORF2 recombinant is the first example of HAstV with this genetic signature.
Fig 2.
Recombinant nature of the type 2c and 2d HAstVs. The lineage 2c strain ITA/2002/PA65R was plotted against the type 2b strain GBR/1993/Oxford/H2 and the type 3a strain DEU/1999/Berlin (A). The lineage 2d strain ITA/2009/PR5142 was plotted against the type 2b strain GBR/1993/Oxford/H2 and the type 1b strain CHN/2008/SH1 (B). Sequences were analyzed with Simplot (14) using a window size of 200 and step size of 20 with gap strip off and Hamming correction on. The recombination breakpoint is shown as a vertical dashed line. The genome organization of HAstV from the 3′ end of ORF1b to the poly(A) tail is also shown.
By reviewing the sequence databases and the literature, there is evidence that 2c viruses were already circulating in the early 1990s in Australia and Europe (3, 21) and that 2d viruses were already circulating in Brazil in 2004 (8) and in Mexico in the late 1980s (24), and, therefore, type 2c and 2d HAstVs reemerged in Italy in 2002 and 2009 (5, 6, 15), respectively, rather than representing completely novel lineages. Investigation systematically of the genetic variability of HAstVs is important to understand if periodic genotype shifts and genetic or antigenic drifts occur and if these changes may fit a model of evolution analogous to those observed for other antigenically or genetically heterogeneous viruses. Also, generation of a database of HAstV sequences of various types and lineages will be important for tracking the geographical or temporal origin of novel strains and for improving or harmonizing the diagnostic strategies for large molecular epidemiological studies.
ACKNOWLEDGMENTS
This investigation was supported by grant “RicercaScientifica FIL 2009” from the University of Parma, Parma, Italy, and by grant MicroMap (PON01_02589).
Footnotes
Published ahead of print 29 August 2012
REFERENCES
- 1.Ahmed SF, et al. 2011. Novel astroviruses in children, Egypt. Emerg. Infect. Dis. 17:2391–2393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bull RA, Tanaka MM, White PA. 2007. Norovirus recombination. J. Gen. Virol. 88:3347–3359 [DOI] [PubMed] [Google Scholar]
- 3.Colomba C, et al. 2006. Viral gastroenteritis in children hospitalised in Sicily, Italy. Eur. J. Clin. Microbiol. Infect. Dis. 25:570–575 [DOI] [PubMed] [Google Scholar]
- 4.Dalton RM, et al. 2002. Astrovirus acute gastroenteritis among children in Madrid, Spain. Pediatr. Infect. Dis. J. 21:1038–1041 [DOI] [PubMed] [Google Scholar]
- 5.De Grazia S, et al. 2011. Surveillance of human astrovirus circulation in Italy 2002–2005: emergence of lineage 2c strains. Clin. Microbiol. Infect. 17:97–101 [DOI] [PubMed] [Google Scholar]
- 6.De Grazia S, et al. 17 May 2012. Nationwide surveillance study of human astrovirus infections in an Italian paediatric population. Epidemiol. Infect. [Epub ahead of print.] doi:org/10.1017/S0950268812000945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dong J, Dong L, Mendez E, Tao Y. 2011. Crystal structure of the human astrovirus capsid spike. Proc. Natl. Acad. Sci. U. S. A. 108:12681–12686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gabbay YB, et al. 2006. Characterization of an astrovirus genotype 2 strain causing an extensive outbreak of gastroenteritis among Maxakali Indians, Southeast Brazil. J. Clin. Virol. 37:287–292 [DOI] [PubMed] [Google Scholar]
- 9.Gabbay YB, et al. 2007. Molecular epidemiology of astrovirus type 1 in Belem, Brazil, as an agent of infantile gastroenteritis, over a period of 18 years (1982–2000): identification of two possible new lineages. Virus Res. 129:166–174 [DOI] [PubMed] [Google Scholar]
- 10.Gabbay YB, et al. 2007. Prevalence of human astrovirus genotypes associated with acute gastroenteritis among children in Belem, Brazil. J. Med. Virol. 79:530–538 [DOI] [PubMed] [Google Scholar]
- 11.Gaggero A, et al. 1998. Prevalence of astrovirus infection among Chilean children with acute gastroenteritis. J. Clin. Microbiol. 36:3691–3693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guix S, et al. 2002. Molecular epidemiology of astrovirus infection in Barcelona, Spain. J. Clin. Microbiol. 40:133–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guo L, et al. 2010. Molecular characterization of astrovirus infection in children with diarrhea in Beijing, 2005–2007. J. Med. Virol. 82:415–423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lole KS, et al. 1999. Full-length human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination. J. Virol. 73:152–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Medici MC, et al. 2012. Molecular detection and epidemiology of astrovirus, bocavirus, and sapovirus in Italian children admitted to hospital with acute gastroenteritis, 2008–2009. J. Med. Virol. 84:643–650 [DOI] [PubMed] [Google Scholar]
- 16.Medina SM, Gutierrez MF, Liprandi F, Ludert JE. 2000. Identification and type distribution of astroviruses among children with gastroenteritis in Colombia and Venezuela. J. Clin. Microbiol. 38:3481–3483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mendez E, Arias CF. 2007. Astroviruses, 5th ed Lippincott Willliams & Wilkins, Philadelphia, PA [Google Scholar]
- 18.Noel JS, Lee TW, Kurtz JB, Glass RI, Monroe SS. 1995. Typing of human astroviruses from clinical isolates by enzyme immunoassay and nucleotide sequencing. J. Clin. Microbiol. 33:797–801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pantin-Jackwood MJ, Spackman E, Woolcock PR. 2006. Phylogenetic analysis of Turkey astroviruses reveals evidence of recombination. Virus Genes 32:187–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pantin-Jackwood MJ, et al. 2011. Molecular characterization of avian astroviruses. Arch. Virol. 156:235–244 [DOI] [PubMed] [Google Scholar]
- 21.Schnagl RD, et al. 2002. Incidence of human astrovirus in central Australia (1995 to 1998) and comparison of deduced serotypes detected from 1981 to 1998. J. Clin. Microbiol. 40:4114–4120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tamura K, et al. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28:2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Victoria M, Carvalho-Costa FA, Heinemann MB, Leite JP, Miagostovich MP. 2007. Genotypes and molecular epidemiology of human astroviruses in hospitalized children with acute gastroenteritis in Rio de Janeiro, Brazil. J. Med. Virol. 79:939–944 [DOI] [PubMed] [Google Scholar]
- 24.Walter JE, et al. 2001. Molecular epidemiology of human astrovirus diarrhea among children from a periurban community of Mexico City. J. Infect. Dis. 183:681–686 [DOI] [PubMed] [Google Scholar]
- 25.Wang QH, et al. 2001. Genetic analysis of the capsid region of astroviruses. J. Med. Virol. 64:245–255 [DOI] [PubMed] [Google Scholar]
- 26.Wolfaardt M, Kiulia NM, Mwenda JM, Taylor MB. 2011. Evidence of a recombinant wild-type human astrovirus strain from a Kenyan child with gastroenteritis. J. Clin. Microbiol. 49:728–731 [DOI] [PMC free article] [PubMed] [Google Scholar]