Abstract
In 1991, multiresistant Escherichia coli O78:H10 strains caused an outbreak of urinary tract infections in Copenhagen, Denmark. The phylogenetic origin, clonal background, and virulence characteristics of the outbreak isolates, and their relationship to nonoutbreak O78:H10 strains according to these traits and resistance profiles, are unknown. Accordingly, we extensively characterized 51 archived E. coli O78:H10 isolates (48 human isolates from seven countries, including 19 Copenhagen outbreak isolates, and 1 each of calf, avian, and unknown-source isolates), collected from 1956 through 2000. E. coli O78:H10 was clonally heterogeneous, comprising one dominant clonal group (61% of isolates, including all 19 outbreak isolates) from ST10 (phylogenetic group A) plus several minor clonal groups (phylogenetic groups A and D). All ST10 isolates, versus 25% of non-ST10 isolates, were identified by molecular methods as enteroaggregative E. coli (EAEC) (P < 0.001). Genes present in >90% of outbreak isolates included fimH (type 1 fimbriae; ubiquitous in E. coli); fyuA, traT, and iutA (associated with extraintestinal pathogenic E. coli [ExPEC]); and sat, pic, aatA, aggR, aggA, ORF61, aaiC, aap, and ORF3 (associated with EAEC). An outbreak isolate was lethal in a murine subcutaneous sepsis model and exhibited characteristic EAEC “stacked brick” adherence to cultured epithelial cells. Thus, the 1991 Copenhagen outbreak was caused by a tight, non-animal-associated subset within a broadly disseminated O78:H10 clonal group (ST10; phylogenetic group A), members of which exhibit both ExPEC and EAEC characteristics, whereas O78:H10 isolates overall are phylogenetically diverse. Whether ST10 O78:H10 EAEC strains are both uropathogenic and diarrheagenic warrants further investigation.
INTRODUCTION
In 1991, a cluster of 18 patients in the Copenhagen area of Denmark experienced predominantly community-acquired urinary tract infection (UTI) caused by multiresistant Escherichia coli O78:H10, the only reported outbreak to date involving this serotype (32). In addition, one patient with diarrhea was identified as carrying the outbreak strain by screening for serotype O78:H10 among 500 E. coli from diarrheal stool specimens from Copenhagen around the time of the outbreak. Notably, O78:H10 was not found among 377 E. coli bacteremia isolates from a university hospital in Copenhagen during the 5 years prior to outbreak (1986 to 1990) (33). The outbreak strain's source was never identified.
Before and after the 1991 Copenhagen outbreak, several other UTI outbreaks involving multiresistant clonal groups of E. coli, including “Clonal Group A” (sequence type [ST] ST69, according to multilocus sequence typing [MLST]), E. coli O15:K52:H1 (MLST clonal complex CC31), and the extended-spectrum-beta-lactamase (ESBL)-associated O25:H4 clonal group (ST131), were reported (21, 25, 27). These three clonal groups share uropathogenic traits corresponding to iha (adhesin-siderophore), sat (secreted autotransporter toxin), iutA (aerobactin system), fyuA (yersiniabactin system), and the pap operon (digalactoside-binding adhesins); each also exhibits additional virulence genes typical of extraintestinal pathogenic E. coli (ExPEC) (16).
The Copenhagen O78:H10 outbreak isolates exhibited the same distinctive resistance profile, i.e., ACSSuTTp (ampicillin, chloramphenicol, streptomycin, sulfonamides, tetracycline, and trimethoprim), as the E. coli O15:K52:H1 strain that caused a yearlong, community-wide outbreak of multiresistant extraintestinal infections in South London in 1986 to 1987, shortly before the Copenhagen outbreak (37). We previously demonstrated that E. coli O15:K52:H1 is clonally homogeneous based on a large number of phenotypic and molecular characteristics (34).
Here, we sought further insights into the basis for the Copenhagen outbreak. Specifically, we hypothesized that the Copenhagen O78:H10 outbreak isolates represented a single clonal strain, which might (i) fulfill molecular criteria for ExPEC (17), (ii) exhibit distinctive virulence-associated traits in comparison with other O78:H10 isolates, (iii) have a food animal source, and (iv) occur sporadically in other locales. To test these hypotheses, we compared the previously described O78:H10 outbreak isolates to archival nonoutbreak O78:H10 isolates according to phylogenetic background, clonality, virulence factors, resistance profiles, and clinical/ecological source.
MATERIALS AND METHODS
Clinical and epidemiological information.
The WHO Collaborating Centre for Reference and Research on Escherichia and Klebsiella at the Staten Serum Institut (SSI) in Copenhagen, Denmark, holds >70,000 E. coli isolates, received from 85 different countries since 1951 from humans (>60,000) and animals (>10,000) for diverse indications and projects. The Centre's E. coli database (1951 to 2009) was searched for serotype O78:H10. In total, 51 unique E. coli O78:H10 isolates, including 48 human isolates originating from 48 patients plus 1 calf isolate, 1 avian isolate, and 1 isolate from an unknown host, all received between 1956 and 2000, were identified. Serotyping was done according to the method of Ørskov and Ørskov (35). Of the isolates, the 19 from the 1991 Danish outbreak were described elsewhere (32). The remaining 32 (9 from Denmark, 8 from Germany, 6 from Switzerland, 3 from Bulgaria, 3 from Chile, 2 from Sweden, and 1 from Argentina) have not been reported previously.
Of the 48 E. coli O78:H10 human clinical isolates, 20 (including 18 of the 19 Copenhagen outbreak isolates) were from urine and 24 from feces (23 of these patients had diarrhea). Two fecal isolates, presumptively representing one strain (same serogroup and biotype), were from the feces of 30 German neonates from a maternity clinic in 1970. One child had diarrhea; no information was available regarding the others. The poultry isolate was from Denmark and the calf isolate from Germany.
Biotyping, epithelial cell adherence, and mouse virulence.
Biotyping was done using 11 carbohydrates: adonitol, dulcitol, sorbitol, xylose, rhamnose, maltose, salicin, inositol, lactose, sucrose, and sorbose (24). One typical outbreak isolate was examined for its pattern of adherence to HEp-2 cells (30) and its lethality in an established mouse model of subcutaneous sepsis, in comparison with positive- and negative-control strains CFT073 and MG1655, respectively (19).
PFGE analysis.
Pulsed-field gel electrophoresis (PFGE) analysis was done according to the PulseNet protocol (43). Pulsotypes were defined at the ≥94% profile similarity level, which approximately corresponds to a ≤3-band difference, suggesting genetic relatedness (47). For isolates from ST10, a dendrogram was inferred according to the unpaired group method within BioNumerics (Applied Maths).
Phylogenetic analysis and MLST.
Classification of major E. coli phylogenetic groups (A, B1, B2, and D) was performed by triplex PCR (6). MLST was performed according to the Achtman scheme (54) using seven housekeeping genes, i.e., adk, fumC, gyrB, icd, mdh, purA, and recA. Procedures, primers, and sequence type (ST) assignment were as described at http://mlst.ucc.ie/mlst/dbs/Ecoli. All seven MLST loci were sequenced for up to two representatives of each pulsotype, as available, since the MLST status of any member of a pulsotype reliably predicts the MLST status of other members of the same pulsotype (J. R. Johnson, unpublished data). Any remaining isolates within multiple-isolate pulsotypes underwent confirmatory sequencing of fumC and adk, which uniformly identified the same alleles at these loci as found in the corresponding pulsotype's full MLST isolate (not shown).
Based on the MLST allele data, minimum-spanning trees were constructed by using eBURST (http://eburst.mlst.net/) (9) (Fig. 1). Clonal groups were defined based on ST. STs differing at only one of seven MLST loci were considered part of the same clonal complex (Table 1). To further elucidate the phylogenetic relationships among the isolates, concatenated MLST sequence data (as directly determined, for most isolates, or partially inferred, for isolates assigned to STs based on pulsotype plus fumC and adk sequence) were aligned by using CLUSTAL-X (48) and then used to infer a phylogram according to maximum parsimony, as implemented within PAUP* (46) (Fig. 2).
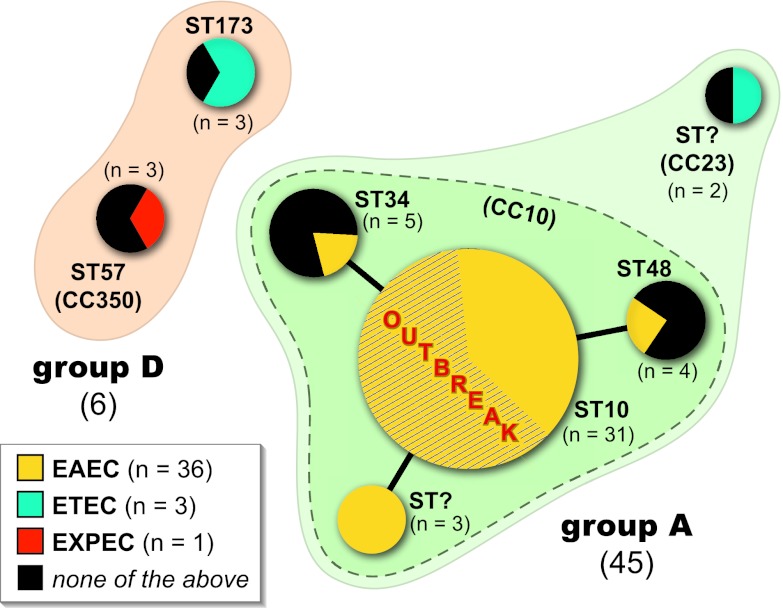
Fig 1.
BURST diagram for 51 O78:H10 Escherichia coli isolates. Each sequence type (ST) is represented by a circle, the size of which is proportional to the number of isolates in the ST. STs belonging to the same clonal complex (CC10) are connected by solid black lines and enclosed within a medium-green-shaded zone with a dashed border. STs belonging to the same major phylogenetic group (group A or D) are enclosed within a pale-tan- or -green-shaded zone with a solid border. Different E. coli pathotypes (EAEC, enteroaggregative E. coli; ETEC, enterotoxigenic E. coli; EXPEC, extraintestinal pathogenic E. coli) are color coded within each ST circle, with the area of the circle that corresponds to coding for a particular pathotype being proportional to the number of isolates of that pathotype within the ST. The 19 Copenhagen outbreak isolates (ST10) are indicated by diagonal hash marks.
Table 1.
Distribution by sequence type, clonal complex, and phylogenetic group of alleles at seven housekeeping gene loci among 51 isolates of Escherichia coli O78:H10a
| ST | CC | Phylogenetic group | No. of isolates | Allele at indicated locusb |
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| adk | fumC | gyrB | icd | mdh | purA | recA | ||||
| ST10c | 10 | A | 31c | 10 | 11 | 4 | 8 | 8 | 8 | 2 |
| ST34 | 10 | A | 5 | 10 | 11 | 4 | 1 | 8 | 8 | 2 |
| ST48 | 10 | A | 4 | 6 | 11 | 4 | 8 | 8 | 8 | 2 |
| Novel | 10 | A | 3 | 10 | 11 | 4 | 8 | 8 | 2 | 2 |
| Novel | 23 | A | 2 | 6 | 11 | 12 | 1 | 20 | 13 | 7 |
| ST57 | 350 | D | 3 | 6 | 31 | 5 | 28 | 1 | 1 | 2 |
| ST173 | None | D | 3 | 6 | 6 | 15 | 16 | 42 | 46 | 7 |
ST, sequence type; CC, clonal complex.
Alleles are numbered according to the Achtman system (http://mlst.ucc.ie/mlst/dbs/Ecoli). Allele numbers shown in bold correspond to ST10 (top row). For 36 isolates, all loci were sequenced. For 15 of the presumptive ST10 isolates (all from a pulsotype containing an ST10 isolate according to full multilocus sequence typing), two loci were sequenced, i.e., adk and fumC.
The 19 Copenhagen outbreak isolates fell within ST10.
Fig 2.
Phylogram and associated bacterial characteristics and source data among 51 isolates of Escherichia coli O78:H10. The phylogram is based on maximum-parsimony analysis of the concatenated multilocus sequence data, with sequence type (ST) ST173 used as the outgroup. The scale represents numbers of nucleotide changes. Horizontal dashed lines separate different STs. Gray shading indicates the 19 Copenhagen outbreak isolates. Red text indicates isolate C555-91, the outbreak isolate used in the adherence assay and mouse sepsis model. Colored squares, presence of virulence genes (color coded by functional category or pathotype), as defined in Table 2. Asterisks identify the presumptive ST10 isolates for which ST status was inferred based on pulsotype and sequence of fumC and adk. Pathotype abbreviations: EAEC, enteroaggregative E. coli; ETEC, enterotoxigenic E. coli; EXPEC, extraintestinal pathogenic E. coli. DEC, diarrheagenic E. coli. Phylotype, major E. coli phylogenetic group. Blank spaces for “host” and “sample type” reflect missing data. The biotypes were negative for adonitol, sorbose, salicin, inositol, and sucrose and positive for dulcitol, sorbitol, xylose, rhamnose, maltose, and lactose, except as follows: A1, rhamnose and lactose negative; A2, rhamnose negative; C, salicin and sucrose positive; D, dulcitol negative and sucrose positive; E, dulcitol negative and salicin positive; F, sorbose positive; G, dulcitol negative; and H, salicin positive.
Virulence genotyping.
Isolates were tested for various extraintestinal and diarrheagenic virulence genes by three different PCR methods, comprising a wide array of virulence markers (n = 61) (Table 2). Testing was done in duplicate using independently prepared boiled lysates of each isolate, together with appropriate positive and negative controls.
Table 2.
Prevalence of significantly distributed virulence genes among 51 outbreak and nonoutbreak isolates of Escherichia coli O78:H10
| Pathotypec | Gened | No. (%) of genesa |
P value,b outbreak isolates vs: |
||||
|---|---|---|---|---|---|---|---|
| Total (n = 51) | Outbreak isolates (n = 19) | Nonoutbreak isolates |
|||||
| All (n = 32) | ST10 only (n = 12) | All nonoutbreak isolates | ST10 nonoutbreak isolates | ||||
| ExPEC | traT | 25 (49) | 17 (90) | 8 (25) | 4 (33) | <0.001 | 0.004 |
| iutA | 31 (61) | 17 (90) | 14 (44) | 8 (67) | 0.002 | ||
| EAEC | aaiC | 34 (67) | 18 (95) | 16 (50) | 11 (92) | 0.002 | |
| aatA | 32 (63) | 18 (95) | 14 (44) | 10 (83) | <0.001 | ||
| aap | 32 (63) | 18 (95) | 14 (44) | 10 (83) | <0.001 | ||
| agg3-agg4C | 13 (26) | 0 (0) | 13 (41) | 3 (25) | 0.0015 | ||
| aggA | 31 (61) | 19 (100) | 12 (38) | 9 (75) | <0.001 | ||
| aggR | 33 (65) | 19 (100) | 14 (44) | 10 (83) | <0.001 | ||
| ORF3 | 32 (63) | 18 (95) | 14 (44) | 9 (75) | <0.001 | ||
| ORF61 | 32 (63) | 19 (100) | 13 (41) | 9 (75) | <0.001 | ||
| Both | astA | 14 (28) | 0 (0) | 14 (44) | 9 (75) | <0.001 | <0.001 |
| sat | 31 (61) | 19 (100) | 12 (38) | 10 (83) | <0.001 | ||
| pic | 31 (61) | 18 (95) | 13 (41) | 8 (67) | <0.001 | ||
Only data for virulence genes that yielded P < 0.05 in at least one comparison are shown. Percent values represent comparisons to column totals.
P values, determined by Fisher's (2-tailed) exact test, are shown where P < 0.05.
ExPEC, extraintestinal pathogenic E. coli; EAEC, enteroaggregative E. coli.
Traits shown in the table correspond to the following genes: aatA (ABC transporter), aaiC (type VI secretion system), aap (dispersin transporter protein), agg3-agg4C (usher for AAF/III and IV), aggA (encodes AFF/I; colonic mucosa adhesin and hemagglutinin), aggR (master regulator), astA (enteroaggregative heat-stable enterotoxin [EAST-1]), iutA (aerobactin receptor), ORF3 (putative ORF3-ORF4a coregulated two-gene cluster with homology to isoprenoid synthesis genes), ORF61 (putative contact hemolysis of erythrocytes), pic (serine protease autotransporter), sat (secreted autotransporter toxin), and traT (serum resistance associated). Traits detected in ≥1 isolate each but not yielding a significant between-group difference (definition [overall prevalence]): air (aggregation and adherence, EAEC [10%]), capU (hexosyltransferase homologue, EAEC [92%]), cvaC (microcin V, ExPEC [2%]), fyuA (yersiniabactin receptor, ExPEC [80%]), hlyF (hemolysin F, ExPEC [2%]), hra (heat-resistant agglutinin, ExPEC [6%]), iha (adhesin-siderophore, ExPEC [2%]), iroN (siderophore receptor, ExPEC [4%]), iss (increased serum survival, ExPEC [2%]), LT1 (heat-labile I enterotoxin, characteristic of enterotoxigenic E. coli [ETEC] [6%]), ompT (outer membrane protease T, ExPEC [8%]), papAH, papC (P fimbriae operon), sepA (extracellular protein [S. flexneri, EAEC [2%]), tsh (temperature-sensitive hemagglutinin, ExPEC [2%), and vat (vacuolating autotransporter, ExPEC [4%). Traits screened for but not detected in any isolate: aafA (AAF/II; colonic mucosa adherence, EAEC), aafC (AAF/II usher, EAEC), afa/dra (Dr family adhesins, ExPEC), agg3A (AAF/III; hemagglutinin, EAEC), agg4A (AAF/IV; colonic mucosa adherence, EAEC), bmaE (M fimbriae, ExPEC), cdtB (cytolethal distending toxin, ExPEC), clpG (fimbrial adhesin CS31A, ExPEC), cnf1 (cytotoxic necrotizing factor, ExPEC), eae (enterocyte attachment and effacement, EPEC and A/EEC), fliC (H7 flagellin, ExPEC), focG (F1C fimbriae, ExPEC), gafD (G fimbriae, ExPEC), hlyD (hemolysin, ExPEC), ibeA (invasion of brain endothelium, ExPEC), ipah (the invasion plasmid antigen H gene sequence, enteroinvasive E. coli [EIEC] and Shigella spp.), ireA (siderophore receptor, ExPEC), kpsMII (group 2 capsule, ExPEC), malX (pathogenicity island marker, ExPEC), pap (P fimbriae operon including papEG and papG alleles I, II, and III; ExPEC), rfc (O4 lipopolysaccharide, ExPEC), sfa/focDE (S and F1C fimbriae, ExPEC), sfaS (S fimbriae, ExPEC), sigA (protease-like homologue, EAEC), AAF/II-IV (aggregative adherence fimbria, EAEC), stx1 and stx2 (toxins of Shiga toxin-producing E. coli, STEC), ST1 (heat-stable I enterotoxin, enterotoxigenic E. coli, ETEC), and usp (uropathogenic-specific protein, ExPEC).
First, established multiplex PCR assays were used to detect 35 markers associated with ExPEC (Table 2) (14, 18, 20, 22). Based on epidemiological and in vivo experimental data, isolates were regarded as ExPEC if positive for ≥2 of 5 classic ExPEC-defining traits, including the presence of papA and/or papC (P fimbriae; counted as one), sfa-foc (S and F1C fimbriae), afa-dra (Dr-binding adhesins), kpsMII (group 2 capsule), and iutA (aerobactin system) (17).
Second, real-time PCR (Light Cycler) was used to detect the following six diarrhea-associated genes (41, 42): stx1 and stx2, characteristic of Shiga toxin-producing E. coli (STEC); eltI and estA (Heat-Labile I and Heat-Stable I enterotoxin), both characteristic of enterotoxigenic E. coli (ETEC); eae, characteristic of enteropathogenic E. coli (EPEC) and attaching and effacing E. coli (A/EEC) (13); and ipaH, characteristic of both enteroinvasive E. coli (EIEC) and Shigella spp. ipaH was detected using the following primer and probes: primers IpaHF (5′-TGGAAAAACTCAGTGCCTCT-3′) and IpaHR (5′-CCAGTCCGTAAATTCATTCT-3′) and probes IpaH-HP-1 (GATAAAGTCAGAACTCTCCATTTTGT-FL) and IpaH-HP-2 ([Red 640]-GATGAGATAGAAGTCTACCTGGCCT-[Ph]). Third, four multiplex PCR assays (3) were used to screen for 20 enteroaggregative E. coli (EAEC)-associated putative virulence genes (aaiC, aafA, aafC, aap, aatA, agg3A, agg4A, agg3-agg4C, aggA, aggR, air, astA, capU, ORF3, ORF61, pet, pic, sat, sepA, and sigA (3). Strains exhibiting ≥1 of aggR, aatA, and aaiC were considered to fulfill molecular criteria for EAEC (4, 5, 8).
The number of virulence markers detected was the virulence gene score. In the MLST-based phylogram, the isolates within each ST were sorted based on their pairwise similarity relationships according to extended virulence profiles (i.e., the presence or absence of all detected markers), using BioNumerics. The virulence genes were then displayed adjacent to the phylogram to illustrate their clonal distribution (Fig. 2).
Susceptibility testing.
Susceptibility to 11 antimicrobial agents (ampicillin, amdinocillin, cefuroxime, gentamicin, tetracycline, chloramphenicol, ciprofloxacin, trimethoprim, nitrofurantoin, streptomycin, and sulfonamides) was determined on 5% Danish blood agar (Statens Serum Institut) using Neo-Sensitabs, according to the instructions of the manufacturer (Rosco). Intermediate isolates were analyzed as resistant. Multiresistance was defined as resistance to ≥5 antimicrobial agents. The number of resistance markers was the resistance score.
Statistical methods.
Comparisons were tested using Fisher's (2-tailed) exact test for proportions and the Mann-Whitney (2-tailed) U test for scores. Statistical significance was defined as P < 0.05.
RESULTS
Phylogenetic group and MLST.
PCR-based phylotyping placed 45 (88%) of the 51 E. coli O78:H10 isolates, including all 19 Copenhagen outbreak isolates, within phylogenetic group A, and 6 (12%) within group D (Table 1 and Fig. 1). According to MLST, one large clonal group, corresponding to ST10, accounted for 31 (69%) of the group A isolates (61% of isolates overall) and included all 19 outbreak isolates. The remaining 20 isolates were divided among six smaller clonal groups (STs), each accounting for between 2 and 5 isolates. Of the 6 non-ST10 STs, 3 were single-locus variants of ST10 and so part of CC10, whereas 1 was from group A but not CC10 and 2 were from separate clonal complexes within group D.
Phylogram.
Phylogenetic relationships among the isolates and their respective STs were visualized by constructing a phylogram from the concatenated MLST sequence data, which allowed associated source data and bacterial characteristics to be displayed within a phylogenetic framework (Fig. 2). In the phylogram, the CC10 isolates formed a fairly tight cluster that included ST10, relative to which the ST48 isolates (group A, non-CC10) were nearest neighbors and the isolates from the two group D STs were outliers.
Locale, year, and host.
Four of the STs were geographically heterogeneous (Fig. 2). The three homogeneous exceptions included ST34 (Denmark, n = 5) and the two novel STs (CC10 [Germany; n = 3] and CC23 [Chile; n = 2]). The three earliest isolates, all collected from humans in Switzerland in 1956, were from ST48 (n = 1) and ST10 (n = 2). The poultry isolate was from Denmark (1981) and corresponded to ST350 (group D), whereas the calf isolate was from Germany (1964) and corresponded to a novel ST within CC10 (group A).
Biotyping and adherence.
The nine observed biotypes differed only with respect to fermentation of rhamnose, dulcitol, salicin, sucrose, and sorbose (among 12 carbohydrates) (Fig. 2). Rhamnose-negative biotypes (A1 and A2) predominated, accounting for 36 isolates (71%) and occurring throughout the study period. Partial phylogenetic segregation of biotypes was evident (Fig. 2). Notably, the 19 Copenhagen outbreak isolates all corresponded to biotype A1, which categorically differentiated them from all other isolates (0%; P < 0.001).
Virulence genotypes and phenotypes.
The 34 virulence genes that were detected in ≥1 isolate also exhibited partial phylogenetic segregation (Fig. 2). Notably, the 19 ST10 outbreak isolates had highly homogeneous virulence profiles that included the following genes (prevalences): fimH, fyuA, sat, aatA, aggR, aggA, and ORF61 (100% each); aaiC, aap, ORF3, and pic (95% each); and traT and iutA (90% each) (Table 2 and Fig. 2).
Despite 18 of the 19 outbreak isolates being from urine, none fulfilled the operational molecular criterion for ExPEC, since their only detected ExPEC-defining gene was iutA. (The study's only ExPEC-qualifying isolate according to this molecular criterion was a nonoutbreak isolate from ST57 [group D] that contained both papAC and sfa/foc [Fig. 1 and 2]). However, in addition to iutA, the outbreak isolates contained fyuA and traT (associated with ExPEC) plus sat and pic (associated with both ExPEC and EAEC). Moreover, in the mouse subcutaneous sepsis model, a typical outbreak isolate (C555-91) was highly lethal, killing 10/10 challenged mice within 48 h, compared with 0/10 for negative-control strain MG1655 (P < 0.001) and 10/10 for positive-control strain CFT073 (P > 0.10).
In contrast, all 19 outbreak isolates exhibited the presence of ≥1 of aggR, aatA, and aaiC, with 18 of 19 outbreak isolates having all three genes, thereby qualifying molecularly as EAEC, compared with only 17 (53%) of the nonoutbreak O78:H10 isolates (P < 0.001) (Table 2 and Fig. 1 and 2). Consistent with this, typical outbreak isolate C555-91 exhibited classic “stacked brick” adherence to cultured epithelial cells, the defining phenotype of EAEC (not shown).
Another 17 isolates also qualified molecularly as EAEC. All were from group A, 12 (71%) were from ST10 proper, and 13 (76%) exhibited biotype A2 (Fig. 1 and 2). Thus, the study's total of 36 EAEC isolates (71% of all isolates) were significantly concentrated within (i) group A (80% EAEC versus 0% of group D isolates, P = 0.001); (ii) ST10 (100% EAEC versus 25% for non-ST10 isolates, P < 0.001); and (iii) biotypes A1 and A2 (92% EAEC versus 20% for other biotypes, P < 0.001).
Overall, the outbreak isolates differed significantly from nonoutbreak isolates with respect to the prevalence of numerous virulence genes, including EAEC-associated genes, with most such differences favoring the outbreak isolates (Table 2 and Fig. 2). Accordingly, the median virulence gene score for the outbreak isolates was 16 (range, 12 to 16) versus only 9 (range, 2 to 19) for nonoutbreak isolates (P = 0.002). Although virulence gene scores did not differ significantly between outbreak and nonoutbreak isolates within ST10, the range of scores was much smaller among outbreak isolates (i.e., 12 to 16 for outbreak versus 4 to 19 for nonoutbreak isolates), consistent with these isolates being more genetically homogeneous.
As for non-EAEC diarrheagenic pathotypes, three nonoutbreak isolates (two from feces, one of unknown origin; from non-CC10 STs from groups A and D) represented enterotoxigenic E. coli (ETEC), since they contained eltI (Fig. 1 and 2). No isolate exhibited genes associated with other E. coli diarrheal pathotypes.
Pulsotypes.
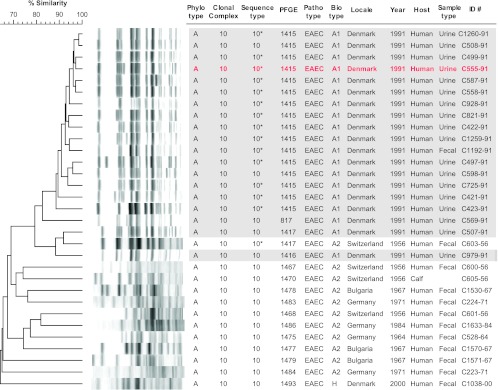
PFGE analysis of the 51 study isolates resolved 35 distinct pulsotypes, as defined at the 94% similarity level, each of which was homogeneous according to ST (not shown). In a PFGE-based dendrogram based on just the ST10 isolates, the 19 Copenhagen outbreak isolates clustered together at approximately the 79% similarity level (Fig. 3). Sixteen (84%) of the outbreak isolates, including the sole fecal isolate (C1192-91), shared the same pulsotype (1415), whereas, of the remaining three, each represented a distinct pulsotype closely related to pulsotype 1415. The only interposed nonoutbreak isolate was a 1956 human fecal isolate from Switzerland (C603-56), which shared pulsotype 1417 with an outlier outbreak isolate. Although this isolate's virulence gene profile resembled that of the outbreak isolates (Fig. 2), its biotype was different (A2 rather than A1) (Fig. 2 and 3).
Fig 3.
XbaI pulsed-field gel electrophoresis profiles for 31 Escherichia coli O78:H10 isolates from sequence type ST10. The tree was inferred according to the unweighted pair group method, using BioNumerics. Gray shading indicates the 19 Copenhagen outbreak isolates. Red text indicates outbreak isolate C555-91, which was used in the adherence assay and mouse sepsis model. Asterisks identify the presumptive ST10 isolates for which ST status was inferred based on pulsotype and sequence of fumC and adk. The 19 Copenhagen outbreak isolates cluster together at approximately the 79% similarity level, with a single interposed nonoutbreak isolate.
Antimicrobial susceptibility.
Of the 51 study isolates, 39 (76%) exhibited resistance to from 1 to 7 (median, 5) of the 10 tested antimicrobial agents, and 19 (37%) were multiresistant (resistant to ≥5 agents); only 24% were fully susceptible. Likewise, resistance to all agents except aminoglycosides, cephalosporins, and fluoroquinolones, was detected. All 19 outbreak isolates (100%) were multiresistant versus 0% of other isolates (P < 0.001), with a median resistance score of 6 versus only 1 for the nonoutbreak isolates (P < 0.001). The signature resistance profile ACSSuTTp occurred in all 19 outbreak isolates versus none of the nonoutbreak isolates (P < 0.001).
DISCUSSION
In this study, we investigated the characteristics of 51 E. coli archival O78:H10 isolates, including 48 human clinical isolates, 1 calf isolate, and 1 poultry isolate, from seven countries, as submitted to the SSI from 1956 through 2000. We sought to determine whether the 1991 Copenhagen multiresistant UTI outbreak (32) was caused by a single clonal strain that fulfills molecular criteria for ExPEC (17), exhibits distinctive virulence-associated traits in comparison with other O78:H10 isolates, has an identifiable food animal source, and has occurred in other locales.
Supporting our clonality hypothesis, we found that 69% of the O78:H10 isolates, including the 19 Copenhagen outbreak isolates, represented a single clonal group within phylogenetic group A, corresponding to ST10. The remaining 15 O78:H10 isolates belonged to six other STs, including 4 within phylogenetic group A (3 being single-locus variants of ST10) and 2 within group D. Moreover, within ST10 proper, the outbreak isolates were distinct from, and more highly homogeneous than, nonoutbreak isolates with respect to resistance profile (more extensive [uniformly ACSSuTTp]), biotype (A1), and PFGE profiles (79% similarity). Thus, the Copenhagen outbreak isolates clearly represented a single clonal strain within ST10, whereas E. coli serotype O78:H10 overall is highly clonally diverse.
Unexpectedly, 100% of the present ST10 O78:H10 isolates, including all 19 Copenhagen outbreak isolates, fulfilled molecular criteria for the diarrheagenic pathotype EAEC, but not for ExPEC, despite containing several ExPEC-associated virulence genes (as discussed below). Additionally, a representative EAEC outbreak isolate (C555-91) exhibited “stacked brick” adherence to cultured epithelial cells, the classic phenotype of EAEC. The finding of EAEC-defining characteristics in the Copenhagen outbreak clone conflicts with those of previously described multiresistant ExPEC clonal groups, including CGA (ST69, group D), E. coli O15:K52:H1 (CC31, group D), and O25:H4 (ST131, group B2), members of which rarely if ever contain EAEC-defining or other diarrheagenic markers (unpublished data). It also conflicts with the conventional understanding that the E. coli strains that cause diarrhea are distinct from those that cause extraintestinal disease, both phylogenetically and according to their discrete repertoires of syndrome-specific virulence genes (23).
Regarding ST10 and EAEC, ST10 is known to be pathotypically heterogeneous (31), comprising EAEC, ETEC (31, 50, 54), ExPEC, and commensal E. coli (2, 49). EAEC is a very common diarrheal pathotype, occurring in multiple lineages (31). Its pathogenic potential is illustrated by volunteer studies, outbreaks, and other epidemiological data (11, 12, 28). Additionally, the 2011 European outbreak of Shiga toxin-producing EAEC highlights the epidemic potential of EAEC (39). However, identification of truly pathogenic EAEC strains remains elusive. Okeke et al. found that ST10 was the most common ST complex among Nigerian children with EAEC, that ST10 was significantly overrepresented among EAEC compared to all other isolates in the database (P < 0.001), and that EAEC ST10 was associated with diarrhea among children 10 months of age or older (31). It is conceivable that the O78:H10 Copenhagen UTI outbreak strain represents one of the ST10 EAEC clones described by Okeke et al., despite the Copenhagen outbreak involving UTI rather than diarrhea.
Whether the present O78:H10 ST10 EAEC isolates can cause diarrhea is unknown. However, 12 of these 13 isolates were recovered from the feces of patients with diarrhea. Moreover, one such isolate originated from an AIDS patient with diarrhea, consistent with the ability of EAEC to cause persistent diarrhea in HIV-infected patients (10). Interestingly, two O78:H10 EAEC fecal isolates from Brazilian children participating in a case-control study of diarrhea were reported in 2005 (51), although both children were in the control group (W. P. Elias, personal communication).
As for the true extraintestinal virulence capability (i.e., ExPEC status) of the present Copenhagen outbreak isolates, three lines of evidence are indicative. First, the isolates were recovered from the urine of patients with symptomatic UTI, implying pathogenicity on epidemiological grounds. Second, they contained five ExPEC-associated virulence genes (fyuA, iutA, pic, traT, and sat). Although only one of these is part of an established molecular definition of ExPEC that requires the presence of ≥2 of papA (and/or papC), sfa-foc, afa-dra, iutA, and kpsMII for an isolate to be classified operationally as ExPEC, this definition provides only an approximate indication of an isolate's extraintestinal virulence potential, as determined experimentally (15, 19). Third, and perhaps most tellingly, a typical outbreak isolate was highly lethal in a mouse model of subcutaneous sepsis. Taken together, these observations suggest that the outbreak clone indeed does represent ExPEC.
We suspect that the outbreak strain's known ExPEC-associated virulence genes, although not fulfilling the standard molecular epidemiological criteria for ExPEC, nonetheless contribute to extraintestinal virulence. Likewise, certain of the strain's EAEC-associated genes also may contribute to its extraintestinal virulence. This concept is supported by in vitro assays (55) demonstrating that (EAEC-associated) aggregative adherence fimbriae variant type 1 (AAF/I) encoded by the aggA gene might facilitate uroepithelial cell adherence (29).
Regarding the commonality of virulence traits between ExPEC and diarrheagenic E. coli, classic uropathogenic traits among certain EAEC fecal isolates have been described previously. Yamamoto et al. reported an EAEC O127a:H2 fecal isolate from a child with diarrhea that adhered avidly to human urethral mucosa, suggesting that its adhesin(s) is related to those of uropathogenic E. coli (UPEC) (55). Likewise, Suzart et al. reported four fecal EAEC isolates, from three patients with and one without diarrhea, carrying hly and pap, classic ExPEC-associated virulence genes (45). Diarrheagenic E. coli and UPEC strains were comparatively characterized in a study from 2010 (40). Amplification assays revealed that 45% and 22% of EAEC and EPEC strains, respectively, carried at least one of the urovirulence sequences. Additionally, among EAEC strains from Nigeria, 10 independent antimicrobial-resistant isolates belonged to the ST69 clonal complex, which includes uropathogenic E. coli “clonal group A” (53).
Conversely, EAEC characteristics among urine isolates from children with UTI have been documented by Park et al. (36). Specifically, Park et al. searched for two EAEC genes (aggR and aap) among E. coli isolates from suprapubic urine specimens of 22 young children with febrile UTI. Nine isolates (41%) carried aggR, the master regulator of EAEC (8). Likewise, the presence of set1 encoding Shigella enterotoxin 1 has been demonstrated in uropathogenic E. coli (44). Finally, Abe et al. found 16 EAEC isolates among 225 uropathogenic E. coli isolates (1), as well as the genes astA, ehly (enterohemolysin), eae, iha, irp2, pet, pic, pilS, and shf, all characteristic of diarrheagenic E.coli.
We were not able to identify a possible food animal source for the Copenhagen outbreak. The single poultry isolate belonged to a different clonal complex than the outbreak isolates, and the calf isolate belonged to an undefined single-locus variant of ST10. Although this isolate's virulence profile resembled that of the outbreak isolates, including the EAEC genes, the isolate differed from the outbreak isolates according to astA status (positive), biotype (D), and PFGE profile (data not shown). Thus, although a potential food or animal origin has been proposed for certain human-associated, multidrug-resistant uropathogenic E. coli clonal groups (26, 38), and a close relationship has been demonstrated between E. coli from retail chicken meat and E. coli causing human UTI (52), no candidate animal source for the Copenhagen outbreak was identified here.
Although we previously reported finding the estA gene, encoding the ETEC heat-stable enterotoxin, in several (ExPEC) O15:K52:H1 clonal group members (34), to our knowledge eltI has not previously been detected among extraintestinal E. coli isolates. Heat-labile toxin (LT) has, however, been demonstrated in several E. coli isolates causing urinary tract infection in pigs (7).
Study strengths include the geographically and temporally diverse set of O78:H10 study isolates, the extensive and complementary phylogenetic, molecular, and phenotypic methods used to characterize them, and the use of in vivo testing to assess the extraintestinal virulence of a typical outbreak isolate. Limitations include the small number of animal isolates, the limited associated clinical data, and the absence of in vivo testing regarding the isolates' diarrheagenic potential.
In conclusion, this report demonstrates that the 1991 Copenhagen O78:H10 outbreak was caused by a fairly genetically and phenotypically homogeneous, human-associated subset within a larger, more broadly disseminated O78:H10 (ST10) clonal group. In contrast, at least six other clonal groups, including 2 from phylogenetic group D, also exhibit serotype O78:H10. Remarkably, 71% of the study isolates, including all 19 outbreak isolates, fulfilled molecular criteria for EAEC. Moreover, an EAEC outbreak isolate exhibited the classic “stacked brick” pattern of adherence to epithelial cells but also contained several ExPEC-associated virulence genes and was highly lethal in a mouse subcutaneous sepsis model. Whether this outbreak strain, and other EAEC isolates that contain urovirulence genes, are both diarrheagenic and extraintestinal pathogenic remains to be determined.
ACKNOWLEDGMENTS
This material is based upon work supported by the Office of Research and Development, Medical Research Service, Department of Veterans Affairs (J.R.J.), and a research grant from Hillerød Hospital.
We thank Susanne Jespersen (Statens Serum Institut) for excellent technical assistance.
Footnotes
Published ahead of print 12 September 2012
REFERENCES
- 1. Abe CM, et al. 2008. Uropathogenic Escherichia coli (UPEC) strains may carry virulence properties of diarrhoeagenic E. coli. FEMS Immunol. Med. Microbiol. 52:397– 406 doi:10.1111/j.1574-695X.2008.00388.x [DOI] [PubMed] [Google Scholar]
- 2. Bert F, et al. 2010. Genetic diversity and virulence profiles of Escherichia coli isolates causing spontaneous bacterial peritonitis and bacteremia in patients with cirrhosis. J. Clin. Microbiol. 48:2709– 2714 doi:10.1128/JCM.00516-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Boisen N, et al. 2012. Genomic characterization of enteroaggregative Escherichia coli from children in Mali. J. Infect. Dis. 205:431– 444 doi:10.1093/infdis/jir757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Boisen N. 2011. Virulence characterization and epidemiology of diarrheagenic enteroaggregative Escherichia coli (EAEC). Ph.D. thesis. Faculty of Health Sciences, University of Copenhagen, Denmark: [Google Scholar]
- 5. Cerna JF, Nataro JP, Estrada-Garcia T. 2003. Multiplex PCR for detection of three plasmid-borne genes of enteroaggregative Escherichia coli strains. J. Clin. Microbiol. 41:2138– 2140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Clermont O, Bonacorsi S, Bingen E. 2000. Rapid and simple determination of the Escherichia coli phylogenetic group. Appl. Environ. Microbiol. 66:4555– 4558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. de Brito BG, Leite DS, Linhares RE, Vidotto MC. 1999. Virulence-associated factors of uropathogenic Escherichia coli strains isolated from pigs. Vet. Microbiol. 65:123– 132 [DOI] [PubMed] [Google Scholar]
- 8. Dudley EG, Thomson NR, Parkhill J, Morin NP, Nataro JP. 2006. Proteomic and microarray characterization of the AggR regulon identifies a pheU pathogenicity island in enteroaggregative Escherichia coli. Mol. Microbiol. 61:1267– 1282 doi:10.1111/j.1365-2958.2006.05281.x [DOI] [PubMed] [Google Scholar]
- 9. Feil EJ, Li BC, Aanensen DM, Hanage WP, Spratt BG. 2004. eBURST: inferring patterns of evolutionary descent among clusters of related bacterial genotypes from multilocus sequence typing data. J. Bacteriol. 186:1518– 1530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gassama-Sow A, et al. 2004. Characterization of pathogenic Escherichia coli in human immunodeficiency virus-related diarrhea in Senegal. J. Infect. Dis. 189:75– 78 doi:10.1086/380489 [DOI] [PubMed] [Google Scholar]
- 11. Huang DB, et al. 2006. Enteroaggregative Escherichia coli is a cause of acute diarrheal illness: a meta-analysis. Clin. Infect. Dis. 43:556– 563 doi:10.1086/505869 [DOI] [PubMed] [Google Scholar]
- 12. Itoh Y, Nagano I, Kunishima M, Ezaki T. 1997. Laboratory investigation of enteroaggregative Escherichia coli O untypeable:H10 associated with a massive outbreak of gastrointestinal illness. J. Clin. Microbiol. 35:2546– 2550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Jerse AE, Yu J, Tall BD, Kaper JB. 1990. A genetic locus of enteropathogenic Escherichia coli necessary for the production of attaching and effacing lesions on tissue culture cells. Proc. Natl. Acad. Sci. U. S. A. 87:7839– 7843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Johnson JR, Brown JJ. 1996. A novel multiply primed polymerase chain reaction assay for identification of variant papG genes encoding the Gal(alpha 1–4)Gal-binding PapG adhesins of Escherichia coli. J. Infect. Dis. 173:920– 926 [DOI] [PubMed] [Google Scholar]
- 15. Johnson JR, et al. 2006. Experimental mouse lethality of Escherichia coli isolates, in relation to accessory traits, phylogenetic group, and ecological source. J. Infect. Dis. 194:1141– 1150 doi:10.1086/507305 [DOI] [PubMed] [Google Scholar]
- 16. Johnson JR, et al. 2009. Epidemic clonal groups of Escherichia coli as a cause of antimicrobial-resistant urinary tract infections in Canada, 2002 to 2004. Antimicrob. Agents Chemother. 53:2733– 2739 doi:10.1128/AAC.00297-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Johnson JR, et al. 2003. Isolation and molecular characterization of nalidixic acid-resistant extraintestinal pathogenic Escherichia coli from retail chicken products. Antimicrob. Agents Chemother. 47:2161– 2168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Johnson JR, Owens KL, Clabots CR, Weissman SJ, Cannon SB. 2006. Phylogenetic relationships among clonal groups of extraintestinal pathogenic Escherichia coli as assessed by multi-locus sequence analysis. Microbes Infect. 8:1702– 1713 doi:10.1016/j.micinf.2006.02.007 [DOI] [PubMed] [Google Scholar]
- 19. Johnson JR, Porter SB, Zhanel G, Kuskowski MA, Denamur E. 2012. Virulence of Escherichia coli clinical isolates in a murine sepsis model in relation to sequence type ST131 status, fluoroquinolone resistance, and virulence genotype. Infect. Immun. 80:1554– 1562 doi:10.1128/IAI.06388-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Johnson JR, Stell AL. 2000. Extended virulence genotypes of Escherichia coli strains from patients with urosepsis in relation to phylogeny and host compromise. J. Infect. Dis. 181:261– 272 doi:10.1086/315217 [DOI] [PubMed] [Google Scholar]
- 21. Johnson JR, et al. 2002. Global molecular epidemiology of the O15:K52:H1 extraintestinal pathogenic Escherichia coli clonal group: evidence of distribution beyond Europe. J. Clin. Microbiol. 40:1913– 1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Johnson JR, et al. 2000. Analysis of the F antigen-specific papA alleles of extraintestinal pathogenic Escherichia coli using a novel multiplex PCR-based assay. Infect. Immun. 68:1587– 1599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kaper JB, Nataro JP, Mobley HL. 2004. Pathogenic Escherichia coli. Nat. Rev. Microbiol. 2:123– 140 doi:10.1038/nrmicro818 [DOI] [PubMed] [Google Scholar]
- 24. Kauffmann F. 1966. The bacteriology of enterobacteriaceae: collected studies of the author and his co-workers. Munksgaard (pr Aarhuus), Cophenhagen, Denmark: [Google Scholar]
- 25. Lau SH, et al. 2008. UK epidemic Escherichia coli strains A-E, with CTX-M-15 beta-lactamase, all belong to the international O25:H4-ST131 clone. J. Antimicrob. Chemother. 62:1241– 1244 doi:10.1093/jac/dkn380 [DOI] [PubMed] [Google Scholar]
- 26. Manges AR, et al. 2007. Retail meat consumption and the acquisition of antimicrobial resistant Escherichia coli causing urinary tract infections: a case-control study. Foodborne Pathog. Dis. 4:419– 431 doi:10.1089/fpd.2007.0026 [DOI] [PubMed] [Google Scholar]
- 27. Manges AR, Tabor H, Tellis P, Vincent C, Tellier PP. 2008. Endemic and epidemic lineages of Escherichia coli that cause urinary tract infections. Emerg. Infect. Dis. 14:1575– 1583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Nataro JP, et al. 1995. Heterogeneity of enteroaggregative Escherichia coli virulence demonstrated in volunteers. J. Infect. Dis. 171:465– 468 [DOI] [PubMed] [Google Scholar]
- 29. Nataro JP, et al. 1992. Aggregative adherence fimbriae I of enteroaggregative Escherichia coli mediate adherence to HEp-2 cells and hemagglutination of human erythrocytes. Infect. Immun. 60:2297– 2304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Okeke IN, Nataro JP. 2001. Enteroaggregative Escherichia coli. Lancet Infect. Dis. i:304– 313 doi:10.1016/S1473-3099(01)00144-X [DOI] [PubMed] [Google Scholar]
- 31. Okeke IN, et al. 2010. Multi-locus sequence typing of enteroaggregative Escherichia coli isolates from Nigerian children uncovers multiple lineages. PLoS One 5:e14093 doi:10.1371/journal.pone.0014093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Olesen B, Kolmos HJ, Orskov F, Orskov I. 1994. Cluster of multiresistant Escherichia coli O78:H10 in greater Copenhagen. Scand. J. Infect. Dis. 26:406– 410 [DOI] [PubMed] [Google Scholar]
- 33. Olesen B, Kolmos HJ, Orskov F, Orskov I. 1995. A comparative study of nosocomial and community-acquired strains of Escherichia coli causing bacteraemia in a Danish University Hospital. J. Hosp. Infect. 31:295– 304 [DOI] [PubMed] [Google Scholar]
- 34. Olesen B, et al. 2009. Three-decade epidemiological analysis of Escherichia coli O15:K52:H1. J. Clin. Microbiol. 47:1857– 1862 doi:10.1128/JCM.00230-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ørskov F, Ørskov I. 1984. Serotyping of Escherichia coli, p 43–112 In Bergan T. (ed), Methods in microbiology, vol. 14 Academic Press, London, United Kingdom [Google Scholar]
- 36. Park HK, et al. 2009. Comparison of Escherichia coli uropathogenic genes (kps, usp and ireA) and enteroaggregative genes (aggR and aap) via multiplex polymerase chain reaction from suprapubic urine specimens of young children with fever. Scand. J. Urol. Nephrol. 43:51– 57 doi:10.1080/00365590802299338 [DOI] [PubMed] [Google Scholar]
- 37. Phillips I, et al. 1988. Epidemic multiresistant Escherichia coli infection in West Lambeth Health District. Lancet i:1038– 1041 [DOI] [PubMed] [Google Scholar]
- 38. Ramchandani M, et al. 2005. Possible animal origin of human-associated, multidrug-resistant, uropathogenic Escherichia coli. Clin. Infect. Dis. 40:251– 257 doi:10.1086/426819 [DOI] [PubMed] [Google Scholar]
- 39. Rasko DA, et al. 2011. Origins of the E. coli strain causing an outbreak of hemolytic-uremic syndrome in Germany. N. Engl. J. Med. 365:709– 717 doi:10.1056/NEJMoa1106920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Regua-Mangia AH, et al. 2010. Molecular characterization of uropathogenic and diarrheagenic Escherichia coli pathotypes. J. Basic Microbiol. 50(Suppl 1):S107–S115 doi:10.1002/jobm.200900364 [DOI] [PubMed] [Google Scholar]
- 41. Reischl U, et al. 2002. Real-time fluorescence PCR assays for detection and characterization of Shiga toxin, intimin, and enterohemolysin genes from Shiga toxin-producing Escherichia coli. J. Clin. Microbiol. 40:2555– 2565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Reischl U, Youssef MT, Wolf H, Hyytia-Trees E, Strockbine NA. 2004. Real-time fluorescence PCR assays for detection and characterization of heat-labile I and heat-stable I enterotoxin genes from enterotoxigenic Escherichia coli. J. Clin. Microbiol. 42:4092– 4100 doi:10.1128/JCM.42.9.4092-4100.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ribot EM, et al. 2006. Standardization of pulsed-field gel electrophoresis protocols for the subtyping of Escherichia coli O157:H7, Salmonella, and Shigella for PulseNet. Foodborne Pathog. Dis. 3:59– 67 doi:10.1089/fpd.2006.3.59 [DOI] [PubMed] [Google Scholar]
- 44. Soto SM, Guiral E, Bosch J, Vila J. 2009. Prevalence of the set-1B and astA genes encoding enterotoxins in uropathogenic Escherichia coli clinical isolates. Microb. Pathog. 47:305– 307 doi:10.1016/j.micpath.2009.09.007 [DOI] [PubMed] [Google Scholar]
- 45. Suzart S, Guth BE, Pedroso MZ, Okafor UM, Gomes TA. 2001. Diversity of surface structures and virulence genetic markers among enteroaggregative Escherichia coli (EAEC) strains with and without the EAEC DNA probe sequence. FEMS Microbiol. Lett. 201:163– 168 [DOI] [PubMed] [Google Scholar]
- 46. Swofford DL. 2002. PAUP*: phylogenetic analysis using parsimony (and other methods). Sinauer Associates, Sunderland, MA [Google Scholar]
- 47. Tenover FC, et al. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233– 2239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876– 4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Trobos M, et al. 2009. Characterization of sulphonamide-resistant Escherichia coli using comparison of sul2 gene sequences and multilocus sequence typing. Microbiology 155:831– 836 doi:10.1099/mic.0.024190-0 [DOI] [PubMed] [Google Scholar]
- 50. Turner SM, et al. 2006. Phylogenetic comparisons reveal multiple acquisitions of the toxin genes by enterotoxigenic Escherichia coli strains of different evolutionary lineages. J. Clin. Microbiol. 44:4528– 4536 doi:10.1128/JCM.01474-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Uber AP, et al. 2006. Enteroaggregative Escherichia coli from humans and animals differ in major phenotypical traits and virulence genes. FEMS Microbiol. Lett. 256:251– 257 doi:10.1111/j.1574-6968.2006.00124.x [DOI] [PubMed] [Google Scholar]
- 52. Vincent C, et al. 2010. Food reservoir for Escherichia coli causing urinary tract infections. Emerg. Infect. Dis. 16:88– 95 doi:10.3201/eid1601.091118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Wallace-Gadsden F, Johnson JR, Wain J, Okeke IN. 2007. Enteroaggregative Escherichia coli related to uropathogenic clonal group A. Emerg. Infect. Dis. 13:757– 760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Wirth T, et al. 2006. Sex and virulence in Escherichia coli: an evolutionary perspective. Mol. Microbiol. 60:1136– 1151 doi:10.1111/j.1365-2958.2006.05172.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Yamamoto T, Endo S, Yokota T, Echeverria P. 1991. Characteristics of adherence of enteroaggregative Escherichia coli to human and animal mucosa. Infect. Immun. 59:3722– 3739 [DOI] [PMC free article] [PubMed] [Google Scholar]