Abstract
A novel eSensor respiratory viral panel (eSensor RVP) multiplexed nucleic acid amplification test (GenMark Diagnostics, Inc., Carlsbad, CA) was compared to laboratory-developed real-time PCR assays for the detection of various respiratory viruses. A total of 250 frozen archived pediatric respiratory specimens previously characterized as either negative or positive for one or more viruses by real-time PCR were examined using the eSensor RVP. Overall agreement between the eSensor RVP and corresponding real-time PCR assays for shared analytes was 99.2% (kappa = 0.96 [95% confidence interval {CI}, 0.94 to 0.98]). The combined positive percent agreement was 95.4% (95% CI, 92.5 to 97.3); the negative percent agreement was 99.7% (95% CI, 99.4 to 99.8). The mean real-time PCR threshold cycle (CT) value for specimens with discordant results was 39.73 (95% CI, 38.03 to 41.43). Detection of coinfections and correct identification of influenza A virus subtypes were comparable between methods. Of note, the eSensor RVP rhinovirus assay was found to be more sensitive and specific than the corresponding rhinovirus real-time PCR. In contrast, the eSensor RVP adenovirus B, C, and E assays demonstrated some cross-reactivity when tested against known adenovirus serotypes representing groups A through F. The eSensor RVP is robust and relatively easy to perform, it involves a unique biosensor technology for target detection, and its multiplexed design allows for efficient and simultaneous interrogation of a single specimen for multiple viruses. Potential drawbacks include a slower turnaround time and the need to manipulate amplified product during the protocol, increasing the possibility of contamination.
INTRODUCTION
Acute viral upper and lower respiratory tract infections are responsible for substantial morbidity in both pediatric and adult populations worldwide. These infections can be severe and even fatal, especially in susceptible infants, older adults, patients with compromised immune systems, and individuals with underlying cardiopulmonary diseases. Rapid and accurate laboratory detection of respiratory viruses plays an important role in clinical management, guiding the appropriate prescription of antivirals, reducing the need for additional diagnostic studies and hospital procedures, and limiting the use of unnecessary antibiotics (2, 4, 5, 6, 10, 19, 31, 35). In addition, a virologic diagnosis is instrumental to efforts focused on prevention of health care-associated infections (3, 9, 23, 24).
PCR has demonstrated superior sensitivity for the detection of respiratory viruses over the more conventional laboratory methods of virus culture and rapid direct antigen detection tests (7, 12, 20, 22, 34). Both laboratory-developed and commercial molecular assays are now available, but the overall complexity of most of these tests has made implementation challenging for many clinical laboratories (18). However, significant recent advances in microfluidics, microelectronics, and microfabrication have allowed the development of simplified molecular systems that can be used by all laboratories regardless of their size, resources, or capacity (14, 29, 30, 32, 36). With this study, we compared one such commercial system, the eSensor XT-8 respiratory viral panel (eSensor RVP; GenMark Diagnostics, Inc., Carlsbad, CA), to laboratory-developed real-time PCR assays for the rapid detection of a number of respiratory viruses. To our knowledge, this is the first reported evaluation of the newly developed eSensor RVP.
MATERIALS AND METHODS
Specimens.
A total of 250 archived respiratory specimens from pediatric patients were used, including 239 (95.6%) nasopharyngeal aspirates, 4 (1.6%) nasopharyngeal swabs, 5 (2.0%) tracheal aspirates, and 2 (0.8%) bronchoalveolar lavage specimens. The samples were originally submitted to the Clinical Virology Laboratory at the Children's Hospital of Philadelphia between January 2007 and January 2012, and multiple single-use aliquots of each specimen were immediately stored frozen at −70°C following processing for clinical testing. Patients from whom specimens had been collected ranged in age from 19 days to 22 years, with a median age of 2.5 years; 67.2% of specimens were from patients under 5 years of age. All samples were previously characterized as positive or negative for one or more respiratory viruses using a panel of laboratory-developed real-time PCR assays for adenovirus; respiratory syncytial virus types A and B; influenza virus type A [including subtype determination for seasonal H1 and H3 and (H1N1)pdm09] and influenza virus type B; parainfluenza virus types 1, 2, and 3; human metapneumovirus; and rhinovirus. The individual threshold cycle (CT) values for positive analytes spanned the reportable range of the real-time PCR assays.
In addition to the clinical specimens tested, 20 characterized adenovirus serotypes representing groups A (type 12), B (types 3, 7, 11, 14, 21, and 35), C (types 1, 2, 5, and 6), D (types 10, 19, 20, 29, 32, and 37), E (type 4), and F (types 40 and 41) were obtained from either the American Type Culture Collection (ATCC), Manassas, VA, or Adriana Kajon, Infectious Diseases Program, Lovelace Respiratory Research Institute, Albuquerque, NM. The different serotypes were grown in lung adenocarcinoma epithelial cells (A549; ATCC CCL-185), diluted to achieve targeted real-time PCR CT values of between 25.00 and 27.99, and then tested by the eSensor RVP. Nineteen characterized enterovirus isolates (echovirus types 4, 6, 9, 11, and 30; coxsackievirus types A13, B1, B2, B3, B4, B5, and B6; enterovirus types 68, 69, 70, and 71; and poliovirus types 1, 2, and 3) were also obtained from the ATCC, grown in primary rhesus monkey kidney cells, and then tested by both real-time PCR and the eSensor RVP.
This work was deemed not to be human subject research and was declared to be exempt by the Institutional Review Board at the Children's Hospital of Philadelphia.
Real-time PCR assays.
Nucleic acids were extracted from 200 μl of each clinical specimen by standard procedures using a MagNA Pure LC automated instrument (Roche Diagnostics, Indianapolis, IN) and corresponding Roche total nucleic acid isolation kit. Individual real-time PCR assays were performed in 50-μl volumes on a 7500 real-time PCR system (Life Technologies/Applied Biosystems, Foster City, CA) using 5 μl of eluted nucleic acid, universal master mixes for either RNA (Ambion AgPath-ID One-Step reverse transcription-PCR master mix; Life Technologies/Applied Biosystems) or DNA (TaqMan universal master mix; Life Technologies/Applied Biosystems), and universal amplification conditions consisting of 1 cycle for 10 min at 45°C and 1 cycle for 10 min at 95°C, followed by 45 two-step cycles of 15 s at 95°C and 45 s at 60°C and TaqMan fluorogenic chemistry for detection. Positive and negative controls were processed with each batch of clinical specimens from extraction of nucleic acids through the detection of amplified products. Negative controls consisted of 1.0 × 106 cells/ml of an uninfected human lung carcinoma cell line (A549 cells; ATCC CCL-185), and positive controls were prepared as a mixture of clinical material from previously positive patients. No-template controls were included in each reaction plate for all sets of primers and probes. Primer and probe sequences targeted conserved regions of the genome for each organism and have been previously published (28). A human albumin gene primer and probe set (28) was used in separate PCRs as an internal control to ensure that samples contained nucleic acid and to exclude the presence of inhibitors. Specimens and controls were considered positive when the generated fluorescence signal at the CT exceeded a defined threshold limit. Specimens that reached the threshold before 38 cycles were considered positive without further testing, and those that reached the threshold at or after 38 cycles but before the last of 45 cycles were considered positive only if, upon duplicate repeat testing of separate aliquots of stored original specimen, at least one of the two repeat tests also reached the threshold before 45 cycles. For certain experiments, the quantity of adenovirus DNA or enterovirus RNA was determined by real-time PCR from a standard curve generated using a set of five nucleic acid standards ranging from 108 to 104 copies/ml or 109 to 105 copies/ml, respectively.
eSensor XT-8 instrument and respiratory viral panel.
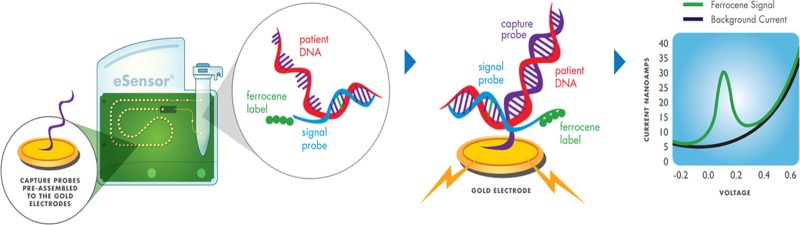
Specimens were tested using the eSensor XT-8 system and corresponding premarket respiratory viral panel kit (GenMark Diagnostics, Inc.) according to the manufacturer's instructions. This panel includes assays for adenovirus groups B, C, and E; coronavirus types 229E, HKU1, OC43, and NL63; influenza A virus (including subtype determination); influenza B virus; human metapneumovirus; parainfluenza virus types 1, 2, 3, and 4; respiratory syncytial virus types A and B; and rhinovirus. Nucleic acids were extracted as described for the real-time PCR assays, but with the addition of 10 μl of bacteriophage MS2 internal control (included in the eSensor RVP kit) to each specimen immediately prior to extraction on the MagNA Pure system. Conventional endpoint PCR assays were performed in 35-μl volumes on a GeneAmp PCR system 9700 (Applied Biosystems) thermal cycler using 5 μl of eluted nucleic acid; kit-supplied multiplex master mix; and amplification conditions consisting of 1 cycle for 30 min at 50°C for reverse transcription and 1 cycle for 15 min at 95°C for initial PCR activation, followed by 40 three-step cycles of 30 s at 94°C, 60 s at 60°C, and 60 s at 72°C for denaturation, annealing, and extension, respectively. After completion of the PCR, 5 μl of exonuclease was added to each reaction mixture and the reaction plate was incubated in the thermal cycler for 20 min at 37°C to create single-stranded DNA by digestion of the double-stranded amplicons, followed by 2 min at 95°C for enzyme inactivation. A total of 100 μl of hybridization buffer containing ferrocene-labeled signal probes specific for the different viral targets was then added to each reaction mixture; if present, target DNA immediately hybridizes to the corresponding signal probes. This was followed by the addition of 125 μl of the mixture of amplified sample and signal hybridization solution into a detection cartridge. Cartridges were then loaded onto the eSensor XT-8 instrument for data acquisition and automated analysis.
Each cartridge has a preprogrammed memory chip that is read by the instrument and contains the test protocol, lot number, and expiration date. The XT-8 instrument then activates a pneumatic pump membrane within each cartridge to propel the mixture of amplified and hybridized sample through a narrow channel across a single-file microarray of preassembled gold-plated electrodes positioned in a serpentine path and coated with single-stranded oligonucleotide capture probes specific for individual viral targets (15, 33). Target DNA-signal probe complexes present in the sample hybridize to the capture probes, bringing the ferrocene label in proximity with the gold electrode. An electrical current is applied to each electrode, and captured target DNA is analyzed by electrochemical detection using voltammetry; a signal of 3 nA or greater for a given analyte is interpreted as positive (Fig. 1). The XT-8 instrument has a modular design with a base module and up to three processing towers that contain eight cartridge slots each, allowing up to 24 samples to be analyzed simultaneously.
Fig 1.
eSensor RVP cartridge and detection technology.
Resolution of discordant results.
When initial results were discordant for a particular virus when a specimen was tested by the eSensor RVP and laboratory-developed real-time PCR assays, the two tests were repeated concurrently on two separate aliquots of the relevant original sample that had been stored frozen at −70°C. The final determination of whether a specimen was concordant or discordant was based on an analysis of the initial and repeat test results obtained by both methods. If the initial test result was positive for a given assay and one or both of the duplicate retests were positive, the final result was reported as positive. Conversely, if the initial test result was negative for a given assay and one or both of the duplicate retests were negative, the final result was reported as negative. Results for viruses not included in the panel of real-time PCR assays but positive by eSensor RVP, i.e., parainfluenza virus type 4 and the coronaviruses, were not considered discordant between the two methods.
Statistical analysis.
The kappa statistic and all confidence intervals (CIs) were calculated using the VassarStats website for statistical computation, http://vassarstats.net/. This site was accessible as of 6 May 2012. Squared correlation coefficients were calculated using Microsoft Excel 2007 software (Microsoft Corporation, Redmond, WA).
RESULTS
Among viruses tested for by both methods, the overall agreement between the eSensor RVP and the corresponding real-time PCR assays was 99.2% (kappa = 0.96 [95% CI, 0.94 to 0.98]). The combined positive percent agreement for analytes shared by the two methods was 95.4% (95% CI, 92.5 to 97.3), and the combined negative percent agreement was 99.7% (95% CI, 99.4 to 99.8). Results by virus are presented in Table 1. Real-time PCR-positive and eSensor RVP-negative discordant results were more frequently obtained for specimens with higher CT values (i.e., lower viral loads) for the real-time PCR assays (Table 2). The mean real-time PCR CT value of all tests with discordant results was 39.73 (95% CI, 38.03 to 41.43). With respect to subtype determination for influenza A virus, the eSensor RVP generated the same subtype result identified by our laboratory-developed real-time PCR subtyping assays in 68 out of the 70 (97.1%) specimens that were positive by both methods for influenza A virus. One specimen that was subtyped as influenza A virus seasonal H1 and another subtyped as influenza A virus (H1N1)pdm09 by our laboratory-developed PCR assays were also positive for influenza A virus by the eSensor RVP but could not be subtyped using this system.
Table 1.
Detection of respiratory viruses by eSensor RVP and real-time PCR assaysa
| Virus | No. of specimens with the following virus detection results by eSensor RVP/PCR: |
Agreement | ||||||
|---|---|---|---|---|---|---|---|---|
| Positive |
Negative |
|||||||
| +/+ (a) | +/− (b) | −/+ (c) | −/− (d) | a/(a + c) (%) | 95% CI | d/(b + d) (%) | 95% CI | |
| IV A | 70 | 0 | 2 | 178 | 70/72 (97.2) | 90.4–99.2 | 178/178 (100) | 97.9–100 |
| H1 | 24b | 0 | 0 | 226 | 24/24 (100) | 86.2–100 | 226/226 (100) | 98.3–100 |
| H3 | 23 | 0 | 1 | 226 | 23/24 (95.8) | 79.8–99.3 | 226/226 (100) | 98.3–100 |
| (H1N1)pdm09 | 23b | 0 | 1 | 226 | 23/24 (95.8) | 79.8–99.3 | 226/226 (100) | 98.3–100 |
| IV B | 22 | 0 | 2 | 226 | 22/24 (91.7) | 74.2–97.8 | 226/226 (100) | 98.3–100 |
| RSV A | 24 | 0 | 0 | 226 | 24/24 (100) | 86.2–100 | 226/226 (100) | 98.3–100 |
| RSV B | 23 | 0 | 1 | 226 | 23/24 (95.8) | 79.8–99.3 | 226/226 (100) | 98.3–100 |
| PIV type 1 | 23 | 0 | 1 | 226 | 23/24 (95.8) | 79.8–99.3 | 226/226 (100) | 98.3–100 |
| PIV type 2 | 24 | 0 | 0 | 226 | 24/24 (100) | 86.2–100 | 226/226 (100) | 98.3–100 |
| PIV type 3 | 22 | 0 | 2 | 226 | 22/24 (91.7) | 74.2–97.8 | 226/226 (100) | 98.3–100 |
| MPV | 23 | 0 | 2 | 225 | 23/25 (92.0) | 75.0–97.8 | 225/225 (100) | 98.3–100 |
| RhV | 34 | 9 | 1 | 206 | 34/35 (97.1) | 85.5–99.5 | 206/215 (95.8) | 92.2–97.8 |
| AdV | 28 | 0 | 3 | 219 | 28/31 (90.3) | 75.1–96.7 | 219/219 (100) | 98.3–100 |
| Total | 293 | 9 | 14 | 2,684 | 293/307 (95.4) | 92.5–97.3 | 2,684/2,693 (99.7) | 99.4–99.8 |
Abbreviations: IV, influenza virus; RSV, respiratory syncytial virus; PIV, parainfluenza virus; MPV, human metapneumovirus; RhV, rhinovirus; AdV, adenovirus.
One influenza A virus H1 subtype and one influenza A virus (H1N1)pdm09 subtype were detected as influenza A virus by eSensor RVP but were not able to be typed.
Table 2.
Performance of eSensor RVP for detection of 12 respiratory viruses based on real-time PCR CT valuesa
| Virus | No. positive by eSensor RVP based on the following real-time PCR CT values/total no. (%): |
||||||
|---|---|---|---|---|---|---|---|
| <20 | 20.00–24.99 | 25.00–29.99 | 30.00–34.99 | 35.00–39.99 | 40.00–44.99 | All | |
| IV A H1 | 3/3 | 6/6 | 6/6 | 5/5 | 4/4 | NA | 24/24 (100) |
| IV A H3 | 3/3 | 6/6 | 6/6 | 5/5 | 2/3 | 1/1 | 23/24 (95.8) |
| IV A (H1N1)pdm09 | 3/3 | 5/5 | 6/6 | 6/6 | 3/3 | 0/1 | 23/24 (95.8) |
| IV B | 3/3 | 5/5 | 6/6 | 5/5 | 3/3 | 0/2 | 22/24 (91.7) |
| RSV A | 4/4 | 5/5 | 6/6 | 5/5 | 3/3 | 1/1 | 24/24 (100) |
| RSV B | 3/3 | 6/6 | 6/6 | 5/5 | 2/3 | 1/1 | 23/24 (95.8) |
| PIV type 1 | 3/3 | 5/5 | 6/6 | 5/5 | 3/3 | 1/2 | 23/24 (95.8) |
| PIV type 2 | 2/2 | 6/6 | 6/6 | 5/5 | 4/4 | 1/1 | 24/24 (100) |
| PIV type 3 | 3/3 | 6/6 | 5/5 | 5/5 | 3/4 | 0/1 | 22/24 (91.7) |
| MPV | 3/3 | 5/5 | 6/6 | 6/6 | 3/3 | 0/2 | 23/25 (92.0) |
| RhV | 3/3 | 5/5 | 6/6 | 5/6 | 11/11 | 4/4 | 34/35 (97.1) |
| AdV | 3/3 | 5/5 | 6/6 | 6/6 | 6/7 | 2/4 | 28/31 (90.3) |
| Total | 36/36 (100) | 65/65 (100) | 71/71 (100) | 63/64 (98.4) | 47/51 (92.2) | 11/20 (55.0) | 293/307 (95.4) |
Abbreviations: IV, influenza virus; RSV, respiratory syncytial virus; PIV, parainfluenza virus; MPV, human metapneumovirus; RhV, rhinovirus; AdV, adenovirus; NA, specimens not available at this concentration of virus.
The eSensor RVP was also highly specific compared to the real-time PCR assays, with differences observed for only 9 out of 2,693 negative determinations; all 9 specimens were reproducibly positive for rhinovirus by eSensor RVP but were repeatedly rhinovirus negative by real-time PCR. To investigate whether the higher number of rhinovirus detections by eSensor RVP truly reflected an enhanced sensitivity or nonspecific detection of other picornaviruses (i.e., enteroviruses), a total of 19 characterized enterovirus types present at high viral loads were tested by both eSensor RVP and the laboratory-developed rhinovirus real-time PCR assay. While all 19 enterovirus strains generated positive results with the rhinovirus real-time PCR assay, none were positive for rhinovirus by eSensor RVP (Table 3).
Table 3.
Comparison of specificity of eSensor RVP and real-time PCR assay for rhinovirusa
| Enterovirus type | Enterovirus real-time PCR result (CT) | Enterovirus real-time PCR no. of copies/ml (log10) | Rhinovirus result |
|
|---|---|---|---|---|
| eSensor RVP (nA) | Real-time PCR (CT) | |||
| Echovirus 4 | Pos (17.04) | 1.04 × 1010 (10.02) | Neg (0.2) | Pos (41.30) |
| Echovirus 6 | Pos (26.68) | 1.46 × 107 (7.16) | Neg (0.1) | Pos (31.84) |
| Echovirus 9 | Pos (17.59) | 7.14 × 109 (9.85) | Neg (0.2) | Pos (33.24) |
| Echovirus 11 | Pos (15.51) | 2.95 × 1010 (10.47) | Neg (0.3) | Pos (28.06) |
| Echovirus 30 | Pos (16.57) | 1.44 × 1010 (10.16) | Neg (0.1) | Pos (29.72) |
| Coxsackievirus A13 | Pos (21.23) | 6.01 × 108 (8.78) | Neg (0.5) | Pos (38.59) |
| Coxsackievirus B1 | Pos (14.75) | 4.97 × 1010 (10.70) | Neg (0.1) | Pos (29.82) |
| Coxsackievirus B2 | Pos (15.83) | 2.38 × 1010 (10.38) | Neg (0.1) | Pos (31.17) |
| Coxsackievirus B3 | Pos (17.18) | 9.46 × 109 (9.98) | Neg (0.2) | Pos (28.55) |
| Coxsackievirus B4 | Pos (15.95) | 2.19 × 1010 (10.34) | Neg (0.1) | Pos (25.48) |
| Coxsackievirus B5 | Pos (15.76) | 2.49 × 1010 (10.40) | Neg (0.2) | Pos (31.52) |
| Coxsackievirus B6 | Pos (17.76) | 6.38 × 109 (9.80) | Neg (0.2) | Pos (30.74) |
| Enterovirus 68 | Pos (17.35) | 8.43 × 109 (9.93) | Neg (0.4) | Pos (26.33) |
| Enterovirus 69 | Pos (17.78) | 6.30 × 109 (9.80) | Neg (0.2) | Pos (31.64) |
| Enterovirus 70 | Pos (19.45) | 2.01 × 109 (9.30) | Neg (1.3) | Pos (26.94) |
| Enterovirus 71 | Pos (17.07) | 1.02 × 1010 (10.01) | Neg (0.1) | Pos (32.08) |
| Poliovirus 1 | Pos (17.38) | 8.26 × 109 (9.92) | Neg (0.2) | Pos (24.10) |
| Poliovirus 2 | Pos (18.66) | 3.46 × 109 (9.54) | Neg (0.2) | Pos (24.80) |
| Poliovirus 3 | Pos (17.97) | 5.53 × 109 (9.74) | Neg (0.2) | Pos (25.91) |
Abbreviations: Pos, positive: Neg, negative.
Of interest, although the qualitative agreement between real-time PCR and eSensor RVP results among clinical samples was excellent, the relationship between real-time PCR CT values and eSensor RVP nA signal strengths was nonlinear and exhibited variation among viruses. The greatest deviation from linearity was seen for detection of rhinoviruses. Squared correlation coefficient (R2) values were 0.87, 0.81, and 0.92 for influenza virus A seasonal H1, seasonal H3, and (H1N1)pdm09, respectively; 0.83 for influenza virus B; 0.79 and 0.91 for respiratory syncytial virus types A and B, respectively; 0.95, 0.93, and 0.85 for parainfluenza virus types 1, 2, and 3, respectively; 0.80 for human metapneumovirus; 0.83 for adenovirus; and 0.35 for rhinovirus.
Forty-seven of the 250 tested clinical specimens (18.8%) were positive for more than one virus when tested with our laboratory-developed real-time PCR assays. The performance of eSensor RVP in the detection of these coinfections is summarized in Table 4. The eSensor RVP identified all expected viruses in 39 of the 47 specimens (83.0%). In the remaining eight samples (seven with two viruses and one with three viruses), one of the analytes detected by real-time PCR was not detected by eSensor RVP. The mean real-time PCR CT value for analytes not detected by eSensor RVP when present as part of coinfections (n = 8) was 40.75 (95% CI, 39.35 to 42.14), which was not significantly different from the mean CT value (38.37; 95% CI, 34.33 to 42.41) obtained for undetected analytes that were not part of coinfections (n = 6).
Table 4.
Comparative detection of coinfections by eSensor RVP and real-time PCR assaysa
| Distinct combinations detected by real-time PCR assays |
Total no. of coinfections | No. of discrepant coinfectionsb | Discrepant analyteb or no. of discrepant analytes | |||
|---|---|---|---|---|---|---|
| Analyte 1 | Analyte 2 | Analyte 3 | Analyte 4 | |||
| IV A H1 | IV B | 1 | 1 | IV B | ||
| IV A H1 | AdV | 1 | 0 | |||
| IV A H3 | MPV | 1 | 0 | |||
| IV A H3 | AdV | 2 | 1 | IV A H3 | ||
| IV A (H1N1)pdm09 | RSV B | 1 | 0 | |||
| IV B | PIV type 3 | 2 | 1 | IV B | ||
| IV B | AdV | 1 | 1 | AdV | ||
| RSV A | PIV type 3 | 1 | 0 | |||
| RSV A | RhV | 2 | 0 | |||
| RSV A | AdV | 2 | 1 | AdV | ||
| RSV B | RhV | 7 | 1 | RSV B | ||
| PIV type 1 | RhV | 3 | 0 | |||
| PIV type 1 | AdV | 1 | 0 | |||
| PIV type 2 | RhV | 1 | 0 | |||
| PIV type 2 | AdV | 1 | 0 | |||
| PIV type 3 | MPV | 1 | 0 | |||
| PIV type 3 | RhV | 1 | 0 | |||
| MPV | RhV | 3 | 1 | MPV | ||
| MPV | AdV | 1 | 0 | |||
| RhV | AdV | 5 | 0 | |||
| Total dual infections | 38 | 7 | 7/76c | |||
| IV A (H1N1)pdm09 | MPV | RhV | 1 | 0 | ||
| RSV A | RhV | AdV | 1 | 0 | ||
| RSV B | PIV type 1 | RhV | 1 | 0 | ||
| RSV B | RhV | AdV | 1 | 0 | ||
| PIV type 1 | RhV | AdV | 1 | 1 | PIV type 1 | |
| PIV type 3 | MPV | AdV | 1 | 0 | ||
| MPV | RhV | AdV | 1 | 0 | ||
| Total triple infections | 7 | 1 | 1/21c | |||
| IV A (H1N1)pdm09 | PIV type 3 | MPV | RhV | 1 | 0 | |
| RSV A | PIV type 2 | RhV | AdV | 1 | 0 | |
| Total quadruple infections | 2 | 0 | 0/8c | |||
| Total coinfections | 47 | 8 | 8/105c | |||
Abbreviations: IV, influenza virus; RSV, respiratory syncytial virus; PIV, parainfluenza virus; MPV, human metapneumovirus; RhV, rhinovirus; AdV, adenovirus.
A discrepant coinfection or discrepant analyte was defined as one detected by real-time PCR but not detected by eSensor RVP.
The denominator represents the expected number of viral detections.
Of the 28 clinical specimens positive for adenovirus by both eSensor RVP and our laboratory-developed real-time PCR assay, 23 of the adenoviruses were typed by the eSensor assays as belonging to group C, 3 were typed as belonging to group B, and 2 were typed as coming from both groups B and C. Though we were unable to independently type these adenoviruses, group C viruses have recently predominated among respiratory adenoviruses in our pediatric population; 69.3% (480/693) of adenoviruses whose groups were determined over 7.5 years from 2001 to 2008 have represented group C (Adriana Kajon, personal communication). The finding of both group B and C adenoviruses in two specimens was thought to be unusual, and additional testing was performed to verify the specificity of the eSensor RVP adenovirus B, C, and E assays. Upon testing 20 well-characterized adenovirus serotypes from groups A to F, the eSensor RVP adenovirus B, C, and E assays demonstrated some cross-reactivity with adenovirus serotypes from other groups; all tested serotypes from groups A through E were called positive for one or more of adenovirus groups B, C, and E by the eSensor RVP, with only the group F enteric adenoviruses (types 40 and 41) not detected at the tested viral loads (Table 5). Six of the 18 (33.3%) eSensor RVP-positive adenovirus serotypes (1 from each of groups B, C, and E and 3 from group D) were positive by more than one of the eSensor RVP adenovirus assays.
Table 5.
Performance of eSensor RVP for detection of characterized adenovirus serotypesa
| Adenovirus group and type | PCR CT value | PCR no. of DNA copies/ml (log10) | eSensor RVP result (nA) |
||
|---|---|---|---|---|---|
| AdV B | AdV C | AdV E | |||
| Group A, ATCC AdV-12 (VR-863) | 26.05 | 3.41 × 107 (7.53) | Pos (25.6) | Neg (0.3) | Neg (0.1) |
| Group B | |||||
| LRRI 3p | 26.16 | 1.75 × 107 (7.24) | Pos (106.7) | Neg (0.4) | Neg (0.1) |
| ATCC AdV-7 (VR-7) | 25.70 | 4.24 × 107 (7.63) | Pos (115.0) | Neg (0.3) | Neg (0.1) |
| ATCC AdV-11 (VR-12) | 26.47 | 2.61 × 107 (7.42) | Pos (30.8) | Neg (0.6) | Neg (0.2) |
| LRRI 14p | 27.20 | 1.65 × 107 (7.22) | Pos (51.5) | Pos (224.9) | Neg (0.3) |
| ATCC AdV-21 (VR-256) | 23.77 | 5.47 × 107 (7.74) | Pos (163.0) | Neg (0.3) | Neg (0.1) |
| ATCC AdV-35 (VR-718) | 28.30 | 6.02 × 106 (6.78) | Pos (35.1) | Neg (0.2) | Neg (0.1) |
| Group C | |||||
| ATCC AdV-1 (VR-1) | 27.87 | 5.98 × 106 (6.78) | Neg (0.7) | Pos (185.6) | Neg (0.2) |
| LRRI 2 | 27.16 | 9.29 × 106 (6.97) | Pos (152.1) | Pos (171.1) | Neg (0.1) |
| LRRI 5p | 27.92 | 1.05 × 107 (7.02) | Neg (0.3) | Pos (166.6) | Neg (0.1) |
| ATCC AdV-6 (VR-6) | 26.84 | 2.07 × 107 (7.32) | Neg (0.8) | Pos (203.4) | Neg (0.1) |
| Group D | |||||
| LRRI 10 | 27.18 | 9.16 × 106 (6.96) | Neg (0.9) | Pos (33.4) | Neg (0.1) |
| ATCC AdV-19 (VR-254) | 25.24 | 2.18 × 107 (7.34) | Neg (1.2) | Pos (45.6) | Neg (0.2) |
| LRRI 20p | 26.37 | 1.06 × 107 (7.03) | Pos (6.5) | Pos (5.4) | Neg (0.1) |
| ATCC AdV-29 (VR-272) | 26.77 | 8.27 × 106 (6.92) | Neg (1.8) | Pos (32.8) | Neg (0.5) |
| ATCC AdV-32 (VR-625) | 27.70 | 7.13 × 106 (6.85) | Pos (5.8) | Pos (17.3) | Neg (0.2) |
| ATCC AdV-37 (VR-929) | 26.41 | 1.57 × 107 (7.20) | Pos (21.1) | Pos (18.9) | Neg (0.1) |
| Group E, ATCC AdV-4 (VR-4) | 24.46 | 3.54 × 107 (7.55) | Pos (115.4) | Neg (0.1) | Pos (94.6) |
| Group F | |||||
| ATCC AdV-40 (VR-931) | 26.89 | 1.17 × 107 (7.07) | Neg (1.5) | Neg (2.5) | Neg (0.1) |
| ATCC AdV-41 (VR-930) | 27.20 | 9.61 × 106 (6.98) | Neg (0.9) | Neg (0.3) | Neg (0.1) |
Abbreviations: ATCC, American Type Culture Collection; LRRI, Lovelace Respiratory Research Institute; AdV, adenovirus; Pos, positive; Neg, negative.
In the 250 clinical specimens examined, a total of 11 detections of viruses unique to the eSensor RVP and not routinely tested for in our laboratory were observed, including parainfluenza virus type 4 in 6 specimens, coronavirus OC43 in 3 specimens, and coronaviruses NL63 and HKU1 in 1 specimen each. These viruses were repeatedly identified in the specimens tested. There were no detections of coronavirus 229E. One of the parainfluenza virus type 4 detections was in a sample that was also positive for this virus using the FilmArray Respiratory Panel from Idaho Technologies, Inc., Salt Lake City, UT (28). Four additional respiratory samples that were positive for parainfluenza virus type 4 by culture and direct fluorescent-antibody testing (a gift from Danny L. Wiedbrauk, Warde Medical Laboratories, Ann Arbor, MI) were also tested and determined to be positive by the eSensor RVP (data not shown).
During our evaluation of the eSensor RVP, there were a total of 6 invalid test cartridge runs out of 399 performed over the 3-month study period for a failure rate of 1.5%. This necessitated repeat testing of the affected sample to obtain valid results in each case. One cartridge would not electronically connect with the instrument, despite multiple attempts in multiple different analyzer slots. In a single cartridge containing a negative-control sample, the internal control failed and adenovirus group C was detected with a measured current signal of 58.9 nA. According to the manufacturer, unexplained false positivity for adenovirus group C accompanied by internal control failure has also been seen in a small percentage of cartridges tested in premarket evaluations by other users (M. Langley, GenMark Diagnostics, personal communication). In the other four failed cartridges, the internal control was negative in the setting of no positive analytes. Of note, the internal control was also negative in 81 of the 393 (20.6%) other cartridge runs performed. Test results from these cartridges were deemed acceptable and were not retested since one or more viral analytes was detected by eSensor RVP, while the internal control was negative.
DISCUSSION
With this study, the eSensor RVP was found to be comparable in sensitivity and specificity to laboratory-developed real-time TaqMan PCR assays for the qualitative detection of a variety of respiratory viruses. Discordant results between the eSensor RVP and comparator PCR assays were minimal and mostly involved viruses that were present in small quantity within tested specimens. The eSensor RVP is relatively easy to perform, involves a unique enclosed cassette and biosensor technology for target detection, can run up to 24 samples using one instrument and three modular towers, and has a multiplexed design that allows for efficient and simultaneous interrogation of a single specimen for multiple viruses. A potential drawback of the eSensor RVP is the need to manipulate amplified product during the protocol, increasing the risk of contamination. However, no known or suspected contamination events occurred during this evaluation.
The rate of correct identification of influenza A virus subtypes using the eSensor RVP was comparable to that using the real-time PCR. There were only 2 (2.9%) of the 70 samples positive for influenza A virus by both methods for which the eSensor RVP detected the presence of influenza A virus but did not make a subtype determination; 1 of these samples had previously been characterized as seasonal H1 and 1 had previously been characterized as (H1N1)pdm09 by our laboratory-developed typing assays. The ability of the eSensor RVP to simultaneously detect and accurately subtype influenza A virus may be a particularly attractive feature to clinical laboratories since results may impact the clinical decision to prescribe appropriate antiviral medications.
The eSensor RVP assays for adenovirus groups B, C, and E demonstrated some cross-reactivity with other adenovirus serotypes; all tested serotypes from groups A, B, C, D, and E were called positive for one or more of the adenovirus groups B, C, and E by the eSensor RVP. However, differentiation between these groups is not routinely necessary for clinical use and may be more important for epidemiologic investigations and in other special circumstances (8, 16, 25). Though we did not directly compare the eSensor RVP with any other commercially available molecular platform for detection of respiratory pathogens, the high positive percent agreement between the eSensor RVP and the adenovirus real-time PCR observed in the current study suggests a greater sensitivity of the eSensor RVP for the genetically heterogeneous adenoviruses than has been reported for some other commercial molecular assays (26, 28).
The eSensor RVP detected rhinovirus in nine samples that were negative for this virus by our laboratory-developed rhinovirus real-time PCR assay. Many assays with high sensitivity for rhinovirus often demonstrate cross-reactivity with other picornaviruses, such as the enteroviruses. Our laboratory-developed rhinovirus real-time PCR assay, using primers and probes originally designed by the Centers for Disease Control and Prevention (CDC), is no exception and is known to be biased toward an enhanced specificity for rhinovirus but has some cross-reactivity with enteroviruses that are present at high viral loads (17). Interestingly, our testing of characterized enterovirus strains demonstrates that the eSensor RVP is most likely not only more sensitive but also more specific than the comparator rhinovirus-biased laboratory-developed real-time PCR assay. With that said, however, the possibility still remains that the results for these nine rhinovirus-positive samples detected by only the eSensor RVP may potentially represent false-positive results due to cross-reactivity with nucleic acid from some other organism that does not represent the picornavirus family. Conversely, the manufacturer has conducted extensive cross-reactivity studies, testing 32 bacterial and 2 yeast isolates and 13 different viruses that are not targets of the eSensor RVP but can be found in the respiratory tract. The eSensor RVP did not cross-react with any of these organisms when examined at very high concentrations (Peter Krein, GenMark Diagnostics, personal communication, and product package insert). Also, the lower limit of detection of the eSensor RVP for rhinovirus is reported by the manufacturer to be quite low at 0.005 50% tissue culture infectious dose (TCID50)/ml.
In our laboratory, small batches of six respiratory specimens are continuously processed and extracted throughout the day and evening shifts and tested with our 10-member respiratory virus panel of laboratory-developed real-time PCR assays. Our standard procedure incorporates the use of robotic systems, including the epMotion 5075 liquid handling workstation (Eppendorf North America, Hauppauge, NY) for the preparation and routine dispensing of master mix formulations into 96-well plates and the Roche MagNA Pure LC instrument for the postelution pipetting of nucleic acids extracted from clinical specimens. The total run time is approximately 3 h 10 min with a hands-on time of 45 min, and the cost of testing six specimens for all 10 respiratory viruses is $334.90, including reagents, supplies, and labor. Using the eSensor RVP protocol and corresponding XT-8 instrument, it takes about 6 h 26 min to test six specimens for 20 respiratory viral targets, with a total hands-on time of 63 min. The list price for a 48-test eSensor RVP kit is $4,320 ($90/test), but several additional reagents and supplies must be provided by the laboratory. Assuming that external negative and positive controls are run with each batch of samples and incorporating the cost of labor, the total cost to run six samples by eSensor RVP in our laboratory would be $837.26. The laboratory would also need to supply its own thermal cycler. However, the overall ease of use of the system and the equation of hands-on time, turnaround time, specimen batch size, and cost may be favorable for laboratories with different work flows, capacities, and expertise to perform molecular testing.
The eSensor RVP and corresponding XT-8 instrument have recently been submitted to the U.S. Food and Drug Administration (FDA) as the first infectious diseases platform produced by the manufacturer for clearance. Molecular assays utilizing this technology and instrumentation and targeting other genetic markers of human disease have previously been licensed by the FDA (1, 11, 13, 21, 27).
ACKNOWLEDGMENTS
We thank Adriana Kajon (Infectious Diseases Program, Lovelace Respiratory Research Institute, Albuquerque, NM) for providing characterized adenovirus strains and typing information and Danny L. Wiedbrauk (Warde Medical Laboratories, Ann Arbor, MI) for providing characterized parainfluenza virus type 4-positive clinical samples.
Footnotes
Published ahead of print 8 August 2012
REFERENCES
- 1. Bernacki SH, et al. 2003. Bioelectronic sensor technology for detection of cystic fibrosis and hereditary hemochromatosis mutations. Arch. Pathol. Lab. Med. 127: 1565–1572 [DOI] [PubMed] [Google Scholar]
- 2. Bonner AB, Monroe KW, Talley LI, Klasner AE, Kimberlin DW. 2003. Impact of the rapid diagnosis of influenza on physician decision-making and patient management in the pediatric emergency department: results of a randomized, prospective, controlled trial. Pediatrics 112: 363–367 [DOI] [PubMed] [Google Scholar]
- 3. Caram LB, et al. 2009. Respiratory syncytial virus outbreak in a long-term care facility detected using reverse transcriptase polymerase chain reaction: an argument for real-time detection methods. J. Am. Geriatr. Soc. 57: 482–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. D'Heilly SJ, Janoff EN, Nichol P, Nichol KL. 2008. Rapid diagnosis of influenza in older adults: influence on clinical care in a routine clinical setting. J. Clin. Virol. 42: 124–128 [DOI] [PubMed] [Google Scholar]
- 5. Esposito S, Marchisio P, Morelli P, Crovari P, Principi N. 2003. Effect of a rapid influenza diagnosis. Arch. Dis. Child. 88: 525–526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Falsey AR, Murata Y, Walsh EE. 2007. Impact of rapid diagnosis on management of adults hospitalized with influenza. Arch. Intern. Med. 167: 354–360 [DOI] [PubMed] [Google Scholar]
- 7. Freymuth F, et al. 2006. Comparison of multiplex PCR assays and conventional techniques for the diagnostic of respiratory virus infections in children admitted to hospital with an acute respiratory illness. J. Med. Virol. 78: 1498–1504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gray GC, et al. 2007. Genotype prevalence and risk factors for severe clinical adenovirus infection, United States, 2004-2006. Clin. Infect. Dis. 45: 1120–1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Harris JA, Huskins WC, Langley JM, Siegel JD, Pediatric Special Interest Group of the Society of Healthcare Epidemiology of America 2007. Health care epidemiology perspective on the October 2006 recommendations of the Subcommittee on Diagnosis and Management of Bronchiolitis. Pediatrics 120: 890–892 [DOI] [PubMed] [Google Scholar]
- 10. Jernigan DB, et al. 2011. Detecting 2009 pandemic influenza A (H1N1) virus infection: availability of diagnostic testing led to rapid pandemic response. Clin. Infect. Dis. 52: S36–S43 [DOI] [PubMed] [Google Scholar]
- 11. Johnson MA, Yoshitomi MJ, Richards CS. 2007. A comparative study of five technologically diverse CFTR testing platforms. J. Mol. Diagn. 9: 401–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kuypers J, et al. 2006. Comparison of real-time PCR assays with fluorescent-antibody assays for diagnosis of respiratory virus infections in children. J. Clin. Microbiol. 44: 2382–2388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lee CC, McMillin GA, Babic N, Melis R, Yeo KT. 2011. Evaluation of a CYP2C19 genotype panel on the GenMark eSensor platform and the comparison to the Autogenomics Infiniti and Luminex CYP2C19 panels. Clin. Chim. Acta 412: 1133–1137 [DOI] [PubMed] [Google Scholar]
- 14. Lee WG, Kim Y, Chung BG, Demirci U, Khademhosseini A. 2010. Nano/microfluidics for diagnosis of infectious diseases in developing countries. Adv. Drug Deliv. Rev. 62: 449–457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lenigk R, et al. 2002. Plastic biochannel hybridization devices: a new concept for microfluidics DNA arrays. Anal. Biochem. 311: 40–49 [DOI] [PubMed] [Google Scholar]
- 16. Lin B, et al. 2004. Use of oligonucleotide microarrays for rapid detection and serotyping of acute respiratory disease-associated adenoviruses. J. Clin. Microbiol. 42: 3232–3239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lu X, et al. 2008. Real-time reverse transcription-PCR assay for comprehensive detection of human rhinoviruses. J. Clin. Microbiol. 46: 533–539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mahony J. 2008. Detection of respiratory viruses by molecular methods. Clin. Microbiol. Rev. 21: 716–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mahony J, et al. 2009. Cost analysis of multiplex PCR testing for diagnosing respiratory virus infections. J. Clin. Microbiol. 47: 2812–2817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mahony J, et al. 2007. Development of a respiratory virus panel test for detection of twenty human respiratory viruses by use of multiplex PCR and a fluid microbead-based assay. J. Clin. Microbiol. 45: 2965–2970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Maurice CB, et al. 2010. Comparison of assay systems for warfarin-related CYP2C9 and VKORC1 genotyping. Clin. Chim. Acta 411: 947–954 [DOI] [PubMed] [Google Scholar]
- 22. Miernyk K, et al. 2011. Performance of a rapid antigen test (Binax NOW® RSV) for diagnosis of respiratory syncytial virus compared with real-time polymerase chain reaction in a pediatric population. J. Clin. Virol. 50: 240–243 [DOI] [PubMed] [Google Scholar]
- 23. Mills JM, Harper J, Broomfield D, Templeton KE. 2011. Rapid testing for respiratory syncytial virus in a paediatric emergency department: benefits for infection control and bed management. J. Hosp. Infect. 77: 248–251 [DOI] [PubMed] [Google Scholar]
- 24. Milstone AM, Perl TM, Valsamakis A. 2012. Epidemiology of respiratory viruses in children admitted to an infant/toddler unit. Am. J. Infect. Control 40: 462–464 [DOI] [PubMed] [Google Scholar]
- 25. Morfin F, et al. 2009. Differential susceptibility of adenovirus clinical isolates to cidofovir and ribavirin is not related to species alone. Antivir. Ther. 14: 55–61 [PubMed] [Google Scholar]
- 26. Pabbaraju K, Tokaryk KL, Wong S, Fox JD. 2008. Comparison of the Luminex xTAG respiratory viral panel with in-house nucleic acid amplification tests for diagnosis of respiratory virus infections. J. Clin. Microbiol. 46: 3056–3062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Pickering JW, McMillin GA, Gedge F, Hill HR, Lyon E. 2004. Flow cytometric assay for genotyping cytochrome p450 2C9 and 2C19: comparison with a microelectronic DNA array. Am. J. Pharmacogenomics 4: 199–207 [DOI] [PubMed] [Google Scholar]
- 28. Pierce VM, Elkan M, Leet M, McGowan KL, Hodinka RL. 2012. Comparison of the Idaho Technology FilmArray system to real-time PCR for detection of respiratory pathogens in children. J. Clin. Microbiol. 50: 364–371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Riedlinger J, et al. 2010. Multicenter evaluation of the BD Max GBS assay for detection of group B streptococci in prenatal vaginal and rectal screening swab specimens from pregnant women. J. Clin. Microbiol. 48: 4239–4241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Schwartz J, Robinson-Dunn B, Makin J, Boyanton BL., Jr 2012. Evaluation of the BD MAX GBS assay to detect Streptococcus group B in LIM broth-enriched antepartum vaginal-rectal specimens. Diagn. Microbiol. Infect. Dis. 73: 97–98 [DOI] [PubMed] [Google Scholar]
- 31. Sharma V, Dowd MD, Slaughter AJ, Simon SD. 2002. Effect of rapid diagnosis of influenza virus type A on the emergency department management of febrile infants and toddlers. Arch. Pediatr. Adolesc. Med. 156: 41–44 [DOI] [PubMed] [Google Scholar]
- 32. Tanriverdi S, Chen L, Chen S. 2010. A rapid and automated sample-to-result HIV load test for near-patient application. J. Infect. Dis. 201: S52–S58 [DOI] [PubMed] [Google Scholar]
- 33. Vernon SD, et al. 2003. Bioelectronic DNA detection of human papillomaviruses using eSensor: a model system for detection of multiple pathogens. BMC Infect. Dis. 3: 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Weinberg GA, et al. 2004. Superiority of reverse-transcription polymerase chain reaction to conventional viral culture in the diagnosis of acute respiratory tract infections in children. J. Infect. Dis. 189: 706–710 [DOI] [PubMed] [Google Scholar]
- 35. Woo PC, Chiu SS, Seto W, Peiris M. 1997. Cost-effectiveness of rapid diagnosis of viral respiratory tract infections in pediatric patients. J. Clin. Microbiol. 35: 1579–1581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Yager P, et al. 2006. Microfluidic diagnostic technologies for global public health. Nature 442: 412–418 [DOI] [PubMed] [Google Scholar]