Abstract
Buruli ulcer (BU) is a necrotizing infection of skin and soft tissue caused by Mycobacterium ulcerans. In Australia, most cases of BU are linked to temperate, coastal Victoria and tropical, northern Queensland, and strains from these regions are distinguishable by variable-number tandem repeat (VNTR) typing. We present an epidemiological investigation of five patients found to have been infected during interstate travel and describe two nucleotide polymorphisms that differentiate M. ulcerans strains from northern Australia.
TEXT
Mycobacterium ulcerans is a toxin-producing pathogen that causes Buruli ulcer (BU), a spectrum of clinical disease that includes ulcers (Fig. 1), nodules, plaques, edematous lesions, and osteomyelitis (20). Australia is the only developed country with significant local transmission of BU. Foci of infection have been described in tropical, far north Queensland (16) and the Capricorn Coast region of central Queensland (6), with sporadic cases also reported from the Northern Territory (NT) (14), Western Australia (WA) (4), and New South Wales (NSW) (12). In temperate, coastal Victoria, where the disease is known as Bairnsdale ulcer, large clusters of cases have occurred on Phillip Island (19) and on the Mornington (10) and Bellarine (3, 11, 18) peninsulas.
Fig 1.
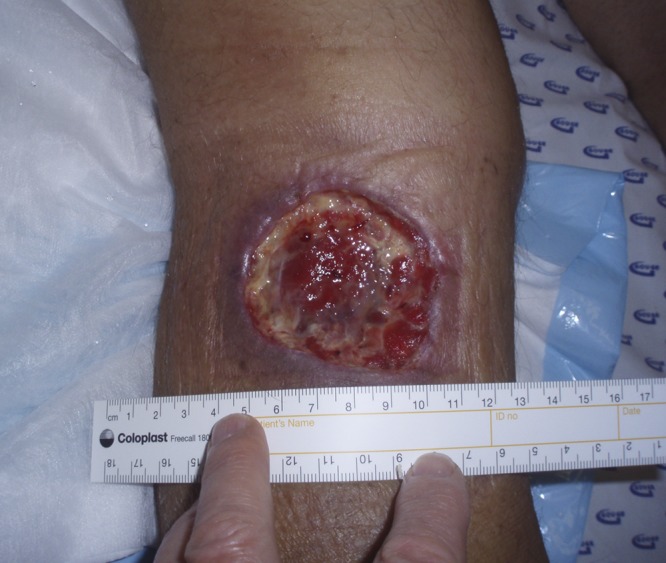
M. ulcerans infection in a 69-year-old male from Victoria acquired during a fishing trip to the Northern Territory (case 1).
Compared to other mycobacteria, such as Mycobacterium tuberculosis, there is little genetic diversity among isolates of M. ulcerans. Nevertheless, variable-number tandem repeat (VNTR) typing is an established method for differentiating M. ulcerans strains from broad geographic regions (2). Only two VNTR profiles have been identified in Australian strains: the Victorian genotype, which is characteristic of M. ulcerans strains from southern Australia (Victoria and NSW), and the Southeast Asian genotype, which is characteristic of M. ulcerans strains from northern Australia (Queensland, NT, and WA), Malaysia, and some strains from Papua New Guinea (2). Although nucleotide sequence differences within some VNTR loci have previously been identified that distinguish northern Australian, Malaysian, and Papua New Guinean isolates with the Southeast Asian genotype (1), no sequence polymorphisms have yet been described that permit discrimination of M. ulcerans strains within northern Australia.
The World Health Organization (WHO) Collaborating Centre for Mycobacterium ulcerans (Western Pacific Region) located at the Victorian Infectious Disease Reference Laboratory (VIDRL) is responsible for recording all cases of BU diagnosed in Australia. In 2011, there was a marked increase in cases of BU in Victoria and Queensland, with 79 and 62 laboratory-confirmed cases recorded, respectively, compared to 33 and four in 2010 (unpublished data). Among those cases diagnosed at VIDRL were five patients who were not residents of an area where BU is known to be endemic but who had a history of interstate/international travel and, in all but one case, had traveled to more than one area where BU is endemic. An investigation was carried out using traditional and molecular epidemiological methods to determine the geographic origin of these patients' infections.
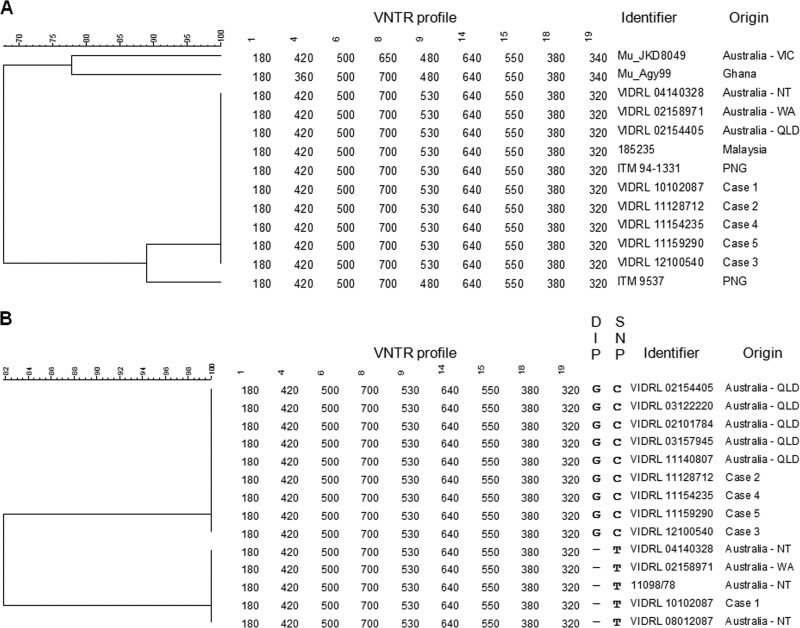
The demographic and clinical characteristics of the five cases are summarized in Table 1. Diagnosis of BU was confirmed in all cases by real-time PCR targeting the M. ulcerans-specific insertion element IS2404 (8) and, in all but one case, isolation of M. ulcerans by culture. Culture could not be performed in case 4, as the entire specimen had been fixed in formalin. The travel history of each patient was reviewed to establish possible exposure locations. All patients had visited at least one area in Victoria (cases 1, 2, and 4), NT (case 1), Queensland (cases 2, 3, 4, and 5), or Africa (case 5) where BU is known to be endemic. VNTR typing (2) was performed using DNA extracted from the patient isolate or, in case 4, from paraffin-embedded fixed-tissue sections, as described previously (8, 12, 13). The PCR products generated from the fixed tissue were sequenced to rule out nonspecific amplification. The VNTR profiles were compared to those of M. ulcerans strains from different geographic regions (Table 2) using BioNumerics version 5.0 (Applied Maths, Belgium). All five strains exhibited the Southeast Asian genotype, indicating that the patients had acquired their infections in northern Australia rather than in areas of Victoria and Africa where BU is endemic (Fig. 2A).
Table 1.
Demographic and clinical characteristics of Buruli ulcer cases diagnosed in travelers to Queensland and the Northern Territory, Australia
| Case no. | Age (yrs) | Sex | Residential addressa | Site and form of disease | Diagnosisb | Treatment | Onset of symptoms | Travel history/possible exposuresa | Origin of infectiona | Incubation period |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 69 | Male | VIC | Left leg ulcer | PCR, culture | Surgery and antibiotics (RIF + MXF) | December 2009 (boil) | VIC (Barwon Heads), July 2009 (<1 h); NT (Darwin, Kakadu, Daly River), July 2009 (10 days) | NT | 5 months |
| 2 | 32 | Male | VIC | Left calf ulcer | PCR, culture, histology | Antibiotics (RIF + CLR) | January 2011 (nontender papule) | QLD (Daintree), October 2010 (5 days); VIC (Point Lonsdale), January 2011 (1 day) | QLD | 3 months |
| 3 | 65 | Female | NSW | Left leg ulcer | PCR, culture, histology | Antibiotics (RIF + CIP) | December 2010 (painless papule) | QLD (Daintree, Mossman), October 2010 (3 weeks) | QLD | 2 months |
| 4 | 64 | Female | WA | Left thigh nonulcerated lesion | PCR | Surgery and antibiotics (RIF + CLR) | April 2011 (itchy lesion) | VIC (Drysdale), January 2011(2 weeks); QLD (Daintree), January 2011 (7 days) | QLD | 3 months |
| 5 | 60 | Female | SA | Right ankle ulcer | PCR, culture | Surgery and antibiotics (RIF + MXF) | June 2011 | South Africa, Mozambique, Zimbabwe, Kenya, and Uganda, 2001–2009 (8 yrs); QLD (Mossman, Port Douglas, Miallo), January 2011 (2–3 weeks) | QLD | 5 months |
NSW, New South Wales; NT, Northern Territory; QLD, Queensland; SA, South Australia; VIC, Victoria; WA, Western Australia.
All cases were confirmed by PCR targeting the M. ulcerans-specific insertion element IS2404. No fresh tissue was available for culture from case 4.
Table 2.
Mycobacterium ulcerans strains used in this study
| Identifier | Alternate identifier | Origin | Yr | Reference |
|---|---|---|---|---|
| Mu_Agy99 | Ga District, Ghana | 1999 | 17 | |
| 185235 | Malaysia | This study | ||
| ITM 94–1331 | 186395 | Papua New Guinea | 2 | |
| VIDRL 71 | Papua New Guinea | 12 | ||
| ITM 9537 | 11878/71 | Papua New Guinea | 2 | |
| Mu_JKD8049 | Mu_04126204 | Victoria, Australia | 2004 | 5 |
| VIDRL 04140328 | Northern Territory, Australia | 2004 | This study | |
| VIDRL 08012087 | Northern Territory, Australia | 2008 | This study | |
| 11098/78 | Northern Territory, Australia | This study | ||
| VIDRL 02101784 | Queensland, Australia | 2002 | This study | |
| VIDRL 02154405 | Queensland, Australia | 2002 | This study | |
| VIDRL 03122220 | Queensland, Australia | 2003 | This study | |
| VIDRL 03157945 | Queensland, Australia | 2003 | This study | |
| VIDRL 11140807 | Queensland, Australia | 2011 | This study | |
| VIDRL 02158971 | Western Australia, Australia | 2002 | This study | |
| VIDRL 10102087 | Case 1 | Northern Territory, Australia | 2010 | This study |
| VIDRL 11128712 | Case 2 | Queensland, Australia | 2011 | This study |
| VIDRL 12100540 | Case 3 | Queensland, Australia | 2011 | This study |
| VIDRL 11154235 | Case 4 | Queensland, Australia | 2011 | This study |
| VIDRL 11159290 | Case 5 | Queensland, Australia | 2011 | This study |
Fig 2.
UPGMA tree showing relationship between M. ulcerans strains from different geographic regions. Panel A shows the VNTR typing results of strains from Africa, Australia, and Southeast Asia. Panel B shows the combined VNTR typing and nucleotide sequencing results of strains from Queensland (QLD), Northern Territory (NT), and Western Australia (WA). Numbers represent the PCR product size (bp) at each of the nine VNTR loci. Letters represent the nucleotide present at the DIP in locus 9 and the SNP in locus 19.
In an attempt to distinguish M. ulcerans strains from different regions of northern Australia, we performed comparative sequence analysis of the VNTR loci in all isolates from northern Australia in our collection. These comprised five isolates from Queensland, three from NT, and one from the only documented case from WA (Table 2). Nucleotide sequencing was performed on both strands as described previously (8), and sequence data were compared using BioNumerics. Two nucleotide sequence polymorphisms (a deletion/insertion polymorphism [DIP] and a single nucleotide polymorphism [SNP]) were identified that differentiated the Queensland strains from the NT/WA strains: a G insertion in the third repeat unit at VNTR locus 9 and a C/T substitution downstream of the second repeat unit at VNTR locus 19 (Table 3). Analysis of these two loci in the five cases under investigation revealed that case 1 clustered with the NT/WA strains and cases 2, 3, 4, and 5 clustered with the Queensland strains (Fig. 2B). Together with the patients' travel histories, this enabled us to conclude that case 1 acquired his infection in NT, while the other four patients acquired their infections in Queensland (Table 1).
Table 3.
Sequence alignment of genomic regions containing polymorphisms that differentiate M. ulcerans strains from northern Australia
| Locus | Origina | Sequence (5′–3′)b | Sequence changec |
|---|---|---|---|
| 9d | QLD | GTGGCGATCGTAAGCTCGGCGCAGCCGGGGTTGCCA | G insertion after nucleotide 2814032 |
| NT/WA | ..........................-......... | ||
| 19e | QLD | GGTACGGTGCACGGTGTGAAGTCGCTCCCGTGCCCA | C/T substitution at nucleotide 4981684 |
| NT/WA | ............................T....... |
QLD, Queensland; NT/WA, Northern Territory/Western Australia.
., identical nucleotide position; -, base deletion.
Nucleotide position relative to M. ulcerans Agy99 complete genome (GenBank accession no. CP000325.1).
Sequence shown represents the third repeat unit in the region amplified by the VNTR locus 9 primers described by Ablordey et al. (2).
Sequence shown begins 50 bp downstream of the second repeat unit in the region amplified by the VNTR 19 primers described by Ablordey et al. (2).
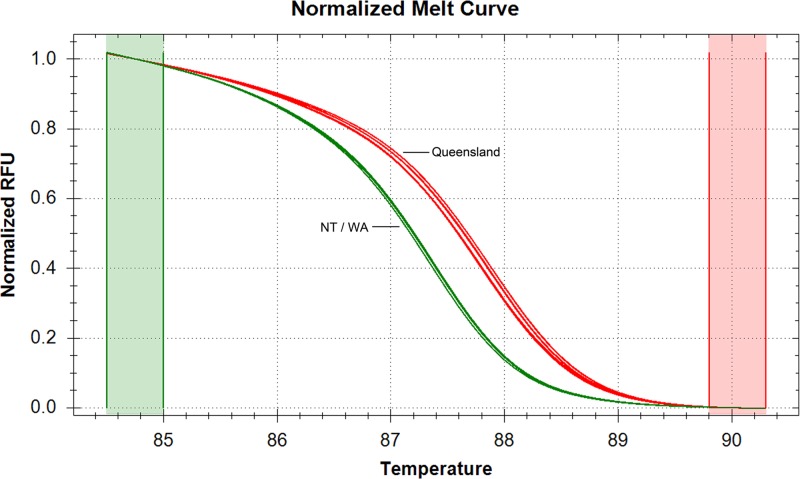
We then attempted to develop high-resolution melting (HRM) assays targeting these two nucleotide polymorphisms. Primers were designed to flank the polymorphic regions at each locus using NCBI Primer-BLAST (http://www.ncbi.nlm.nih.gov/tools/primer-blast/) and Primer3 (http://frodo.wi.mit.edu/). To identify the SNP at locus 19, PCR was performed in 20-μl reaction mixtures containing 1 μl of genomic DNA (extracted from the isolate or, in case 4, directly from the fixed tissue), 1× precision melt supermix (Bio-Rad Laboratories, Hercules, CA), and 200 nM each primer (MUVNTR19SNP-F, 5′-ACGGTACGGTGCACGGTGTG-3′; MUVNTR19SNP-R, 5′-TGTCGCGGCAACCAATGCGA-3′). Amplification and HRM analysis was performed on the Bio-Rad CFX96 real-time system (Bio-Rad) using the following PCR and HRM cycling parameters: 1 cycle of 95°C for 5 min; 45 cycles of 95°C for 10 s and 60°C for 30 s; and 65 to 95°C in 0.5°C increments for 10 s/step. The HRM curve was analyzed using the Bio-Rad precision melt analysis software. All 14 strains from northern Australia listed in Table 2 were tested (nine from Queensland, four from NT, and one from WA). There was a 0.5°C difference in the melting temperature of the two sequence variants which clearly differentiated the Queensland and NT/WA genotypes (Fig. 3). The HRM and sequencing results were in agreement for all samples tested, indicating 100% concordance between the HRM assay and nucleotide sequencing and demonstrating that the HRM method can be applied to DNA extracted from clinical specimens. Despite experimenting with different primer sequences, reaction conditions, and cycling parameters, an assay to detect the DIP at locus 9 could not be developed. We presume that this is due to the DIP being located in a repeat unit, which hampered amplification of the region of interest.
Fig 3.
High-resolution melt curves of M. ulcerans strains from northern Australia demonstrating the difference in melting temperature caused by the SNP at VNTR locus 19. The Queensland (QLD) genotype (C nucleotide) is shown in red, and the Northern Territory (NT)/Western Australia (WA) genotype (T nucleotide) is shown in green.
The five cases presented here are noteworthy for several reasons. First, they highlight the value of nucleotide polymorphism typing as an additional tool to VNTR typing for determining the geographic origin of a patient's infection and for better understanding the genetic diversity and epidemiology of M. ulcerans in Australia (Fig. 4). Although restriction fragment length polymorphism (RFLP) typing also provides increased discrimination of Australian strains (7, 9), it requires large amounts of high-quality DNA, which is time-consuming and labor-intensive or may not be possible if there is no fresh specimen available for culture (as in case 4). In contrast, PCR-based typing methods are rapid and in some cases can be performed on DNA extracted from clinical specimens (provided there is sufficient M. ulcerans DNA in the sample). SNP typing has been successfully used to differentiate M. ulcerans patient isolates from different regions in Ghana (15). We have now identified two nucleotide polymorphisms that permit discrimination of M. ulcerans strains found in northern Australia that were previously indistinguishable on the basis of VNTR typing alone and have developed a high-throughput HRM assay for rapid detection of the SNP at locus 19. It is hoped that whole-genome sequencing will reveal additional SNPs that will provide increased discrimination of M. ulcerans strains from different geographic regions of Australia (5).
Fig 4.
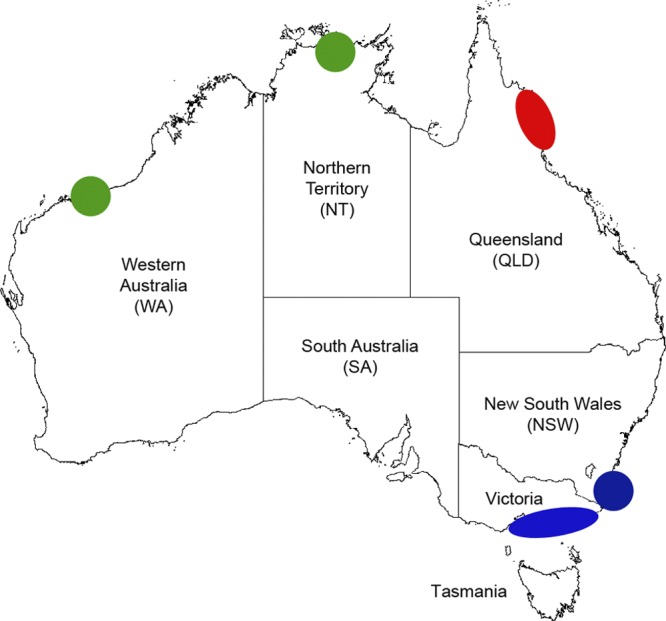
Map of Australia showing distribution of combined VNTR/nucleotide polymorphism M. ulcerans genotypes. Each shape represents the geographic region and genotype of at least one confirmed case of locally acquired M. ulcerans infection: green, Western Australia (WA)/Northern Territory (NT) genotype; red, Queensland genotype; blue, Victoria/New South Wales (NSW) genotype.
Second, cases of BU in travelers to Queensland and NT have, until now, been extremely rare. In contrast to Victoria, where cases of BU in visitors to areas where BU is endemic are common (11), to date almost all patients with M. ulcerans infection in Queensland and NT have been permanent residents of those regions. In a review of 92 cases of M. ulcerans infection in far north Queensland over a 45-year period from 1964 to 2008, only four patients were nonresidents (16). The reason for the recent increase in nonresident cases is not clear, but it is possible that the environmental and/or climatic conditions that might have led to the marked increase in disease in permanent residents of far north Queensland in 2011 also resulted in the increased likelihood of travelers to these regions acquiring the infection.
Third, as the mode of transmission of M. ulcerans is not completely understood, there is currently no way of knowing when most patients have been exposed to the pathogen. This means that the incubation period of BU generally cannot be accurately calculated. However, in cases such as these where it can be clearly demonstrated that the patient acquired her/his infection during a short visit to an area where BU is endemic, the timing of exposure is clear. All five cases acquired the infection during the wet season, and the incubation periods (that is, the time between exposure to M. ulcerans and the first appearance of clinical symptoms) ranged from 2 to 5 months (Table 1), which is consistent with previous estimates of 3 to 7 months in other Australian travelers (12). The fact that some patients visited the area of BU endemicity only for a few days also demonstrates that the duration of exposure may be short.
Finally, these cases reinforce the importance of including M. ulcerans disease in the differential diagnosis of any patient presenting with a nonhealing skin lesion. The regions of Queensland and Victoria where BU is endemic receive large numbers of international visitors, and it is possible that cases of BU may have presented in areas of the world unfamiliar with the disease. Although rarely fatal, BU can cause serious morbidity and result in permanent disability if not detected and treated early (21). Public health interventions to prevent BU are limited, as the mode of transmission is not well understood and there is currently no vaccine available. Therefore, early diagnosis is essential for minimizing the severity of disease and improving patient outcomes.
ACKNOWLEDGMENTS
This study was funded in part by a Victorian Government Department of Health Public Health Grant. We also wish to acknowledge Dallas Wilson for assistance with preparation of figures.
We thank Lynne Brown for providing information on cases 1 and 2, Timothy Stinear for providing sequence data, Peter Jelfs for providing an isolate for case 3, David Dawson for providing isolates from Queensland patients, and Richard Gair, Juliette Esmonde, Sushil Pandey and Chris Coulter for providing figures on Queensland cases in 2011. We also wish to acknowledge Dallas Wilson for assistance with preparation of figures.
Footnotes
Published ahead of print 8 August 2012
REFERENCES
- 1.Ablordey A, Hilty M, Stragier P, Swings J, Portaels F. 2005. Comparative nucleotide sequence analysis of polymorphic variable-number tandem-repeat loci in Mycobacterium ulcerans. J. Clin. Microbiol. 43:5281–5284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ablordey A, et al. 2005. Multilocus variable-number tandem repeat typing of Mycobacterium ulcerans. J. Clin. Microbiol. 43:1546–1551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boyd SC, et al. 2012. Epidemiology, clinical features and diagnosis of Mycobacterium ulcerans in an Australian population. Med. J. Aust. 196:341–344 [DOI] [PubMed] [Google Scholar]
- 4.Clarke B, Haverkort F, Keehner T, Lavender C, Johnson P. 2012. The first two cases of Mycobacterium ulcerans infection diagnosed in Western Australia. A new geographical focus of the Bairnsdale ulcer?, poster 26. Australasian Soc. Infect. Dis. (ASID) Annu. Sci. Meet., Fremantle, Western Australia, Australia. [Google Scholar]
- 5.Doig KD, et al. 2012. On the origin of Mycobacterium ulcerans, the causative agent of Buruli ulcer. BMC Genomics 13:258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Francis G, Whitby M, Woods M. 2006. Mycobacterium ulcerans infection: a rediscovered focus in the Capricorn Coast region of central Queensland. Med. J. Aust. 185:179–180 [DOI] [PubMed] [Google Scholar]
- 7.Fyfe J, Globan M, Lavender C, Johnson P, Stinear T. 2006. Molecular characterization of Mycobacterium ulcerans strains from different geographical locations in Australia and southeast Asia. 2006 WHO Annual Meeting on Buruli Ulcer, Geneva, Switzerland: http://www.who.int/buruli/events/Fyfe_ENG.pdf. [Google Scholar]
- 8.Fyfe JA, et al. 2007. Development and application of two multiplex real-time PCR assays for the detection of Mycobacterium ulcerans in clinical and environmental samples. Appl. Environ. Microbiol. 73:4733–4740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jackson K, Edwards R, Leslie D, Hayman J. 1995. Molecular method for typing Mycobacterium ulcerans. J. Clin. Microbiol. 33:2250–2253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Johnson PD, Veitch MG, Leslie DE, Flood PE, Hayman JA. 1996. The emergence of Mycobacterium ulcerans infection near Melbourne. Med. J. Aust. 164:76–78 [DOI] [PubMed] [Google Scholar]
- 11.Johnson PDR, et al. 2007. Mycobacterium ulcerans in mosquitoes captured during an outbreak of Buruli ulcer, southeastern Australia. Emerg. Infect. Dis. 13:1653–1660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lavender C, et al. 2007. First case of Mycobacterium ulcerans disease (Bairnsdale or Buruli ulcer) acquired in New South Wales. Med. J. Aust. 186:62–63 [DOI] [PubMed] [Google Scholar]
- 13.Lavender CJ, et al. 2008. Evaluation of VNTR typing for the identification of Mycobacterium ulcerans in environmental samples from Victoria, Australia. FEMS Microbiol. Lett. 287:250–255 [DOI] [PubMed] [Google Scholar]
- 14.Radford A. 1975. Mycobacterium ulcerans in Australia. Aust. N. Z. J. Med. 5:162–169 [DOI] [PubMed] [Google Scholar]
- 15.Roltgen K, et al. 2010. Single nucleotide polymorphism typing of Mycobacterium ulcerans reveals focal transmission of Buruli ulcer in a highly endemic region of Ghana. PLoS Negl. Trop. Dis. 4:e751 doi:10.1371/journal.pntd.0000751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Steffen CM, Smith M, McBride WJ. 2010. Mycobacterium ulcerans infection in North Queensland: the ‘Daintree ulcer’. ANZ J. Surg. 80:732–736 [DOI] [PubMed] [Google Scholar]
- 17.Stinear T, et al. 2007. Reductive evolution and niche-adaptation inferred from the genome of Mycobacterium ulcerans, the causative agent of Buruli ulcer. Genome Res. 17:192–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tiong A. 2005. The epidemiology of a cluster of Mycobacterium ulcerans infections in Point Lonsdale. Vic. Infect. Dis. Bull. 8:2–4 [Google Scholar]
- 19.Veitch M, et al. 1997. A large localized outbreak of Mycobacterium ulcerans infection on a temperate southern Australian island. Epidemiol. Infect. 119:313–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Walsh DS, Portaels F, Meyers WM. 2011. Buruli ulcer: advances in understanding Mycobacterium ulcerans infection. Dermatol. Clin. 29:1–8 [DOI] [PubMed] [Google Scholar]
- 21.World Health Organization 2008. Buruli ulcer: progress report, 2004–2008. Wkly. Epidemiol. Rec. 83:145–156 [PubMed] [Google Scholar]