Abstract
The main goal of this study was to determine the prevalence of methicillin-resistant Staphylococcus aureus (MRSA), particularly livestock-associated MRSA (LA-MRSA) in pigs and pork. The genotypic relatedness of isolates on the farm, at slaughter, and at the retail level was assessed. Paired nasal and perianal swab samples were collected from 10 cohorts of market-age pigs (24 pigs per cohort) and carcasses at slaughterhouse, and pork samples were collected at retail. Staphylococci were isolated using selective enrichment method. Isolates were tested for antimicrobial resistance by broth microdilution. Duplex PCR was used to confirm MRSA using species-specific (nuc) and methicillin resistance (mecA) genes. The clonal relatedness of isolates was determined using pulsed-field gel electrophoresis (PFGE), Staphylococcus protein A (spa) typing, multilocus sequence typing (MLST), and staphylococcal cassette chromosome mec element (SCCmec) typing. MRSA was detected in 5 of the 10 cohorts (50%), with the prevalence ranging from 0% to 12.5% per cohort. Of all the pigs sampled on the farm before they went to market, 3% (7/240) were MRSA positive. A higher prevalence of MRSA was detected at holding pens at the slaughterhouse (11% [27/240]). MRSA was also detected in 2% (4/235) of the carcasses and 4% (5/135) of the retail pork. While the isolates appear predominantly to be highly clonal, PFGE had a relatively higher discriminatory power (discriminatory index [DI] = 0.624). Four genotypic clusters were identified by PFGE; of the four clusters, clonal type B was predominant across the farm-to-retail continuum. MLST findings revealed that sequence type 5 (ST5) was the most predominant subtype (32/50). The livestock-associated MRSA (clonal complex 398 [CC398] or sequence type 398 [ST398]) was the second common type (12/50) and was detected at all stages from farm to retail. Nine of the 50 (18%) MRSA isolates belonged to spa type 539/t034 that were of ST398 based on MLST. The results of this study confirm that MRSA, including LA-MRSA, is common in herds of swine in Ohio and hereby shown to persist in the farm to processing and retail continuum.
INTRODUCTION
Methicillin-resistant Staphylococcus aureus (MRSA) primarily causes human disease, and until recently, food animals had not been considered important sources of infection. Among food animals, pigs have been implicated as one source of potential infections to humans, including farmers, slaughterhouse workers, and veterinarians who are in frequent contact with MRSA-colonized pigs (15, 23, 31, 36). Transmission of MRSA from colonized pigs to humans has been reported in The Netherlands (31). MRSA sequence type 398 (ST398) or clonal complex 398 (CC398) based on multilocus sequence typing (MLST), also referred to as livestock-associated MRSA (LA-MRSA), from pigs, pig farmers, and the environment has been reported in a number of European countries (5, 8, 9, 10, 14, 19, 28, 31) and North America (18, 22, 27, 34, 35).
The occurrence of MRSA in food products and the significance of food products in the transmission of MRSA in humans are unknown. One of the debatable issues is whether carcasses obtained from MRSA-colonized pigs could cross-contaminate pork meat and be a source of human infection (8, 36). Few studies from Europe (8) and North America (3, 26, 34) reported the occurrence of MRSA from retail meat. Other studies in North America also demonstrated the occurrence of MRSA in pigs and farm workers (18, 27, 35). Besides these limited investigations, data on the occurrence and prevalence of MRSA in pigs on the farm and at slaughter and carriage in the farm-to-table continuum in the United States are limited. In addition, it is not known how much of the retail meat contamination could be attributed to MRSA strains identified from pigs on the farm or at lairage (holding pens in slaughterhouses) or from the carcass immediately after slaughter. This serial cross-sectional study was conducted to determine the occurrence and persistence of MRSA among herds of swine on farms, at lairage, on carcasses and retail pork and compare the phenotypic and genotypic relatedness of MRSA isolates recovered across the pork production chain.
(Some of the findings in this study were presented at the Conference of Research Workers in Animal Diseases [CRWAD], 5 to 7 December 2010, Chicago, IL, and at the 9th International Conference on the Epidemiology and Control of Biological, Chemical and Physical Hazards in Pigs and Pork, SafePork Conference, 19 to 22 June 2011, Maastricht, The Netherlands.)
MATERIALS AND METHODS
Study design and sample collections.
A serial cross-sectional sampling design was used on market-age pigs on 10 farms in Ohio. The 10 farms were identified, and the factors used in consideration for recruitment in this study were specific slaughterhouse locations and associated retail outlet used where tracking of products was feasible. We collected paired nasal and perianal swabs from selected pigs (24 pigs from different pens on each farm) from a total of 10 farms (n = 480). For each cohort, on day 1, paired nasal swabs from anterior nares and perianal swabs were collected from each study pig on the farm. On day 2, the same batch of pigs was monitored and sampled at lairage prior to stunning. On the same day, matching carcass swab samples (24 per farm) were then collected from the same batch of slaughtered pigs at the postevisceration stage and before chilling (n = 235; five carcasses were missed for various reasons). On the third day, we followed the processed meat products with specific identification marks that originated from the same batch of pigs sampled on the farm and at the slaughterhouse. A total of 135 retail pork samples (n = 12 to 15 per batch) were collected from retail stores within 24 h after processing on the day of product arrival at the retail outlet.
Isolation and identification.
Nasal and perianal swab samples collected aseptically using sterile swab applicators were transported in liquid Stuart's medium (Becton Dickinson [BD], Franklin Lakes, NJ), kept at 4°C during transportation, and processed immediately upon arrival at the laboratory. Each swab sample was inoculated into 5 ml of Mueller-Hinton (MH) broth (BD) with 6.5% NaCl. Carcass swabs (hydrated sponge; 3M, Saint Paul, MN) were cultured in 90 ml of MH broth with 6.5% NaCl, and retail pork samples (25 g) were added to 225 ml of MH broth and incubated overnight at 37°C. On the second day, a loopful of the broth was inoculated onto oxacillin resistance screening agar (ORSA) (Oxoid, Lenexa, KS). The plates were incubated for 24 to 48 h at 37°C. Presumptive colonies were further confirmed by performing the catalase test, tube coagulase test, and S. aureus latex agglutination assay (Pastorex Staph-Plus; Bio-Rad, Hercules, CA). Presumptive S. aureus isolates were stored at −80°C for further phenotypic and genotypic testing. From each of the positive samples, up to three colonies were preserved for further characterization. Identification of the Staphylococcus species was performed at the USDA-ARS, Bacterial Epidemiology and Antimicrobial Resistance Research (BEAR) Laboratory, Athens, GA, using the Vitek 2 system (bioMérieux, Durham, NC) and the Vitek 2 Gram-positive identification cards according to the manufacturer's directions.
Antimicrobial susceptibility testing.
The antimicrobial susceptibility of all S. aureus isolates was tested at the USDA-ARS BEAR Laboratory. MICs (in micrograms per milliliter) for S. aureus were determined by the broth microdilution panels using the Sensititre Gram-Positive Plate GPN3F semiautomated antimicrobial susceptibility system (Trek Diagnostic Systems, Inc., Cleveland, OH). MIC values were manually recorded by Sensitouch where Clinical and Laboratory Standards Institute (CLSI) performance standards were used to determine resistance (6, 7). Results were analyzed using WHONET 5.4 (www.who.int/drugresistance/whonetsoftware) to determine resistance profiles. The antimicrobial agents and breakpoints were as follows: ampicillin (Am), ≥0.5 μg/ml; ceftriaxone (Cf), ≥64 μg/ml; ciprofloxacin (Cp), ≥4 μg/ml; clindamycin (Ca), ≥4 μg/ml; erythromycin (Er), ≥8 μg/ml; gatifloxacin (Gt), ≥2 μg/ml; gentamicin (Gm), ≥16 μg/ml; levofloxacin (Lv), ≥4 μg/ml; linezolid (Ln), ≥8 μg/ml; oxacillin (Ox), ≥4 μg/ml; penicillin G (Pn), ≥0.25 μg/ml; quinupristin-dalfopristin (Qd), ≥4 μg/ml; rifampin (Rif), ≥4 μg/ml; tetracycline (Te), ≥16 μg/ml; trimethoprim-sulfamethoxazole (T/S), ≥4/76 μg/ml; and vancomycin (Vn), ≥16 μg/ml. Daptomycin (Dp) (range, 0.25 to 8 μg/ml) and streptomycin (St) (≥1,000 μg/ml) were also tested; resistance breakpoints for these two antimicrobial agents have not been established by CLSI. S. aureus ATCC 29213 was used as a quality control strain.
Molecular characterization and genotyping.
S. aureus isolates were further confirmed as MRSA using a duplex PCR assay by detecting mecA (methicillin-resistant staphylococcus-specific) and nuc (S. aureus-specific) genes. From a total of 99 confirmed MRSA isolates, 50 were systematically chosen for further genotyping by pulsed-field gel electrophoresis (PFGE), multilocus sequence typing (MLST), Staphylococcus protein A (spa) typing, and staphylococcal cassette chromosome mec element (SCCmec) typing. The isolates were selected to represent farm sources (farms 1 to 10), sample type (nasal, perianal, carcass swab, and retail meat), stages of sampling (on the farm, at lairage, carcass prechilling, and retail store), and antimicrobial resistance profiling.
SCCmec typing.
Genomic DNA was extracted using DNeasy blood and tissue kits (Qiagen, Valencia, CA), and PCRs were performed using Illustra puReTaq ready-to-go PCR beads (GE Healthcare Life Sciences, Piscataway, NJ). Multiplex PCR methods (designated M-PCR1 and M-PCR2) and primers described by Kondo et al. (20) were used to characterize staphylococcal cassette chromosome mec types and mecA gene carriage in S. aureus isolates. These two multiplex methods identify multiple ccr complexes (types 1 through 5) and mec gene complexes (classes A, B, and C), allowing us to assign SCCmec types I through IX following the recommendations of the International Working Group on the Classification of Staphylococcal Cassette Chromosome Elements (IWG-SCC) (17). MRSA positive-control strains N315, 85/2082, CA05, 8/6-3P, 81/108, WIS, and HDE288 were used to validate the multiplex methods and were included as positive controls in every PCR run.
PFGE.
Pulsed-field gel electrophoresis was conducted following a modified protocol as described previously (25). As ST398 strains are nontypeable by PFGE using the standard SmaI macrorestriction enzyme, an isoschizomer enzyme, Cfr9I, which has the same recognition site as SmaI but cuts a different location within the site, was used in this study (1, 4). In brief, isolates were grown overnight at 37°C on MH agar plates (BD). Bacterial cell suspensions were prepared in tubes containing 2 ml of cell suspension buffer (CSB). Each of the adjusted cell suspensions (optical density at 610 nm of 1.3 to 1.4) was transferred to a labeled microcentrifuge tube containing 2 μl lysostaphin (2 mg/ml) and mixed gently. Agarose-embedded cells were then lysed with cell lysis buffer (CLB) (10 mM Tris-HCl [pH 7.2], 50 mM EDTA, 50 mM NaCl, 0.2% deoxycholate, 0.5% N-laurylsarcosine, 1% sarcosyl [Sigma, St. Louis, MO], 20 mg/ml proteinase K). Slices of the plugs were digested with 40 U of Cfr9I restriction enzyme (Fermentas Inc., Glen Burnie, MD) overnight at 37°C. The DNA fragments were then separated using the CHEF-DR III PFGE system (Bio-Rad, Hercules, CA) with the following conditions and reagents: 1% SeaKem Gold agarose (FMC BioProducts, Rockland, ME) in 0.5% Tris-borate-EDTA buffer, temperature of 14°C, voltage of 6 V/cm, run time of 19 h, with an initial switch time of 5.3 s and a final switch time of 34.9 s. Salmonella enterica serovar Braenderup H9812, the PulseNet “universal” standard marker strain, was the molecular reference marker. PFGE images (stained with ethidium bromide) were then analyzed with BioNumerics software version 4.6 (Applied Maths Scientific Software Development, Sint-Martens-Latem, Belgium) with a position tolerance of 2.0% and optimization of 1.5%.
spa typing.
Staphylococcus protein A (spa) repeat region typing was performed by PCR in a total volume of 50 μl containing approximately 100 ng of cleaned DNA, 5 pmol of each primer, 25 μl of 2-fold-concentrated HotStar Taq master mix (catalog no. 203445; Qiagen GmbH, Hilden, Germany), and an additional 7.5 nmol of MgCl2. Thermal cycling conditions included an initial denaturation step (15 min at 95°C), followed by 35 cycles of PCR, with 1 cycle consisting of denaturation (30 s at 94°C), annealing (30 s at 55°C), and extension (30 s at 72°C), with a single final extension step (5 min at 72°C). Standard desalted primers spa-F (F stands for forward) (5′-GAA CAA CGT AAC GGC TTC ATC C-3′) and spa-R (R stands for reverse) (5′-GCT TTT GCA ATG TCA TTT ACT G-3′) were used for amplification. The PCR product was purified by ExoSAP-IT (catalog no. 78201 1 ML; Affymetrix, CA) according to the instructions of the manufacturer. One amplicon that yielded two distinct bands was extracted from the gel separately and purified by QIAquick gel extraction kit (catalog no. 20874; Qiagen Science, MA) following the manufacturer's instructions. The purified amplicons were sequenced using standard Sanger sequencing at the Biomedical Genomics Center (BMGC) of the University of Minnesota using a primer concentration of 3.2 pmol and approximately 20 ng of amplified DNA as the template. Sequences were edited and assembled by using the software Sequencher 5.0. spa types were obtained using the sequence comparisons against egenomics (eGenomics, New York, NY) and Ridom (http://www.spaserver.ridom.de) databases.
MLST.
The 50 isolates were further genotyped using multilocus sequence typing following an established protocol (11). Briefly, specific regions (400 to 450 bp) of seven housekeeping genes, including arcC, aroE, glpF, gmk, pta, tpi, and yqiL, were amplified and sequenced using an eight capillary electrophoresis and CEQ-8000 genetic analysis system (Beckman Coulter, Palo Alto, CA). Allelic profiles and sequence types were generated once the specific complete edited sequences were submitted to the MLST global database at http://saureus.mlst.net.
Analysis of discriminatory power of the genotyping methods.
We applied Simpson's index of diversity to compare the relative discriminatory power of the genotyping approaches used in subtyping the 50 systematically selected isolates. The genotyping methods evaluated include SCCmec, PFGE, spa, and MLST. The discriminatory index (DI) of each of the methods was calculated using the equation shown below as recommended previously (16):
where DI is the discriminatory index, N is the total number of isolates in the sample population, S is the total number of types described, and nj is the number of isolates belonging to the jth type. The DI value (ranges between 0.0 and 1.0) shows the probability that two unrelated strains sampled from the test population will be placed into different types. The higher the DI value, the more discriminatory the method is.
RESULTS
MRSA in pigs on the farm and at lairage.
Of the total 10 cohorts of pigs included in this study, one or more pigs tested MRSA positive on the farm in three of the cohorts (cohorts 1, 3, and 6) and at lairage in five of the cohorts (Table 1). Of all the pigs examined on the farm, 3% (7/240) ranging from 0% to 12.5% of the samples tested were MRSA positive and prevalence at lairage was higher (11% [27/240]). MRSA was detected in 5 of the 10 herds included in the study (herds 1, 2, 3, 6, and 9) (Table 1). The prevalence of MRSA at lairage ranged from 0% to 54% per cohort. In addition, the proportion of MRSA-positive samples was relatively higher in nasal swabs (3% [6/240]) than in perianal swabs (<1% [2/240]) and at lairage (8% [20/240] for nasal swabs compared to 5% [12/240] for perianal samples).
Table 1.
Occurrence and prevalence of MRSA in pigs on the farm, at lairage, and from carcass swabs and retail pork
| Cohort no. | No. (%) of MRSA-positive pigs examined |
No. (%) of MRSA-positive carcass swabs examined (n = 235) | No. (%) of MRSA-positive retail pork samples examined (n = 135) | |
|---|---|---|---|---|
| On the farm (n = 240) | At lairage (n = 240) | |||
| 1 | 3/24 (12.5) | 13/24 (54.2) | 0/24 | 1/15 (6.7) |
| 2 | 0/24 | 1/24 (4.2) | 0/24 | 0/15 |
| 3 | 3/24 (12.5) | 11/24 (45.8) | 3/23 (13) | 2/15 (13.3) |
| 4 | 0/24 | 0/24 | 0/23 | 0/12 |
| 5 | 0/24 | 0/24 | 1/24 (4.2) | 0/14 |
| 6 | 1/24 (4.2) | 1/24 (4.2) | 0/24 | 0/12 |
| 7 | 0/24 | 0/24 | 0/24 | 0/12 |
| 8 | 0/24 | 0/24 | 0/23 | 0/15 |
| 9 | 0/24 | 1/24 (4.2) | 0/23 | 2/12 (16.7) |
| 10 | 0/24 | 0/24 | 0/23 | 0/13 |
| All herds (total)a | 7/240 (2.9) | 27/240 (11.3) | 4/235 (1.7) | 5/135 (3.7) |
The total number of MRSA-positive samples from pigs on the farm, at lairage, and from carcass swabs and retail pork is 43. However, the total number of isolates is 99 (as indicated in the text), since we collected paired nasal and perianal swabs from each pig and in addition from each of the positive samples, up to three colonies were picked for further characterization.
MRSA in carcass swabs.
We collected carcass swab samples that originated from the same cohorts of pigs at the postevisceration stage and before chilling. Of the 235 carcass swabs examined (five carcasses were missed for various reasons), 4 (2%) were MRSA positive (Table 1.). In three of the positive carcass swabs, the carcasses originated from pigs that belonged to cohort (herd) 3, which had a relatively higher proportion of MRSA-positive pigs. The fourth MRSA-positive carcass swab originated from pigs that belonged to herd 5; however, pigs in this particular herd were MRSA negative on the farm and at the lairage stage. On the other hand, in three of the herds (herds 1, 6, and 9) in which MRSA-positive pigs were detected on the farm and at lairage, MRSA was not detected in any of the carcasses.
MRSA in retail pork samples.
We monitored the same cohort of carcasses to the retail level, and a total of 135 pork samples were examined of which 5 (4%) tested MRSA positive. These positive samples originated from herds 1, 3, and 9 (Table 1). In herd 1, one or more MRSA-positive samples were detected in pigs on the farm and at lairage and retail pork; however, all carcass swabs (n = 24) taken from pigs in this herd were MRSA negative. Conversely, in herd 3, MRSA was detected across all sampling stages (on the farm, at lairage, carcass swabs, and retail pork). The third group of retail pork samples in which MRSA was detected originated from herd 9 in which one of the pigs was MRSA positive at lairage. However, MRSA was not detected among the carcass swabs from this cohort.
Antimicrobial resistance patterns (R-types).
MRSA isolates recovered from various stages of sampling exhibited multidrug-resistant (MDR) patterns, with resistance from 3 to up to 11 antimicrobial agents. The main resistance patterns exhibited included R-types AmCaErGmOxPnTe (46%) and AmCaOxPnTe (20%) (data not shown). These results are expected, as we utilized an oxacillin selective agar for isolation and identification. All MRSA isolates tested were susceptible to ciprofloxacin, daptomycin, linezolid, rifampin, and vancomycin.
Discriminatory power of genotyping methods.
The DI value of the genotyping methods used in this study was evaluated. Overall, while the isolates appear predominantly to be highly clonal, PFGE had a relatively higher discriminatory power among the methods evaluated. PFGE showed a DI value of 0.624. MLST showed a DI value of 0.538, and spa typing showed a DI value of 0.586. SCCmec typing of the isolates showed that only 12 of the 50 isolates conform to currently known types, while the remaining 38 appear to be of novel types. Therefore, we did not calculate the DI value of this method. Similarly, Ridom spa typing did not conform with most of the isolates in this study. Instead, we adopted a spa typing interpretation approach that is often used in isolates of animal origin. Using this typing, most of the isolates conformed with known types, and the discriminatory power of this approach was slightly higher than that of MLST as shown above.
Genotyping of MRSA isolates.
Of the total MRSA isolates tested for SCCmec types (n = 99), types II (7%), IV (5.1%), and V (16%) and nontypeable (4%) were detected. Although the majority of the MRSA isolates (67%) had identifiable ccr gene and mec gene complexes, the combinations we found did not match the currently reported types, suggesting that a new SCCmec type(s) might be circulating in the porcine isolates.
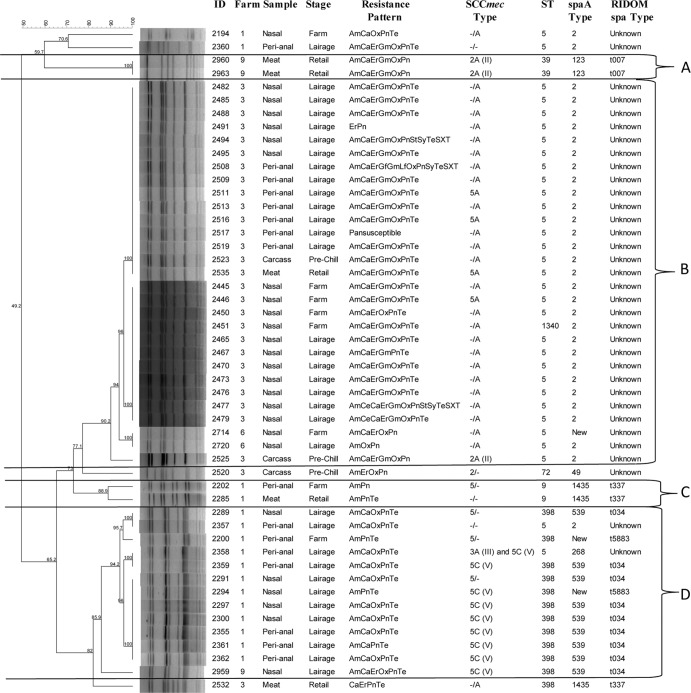
spa typing results (Fig. 1) showed that among the 50 MRSA isolates characterized, spa 2/unknown, 539/t034, and 1435/t37 MRSA were the main clones identified. The 539/t034 and 1435/t337 MRSA isolates have been previously shown to be highly associated with the LA-MRSA (CC/ST398). These isolates were detected among isolates recovered from pigs and retail pork. Overall, from the 50 isolates genotyped by PFGE, four genotypic clusters (A, B, C, and D) were detected (Fig. 1). Cluster B was the predominant cluster containing the majority (29 of 50 [58%]) of isolates and belonged to spa type 2/unknown and sequence type 5 (ST5) MRSA. The majority of these isolates in this cluster (27 of 29) originated from cohort 3, indicating clonal dissemination of the isolates on the farm, at lairage, and persistence during processing (carcasses and retail stores). The remaining two isolates originated from cohort 6 from pigs on the farm and at lairage. The majority (62%) of the isolates in this group had a similar R-type (AmCaErGmOxPnTe). Cluster C consisted of isolates from pigs on the farm (isolate 2202) and retail meat (isolate 2285) which originated from the same herd (herd 1) and belonged to the same sequence type (ST9) and spa type (1435/t337). Cluster D included isolates from pigs on the farm and at lairage, and the majority belonged to the same sequence type (ST398), spa type (539/t034), and SCCmec type (V), the typical livestock-associated MRSA (LA-MRSA) that has been reported in other regions, primarily in Europe.
Fig 1.
Dendrogram of Cfr9I macrorestriction PFGE profiles of representative MRSA isolates recovered from pigs on the farm, at lairage, from carcass swabs and retail pork samples, and association with sources (farms 1 to 10), sample type (nasal, perianal, carcass swab, retail meat), stage of sampling (location of sampling [on the farm, at lairage, carcass prechilling, and retail store]), antimicrobial resistance pattern, and SCCmec type. In the SCCmec type definition, we used a combination of the ccr type and mec class following the recommendations of the International Group on the Classification of Staphylococcal Cassette Chromosome Elements (IWG-SCC), and the − sign refers to the absence of amplification of the ccr gene complex and/or the mec gene complex by the multiplex PCR we used. Abbreviations: ID, identification; Am, ampicillin; Ce, ceftriaxone; Ca, clindamycin; Er, erythromycin; Gf, gatifloxacin; Gm, gentamicin; Lf, levofloxacin; Ox, oxacillin; Pn, penicillin G; St, streptomycin; Sy, Synercid (quinupristin and dalfopristin); Te, tetracycline; TeSXT, trimethoprim-sulfamethoxazole.
MLST genotyping showed that the 50 isolates belonged to six sequence types: ST5 (n = 32), ST398 (n = 12), ST9 (n = 2), ST39 (n = 2), and ST72 (n = 1) and ST1340 (n = 1). Isolates in PFGE cluster B genotyped by MLST belonged to ST5 (n = 28) and spa type 2/unknown (Fig. 1). PFGE cluster C consisted of two genotypically indistinguishable isolates from pigs on the farm (isolate 2202) and retail pork (isolate 2285). These isolates originated from farm 1 and shared a similar antimicrobial resistance pattern (R-type AmPn/Te), belonged to ST9 and spa type 1435/t337. ST398, the livestock-associated MRSA previously reported commonly from pigs mainly in European countries, was the second common strain (n = 12) and had the spa type 539/t034 (n = 9), 1435/t337 (n = 1), and new/t5883 (n = 2). ST398 was detected from three herds (herds 1, 3, and 9), and isolates were recovered from pigs on the farm (herd 1), at lairage (herds 1 and 9), and from retail pork (herd 3). ST5 (n = 9) was detected from two herds (herds 3 and 6), and isolates were recovered from pigs on the farm (n = 2), at lairage (n = 5), from a carcass (n = 1) and retail pork (n = 1). ST9 (n = 2) with spa type 1435/t337 was found in pigs on the farm and from retail pork (herd 1). The two isolates of ST39 and spa type 123/t007 were detected only from retail meat, perhaps implying that the strain is acquired during the processing, packaging, or later stages in handling products. ST72 and spa type 49/unknown (n = 1) was found from the carcass only.
DISCUSSION
Similar to reports in other countries, the findings in the current study further confirm that MRSA is present in swine herds in the United States. Among the 10 herds included in this study, at least one MRSA-colonized pig was detected in three of the herds (30%) sampled on the farm and five of the herds (50%) sampled at holding pens in slaughterhouses (lairage). The higher proportion of MRSA-positive cohorts as well as pigs at lairage in the slaughterhouses implies the presence of other risk factors contributing to an increased proportion of contaminated or colonized pigs during transportation or lairage.
Our findings were concordant with the results of a recent study in The Netherlands (5) which reported that pigs can become colonized with MRSA in the short period of time during transportation from the farm to the slaughterhouse. MRSA has previously been detected in swine transportation trucks, at lairage, in slaughter areas, in pork production shower facilities, and from farm and slaughterhouse personnel (2, 5, 9, 22, 29). Broens and colleagues (5) followed individual pigs from the farm to slaughter, indicated that many of the pigs that tested MRSA negative on the farm became MRSA positive during transportation from the farm to the slaughterhouse, and suggested that sources of colonization of pigs with MRSA could be mixing of pigs from different farms during the time between loading and stunning, contaminated trucks and lairage, or personnel (including truck drivers and/or slaughterhouse personnel). The same study (5) reported that the proportion of MRSA-positive pigs increased by 21% in those herds transported in trucks that were MRSA positive, whereas no MRSA was detected in those pigs transported in trucks that were MRSA negative. This indicated the role of transportation trucks and other related activities during transportation and holding at lairage as sources of colonization of pigs with MRSA.
A study by van Cleef and colleagues (29) estimated the occurrence of MRSA in slaughterhouse environments and workers and found that the status of MRSA of the environmental samples such as lairage and slaughter areas of pigs correlated well with the MRSA status of humans working in the slaughterhouse. Beneke et al. (2) also reported a 12% prevalence of MRSA in environmental samples collected from a large pig slaughterhouse with an integrated pork-processing unit. Leedom Larson and colleagues (22) detected MRSA in pork production shower facilities in two commercial swine production systems (3% and 26%) known to harbor MRSA-positive pigs. These studies underlined the role of the environment as a source of MRSA in the commercial pig production chain. In our study, in two of the herds (herds 2 and 9), none of the samples taken from pigs on the farm were MRSA positive, whereas MRSA was detected in pigs belonging to same herds sampled at lairage. These data support the findings of previous studies (2, 5, 29) on the potential role of the environment as a source of MRSA to pigs. We did not sample any of the transportation trucks, lairage, slaughter areas, or personnel associated with transportation and slaughtering of pigs, so it is difficult to draw any conclusion from this study on the role of the environment as a source of colonization of pigs with MRSA during transportation and holding at lairage, carcass, and retail meat contaminations.
We also noted that even though the proportion of MRSA-positive samples was relatively higher in nasal swabs collected from the same batch of pigs at lairage, 5% of paired perianal samples collected at lairage were MRSA positive. This suggests the occurrence of MRSA in the perianal region of finisher pigs and its potential implications in contaminating carcasses during the slaughtering process.
We found 2% of the carcasses positive for MRSA after evisceration. A previous study (2) reported a higher prevalence of 6% of MRSA in pig carcasses. Both studies confirm the occurrence of contamination of pig carcasses with MRSA during the slaughtering process. In our study, in herd 3, MRSA was also detected in pigs on the farm (13%) and at lairage (46%); 3 of the 24 (13%) carcasses from the same herd tested MRSA positive, illustrating the occurrence and persistence of MRSA at all stages of the pork production chain in this particular herd. In herd 5, none of the samples collected from pigs on the farm and at lairage were MRSA positive and one of the carcasses from this herd tested MRSA positive, suggesting the presence of other sources of MRSA contamination in the slaughtering process. It is also important to note that in four of the five herds (herds 1, 2, 6, and 9) in which MRSA was detected in pigs at lairage, none of the carcass swab samples tested MRSA positive. Since we did not sample carcasses before and after washing the carcasses, no conclusion can be drawn on the role of hygienic measures playing a role in minimizing contamination of carcasses from MRSA-colonized pigs. Further research is required to elucidate the role of hygienic measures in minimizing carcass contaminations with MRSA.
Of the retail meat samples examined, 4% (5/135) were contaminated with MRSA, which was in agreement with previous reports on MRSA in retail pork (26, 34). The five MRSA-positive retail meat samples originated from pig herds (herds 1, 3, and 9) in which MRSA was detected either in pigs on the farm and/or at lairage or carcass swabs. In one of the herds (herd 3), we detected MRSA across all sampling points (Table 1), and the majority of the isolates that originated from this herd showed similar phenotypic (antimicrobial resistance pattern AmCaErGmOxPn) and genotypic characteristics (PFGE, spa type, and MLST).
Genotypic findings overall showed the high clonal nature of the isolates identified from the 10 cohorts. In general, clustering of isolates is expected mainly because pigs are kept in intensive production systems. While the clonality of the isolates was an expected phenomenon, it was interesting to see the variation in the discriminatory power of each of the methods. Overall, Cfr9I PFGE had the highest DI value, followed by spa typing and MLST. PFGE, MLST SCCmec typing, and staphylococcal protein A (spa) sequence typing have been considered the traditional genotyping tools for MRSA characterization of which Cfr9I PFGE is a highly discriminatory method (1, 4).
Representative isolates recovered from pigs on the farm, at lairage, and from carcass swabs and retail meat (herd 3) and genotyped by MLST and spa typing indicated that the tested isolates from this herd belonged to ST5 and spa types 2/unknown. Even though we did not sample any farm worker in this study, other studies have shown that ST5 has previously been associated with human infections (12) but has not been implicated to be specifically associated with pigs, unlike ST398. A recent study from the United States identified this sequence type (ST5) from backyard pigs (13).
We identified MRSA from pigs on the farm and at lairage and subsequently from carcass swabs and retail meat. However, detection of MRSA isolates from different sampling stages by itself does not necessarily indicate that pigs are the only sources for the meat contamination. Therefore, attempts were made to determine how much of the retail meat contamination could be attributed to strains identified from pigs on the farm and at lairage or from carcass swabs at slaughter through comparisons of phenotypic and genotypic relatedness of isolates recovered from various sampling points. The results indicated the presence of phenotypic and genotypically related isolates among isolates recovered from pigs, carcasses, and retail pork as depicted in the Cfr9I PFGE dendrogram. As expected, many of the isolates recovered from pigs on the farm and at lairage from the same herd (herds 1 and 3) were more clonal and of the same sequence type compared to isolates recovered from different herds. However, the SCCmec types, even within the same cluster were often different. This finding is expected, as the SCC targets a specific chromosomal region unlike the other genotyping methods and thus is not suitable to determine overall clonality of isolates compared to PFGE, spa typing, and MLST. Other studies have also shown that MRSA strains with identical spa types can carry different SCCmec elements (30).
Even though relatively few MRSA isolates were genotyped by MLST in our study (n = 50), ST398 with spa type 539/t034 and 1435/t337 was identified in three of the five MRSA-positive herds from pigs on the farm and at lairage (herd 1), pigs at lairage (herd 9), and retail meat (from herd 3), indicating the common occurrence of MRSA ST398 from the farm to the retail level in the study area. Eight of the 12 ST398 strains carried SCCmec type V (5C [Fig. 1]), which is often associated with this clonal type. It has been reported that MRSA ST398 carrying an SCCmec of type V has been the predominant livestock-associated MRSA clone in Europe (1, 28), and a previous study of pigs and farm workers in Midwestern United States showed that the ST398 isolates were SCCmec type V (27).
A previous study (27) detected ST398 in pigs on the farm and farm workers in Iowa and Illinois. In our study, we report the occurrence of MRSA ST398 not only in pigs on the farm and at lairage but also in retail pork products. It is well-known that ST398 has been the predominant strain identified among pigs in Europe and North America. In this study, in addition to ST398, we also identified other sequence types, including ST5 and ST9 among U.S. pigs on the farm and at lairage, carcass, and/or retail pork. ST5 has been reported recently from U.S. retail pork (33). Khanna and colleagues (18) from Canada reported both ST398 and ST5 in pigs on the farm and farm workers. ST9 has been shown to be the predominant strain in Asian pigs (32) compared to Europe and North America where LA-MRSA belong predominantly to CC398 (14). Other less common MLST sequence types in pigs identified included ST72 from carcasses and ST39 from retail pork. ST72 MRSA clone has been previously reported from pork and beef (24) and from both community surveillance and hospital-acquired MRSA outbreaks elsewhere (21). We would also like to note that the 50 MRSA isolates characterized using genotypic approaches were also tested further for the carriage of staphylococcal enterotoxins (staphylococcal enterotoxin A [SEA], SEB, SEC, SED, and SEE) genes, and none of them were positive.
In summary, this study showed that MRSA is detected at different stages of the pork production chain and clearly shows its persistence from farm to retail levels and also the acquisition of strains along the food production and processing chain. Although we have not typed all MRSA isolates recovered in this study, we identified MRSA ST398 clones colonizing U.S. pigs on the farm and at lairage. Contamination of retail meat with ST398 and other clones that have previously been shown to be of significance to public health, including ST5, ST9, ST39, and ST72, shows the presence of diverse MRSA clones across the pork production chain and that pigs and pork could be important players in MRSA dissemination. In the present study, we examined a limited number of cohorts of pigs, with limited geographic representation, and therefore, the findings cannot be generalized.
ACKNOWLEDGMENTS
This study was financially supported by the National Pork Board (NPB6002-1611).
We thank the pig farmers and the slaughterhouse and retail shop personnel for their kind cooperation during sampling in the study period. We are grateful to T. Smith at the University of Iowa and P. Davies at the University of Minnesota for subject matter consultation. We thank T. Ito and K. Hiramatsu for providing reference strains for SCCmec typing.
Footnotes
Published ahead of print 12 September 2012
REFERENCES
- 1. Argudín MA, et al. 2010. High heterogeneity within methicillin-resistant Staphylococcus aureus ST398 isolates, defined by Cfr9I macrorestriction–pulsed-field gel electrophoresis profiles and spa and SCCmec types. Appl. Environ. Microbiol. 76:652–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Beneke B, et al. 2011. Prevalence of methicillin-resistant Staphylococcus aureus in a fresh meat pork production chain. J. Food Prot. 74:126–129 [DOI] [PubMed] [Google Scholar]
- 3. Bhargava K, et al. 2011. Methicillin-resistant Staphylococcus aureus in retail meat, Detroit, Michigan, USA. Emerg. Infect. Dis. 17:1135–1137 (Letter.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bosch T, et al. 2010. PFGE diversity within the methicillin-resistant Staphylococcus aureus clonal lineage ST398. BMC Microbiol. 10:40 doi:10.1186/1471-2180-10-40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Broens EM, Graat EA, Van Der Wolf PJ, Van De Giessen AW, De Jong MC. 2011. Transmission of methicillin-resistant Staphylococcus aureus among pigs during transportation from farm to abattoir. Vet. J. 189:302–305 [DOI] [PubMed] [Google Scholar]
- 6. Clinical and Laboratory Standards Institute (CLSI) 2006. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, 7th ed. Approved standard M7-A7 Clinical and Laboratory Standards Institute (CLSI), Villanova, PA [Google Scholar]
- 7. Clinical and Laboratory Standards Institute (CLSI) 2011. Performance standards for antimicrobial susceptibility testing; twenty-first informational supplement. CLSI document M100-S21 Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 8. de Boer E, et al. 2009. Prevalence of methicillin-resistant Staphylococcus aureus in meat. Int. J. Food Microbiol. 134:52–56 [DOI] [PubMed] [Google Scholar]
- 9. de Neeling AJ, et al. 2007. High prevalence of methicillin-resistant Staphylococcus aureus in pigs. Vet. Microbiol. 122:366–372 [DOI] [PubMed] [Google Scholar]
- 10. Denis O, et al. 2009. Methicillin-resistant Staphylococcus aureus ST398 in swine farm personnel, Belgium. Emerg. Infect. Dis. 15:1098–1101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Enright MC, Day NP, Davies CE, Peacock SJ, Spratt BG. 2000. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J. Clin. Microbiol. 38:1008–1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Feßler AT, et al. 2011. Characterization of methicillin-resistant Staphylococcus aureus isolates from food and food products of poultry origin in Germany. Appl. Environ. Microbiol. 77:7151–7157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gordoncillo MJ, et al. 2012. Detection of methicillin-resistant Staphylococcus aureus (MRSA) in backyard pigs and their owners, Michigan, USA. Zoonoses Public Health 59:212–216 [DOI] [PubMed] [Google Scholar]
- 14. Graveland H, Duim B, van Duijkeren E, Heederika D, Wagenaar JA. 2011. Livestock-associated methicillin-resistant Staphylococcus aureus in animals and humans. Int. J. Med. Microbiol. 301:630–634 [DOI] [PubMed] [Google Scholar]
- 15. Huijsdens XW, et al. 2006. Community-acquired MRSA and pig-farming. Ann. Clin. Microbiol. Antimicrob. 5:26 doi:10.1186/1476-0711-5-26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hunter PR, Gaston MA. 1988. Numerical index of the discriminatory ability of typing systems: an application of Simpson's index of diversity. J. Clin. Microbiol. 26:2465–2466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. International Working Group on the Classification of Staphylococcal Cassette Chromosome Elements (IWG-SCC) 2009. Classification of staphylococcal cassette chromosome mec (SCCmec): guidelines for reporting novel SCCmec elements. Antimicrob. Agents Chemother. 53:4961–4967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Khanna T, Friendship R, Dewey C, Weese JS. 2008. Methicillin-resistant Staphylococcus aureus colonization in pigs and pig farmers. Vet. Microbiol. 128:298–303 [DOI] [PubMed] [Google Scholar]
- 19. Köck R, et al. 2009. Prevalence and molecular characteristics of methicillin-resistant Staphylococcus aureus (MRSA) among pigs on German farms and import of livestock-related MRSA into hospitals. Eur. J. Clin. Microbiol. Infect. Dis. 28:1375–1382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kondo Y, et al. 2007. Combination of multiplex PCRs for staphylococcal cassette chromosome mec type assignment: rapid identification system for mec, ccr, and major differences in junkyard regions. Antimicrob. Agents Chemother. 51:264–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lee J, et al. 2011. Molecular characterization of methicillin-resistant Staphylococcus aureus obtained from the anterior nares of healthy Korean children attending daycare centers. Int. J. Infect. Dis. 15:e558–e563 [DOI] [PubMed] [Google Scholar]
- 22. Leedom Larson KR, et al. 2011. Methicillin-resistant Staphylococcus aureus in pork production shower facilities. Appl. Environ. Microbiol. 77:696–698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lewis HC, et al. 2008. Pigs as source of methicillin-resistant Staphylococcus aureus CC398 infections in humans, Denmark. Emerg. Infect. Dis. 14:1383–1389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lim SK, et al. 2010. Prevalence and characterization of methicillin-resistant Staphylococcus aureus in raw meat in Korea. J. Microbiol. Biotechnol. 20:775–778 [PubMed] [Google Scholar]
- 25. Mulvey MR, et al. 2001. Development of a Canadian standardized protocol for subtyping methicillin-resistant Staphylococcus aureus using pulsed-field gel electrophoresis. Canadian Committee for the Standardization of Molecular Methods. J. Clin. Microbiol. 10:3481–3485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Pu S, Han F, Ge B. 2009. Isolation and characterization of methicillin-resistant Staphylococcus aureus strains from Louisiana retail meats. Appl. Environ. Microbiol. 75:265–267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Smith TC, et al. 2009. Methicillin-resistant Staphylococcus aureus (MRSA) strain ST398 is present in Midwestern U.S. swine and swine workers. PLoS One 4:e4258 doi:10.1371/journal.pone.0004258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Tenhagen B-A, et al. 2009. Prevalence of MRSA types in slaughter pigs in different German abattoirs. Vet. Rec. 165:589–593 [DOI] [PubMed] [Google Scholar]
- 29. van Cleef BA, et al. 2010. High prevalence of nasal MRSA carriage in slaughterhouse workers in contact with live pigs in The Netherlands. Epidemiol. Infect. 138:756–763 [DOI] [PubMed] [Google Scholar]
- 30. van Duijkeren E, et al. 2008. Transmission of methicillin-resistant Staphylococcus aureus strains between different kinds of pig farms. Vet. Microbiol. 126:383–389 [DOI] [PubMed] [Google Scholar]
- 31. Voss A, Loeffen F, Bakker J, Klaassen C, Wulf M. 2005. Methicillin-resistant Staphylococcus aureus in pig farming. Emerg. Infect. Dis. 11:1965–1966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wagenaar JA, et al. 2009. Unexpected sequence types in livestock associated methicillin-resistant Staphylococcus aureus (MRSA): MRSA ST9 and a single locus variant of ST9 in pig farming in China. Vet. Microbiol. 139:405–409 [DOI] [PubMed] [Google Scholar]
- 33. Waters AE, et al. 2011. Multidrug-resistant Staphylococcus aureus in US meat and poultry. Clin. Infect. Dis. 52:1227–1230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Weese JS, Reid-Smith R, Rousseau J, Avery B. 2010. Methicillin-resistant Staphylococcus aureus (MRSA) contamination of retail pork. Can. Vet. J. 51:749–752 [PMC free article] [PubMed] [Google Scholar]
- 35. Weese JS, Rousseau J, Deckert A, Gow S, Reid-Smith RJ. 2011. Clostridium difficile and methicillin-resistant Staphylococcus aureus shedding by slaughter-age pigs. BMC Vet. Res. 7:41 doi:10.1186/1746-6148-7-41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wulf MW, et al. 2008. MRSA carriage in healthcare personnel in contact with farm animals. J. Hosp. Infect. 70:186–190 [DOI] [PubMed] [Google Scholar]