Abstract
Ninety-seven animal, human, and dairy Streptococcus porcinus or Streptococcus pseudoporcinus isolates in the CDC Streptococcus strain collection were evaluated on the basis of DNA-DNA reassociation, 16S rRNA and rpoB gene sequencing, conventional biochemical and Rapid ID 32 Strep identification methods, and antimicrobial susceptibility testing to determine their taxonomic status, characteristics for species differentiation, antimicrobial susceptibility, and relevance of clinical source. Nineteen of the 97 isolates (1 human, 18 swine) were identified as S. porcinus. The remaining 72 human isolates and 6 dairy isolates were identified as S. pseudoporcinus. The use of 16S rRNA or rpoB gene sequencing was required to differentiate S. porcinus from S. pseudoporcinus. The human and dairy S. pseudoporcinus isolates were biochemically distinct from each other as well as distinct by 16S rRNA and rpoB gene sequencing. Therefore, we propose the subspecies denominations S. pseudoporcinus subsp. hominis subsp. nov. for the human isolates and S. pseudoporcinus subsp. lactis subsp. nov. for the dairy isolates. Most strains were susceptible to the antimicrobials tested, with the exception of tetracycline. Two strains of each species were also resistant to clindamycin and erythromycin and carried the erm(A) (S. pseudoporcinus) or the erm(B) (S. porcinus) gene. S. porcinus was identified from a single human isolate recovered from a wound in an abattoir worker. S. pseudoporcinus was primarily isolated from the genitourinary tract of women but was also associated with blood, placental, and wound infections. Isolates reacting with group B antiserum and demonstrating wide beta-hemolysis should be suspected of being S. pseudoporcinus and not S. agalactiae.
INTRODUCTION
Due to the advancement and use of genetic sequence markers, the taxonomic standing of many microorganisms characterized in the past has been reevaluated. A recent example involves isolates that were previously identified as Streptococcus porcinus. This species was originally described in 1984 (8) as a pathogen associated with swine. The isolation from human sources of strains with nearly identical phenotypic characteristics was later described (10, 14, 22). A more recent publication describing a study that used 16S rRNA gene sequencing revealed that isolates primarily from the genital tract of female patients were significantly different (greater than 2.1% dissimilarity) and constituted a separate species, named Streptococcus pseudoporcinus (3). Consequently, we decided to reevaluate the identification of S. porcinus isolates from our previous studies (10, 14, 32) and more recent isolates included in the CDC Streptococcus Laboratory collection. The goal of this study was to determine the association of S. porcinus and S. pseudoporcinus isolates with different sources, analyze genetic and phenotype characteristics useful for their differentiation and precise identification, and examine potential differences in antimicrobial susceptibility patterns. Accordingly, isolates were evaluated by 16S rRNA and rpoB gene sequencing, DNA-DNA reassociation experiments, conventional biochemical testing, Rapid ID 32 Strep panels, and antimicrobial susceptibility patterns.
MATERIALS AND METHODS
Bacterial strains.
We included in the present study a total of 97 strains belonging to the S. porcinus/S. pseudoporcinus complex, comprising most (n = 42) of the strains described in previous publications of our group (10, 14, 32), 2 additional group E reference strains, and 53 additional isolates obtained between 1984 and 2010. From the last group, 22 strains were received from the collection of the Wadsworth Center, New York State Department of Health, and 3 strains were recovered from dairy products that were cultured during the investigation to elucidate a nephritis outbreak in a Brazilian city (2). The type strains of several other streptococcal species were also included for comparative purposes.
Phenotypic characteristics.
The isolates were tested for their phenotypic characteristics by using a conventional biochemical identification schema (13, 12) and the Rapid ID 32 Strep system (18) (bioMérieux, Inc.). Serogroups were determined by using the Lancefield method and antisera for groups B, E, P, U, V, NG1, NG2, and NG3, as previously described (10, 14, 16). Reactivity with group B antiserum was also evaluated using a PathoDX Strep grouping latex agglutination test kit (Remel, Lenexa, KS), in accordance with the manufacturer's package insert.
Antimicrobial susceptibility testing.
MICs were determined by using a broth microdilution panel method according to the manufacturer's instructions (PML Microbiologicals, Wilsonville, Oregon). The following antibiotics were tested: ampicillin, cefotaxime, clindamycin, erythromycin, levofloxacin, penicillin, tetracycline, and vancomycin. The MIC interpretive standards for beta-hemolytic streptococci were used (7).
Determination of macrolide resistance phenotypes and genotypes.
Macrolide resistance phenotypes were determined by using a modification of the double-disk test described by Seppälä et al. (24). Disks (Oxoid Ltd., Basingstoke, United Kingdom) of erythromycin (15 mg) and clindamycin (2 mg) were placed 12 mm apart on Mueller-Hinton agar (BBL Microbiology Systems, Cockeysville, MD) supplemented with 5% sheep blood which had been inoculated with a swab dipped into a bacterial suspension with a turbidity equivalent to a 0.5 McFarland standard. Resistant isolates were evaluated for the presence of macrolide resistance genes by PCR on the basis of previous recommendations (11, 29). Briefly, one loopful from an overnight culture on 5% sheep blood agar was suspended in 100 μl of TE buffer (10 mM Tris-HCl, 1 mM EDTA, pH 8.0) containing 0.1 mg of lysozyme and 5 U of mutanolysin. After 30 min incubation at 37°C, the suspensions were heated at 100°C for 5 min and the lysates were stored at −20°C. PCR conditions were as described by Sutcliffe et al. (29). The PCRs were done separately for each erythromycin-resistant determinant, with 4 mM magnesium used for both the erm(A) and mef primer sets and 2 mM used for the erm(B) primer set.
16S rRNA gene sequencing.
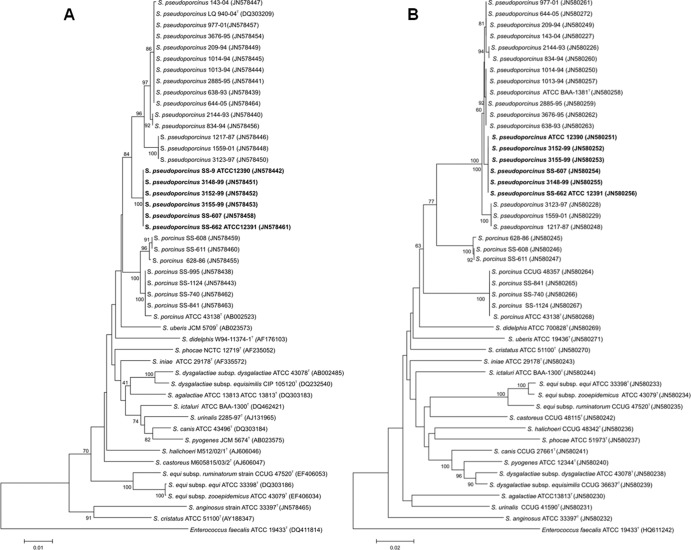
Amplification and sequencing of the 16S rRNA gene were performed as previously described (25, 26). Sequencing reaction products were purified with Centri-Sep plates (Princeton Separations, Princeton, NJ). Reaction mixtures were electrophoresed on an ABI 3130 or 3730 apparatus using POP-7 polymer (Applied Biosystems). Chromatograms were assembled and analyzed in Seqmerge (Wisconsin Package, version 10.3; Accelrys Inc., San Diego, CA). The consensus sequences were aligned by using the CLUSTAL W program (33) and trimmed to 1,419 bp to create a phylogenetic tree (Fig. 1A). Evolutionary analyses were conducted in the MEGA5 program (31). The evolutionary history was inferred using the neighbor-joining method (23). The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1,000 replicates) was determined (17). The evolutionary distances were computed using the maximum composite likelihood method (30).
Fig 1.
Phylogenetic trees from 16S rRNA gene (A) and rpoB gene (B) sequence data of representative strains belonging to the Streptococcus porcinus/Streptococcus pseudoporcinus complex and other select species of Streptococcus. GenBank accession numbers are given in parentheses. The evolutionary history was inferred by using the neighbor-joining method. The associated taxa clustered together in 60% or more of the 1,000 replicate trees. Enterococcus faecalis was used as the outgroup sequence. The scale bar indicates the units of the number of base substitutions per site.
rpoB gene sequencing.
Approximately 1,200 bp of the RNA polymerase beta subunit (rpoB) gene from each isolate was amplified with HotStarTaq (Qiagen, Valencia, CA) using primers UnivrpoB3F (5′-ATGGGNDCGNAAYATGCA) and UnivrpoB23R (5′-GAYATGGAYGTNTGYGC) in a 50-μl PCR mixture. The thermal cycling conditions were 95°C for 5 min; 15 cycles at 94°C for 1 min, 55°C for 1 min, and 72°C for 1 min with a decrease of the annealing temperature by 1°C at each cycle; 25 cycles of 94°C for 1 min, 50°C for 1 min, and 72°C for 1 min; 72°C for 10 min; and a final hold at 4°C. The PCR amplicons were purified through a Nucleospin column (Clontech) and eluted in 120 μl PCR-grade water. Cycle sequencing was performed as described above for the 16S rRNA gene but instead using primers UnivrpoB3F, UnivrpoB23R, UnivrpoBseq1 (5′-GGNGAYAARNTNKSNRR), and UnivrpoBseq2 (5′-YYNSMNANYTTRTCNCC). Raw sequence data were assembled in Geneious (version 5.4) software (9), and a neighbor-joining tree was created in MEGA5 using an alignment of 1,149 bp (Fig. 1B).
DNA-DNA reassociation experiments.
The cultures were grown in 2-liter flasks containing 1 liter Todd-Hewitt broth and incubated at 35°C for 18 h to 24 h on a rotary shaker. The cells were centrifuged to obtain a pellet, and total DNA was isolated with phenol-chloroform extraction and purified. DNA-DNA reassociation experiments were performed as previously described (4, 27). The temperatures used for DNA reassociation were 55°C (optimal conditions) and 70°C (stringent conditions).
Nucleotide sequence accession numbers.
The 16S rRNA and rpoB gene sequences for strains belonging to the S. porcinus/S. pseudoporcinus complex generated during this study have been deposited in GenBank. The accession numbers are shown in Fig. 1. The sequences of the 16S rRNA genes determined in this study have GenBank accession numbers JN578438 to JN578465. The partial sequences of the rpoB genes determined in this study have GenBank accession numbers JN580226 to JN580272.
RESULTS AND DISCUSSION
16S rRNA gene and rpoB gene sequence analyses.
The results of comparative 16S rRNA and rpoB gene sequence analysis for the isolates were confirmed with DNA-DNA reassociation experiments and are shown in Fig. 1A and B. Of the 97 isolates, 19 showed greater than 99% 16S rRNA gene sequence identity to the type strain of S. porcinus (GenBank accession number AB002523), and 78 isolates had greater than 98% sequence identity to S. pseudoporcinus (GenBank accession number DQ303209). However, six of these S. pseudoporcinus isolates, including SS-662 (ATCC 12391, a dairy isolate), formed a separate clade on the dendrogram (Fig. 1A) and differed significantly from other S. pseudoporcinus isolates on the basis of source and biochemical data. The sequence from SS-662 was 98.8% identical to the 16S rRNA gene sequence of the S. pseudoporcinus type strain and 98.3% identical to that of the S. porcinus type strain, values which exceed the criteria recommended for species identification (6). The identification of the dairy isolates as S. pseudoporcinus was possible on the basis of a signature sequence in the first 100 nucleotides of the 16S rRNA gene. All S. pseudoporcinus 16S rRNA gene sequences were identical and differed by 7 nucleotides from the S. porcinus sequence (region from positions 61 to 91 of the Escherichia coli 16S rRNA gene sequence, GenBank accession number X80725). The sequences are as follows: TGAGGTCTGGTGCTTGCACTAGACCAAG for S. pseudoporcinus and AGAGGACAGGTGCTTGCACCAGTCTAAT for S. porcinus. The S. pseudoporcinus dairy isolates could also be distinguished from the S. pseudoporcinus human isolates by a signature sequence in the region from positions 175 to 218 of the E. coli 16S rRNA gene sequence, GenBank accession number X80725. The sequences are as follows: CAATAGAGTACACATGTACTTAATTTAAAAGGGGCAACTGCTC for dairy isolates and GACTGGGGTTCACATGAACCCGAGTTAAAAGGAGCAAAAGCTT for human isolates. DNA-DNA hybridization or sequencing of an additional genetic target, the rpoB gene, was required for unambiguous identification of the dairy isolates as S. pseudoporcinus.
Overall, the rpoB gene sequencing results corroborated those obtained by 16S rRNA gene sequencing. Comparative analysis of the rpoB gene sequencing results is shown in Fig. 1B. The rpoB gene, however, is a more powerful tool to distinguish these two difficult-to-separate species. The rpoB sequence of S. pseudoporcinus strain SS-662 was 99.2% identical to that of S. pseudoporcinus and 89.6% identical to that of S. porcinus. In both the 16S rRNA and rpoB trees, the six S. pseudoporcinus dairy isolates, including SS-662, were strongly grouped together in a clade (100% bootstrap values). To the best of our knowledge, this is the first report on the application of rpoB gene sequencing for the study of microorganisms included in the S. porcinus/S. pseudoporcinus complex.
In a publication prior to the species description of S. pseudoporcinus, restriction fragment length polymorphism (RFLP) of the 16S rRNA gene with the enzyme BpiI was used to distinguish S. porcinus (group E, P, U, V, NG1, NG2, and NG3) strains from other Streptococcus species (1). In silico analysis of the 16S rRNA gene sequences for all S. pseudoporcinus strains in our study revealed that none of these isolates has the restriction enzyme site BpiI. Analysis of the 16S rRNA gene sequence of all 19 S. porcinus isolates in our study confirmed the presence of the BpiI restriction site. Thus, the BpiI RFLP method may be useful to distinguish between S. porcinus and S. pseudoporcinus and should be evaluated.
DNA-DNA reassociation experiments.
The type strains for S. porcinus (ATCC 43138) and S. pseudoporcinus (ATCC BAA-1381), as well as SS-662 (ATCC 12391, a select strain representative of the dairy isolates), were labeled with 32P for the DNA reassociation studies and hybridized with representative strains, as shown in Table 1. The representative group of strains was selected for inclusion in DNA-DNA reassociation studies on the basis of differences in 16S rRNA gene sequences, differences in source, and/or slight differences in biochemical patterns. Two strains are considered the same species if their DNA-DNA relatedness is 70% or higher at the optimal reassociation temperature and 55% or more at the stringent temperature with 5% divergence or less (34). The 6 S. porcinus isolates and human clinical isolate 628-86 were confirmed to be S. porcinus. S. porcinus ATCC 43183T was less than 58% related at the optimal temperature and less than 33% related at the stringent temperature and showed 5.0% or greater divergence from S. pseudoporcinus type strain ATCC BAA-1381 and reference dairy strain SS-662 (ATCC 12391), indicating that these were a different species. S. pseudoporcinus ATCC BAA-1381T showed 24% or less relatedness when hybridized against the type strains of other phylogenetically related species of Streptococcus. In reciprocal experiments, DNA studies with labeled S. pseudoporcinus ATCC BAA-1381T showed greater than 70% relatedness at the optimal and stringent temperatures to all 11 S. pseudoporcinus clinical isolates and the SS-662 dairy isolate and showed 24% or less relatedness to the other Streptococcus species tested. In addition, in experiments with labeled strain SS-662, all 5 additional dairy isolates and S. pseudoporcinus ATCC BAA-1381T were greater than 79% related at the optimal and stringent temperatures with less that 2% divergence. Strain SS-662 (ATCC 12391) was 48% related to S. porcinus ATCC 43183T with a divergence of 9.0%, indicating that it is not S. porcinus. Results of DNA-DNA reassociation with SS-662 against other Streptococcus species revealed 20% or less relatedness.
Table 1.
Levels of DNA relatedness between Streptococcus pseudoporcinus human isolates, S. pseudoporcinus dairy isolates, Streptococcus porcinus, and other selected species of Streptococcus
| Strain source of unlabeled DNA | Result with labeled DNA froma: |
||||||||
|---|---|---|---|---|---|---|---|---|---|
|
S. pseudoporcinus ATCC BAA-1381T (human isolate) |
S. pseudoporcinus ATCC 12391 (SS-662) (dairy isolate) |
S. porcinus ATCC 43138T |
|||||||
| RBR at 55°C | % D | RBR at 70°C | RBR at 55°C | % D | RBR at 70°C | RBR at 55°C | % D | RBR at 70°C | |
| S. porcinus/S. pseudoporcinus reference strains | |||||||||
| S. pseudoporcinus ATCC BAA-1381T (SS-1796) | 100 | 0.0 | 100 | 79 | 2.0 | 79 | 60 | 5.0 | 33 |
| S. pseudoporcinus dairy ATCC 12391T (SS-662) | 90 | 2.0 | 70 | 100 | 0.0 | 100 | 58 | 6.0 | 18 |
| S. porcinus ATCC 43138T (SS-1029) | 58 | 7.0 | 20 | 48 | 9.0 | 25 | 100 | 0.0 | 100 |
| S. porcinus/S. pseudoporcinus test strainsb | |||||||||
| S. pseudoporcinus human isolates (11 strains) | 83–100 | 0.0–1.5 | 71–100 | NP | NP | NP | NP | NP | NP |
| S. pseudoporcinus dairy isolates (5 strains) | NP | NP | NP | 90–100 | 0.0–0.5 | 85–100 | NP | NP | NP |
| S. porcinus animal isolates (6 strains) | NP | NP | NP | NP | NP | NP | 83–100 | 0.5–4.5 | 83–92 |
| S. porcinus human isolate (1 strain) | NP | NP | NP | NP | NP | NP | 97 | 1.5 | 92 |
| Reference strains of other species | |||||||||
| S. iniae ATCC 29178T | 20 | NP | NP | 19 | NP | NP | 19 | NP | NP |
| S. uberis ATCC 19436T | 17 | NP | NP | 17 | NP | NP | 16 | NP | NP |
| S. cristatus ATCC 51100T | 24 | NP | NP | 24 | NP | NP | 23 | NP | NP |
| S. halichoeri CCUG 48342T | 3 | NP | NP | 4 | NP | NP | 6 | NP | NP |
| S. didelphis ATCC 700828T | 19 | NP | NP | 20 | NP | NP | 20 | NP | NP |
| S. agalactiae ATCC 13813T | 17 | NP | NP | 18 | NP | NP | 19 | NP | NP |
| S. pyogenes ATCC 12344T | 18 | NP | NP | 18 | NP | NP | 19 | NP | NP |
| S. dysgalactiae subsp. dysgalactiae ATCC 43078T | 19 | NP | NP | 20 | NP | NP | 20 | NP | NP |
| S. dysgalactiae subsp. equisimilis CCUG 36637 | 21 | NP | NP | 21 | NP | NP | 20 | NP | NP |
| S. equi subsp. equi ATCC 33398T | 17 | NP | NP | 18 | NP | NP | 20 | NP | NP |
| S. equi subsp. zooepidemicus ATCC 43079T | 18 | NP | NP | 18 | NP | NP | 19 | NP | NP |
| S. equi subsp. ruminatorum CCUG 47520T | 10 | NP | NP | 11 | NP | NP | 11 | NP | NP |
| S. canis ATCC 43496T | 21 | NP | NP | 23 | NP | NP | 21 | NP | NP |
| S. anginosus ATCC 33397T | 11 | NP | NP | 10 | NP | NP | 11 | NP | NP |
| S. phocae ATCC 51973T | 8 | NP | NP | 9 | NP | NP | 8 | NP | NP |
RBR, relative binding ratio; % D, percent divergence, calculated to the nearest 0.5%; NP, not performed.
Numbers represent average values of the results obtained for the number of strains indicated in parentheses for each species or category.
Phenotypic characteristics.
Very few biochemical differences were observed between S. porcinus and S. pseudoporcinus, as shown in Table 2. Using conventional biochemical testing, hippurate hydrolysis was the key test in differentiating S. porcinus from S. pseudoporcinus. The six dairy source isolates were distinguished from both S. porcinus and S. pseudoporcinus by their inability to grow in 6.5% NaCl and to acidify pyruvate broth. The 100% positive results for Vogues-Proskauer and lactose tests observed for the dairy isolates are more consistent with S. porcinus (84% positive compared to 18% positive for S. pseudoporcinus). It is noteworthy that the different methodologies used gave different results for the hippurate hydrolysis test. This test is based on the bacterium's ability to hydrolyze hippurate to glycine and benzoic acid. The conventional tube method uses ferric chloride to form a precipitate with benzoic acid (15). The method on the Rapid ID 32 Strep panel uses ninhydrin to detect glycine (20). The ninhydrin method appears to be more sensitive, as the majority of S. porcinus and S. pseudoporcinus isolates were positive when tested by the Rapid ID 32 Strep system. This finding is consistent with that described in a previous publication describing a study that used several of the same strains (10). The Rapid ID 32 Strep system was not very useful in separating these species, as even fewer differences than the number obtained with the traditional biochemical method were observed. Again, it is noteworthy that with this method, 68% of S. porcinus isolates were hippurate positive with ninhydrin; however, all tested negative using the conventional method with ferric chloride. Using the Rapid ID 32 Strep system, all 19 S. porcinus isolates were identified as S. porcinus, with 18 strains having a confidence level of greater than 90%. All 72 S. pseudoporcinus strains were identified as S. porcinus with a confidence level of greater than 90%. Three S. pseudoporcinus dairy isolates were identified as S. porcinus, but with a confidence level of 81.7%, and the other three were identified as S. uberis with a confidence level of 74.1%.
Table 2.
Phenotypic characteristics of Streptococcus pseudoporcinus and Streptococcus porcinus isolatesa
| Test and phenotypic characteristic |
S. pseudoporcinus human isolates (total n = 72) |
S. pseudoporcinus dairy strains (total n = 6) |
S. porcinus (total n = 19) |
||||||
|---|---|---|---|---|---|---|---|---|---|
| No. | % | RXN | No. | % | RXN | No. | % | RXN | |
| Conventional tests | |||||||||
| Pyrase | 59 | 83 | V | 6 (1)b | 100 | + | 17 (6) | 89 | V |
| Bile esculin | 30 | 41 | V | 0 | 0 | − | 1 | 5 | − |
| 6.5% NaCl | 72 | 100 | + | 0 | 0 | − | 19 | 100 | + |
| Esculin | 72 | 100 | + | 6 | 100 | + | 19 | 100 | + |
| Arginine | 72 | 100 | + | 6 | 100 | + | 19 | 100 | + |
| Hippurate | 67 | 93 | + | 0 | 0 | − | 0 | 0 | − |
| Pyruvate | 60 | 83 | V | 0 | 0 | − | 14 | 74 | V |
| Voges-Proskauer | 13 | 18 | V | 6 | 100 | + | 16 (5) | 84 | V |
| CAMP | 70 | 97 | + | 6 | 100 | + | 19 | 100 | + |
| Acid from: | |||||||||
| Glycerol | 52 | 72 | V | 0 | 0 | − | 1 | 5 | − |
| Lactose | 2 | 3 | − | 6 | 100 | + | 6 | 32 | V |
| Maltose | 72 | 100 | + | 6 | 100 | + | 19 | 100 | + |
| Mannitol | 72 | 100 | + | 6 | 100 | + | 19 | 100 | + |
| Ribose | 72 | 100 | + | 6 | 100 | + | 19 | 100 | + |
| Sorbitol | 72 | 100 | + | 6 | 100 | + | 19 | 100 | + |
| Sucrose | 72 | 100 | + | 6 | 100 | + | 19 | 100 | + |
| Trehalose | 72 | 100 | + | 6 | 100 | + | 19 | 100 | + |
| MGPc | 69 | 97 | + | 6 | 100 | + | 14 | 74 | V |
| Rapid ID 32 Strep testsd | |||||||||
| α-Galactosidase | 40 | 56 | V | 0 | 0 | − | 16 | 84 | V |
| Lactose | 0 | 0 | − | 6 | 100 | + | 9 | 43 | V |
| Hippurate | 68 | 94 | + | 6 | 100 | + | 13 | 68 | V |
| Pullulan | 70 | 97 | + | 0 | 0 | − | 17 | 89 | V |
| M-β-Glue | 5 | 7 | − | 0 | 0 | − | 12 | 63 | V |
No., number of strains with positive results; RXN, expected reaction (+, 90% or more of the strains were positive; −, 90% or more of the strains were negative; V, variable [11 to 89% of the strains were positive]).
Numbers in parentheses indicate the number of strains with weak reactions.
MGP, methyl-α-d-glucopyranoside.
Only differential tests are listed.
M-β-Glu, methyl-β-d-glucopyranoside.
Information available in the literature was reviewed (10, 13, 32), and more recent isolates were tested to analyze the serogroup data for S. porcinus and S. pseudoporcinus. The animal S. porcinus strains SS-995 and SS-996 cross-reacted with CDC group B antisera as well as with several commercial kits. In addition, the clinical isolate 1256-95, described as reacting with all commercial group B antisera, has subsequently been identified as S. pseudoporcinus. A noteworthy finding was that only two of the S. porcinus strains tested reacted with NG1 antisera, whereas almost all S. pseudoporcinus strains reacted. Interestingly, NG1 group antiserum was derived from S. porcinus SS-995, an animal isolate. Five of the six S. pseudoporcinus dairy isolates reacted with group E antisera, one isolate was nongroupable, and none reacted with group B or NG1 antisera. This would indicate that antigenic entities are shared between S. porcinus, S. pseudoporcinus, and group B Streptococcus agalactiae. While serogroup may be helpful with identification, it would not be conclusive in identifying S. pseudoporcinus.
Current testing with the Remel PathoDx Strep group B latex reagent revealed that one S. porcinus isolate (5%) reacted strongly (greater than 2+ agglutination), 74% (n = 14) showed a weak reaction (grainy agglutination), and 21% (n = 4) did not react. Ten (14%) of the S. pseudoporcinus strains reacted strongly, 69% (n = 50) reacted weakly, and 17% (n = 12) gave no reaction. None of the S. pseudoporcinus dairy isolates reacted with the PathoDx group B reagent. In addition, two S. porcinus isolates and all the human S. pseudoporcinus isolates reacted with NG1 antisera. None of the six S. pseudoporcinus dairy isolates reacted with NG1 antisera. These six dairy isolates demonstrated serogroup E in previous studies (10). The two additional serogroup E reference strains isolated from pigs, SS-608 and SS-611 (which were not included in the earlier studies), were retested with the current methodology and identified as S. porcinus by 16S rRNA and rpoB gene sequencing and DNA-DNA reassociation. This finding is somewhat expected, since serogroup is often not species specific, and other species of Streptococcus (e.g., S. uberis) also react with serogroup E antisera (35). On the basis of results of this study and previous studies, S. porcinus may possess Lancefield group B, E, P, U, V, NG1, NG2, or NG3 or no group antigen; human S. pseudoporcinus may possess B, P, or NG1 or no group antigen; and dairy S. pseudoporcinus isolates may possess E or no group antigen. Isolates reacting with group B antiserum and demonstrating wide zones of beta hemolysis should be suspected of being S. pseudoporcinus and not S. agalactiae.
Antimicrobial susceptibility.
There was little difference in the susceptibility of S. porcinus and S. pseudoporcinus strains to the antimicrobials tested, as shown in Table 3. A few strains of both species showed resistance to both clindamycin and erythromycin, and more than half of all the isolates were resistant to tetracycline. The finding of high percentages of resistance to tetracycline is in agreement with previously published studies (10, 19). Two S. porcinus isolates showed multiresistance to clindamycin, erythromycin, and tetracycline. The constitutive macrolide, lincosamide, streptogramin B (cMLS) phenotype and the erm(B) gene were detected in these two erythromycin-resistant S. porcinus isolates. Over 72% of the S. pseudoporcinus strains were resistant to tetracycline. Two S. pseudoporcinus strains also showed resistance to both clindamycin and erythromycin, indicative of the cMLS phenotype. The erm(A) gene was detected in these two S. pseudoporcinus isolates. This is the first report of the presence of the erm genes among strains of S. porcinus and S. pseudoporcinus. The six S. pseudoporcinus dairy isolates, isolated from milk and cheese, were very susceptible to all antibiotics.
Table 3.
Antimicrobial susceptibility of 72 Streptococcus pseudoporcinus human isolates, 6 S. pseudoporcinus dairy isolates, and 19 Streptococcus porcinus isolates
| Antimicrobial agent |
S. pseudoporcinus human isolates (n = 72) |
S. pseudoporcinus dairy isolates (n = 6) |
S. porcinus (n = 19) |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MIC (μg/ml) |
No. of isolates susceptible (resistant)a | MIC (μg/ml) |
No. of isolates susceptible | MIC (μg/ml) |
No. of isolates susceptible (resistant) | |||||||
| Range | 50% | 90% | Range | 50% | 90% | Range | 50% | 90% | ||||
| Ampicillin | 0.03–0.06 | 0.06 | 0.06 | 72 | 0.015–.03 | 0.03 | 0.03 | 6 | 0.03–0.06 | 0.06 | 0.06 | 19 |
| Cefotaxime | 0.03–0.06 | 0.03 | 0.06 | 72 | 0.015–0.015 | 0.015 | 0.015 | 6 | 0.03–0.06 | 0.03 | 0.06 | 19 |
| Clindamycin | 0.03–>32 | 0.06 | 0.12 | 70 (2b) | 0.03–0.03 | 0.03 | 0.03 | 6 | 0.06–>32 | 0.06 | 0.06 | 17 (2c) |
| Erythromycin | 0.008–>32 | 0.06 | 0.12 | 70 (2b) | 0.03–0.03 | 0.03 | 0.03 | 6 | 0.03–>32 | 0.06 | >32 | 17 (2c) |
| Levofloxacin | 0.5–1 | 1 | 1 | 72 | 0.05–0.5 | 0.5 | 0.5 | 6 | 0.5–1 | 1 | 1 | 19 |
| Penicillin | 0.008–0.03 | 0.015 | 0.015 | 72 | 0.008–0.008 | 0.008 | 0.008 | 6 | 0.008–0.03 | 0.03 | 0.03 | 19 |
| Tetracycline | 1–>16 | >16 | >16 | 20 (52) | 1–1 | 1 | 1 | 6 | 1–>16 | 1 | >16 | 10 (9) |
| Vancomycin | 0.25–1 | 0.5 | 0.5 | 72 | 0.5–1.0 | 0.5 | 1 | 6 | 0.25–1 | 1 | 1 | |
CLSI interpretive standards for large-colony beta-hemolytic Streptococcus were used.
These two S. pseudoporcinus isolates carried the erm(A) gene.
These two S. porcinus isolates carried the erm(B) gene.
Sources and clinical significance.
Analysis of historical records of the S. porcinus isolates confirmed only one S. porcinus strain that was associated with human clinical disease. This strain was isolated from a skin lesion of a 54-year-old male abattoir worker, and zoonotic transmission is presumed. The remaining 18 S. porcinus isolates were from porcine sources. S. pseudoporcinus was not identified among the swine isolates in our collection.
Seventy-eight isolates were identified as S. pseudoporcinus on the basis of 16S rRNA and rpoB gene sequencing and DNA-DNA hybridization. The majority (n = 72) of the S. pseudoporcinus isolates were obtained from clinical human sources and 6 were from dairy sources. The demographic data for the patients and the sources of isolation for 59 of the 72 clinical S. pseudoporcinus isolates are shown in Table 4. Of the 56 isolates of which the source was provided, 78% (44/56) were recovered from the reproductive tract or urine of females aged from newborn to 56 years. We were not provided with patient nationality and cannot comment on the findings of a previous study (19) linking the isolation of S. pseudoporcinus isolates to patients originating from the Caribbean and sub-Saharan Africa. The gender was not provided for the source of 21 of the 72 isolates, so the percentage of isolates from males could be slightly higher. A recent report describes the isolation of S. pseudoporcinus from a thumb infection of a 33-year-old male (21), which is in agreement with our finding that S. pseudoporcinus infection in males is typically associated with wound infections (Table 4). Blood was listed as the source for 5 isolates (4 from females and 1 from an individual whose gender was not provided). However, in a recent population-based study of invasive disease due to beta-hemolytic streptococci other than groups A and B (5), the CDC Streptococcus Laboratory did not identify any S. porcinus or S. pseudoporcinus isolates. Either the incidence of S. pseudoporcinus associated with invasive disease is very low in the general population, or these isolates were misidentified as S. agalactiae on the basis of their occasional cross-reactivity with group B antiserum (28). We have not retested group B isolates in our population-based study collection to answer this question.
Table 4.
Sources and patient characteristics of 78 Streptococcus pseudoporcinus and 19 Streptococcus porcinus isolates included in the present study
| Source or characteristica | S. pseudoporcinusb (n = 78) | S. porcinusc (n = 19) |
|---|---|---|
| No. of isolates from: | ||
| Animals | 0 | 18 |
| Porcines | 0 | 18 |
| Dairy products | 6 | 0 |
| Milk | 5 | 0 |
| Cheese | 1 | 0 |
| Humans | 73 | 1 |
| No. of human subjects with characteristic/total no. (%) | ||
| Gender (n = 53/59, 90%) | ||
| Male | 3/53 (5.7) | 1/19 (5.3) |
| Female | 50/53 (94.3) | |
| Range (mean) age (yr) by gender (n = 28/59, 47%) | ||
| Male (3/28) | 41–47 (43) | 54 |
| Female (25/28) | Newborn–56 (30) | |
| Clinical source of isolates (n = 55/59, 93%) | ||
| Blood | 5/55 (9.1) | |
| Wound | 7/55 (12.7) | 1 |
| Placenta/umbilicus | 8/55 (14.5) | |
| Urine | 4/55 (7.4) | |
| Cervix | 4/55 (7.3) | |
| Genital | 25/55 (45.4) |
No information (sex, age, or source) was provided for 13/72 (18%) of S. pseudoporcinus human isolates; for the remaining 59/72, at least one piece of information was provided.
All three S. pseudoporcinus isolates from males were from wounds (sinus, scrotum, and no specific anatomic location provided).
The one human S. porcinus isolate was from the wound of a male abattoir worker.
We have applied a polyphasic approach for the characterization of strains belonging to the S. porcinus/S. pseudoporcinus complex. Historical and recent isolates were examined to determine methods for the identification and determination of the antimicrobial susceptibility and occurrence of S. porcinus and S. pseudoporcinus isolates from human, animal, and dairy sources. Sequencing of the rpoB gene was used for the first time to identify these difficult-to-differentiate microorganisms. In spite of nearly identical phenotypic characteristics, DNA-DNA reassociation studies as well 16S rRNA and rpoB gene sequencing confirm that S. porcinus and S. pseudoporcinus are clearly two distinct species. Isolates from human sources were predominantly identified as S. pseudoporcinus, while isolates from animals were identified as S. porcinus. Only one human isolate was identified as S. porcinus. In addition, dairy isolates from bovine milk and cheese were identified as S. pseudoporcinus, and biochemical reactions and source differences in conjunction with molecular data showed that they form distinct subclusters, suggesting that these strains may represent a S. pseudoporcinus subspecies.
On the basis of these findings, we propose to subdivide the species S. pseudoporcinus into two subspecies. For the human isolates that usually grow in the presence of 6.5% NaCl, are positive for hippurate hydrolysis and pyruvate tests, and do not produce acids from lactose, we propose the denomination S. pseudoporcinus subsp. hominis subsp. nov. The second subspecies, for which we propose the denomination S. pseudoporcinus subsp. lactis subsp. nov., accommodates the dairy isolates that do not grow in the presence of 6.5% NaCl, are negative for hippurate hydrolysis and pyruvate tests, and produce acids from lactose.
ACKNOWLEDGMENTS
We thank Shantia Warren and Tim Bailiff for their laboratory support.
L.M.T. and V.L.C.M. were supported in part by grants from Brazilian government agencies (FAPERJ and CNPq).
Footnotes
Published ahead of print 29 August 2012
REFERENCES
- 1.Abdulmawjood A, Weiβ R, Lämmler C. 1998. Species identification of Streptococcus porcinus by restriction fragment length polymorphism analysis of 16S ribosomal DNA. Res. Vet. Sci. 65:85–86 [DOI] [PubMed] [Google Scholar]
- 2.Balter S, et al. 2000. Epidemic nephritis in Nova Serrana, Brazil. Lancet 355:1776–1780 [DOI] [PubMed] [Google Scholar]
- 3.Bekal S, Gaudreau C, Laurence RA, Simoneau E, Raynal L. 2006. Streptococcus pseudoporcinus sp. nov., a novel species isolated from the genitourinary tract of women. J. Clin. Microbiol. 44:2584–2586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brenner DJ, McWhorter AC, Leete Knutson JK, Steigerwalt AG. 1982. Escherichia vulneris: a new species of Enterobacteriaceae associated with human wounds. J. Clin. Microbiol. 15:1133–1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Broyles LN, et al. 2009. Population-based study of invasive disease due to β-hemolytic streptococci of groups other than A and B. Clin. Infect. Dis. 48:706–712 [DOI] [PubMed] [Google Scholar]
- 6.Clarridge JE., III 2004. Impact of 16S rRNA gene sequence analysis for identification of bacteria on clinical microbiology and infectious diseases. Clin. Microbiol. Rev. 17:840–862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clinical and Laboratory Standards Institute 2011. Performance standards for antimicrobial susceptibility testing. M100-S21. Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 8.Collins MD, Farrow JAE, Katie V, Kandler O. 1984. Taxonomic studies on streptococci of serological groups E, P, U, and V: a description of Streptococcus porcinus sp. nov. Syst. Appl. Microbiol. 5:402–413 [Google Scholar]
- 9.Drummond AJ, et al. 2011. Geneious v5.4. Biomatters Ltd., Auckland, New Zealand [Google Scholar]
- 10.Duarte RS, Barros RR, Facklam RR, Teixeira LM. 2005. Phenotypic and genotypic characteristics of Streptococcus porcinus isolated from human sources. J. Clin. Microbiol. 43:4592–4601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Duarte RS, Bellei BC, Miranda OP, Brito MAVP, Teixeira LM. 2005. Distribution of antimicrobial resistance and virulence-related genes among Brazilian group B streptococci recovered from bovine and human sources. Antimicrob. Agents Chemother. 49:97–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Facklam RR. 2002. What happened to the streptococci: overview of taxonomic and nomenclature changes. Clin. Microbiol. Rev. 15:613–630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Facklam RR, Elliot JA. 1995. Identification, classification and clinical relevance of catalase-negative, gram-positive cocci, excluding the streptococci and enterococci. Clin. Microbiol. Rev. 8:479–495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Facklam R, Elliot J, Pigott N, Franklin AR. 1995. Identification of Streptococcus porcinus from human sources. J. Clin. Microbiol. 33:385–388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Facklam RR, Padula JF, Thacker LG, Wortham EC, Sconyers BJ. 1974. Presumptive identification of group A, B, and D streptococci. Appl. Microbiol. 27:107–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Facklam RR, Washington JA., II 1991. Streptococcus and related catalase-negative, gram-positive cocci, p 238–257 In Balows A, Hausler WJ, Jr, Herrmann KL, Isenberg HD, Shadomy HJ. (ed), Manual of clinical microbiology, 5th ed. ASM Press, Washington, DC [Google Scholar]
- 17.Felsenstein J. 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791 [DOI] [PubMed] [Google Scholar]
- 18.Freney J, et al. 1992. Description and evaluation of the semiautomated 4-h Rapid ID 32 Strep method for identification of streptococci and members of related genera. J. Clin. Microbiol. 30:2657–2661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gaudreau C, et al. 2007. Epidemiological, biochemical and antimicrobial susceptibility characteristics of Streptococcus pseudoporcinus isolated in Quebec, Canada, from 1997 to 2006. J. Med. Microbiol. 56:1620–1624 [DOI] [PubMed] [Google Scholar]
- 20.Hwang MN, Ederer GM. 1975. Rapid hippurate hydrolysis method for presumptive identification of group B streptococci. J. Clin. Microbiol. 1:114–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mahlen SD, Clarridge JE., III 2009. Thumb infection caused by Streptococcus pseudoporcinus. J. Clin. Microbiol. 47:3041–3042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Martin C, Fermeaux V, Eyraud JL, Aubard Y. 2004. Streptococcus porcinus as a cause of spontaneous preterm human stillbirth. J. Clin. Microbiol. 42:4396–4398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Saitou N, Nei M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406–425 [DOI] [PubMed] [Google Scholar]
- 24.Seppälä H, Nissinen A, Yu Q, Huovinen P. 1993. Three different phenotypes of erythromycin-resistant Streptococcus pyogenes in Finland. J. Antimicrob. Chemother. 32:885–891 [DOI] [PubMed] [Google Scholar]
- 25.Shewmaker PL, et al. 2007. Streptococcus ictaluri sp. nov., isolated from channel catfish Ictalurus punctatus broodstock. Int. J. Syst. Evol. Microbiol. 57:1603–1606 [DOI] [PubMed] [Google Scholar]
- 26.Shewmaker PL, et al. 2004. Vagococcus carniphilus sp. nov., isolated from ground beef. Int. J. Syst. Evol. Microbiol. 54:1505–1510 [DOI] [PubMed] [Google Scholar]
- 27.Shewmaker PL, Steigerwalt AG, Shealey L, Weyant R, Facklam RR. 2001. DNA relatedness, phenotypic characteristics, and antimicrobial susceptibilities of Globicatella sanguinis strains. J. Clin. Microbiol. 39:4052–4056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stoner KA, Rabe LK, Austin MN, Meyn LA, Hillier SL. 2011. Incidence and epidemiology of Streptococcus pseudoporcinus in the genital tract. J. Clin. Microbiol. 49:883–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sutcliffe J, Grebe T, Tait-Kamradt A, Wondrack L. 1996. Detection of erythromycin-resistant determinants by PCR. Antimicrob. Agents Chemother. 40:2562–2566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tamura K, Nei M, Kumar S. 2004. Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc. Natl. Acad. Sci. U. S. A. 101:11030–11035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tamura K, et al. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28:2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thompson T, Facklam R. 1997. Cross-reactions of reagents from streptococcal grouping kits with Streptococcus porcinus. J. Clin. Microbiol. 35:1885–1886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thompson JD, Higgins DG, Gibson TJ. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wayne LG, et al. 1987. International Committee on Systematic Bacteriology. Report of the Ad Hoc Committee on Reconciliation of Approaches to Bacterial Systematics. Int. J. Syst. Bacteriol. 37:463–464 [Google Scholar]
- 35.Wessman GE. 1986. Biology of the group E streptococci: a review. Vet. Microbiol. 12:297–328 [DOI] [PubMed] [Google Scholar]