Abstract
The Candida haemulonii species complex is currently known as C. haemulonii groups I and II. Here we describe C. haemulonii group II as a new species, Candida duobushaemulonii sp. nov., and C. haemulonii var. vulnera as new a variety of C. haemulonii group I using phenotypic and molecular methods. These taxa and other relatives of C. haemulonii (i.e., Candida auris and Candida pseudohaemulonii) cannot be differentiated by the commercial methods now used for yeast identification. Four isolates (C. haemulonii var. vulnera) differed from the other isolates of C. haemulonii in the sequence of the internal transcribed spacer (ITS) regions of the nuclear rRNA gene operon. The new species and the new variety have a multiresistant antifungal profile, which includes high MICs of amphotericin B (geometric mean MIC, 1.18 mg/liter for C. haemulonii var. vulnera and 2 mg/liter for C. duobushaemulonii sp. nov) and cross-resistance to azole compounds. Identification of these species should be based on molecular methods, such as sequence analysis of ITS regions and matrix-assisted laser desorption ionization–time of flight mass spectrometry.
INTRODUCTION
Candida and Aspergillus species are the most common causes of invasive fungal infections in immunocompromised individuals, but besides these fungi, many other yeast species and filamentous fungi can be pathogenic in such individuals (7). The list of reported species that cause human infection is constantly growing, partly because of recent advances in molecular tools and diagnostics. Thus, new clinically relevant species such as Candida metapsilosis, Candida orthopsilosis, Candida bracariensis, and Candida nivariensis, have been described recently (1, 5, 39).
Candida haemulonii (van Uden and Kolipinsky) S. A. Meyer and D. Yarrow (41) (syn. Torulopsis haemulonii) is one of the rare yeast species that can be isolated from human clinical sources. The species originally described was from the gut of a blue-striped grunt fish (Haemulon scirus) in 1962 (40). The first isolation of this yeast from a human, i.e., from the blood of a patient with renal failure, was reported by Lavarde et al. (22). Since then, several cases of infections due to this yeast have been described in the literature, varying from superficial to deep infections. Cases of catheter-related fungemia (18), bloodstream infections (30, 34), and osteitis (6) and outbreaks in intensive care units (16) have been reported recently. The species has also been isolated from toenails of diabetic patients (13). Noteworthy is the susceptibility profile of this yeast, which shows high MICs of amphotericin B (AMB) and fluconazole (FLC) (ranges, 0.5 to 32 and 4 to >64 mg/liter, respectively), which can hinder the management of patients with deep infections caused by this yeast. This antifungal profile has often been associated with clinical failure (6, 16, 17, 30, 34).
The C. haemulonii species complex was further studied by Lehman et al. in 1993 (24). They studied 25 strains from different geographic origins and clinical sources and described two genetically distinct C. haemulonii groups, I and II. This classification was based on isozyme profiles, DNA reassociation experiments, and physiological characteristics.
In recent years, two species related to C. haemulonii have been described, namely, Candida pseudohaemulonii and Candida auris, which are phylogenetically closely related to C. haemulonii in the Metschnikowiaceae clade (20). In 2006, Sugita et al. described C. pseudohaemulonii, which was isolated from the blood of a Thai patient. This species is as resistant to AMB and azole agents as are the two genetic groups of C. haemulonii (38). The second related species, C. auris, was described in 2009 by Satoh et al. and isolated from the external ear canal of an inpatient in a Japanese hospital. Using sequence analysis of the D1/D2 domain of the 26S rRNA gene and the internal transcribed spacer (ITS) regions of the nuclear rRNA gene operon, it was found that the strain represents a new species with a close phylogenetic relationship to C. haemulonii (35). In 2009, 15 isolates of C. auris were recovered from the ear canals of patients suffering from chronic otitis media in South Korea. All of these isolates showed a reduced susceptibility to AMB and azole compounds (17). Therefore, it is important to identify these species correctly in order to provide optimal patient care.
Recently, C. haemulonii and closely related species have caused outbreaks in South Korea and Kuwait (16, 17). The reasons for their emergence are not clear, but they may be related to selective pressure as a result of the commonly applied FLC or AMB therapy. C. haemulonii and C. pseudohaemulonii were isolated from patients with central venous catheter-related fungemia, whereas the C. auris strains were isolated from the ear canals of inpatients (28). Moreover, the first three cases of bloodstream infection due to C. auris have been described recently (23).
In the present study, the phylogenetic relationships among 30 isolates of the C. haemulonii complex from the collections of the Centraalbureau voor Schimmelcultures (CBS) Fungal Biodiversity Centre (CBS-KNAW), Utrecht, The Netherlands, and the Mycology Reference Laboratory of the National Center of Microbiology, CNM-ISCIII, Majadahonda, Madrid, Spain, were studied and compared with known C. auris and C. pseudohaemulonii isolates. Four strains of the latter species from South Korea were included. The profile of susceptibility of 30 isolates of the C. haemulonii complex to nine antifungal compounds was assessed to establish a correct antifungal susceptibility profile of these species, as well as their ability to produce biofilms. Identification of all species by amplified fragment length polymorphism (AFLP) analyses, ITS sequence data, and matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) is possible.
(This work was partially presented at the 22nd ECCMID 2012 in London, England.)
MATERIALS AND METHODS
C. haemulonii group I and group II isolates.
The 30 isolates of C. haemulonii used in this study are listed in Table 1. These include the type strain of C. haemulonii group I (CBS 5149T) and two representative isolates of C. haemulonii group II (CBS 7798 and CBS 7799). Isolates from animal and environmental sources were included to make the sampling more robust. Four strains of the two closely related species C. auris and C. pseudohaemulonii (including type strain CBS 10004T) were included for comparison.
Table 1.
Isolates and reference strains of C. haemulonii, C. pseudohaemulonii, and C. auris used in this study
| Isolate | Species | Origin | Location |
|---|---|---|---|
| CNM-CL7256 | C. haemulonii var. vulnera | Toenail | Alicante, Spain |
| CNM-CL7462 | C. haemulonii var. vulnera | Patient (source unknown) | Spain |
| CNM-CL4642 | C. haemulonii | Patient (source unknown) | Argentina |
| CNM-CL3458 | C. haemulonii | Blood | Argentina |
| CNM-CL7829W | C. duobushaemulonii | Blood | Spain |
| CNM-CL7239T | C. haemulonii var. vulnera | Skin wound | León, Spain |
| CNM-CL7829P | C. duobushaemulonii | Blood | Spain |
| CNM-CL6800 | C. haemulonii | Skin wound | Elche, Spain |
| CNM-CL7793 | C. haemulonii | Blood | Bilbao, Spain |
| CNM-CL4640 | C. haemulonii | Patient (source unknown) | Argentina |
| CNM-CL7073 | C. haemulonii var. vulnera | Skin wound | Alicante, Spain |
| CNM-CL4641 | C. haemulonii | Patient (source unknown) | Argentina |
| CBS 7800 | C. duobushaemulonii | Foot ulcer | Tennessee |
| CBS 7799 | C. duobushaemulonii | Foot ulcer | Georgia |
| CBS 5149T | C. haemulonii | Gut of Haemulon scirus (fish) | Florida |
| CBS 9754 | C. duobushaemulonii | Pyrrhocois apterus (insect) | Ulm, Germany |
| CBS 7801 | C. haemulonii | Toenail | Hawaii |
| CBS 5150 | C. haemulonii | Seawater | Lisbon, Portugal |
| CBS 6590 | C. haemulonii | Patient (source unknown) | France |
| CBS 5468 | C. haemulonii | Seawater | Brazil |
| CBS 6915 | C. duobushaemulonii | Unknown | Unknown |
| CBS 7798T | C. duobushaemulonii | Foot ulcer | Alabama |
| CBS 7802 | C. haemulonii | Foot ulcer | Rhode Island |
| CBS 6332 | C. haemulonii | Blood | Kuwait |
| CBS 10973 | C. haemulonii | Blood | Kuwait |
| CBS 10972 | C. haemulonii | Blood | Kuwait |
| CBS 10971 | C. haemulonii | Blood | Kuwait |
| CBS 10970 | C. haemulonii | Blood | Kuwait |
| CBS 10969 | C. haemulonii | Blood | Kuwait |
| CBS 10968 | C. haemulonii | Blood | Kuwait |
| CBS 10004T | C. pseudohaemulonii | Blood | Thailand |
| KCTC-17807 | C. pseudohaemulonii | Blood | South Korea |
| KCTC-17808 | C. pseudohaemulonii | Blood | South Korea |
| KCTC-17809 | C. auris | Ear | South Korea |
| KCTC-17810 | C. auris | Ear | South Korea |
a A superscript T indicates the type strain.
DNA extraction.
Genomic DNA was extracted from yeasts grown in glucose yeast peptone agar medium (GYPA) at 25°C. DNA extraction was performed by using the protocol described by Bolano et al., with slight modifications (2). After 2 days of incubation, a suspension of cells was added to 150 μl of sterile sand, 750 μl of lysis buffer, and 750 μl of phenol-chloroform (1:1, pH 8.0). This suspension was shaken at 2,500 rpm for 3 min, and the crude extract was centrifuged at 17,000 × g for 15 min at 4°C. The 700 μl of supernatant was transferred into a 1.5-ml Eppendorf tube. An equal volume of ice-cold 96% ethanol and 100 μl of 3 M ice-cold sodium acetate was added to the supernatant. The solution was mixed and stored for 30 to 60 min at −20°C. The DNA was pelleted at 17,000 × g for 10 min at 4°C. The supernatant was discarded, and the pellet was air dried. This pellet was resuspended in 100 μl of preheated Tris-EDTA buffer and incubated at 37°C and 65°C for at least 10 min.
Amplification and nucleotide sequence determination.
The ITS and the D1/D2 regions of the ribosomal DNA were sequenced. The primer set V9-G/LR3-R was used to obtain the amplicon (11). For sequence analyses, primers ITS1 and ITS4 were used for the ITS region and NL1 (21) and LR3-R were used for the D1/D2 domain (Assembling the Fungal Tree of Life website, http://aftol.org/). The sequencing PCR was performed as follows. For each reaction mixture, 17.75 μl of MilliQ water, 0.75 μl of MgCl2 (50 nM), 2.5 μl of PCR buffer 10x, 1.9 μl of a 1 mM deoxynucleoside triphosphate (dNTP) mixture, 0.5 μl of primer V9-F (10 pmol/μl), 0.5 μl of primer RLR3R-R (10 pmol/μl), 0.1 of Taq polymerase (5 U/μl), and 1 μl of DNA solution were used. The amplification reactions were done using a first cycle of denaturation for 5 min at 96°C, followed by 35 cycles of denaturation at 96°C for 30 s, annealing at 52°C for 30 s, and elongation at 72°C for 1 min, with a final extension step of 5 min at 72°C.
The RPB1 (which encodes the largest subunit of RNA polymerase II) and RPB2 (which encodes the second largest subunit of RNA polymerase II) genes were analyzed as follows. For RPB2, the conditions and primers used for amplification and sequencing were described by Liu et al. (26). The amplicon was obtained with primers RBP2-5F (5′-GAYGAYMGWGATCAYTTYGG-3′) and RPB2-7Cr (5′-CCCATRGCTTGYTTRCCCAT-3′). Because of the length of the amplicon (approximately 1 kb), the sequencing reaction was carried out with primers RPB2-5f and RPB2-7Cr and also with primers RPB26F (5′-TGGGGKWTGGTYTGYCCTGC-3′) and RPB26R (5′-GCAGGRACCAWMCCCCA-3′).
For the RPB1 gene, primers RPB1af (5′-GARTGYCCDGGDCAYTTYGG-3′) and RPB1Cr (5′-CCNGCDATNTCRTTRTCCATRTA-3′) were used (Assembling the Fungal Tree of Life website, http://aftol.org/). A PCR mixture contained 17.75 μl of MilliQ water, 0.75 μl of MgCl2 (50 nM), 2.5 μl of PCR buffer 10x, 1.9 μl of a 1 mM dNTP mixture, 0.5 μl of primer RPB1af (10 pmol/μl), 0.5 μl of primer RPB1Cr (10 pmol/μl), 0.1 μl of Taq polymerase (5 U/μl), and 1 μl of DNA solution. The following amplification conditions were used: a first cycle of denaturation for 5 min at 94°C, followed by 36 cycles of denaturation at 94°C for 50 s, annealing at 52°C for 60 s, and elongation at 72°C for 1 min, with a final extension step of 7 min at 72°C. For the sequencing reaction of the RPB1 gene, the following volumes were used per reaction mixture: 4 μl of MilliQ water, 3 μl of dilution buffer, 0.5 μl of primer (10 pmol/μl), 1 μl of BigDye version 3.1, and 1 μl of diluted amplicon solution. The sequencing PCR conditions used were a first cycle of denaturation for 1 min at 95°C, followed by 30 cycles of denaturation at 95°C for 10 s, annealing at 50°C for 5 s, and elongation at 60°C for 4 min.
Sequencing products were purified with Sephadex (Amersham Pharmacia, The Netherlands). Both strands of purified gene fragments were sequenced at the Uppsala Genome Centre (Uppsala, Sweden). Sequences were assembled and edited with SeqMan II software (DNAStar Inc., Madison, WI) and aligned with MegAlign (DNAStar). The sequences were visually corrected. All phylogenetic analyses used maximum likelihood with 2,000 bootstrap simulations and were conducted with InfoQuest FP software, version 4.50 (Bio-Rad Laboratories, Madrid, Spain).
Identification by MALDI-TOF MS.
For identification by MALDI-TOF MS, the full extraction method (ethanol-formic acid [FA]) of Marklein et al. was used, with slight modifications (27). Strains were subcultured on Sabouraud dextrose agar plates and incubated for ≤24 h at 30°C. An Eppendorf tube (1.5-ml volume) was filled with 0.3 ml of MilliQ water and 2 μl of yeast biomass (with a 1-μl sterile inoculation loop [Greiner Bio-One]). After vortexing, 0.9 ml of absolute ethanol was added and mixed vigorously for 1 min. Following centrifugation (17,000 × g, 2 min), the supernatant was discarded and the residual ethanol was removed by pipetting after a second centrifugation step, followed by air drying of the pellets at room temperature. The volume of 70% FA (Fluka, Zwijndrecht, The Netherlands) was then adjusted to the pellet size (usually 30 to 40 μl) and the material was first detached by pipetting and mixed by vortexing until the pellet was completely dissolved in FA. Then a volume of pure acetonitrile (ACN, Sigma-Aldrich, Zwijndrecht, The Netherlands) equal to the volume of FA was added, mixed by vortexing for 5 min, and then centrifuged (17,000 × g, 2 min). A 1-μl volume of the supernatant was pipetted onto a clean, polished steel target plate (Bruker Daltonik, Bremen, Germany). For each strain tested, two spots were prepared. Once the spotted material was air dried at room temperature, it was overlaid with 1 μl of HCCA matrix solution (α-cyano-4-hydroxycinnamic acid at 10 mg/ml; Bruker Daltonik, Bremen, Germany) dissolved in 50% (vol/vol) CAN–2.5% (vol/vol) trifluoroacetic acid–47.5% MilliQ water. As a positive control, 1 μl of Bacterial Test Standard solution (Bruker Daltonik, Bremen, Germany) was spotted twice and overlaid with HCCA matrix solution. Automatic runs were performed using flexControl version 3.3.108.0. Tested strains were identified by the MALDI Biotyper and MALDI Biotyper RTC software 3.0.
The current Bruker Daltonik database contains 4,110 main spectra (MSP). Ethanol extracts from 600 different yeast species of CBS-KNAW, including the set of strains of the C. haemulonii complex, were prepared and sent to Bruker Daltonik (Bremen, Germany) for MSP library creation according to the Bruker internal database creation standard operating procedures. This library was later uploaded at MALDI Biotyper 3.0 as an in-house CBS-KNAW library. This library could be used in combination with the Bruker database or as a stand-alone library.
For the MALDI-TOF MS-based recognition and classification of tested strains, MALDI Biotyper software 3.0 was used. In accordance with the manufacturer's specifications, the log (score) values demonstrating secure genus and species identification (≥2.0), secure genus identification (1.70 to 1.99), and no reliable identification (<1.7) were used. Bruker flex analysis 3.3.75.0 allowed visualization of the mass spectra acquired. ClinProTools 3.0 (Bruker Daltonik, Bremen, Germany) was used to generate artificial gel views of spectral peak intensities to search for differentiating peaks of these spectra and principal-component analysis clustering. Dendrograms were generated by using the respective functionality of the MALDI Biotyper 3.0 offline client. For the creation of dendrograms, the settings used were correlation as distance measure, ward linkage algorithm, and maximal clustering as 4 (four) and 0 (zero) as the maximal number of top-level nodes, and no cutoff (Co) values were used.
AFLP experiments.
AFLP reactions were performed as described by Borst et al., with some minor modifications (3). In the second PCR, the selective primer MseI-G was used. Selective products were run at the Uppsala Genome Center (Uppsala, Sweden) using GeneScan-500 (6-carboxy-X-rhodamine labeled) as an internal size standard. The data were analyzed with the BioNumerics software package, version 4.61 (Applied Maths, Sint-Martens-Latem, Belgium) using the curve-based cosine similarity coefficient in combination with single-linkage cluster analysis to create the dendrogram.
Physiological tests.
Morphological, biochemical, and physiological characteristics were examined as described by Kurtzman et al. (20). Fermentation, carbon assimilation, and nitrogen assimilation tests were done for 11 isolates that represented the three clades (i.e., CBS 5149, CBS 5150, CBS 7800, CBS 7799, CBS 7798, CNM-CL4640, CBS 6332, CNM-CL7239, CNM-CL7462, CNM-CL7256, and CNM-CL7073). Growth at different temperatures (25, 30, 35, 37, 40, and 42°C), in cycloheximide (0.01, 0.1%), on glucose (50 and 60%), and in NaCl (10, 16%) was also tested. Starch production, urease activity, the diazonium blue B staining reaction, and growth without vitamins were also investigated.
Morphology and mating.
Thirteen isolates (CBS 5149, CBS 5150, CBS 7800, CBS 7799, CBS 7798, CNM-CL4640, CBS 6332, CNM-CL7239, CNM-CL7462, CNM-CL7256, CNM-CL7073, CBS 10913, and CBS 10004) were inoculated onto 5% yeast malt agar (YMA), GYPA, and morphology agar (MoA; Difco) and incubated for 12 days at 25°C to study colony morphology. To investigate cell morphology, strains were inoculated onto MoA plates and into yeast nitrogen base (YNB) medium containing 5% glucose. Mating experiments were carried out with McClary acetate agar. Isolates in the same phylogenetic cluster were mixed two by two as follows: C. haemulonii group II, CBS 7799-CBS 7798, CBS 7799-CBS 7799, CBS 7799-CBS 7800, CBS 7800-CBS 7800, CBS 7800-CBS 7798, and CBS 7798-CBS 7798; four Spanish isolates of C. haemulonii with different ITS sequences, CNM-CL7256–CNM-CL7256, CNM-CL7256–CNM-CL7239, CNM-CL7256–CNM-CL6332, CNM-CL7256–CNM-CL7073, CNM-CL7256–CNM-CL7462, CNM-CL7239–CNM-CL7239, CNM-CL7239–CNM-CL6332, CNM-CL7239–CNM-CL7073, CNM-CL7239–CNM-CL7462, CNM-CL6332–CNM-CL6332, CNM-CL6332–CNM-CL7462, CNM-CL7073–CNM-CL7073, CNM-CL7073–CNM-CL7462, and CNM-CL7462–CNM-CL7462. Plates were incubated at 25°C for 40 days and viewed weekly by microscopy to check for the formation of ascospores.
Susceptibility tests.
Susceptibility tests were done as recommended by the Antifungal Susceptibility Testing Subcommittee of the European Committee on Antibiotic Susceptibility Testing (EUCAST) for fermentative yeasts (9). All of the antifungal tests were evaluated after 48 h. Differences between 24-h and 48-h MIC values were no more than 1 2-fold dilution (data not shown). Slow growth of C. haemulonii strains has been described before (16). The antifungal agents used were AMB, flucytosine (5-FC; Sigma-Aldrich), FLC (Pfizer S.A., Madrid, Spain), itraconazole (ITC; Sigma-Aldrich), voriconazole (VRC; Pfizer S.A.), caspofungin (CAS; Merck & Co., Inc.), micafungin (MCF; Astellas Pharma, Inc., Tokyo, Japan), anidulafungin (ANF; Pfizer S.A), and posaconazole (PSZ; Merck & Co., Inc., Rahway, NJ). Interpretative breakpoints proposed by EUCAST for FLC were used (32, 33). For AMB, VRC, ITC, and PSZ, the breakpoints were defined on the basis of the wild-type distribution of MICs determined by the EUCAST method (epidemiological cut off) and on pharmacokinetic/pharmacodynamic and bibliographic data (8, 10, 31). In the case of echinocandins, breakpoints proposed by the EUCAST were used to interpret the susceptibility results (http://www.eucast.org/clinical_breakpoints/). Statistical and descriptive analyses of the MICs were also done, including the geometric mean (GM), range, and MIC90. The significance of the differences between MIC values was determined by analysis of variance (Bonferroni post hoc test) (PASW statistics 18; IBM Software, Madrid, Spain). A P value of <0.01 was regarded as statistically significant.
Biofilm formation.
Isolates of the C. haemulonii complex (Table 1) were used for biofilm formation. Additionally, strains of other Candida species that are known to form biofilms were selected for comparison, namely, C. albicans CBS 8758 (= SC 5314), C. glabrata CBS 861, C. dubliniensis CBS 7987, C. krusei CBS 573, and C. tropicalis CBS 8072. The standard protocol of Li et al. (25), with minor modifications, as reported by Kolecka et al. (19), was used. The ability to form biofilm was quantified by the crystal violet staining method according to Jin et al. (15). Final results were expressed as an average of three independent experiments ± the standard deviation (SD) where biofilms of each strain tested were cultivated in four parallel wells. The presence of biofilms was evaluated according to the Co value calculated from the average optical density at 600 nm (OD600) of the three measurements of the negative control plus 3 times the SD as suggested by Holá et al. (14). Biofilm formation by each isolate was scored as negative (OD600 = <0.111 [Co value]), weak (OD600 = Co − [2 × Co]), intermediate (OD600 = [2 × Co] − [3 × Co]), or strong (OD600 = >3 × Co).
Nucleotide sequence accession numbers.
The sequences obtained during this study were deposited in GenBank under the accession numbers listed in Table 2.
Table 2.
Isolates used in this study and GenBank accession numbers of the genes sequenced
NS, not sequenced.
RESULTS
Phylogeny.
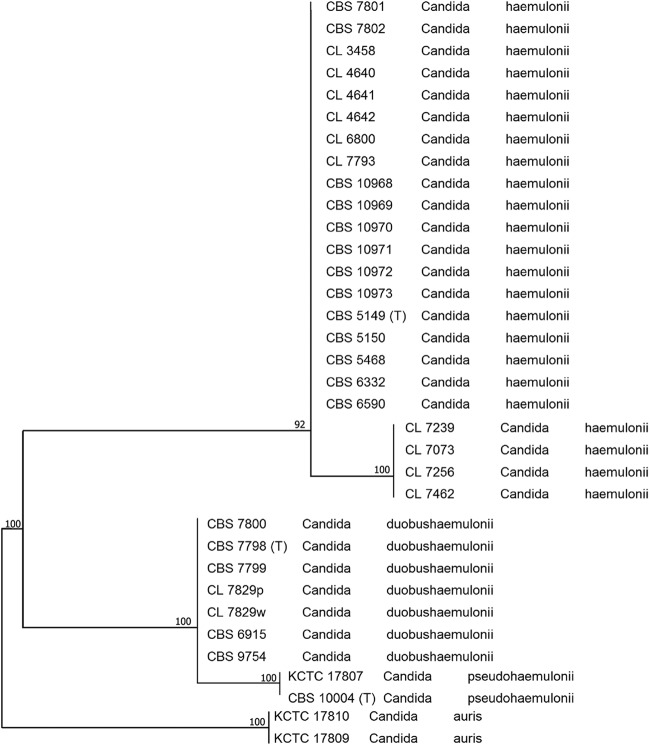
Two main clusters of the strains of the C. haemulonii complex were defined by sequence analysis of the four genes studied (ITS, D1/D2, RPB1, and RPB2). Twenty-three isolates were assigned to the most commonly encountered group, C. haemulonii group I, and seven isolates were assigned to C. haemulonii group II. In the ITS phylogenic tree (Fig. 1), four main clusters could be distinguished. The first cluster contained 19 C. haemulonii group I isolates and 4 isolates (CNM-CL7256, CNM-CL7462, CNM-CL7239, CNM-CL7073) that differ from the other 19 isolates showing 96% similarity to typical isolates of C. haemulonii group I. The percentages of similarity between the four isolates and the C. haemulonii type strain, CBS 5149T, were as follows: 96.51% for CNM-CL7239 and CNM-CL7256, 96.31% for CNM-CL7462, and 96.43% for CNM-CL7073. The second cluster is formed by the seven isolates belonging to C. haemulonii group II. The third and fourth clusters contained C. pseudohaemulonii and C. auris isolates, respectively. The last two species were included for comparison.
Fig 1.
Phylogenetic tree of isolates of the C. haemulonii complex obtained by using maximum-likelihood phylogenetic analyses and 2,000 bootstrap simulations based on ITS sequences.
D1/D2, RPB1, and RPB2 phylogenic studies also revealed four main clusters. The first cluster included 23 C. haemulonii group I isolates, and the other cluster is formed by the 7 isolates belonging to C. haemulonii group II (see Fig. S1 to S3 in the supplemental material). C. pseudohaemulonii and C. auris isolates formed the third and fourth clusters, respectively. Strain KCTC 17807 was not included in the RPB1 phylogenic tree because no good sequence of the RPB1 gene was obtained, despite several tries. The sequence of the RPB2 gene obtained for strain CNM-CL7073 was too short to be aligned with the other sequences, and hence, this strain was not included in the RPB2 phylogenic tree. Phylogenetic trees of these genes (see Fig. S1 to S3 in the supplemental material) indicated good bootstrap support (>90%) for all clusters.
The similarities between the type strain of C. haemulonii group I (CBS 5149T) and the reference strain of C. haemulonii group II (CBS 7798) were 89.04% for ITS, 90.93% for D1/D2, 86.77% for RPB1, and 87.43% for RPB2.
Physiological tests, mating experiments, and biofilms.
Some differences occurred in the fermentation tests, the carbon assimilation tests, growth at different temperatures, and growth in 60% glucose among C. haemulonii group II, C. haemulonii group I, and the C. haemulonii strains that differed in their ITS sequences (see Table S1 in the supplemental material). C. haemulonii group II is able to ferment raffinose, and it has differential patterns of carbon compound assimilation, growth temperatures, and growth in 60% glucose (Table 3). Differences between the two clades of the C. haemulonii complex (C. haemulonii type I and type II) are supported by their patterns of utilization of eight carbon compounds, namely, l-sorbose, arbutin, l-arabinose, l-arabinose, l-rhamnose, melezitose, inulin, and ethanol. The raffinose fermentation profiles differed as well (positive for C. haemulonii type II), as did growth at 37°C (negative for C. haemulonii type I) and growth in 60% glucose (positive for C. haemulonii type II). None of the mating experiments carried out gave any indication of the presence of a sexual cycle.
Table 3.
Differential characteristics of the taxa of the C. haemulonii complexa
| Characteristic | C. haemulonii CBS 5149, CBS 5150, CNM-CL4640, CBS 6332 | C. duobushaemulonii CBS 7798, CBS 7799, CBS 7800 | C. haemulonii var. vulnera CNM-CL7239, CNM-CL7462, CNM-CL7256, CNM-CL7073 | C. aurisb | C. pseudohaemuloniib |
|---|---|---|---|---|---|
| Fermentation of: | |||||
| Raffinose | − | + | − | − | − |
| Sucrose | + | + | + | + | − |
| Assimilation of: | |||||
| l−Sorbose | − | + | − | − | + |
| Arbutin | − | + | − | ND | ND |
| l-Arabinose | − | W | − | − | V |
| d-Arabinose | − | V | − | − | − |
| l-Rhamnose | + | W | + | − | + |
| Melezitose | + | + | D | + | + |
| Inulin | − | + | W | W | − |
| Ethanol | D | W/D | + | − | D |
| d-Galactose | + | + | + | − | + |
| Methanol | W/D | W/D | W/D | − | − |
| Succinate | + | + | + | − | + |
| d-Gluconate | + | + | + | − | + |
| Xylitol | W/D | + | + | − | + |
| Glycerol | + | + | + | − | + |
| Growth at: | |||||
| 37°C | − | + | + | + | + |
| 40°C | − | − | − | + | − |
| Growth in: | |||||
| 60% glucose | − | + | − | − | ND |
| Vitamin-free medium | − | − | − | + | ND |
Biofilms that developed were quantified after 48 h (see Table S2 in the supplemental material). In general, strains of the C. haemulonii species complex do not form well-developed biofilms in vitro in YNB medium supplemented with 50 mM glucose.
Susceptibility tests.
The 30 isolates were tested for susceptibility to nine antifungal compounds (see Materials and Methods). The MICs of AMB ranged from 0.25 to >16 mg/liter for all 30 isolates of the C. haemulonii complex. C. haemulonii group II and the four strains of C. haemulonii that showed deviant ITS sequences had high MICs of AMB (GM, 2.0 mg/liter and 1.18 mg/liter, respectively) and azoles (Table 4). Statistically significantly higher AMB MICs were found for C. duobushaemulonii (P = 0.008).
Table 4.
Antifungal susceptibility profiles of the 30 isolates in this study
| Strain or parameter | MIC (mg/liter) |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| AMB | 5-FC | FLC | ITC | VRC | PSZ | CAS | MCF | ANF | |
| C. haemulonii var. vulnera | |||||||||
| CNM-CL7239 | 1 | <0.12 | >64 | >8 | >8 | >8 | >16a | >16a | >16a |
| CNM-CL7256 | 2 | 0.5 | >64 | >8 | >8 | >8 | >16a | 0.12 | 0.06 |
| CNM-CL7073 | 1 | 0.25 | >64 | >8 | >8 | >8 | 0.5a | 0.12 | 0.06 |
| CNM-CL7462 | 1 | 0.5 | >64 | >8 | >8 | >8 | >16a | 0.06 | <0.03 |
| GM (range) | 1.18 (1.0–2.0) | 0.24 (0.06–0.5) | 128 (>64) | 16 (>8) | 16 (>8) | 16 (>8) | 11.31 (0.5–16) | 0.40 (0.06–>16) | 0.20 (<0.03–>16) |
| MIC90 | 1.7 | 0.5 | 128 | 16 | 16 | 16 | 32 | 22.436 | 22.418 |
| C. duobushaemuloni | |||||||||
| CBS 7798 | 4 | 0.5 | >64 | 4 | >8 | >8 | >16a | 0.06 | 4 |
| CBS 6915 | 2 | 0.12 | 8 | 0.12 | 0.25 | 0.015 | >16a | 0.06 | 0.03 |
| CBS 7800 | 0.25 | 0.25 | >64 | >8 | >8 | >8 | >16 | 0.06 | 0.25 |
| CBS 7799 | 1 | 2 | >64 | >8 | >8 | >8 | 16 | 0.06 | 0.06 |
| CBS 9754 | 1 | 0.25 | >64 | >8 | >8 | >8 | 1 | 0.12 | 0.06 |
| CNM-CL7829W | >16 | <0.125 | >64 | >8 | >8 | >8 | 0.5 | 0.06 | <0.03 |
| CNM-CL7829P | 2 | 0.25 | 16 | 0.06 | 0.12 | 0.03 | 0.5 | 0.06 | <0.03 |
| GM (range) | 2 (0.25–32) | 0.27 (0.06–2) | 64 (8–>64) | 2.93 (0.06–>8) | 4.39 (0.12–>8) | 2.18 (0.015–>8) | 5.38 (0.5–>16) | 0.06 (0.06–0.12) | 0.08 (<0.03–4) |
| MIC90 | 15.2 | 1.1 | 128 | 16 | 16 | 16 | 32 | 0.084 | 1.75 |
| C. haemulonii | |||||||||
| CBS 7802 | 1 | 0.25 | >64 | >8 | >8 | >8 | >16a | >16a | 0.25 |
| CBS 7801 | 1 | 0.25 | >64 | >8 | >8 | >8 | 0.25a | 0.12a | <0.03 |
| CBS 6332 | 0.25 | 0.5 | >64 | >8 | >8 | >8 | >16 | >16a | 4 |
| CBS 10970 | 1 | 1 | >64 | >8 | >8 | >8 | 0.25a | 0.12a | <0.03 |
| CBS 6590 | 0.5 | <0.12 | >64 | >8 | >8 | >8 | >16a | 0.06 | <0.03 |
| CBS 5150 | 0.5 | <0.12 | >64 | >8 | >8 | >8 | >16a | 0.06 | <0.03 |
| CBS 5468 | 0.5 | 0.25 | >64 | >8 | >8 | >8 | >16a | >16a | >16a |
| CBS 5149 | 0.25 | <0.12 | >64 | >8 | >8 | 4 | 0.25 | <0.03 | <0.03 |
| CBS 10972 | 0.5 | 0.5 | >64 | >8 | >8 | >8 | >16a | 0.06 | <0.03 |
| CBS 10971 | 0.5 | 1 | >64 | >8 | >8 | 8 | >16a | 0.06 | <0.03 |
| CBS 10969 | 0.5 | 1 | >64 | >8 | >8 | 8 | >16a | 0.06 | <0.03 |
| CBS 10968 | 0.5 | 2 | >64 | >8 | >8 | >8 | >16a | 0.06 | <0.03 |
| CBS 10973 | 0.5 | 0.25 | >64 | >8 | >8 | >8 | >16a | 0.06 | 0.06 |
| CNM-CL7793 | 2 | 0.25 | >64 | >8 | >8 | >8 | >16a | 0.06 | <0.03 |
| CNM-CL3458 | 0.5 | <0.12 | >64 | >8 | >8 | >8 | >16a | 0.25 | 0.12 |
| CNM-CL4640 | 0.25 | <0.12 | >64 | >8 | >8 | 0.03 | 8a | <0.03 | <0.03 |
| CNM-CL4641 | 1 | <0.12 | >64 | >8 | >8 | >8 | >16 | >16 | >16 |
| CNM-CL4642 | 0.5 | 0.25 | >64 | >8 | >8 | >8 | 0.5 | 0.06 | <0.03 |
| CNM-CL6800 | 1 | 0.12 | >64 | >8 | >8 | >8 | >16a | 0.06 | <0.03 |
| GM (range) | 0.57 (0.25–2) | 0.22 (0.06–2) | 128 (>64) | 16 (>8) | 16 (>8) | 10.68 (0.03–>8) | 11.10 (0.25–>16) | 0.17 (<0.03–>16) | 0.06 (<0.03–>16) |
| MIC90 | 1 | 1 | 128 | 16 | 16 | 16 | 32 | 32 | 9.6 |
Strain that showed some kind of paradoxical growth effect.
All of the isolates of the new variety, 6 out of 7 isolates of C. haemulonii group II, and 6 out of 19 isolates of C. haemulonii group I showed high MICs of AMB (Table 4). The MIC ranges of FLC, ITC, VRC, and PSZ for all 30 isolates were 8 to >64 mg/liter, 0.06 to >8 mg/liter, 0.12 to >8 mg/liter, <0.015 to >8 mg/liter, and 0.015 to >8 mg/liter, respectively. It is noteworthy that 28 out of 30 isolates could be considered cross-resistant to azoles. Only 2 out of 30 isolates with FLC MICs of >4 mg/liter showed low MICs of the other azole compounds (Table 4). In general terms, there were subtle observed differences in azole MICs between the groups of strains tested, showing higher MICs for C. haemulonii than for the other two groups. In some cases (MICs of ITC and VRC for C. haemulonii and C. duobushaemulonii), the differences were significant or close to significance (P = 0.009 and P = 0.028, respectively).
Strains with high MICs of CAS, ANF, and MCF were found as well. Three out of four isolates of the ITS variant of C. haemulonii showed high MICs of CAS, and one of them (CNM-CL7239) showed high MICs of all of the echinocandins studied. Fifteen out of 19 isolates of C. haemulonii showed high MICs of CAS, and 4 out of these 15 isolates showed the same profile for all echinocandins. With respect to C. haemulonii group II, four out of seven isolates showed high MICs of CAS. However, this susceptibility profile was totally different for ANF or MCF (Table 4).
Identification of species.
Representatives of C. haemulonii and C. duobushaemulonii can be identified by ITS sequencing, which gives reliable identification of C. haemulonii, the ITS variant of C. haemulonii, C. duobushaemulonii sp. nov., C. auris, and C. pseudohaemulonii (Fig. 1).
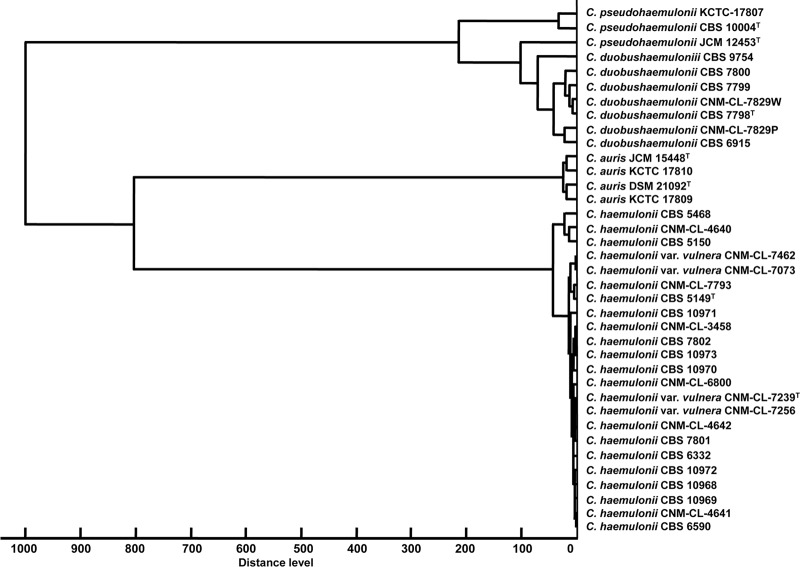
MALDI-TOF MS also identified all of the species, but here the ITS variant of C. haemulonii clustered among typical C. haemulonii strains (Fig. 2). MALDI-TOF MS correctly identified 32 (86.5%) of 37 isolates with log scores of ≥2.0 for at least one spot and 5 (13.5%) of 37 with log scores of 1.700 to 1.999 on one spot (see Table S3 in the supplemental material). The artificial gel view acquired from the type strains illustrated differences in mass spectral peak positions and intensities which are species unique (see Fig. S4 in the supplemental material). The technical variation between duplicates of the same strains obtained from different culture collections are illustrated by C. pseudohaemulonii CBS 1004T and JCM 12453T and C. auris JCM 15448T and DSM 21092T.
Fig 2.
Dendrogram clustering the MALDI-TOF MSP obtained from at least 20 mass spectra of strains belonging to the C. haemulonii complex species and related species. C. auris JCM 15448T and DSM 21092T and C. pseudohaemulonii JCM 12453T were added to make the sampling in MALDI-TOF MS more robust.
The AFLP experiments showed the same strain grouping as the MALDI-TOF MS analyses (Fig. 3), and here the strains of C. haemulonii with a deviating ITS sequence could not be distinguished from typical strains of C. haemulonii.
Fig 3.
AFLP patterns of representative isolates of the C. haemulonii complex and related species.
Taxonomy.
Candida duobushaemulonii sp. nov. E. Cendejas-Bueno, A. Kolecka, A. Alastruey-Izquierdo, A. Gómez-López, M. Cuenca-Estrella, and T. Boekhout.
MycoBank: “Candida duobushaemulonii” (MB 800921).
Etymology: the epithet duobushaemulonii “haemulonii two” refers to the previous name by which this species was known, C. haemulonii group II.
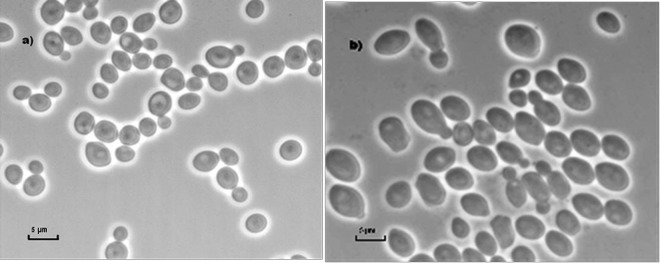
In 5% glucose liquid broth, at 25°C for 72 h, cells are subspherical to ellipsoidal, 2.5 to 5 μm by 2 to 3.5 μm (Fig. 4), and occur singly, in pairs, or in small clusters, with multipolar or unipolar budding. No filaments or hyphae are formed on MoA. On GYPA and YMA after 12 days of growth, the colonies are about 1 cm in diameter, creamy white, and smooth, with a central ring and weak grooves that run from the center of the colony to the entire margin. For descriptions of its biochemical and physiological features, see Table 3; see also Table S1 in the supplemental material. The holotype, CBS 7798 (CNM-CL9157), was isolated from a human foot ulcer in Alabama. Other representative isolates are CBS 7800 (CNM-CL9179; foot ulcer, United States), CBS 7799 (CNM-CL9178; foot ulcer, United States), CBS 9754 (CNM-CL9165; insect, Germany), CBS 6915 (CNM-CL9167; source unknown, country unknown), CNM-CL7829P, and CNM-CL7829W (blood, Spain). These strains have also been deposited in the Centro Nacional de Microbiología (CNM) collection.
Fig 4.
Cell morphology of C. duobushaemulonii sp. nov. on MoA (a) and in 5% glucose liquid broth (b).
Candida haemulonii var. vulnera var. nov. E. Cendejas-Bueno, A. Kolecka, A. Alastruey-Izquierdo, A. Gómez-López, M. Cuenca-Estrella, and T. Boekhout.
MycoBank: “Candida haemulonii vulnera” (MB 800922).
Etymology: the epithet vulnera “wound” refers to a clinical sample from which the type strain was isolated.
Differs from the typical variety (C. haemulonii) on the basis of phenotypic, susceptibility, and molecular features, namely, delayed assimilation of melezitose, weak growth on inulin, growth on ethanol and xylitol, and growth at 37°C. The AMB MIC is higher than that for C. haemulonii, and the ITS sequence differs from the typical C. haemulonii ITS sequence, showing 96% similarity to typical isolates of C. haemulonii group I.
The holotype, CNM-CL7239, was isolated from a human skin wound in León, Spain. This strain has been deposited in the Collection of the CNM Mycology Department. Other isolates are CNM-CL-7256 (toenail, Alicante, Spain), CNM-CL-7462 (source unknown from a human clinical sample, Spain), and CNM-CL-7073 (skin wound, Alicante, Spain). They have also been deposited in the CBS collection under the numbers CBS 12439 (CNM-CL7239), CBS 12436 (CNM-CL7256), CBS 12437 (CNM-CL 7073), and CBS 12438 (CNM-CL 7462).
DISCUSSION
Because of advances in molecular taxonomy, many species of yeasts that can cause infections in humans have been described recently. Some of these new pathogens (e.g., C. orthopsilosis, C. metapsilosis, C. nivariensis, and C. bracarensis) have been well characterized by molecular methods, such as PCR-based procedures and sequence analysis (amplification of ITS regions and the D1/D2 domain and analysis of sequence polymorphisms) (1, 35, 38, 39). Several reports have addressed the difficulty of identifying rare yeast isolates to the species level by conventional methods, since they are highly dependent on variables such as the growth medium and temperature (4). In addition, databases of commercial identification systems are limited to the species commonly found in the clinic, and in general terms, their use is time-consuming. On the other hand, molecular methods based on DNA sequencing resulted in the improved characterization of strains (4). Because of the emergence of resistant yeast pathogens, it is important that the available identification methods provide the highest possible degree of precision. Furthermore, providing reliable antifungal susceptibility profiles of these rare pathogenic yeasts will improve the management of patients with fungal infections due to these organisms.
In 1993, Lehman et al. described two different groups among C. haemulonii isolates (24) and concluded that these isolates represented a species complex. This classification is still in place today. In this study, we demonstrated that the C. haemulonii complex comprises two species, C. haemulonii group I and C. haemulonii group II, using phenotypic and molecular methods. They differ from each other in the four genes sequenced, some phenotypic features, and MALDI-TOF MS and AFLP profiles. Surprisingly, we found four C. haemulonii group I isolates that differ in the ITS gene sequences (96% similarity to the C. haemulonii group I type strain ITS sequences), whereas the other three genes studied were identical. The four strains also differ in some physiological features from C. haemulonii group I, including assimilation of melezitose and inulin and growth at 37°C. However, they did not differ by proteomics and AFLP analysis, and hence, we concluded that these four isolates represent a variety of C. haemulonii.
Only one study included a strain of C. haemulonii CBS 5149T (type strain) for MALDI-TOF MS testing (29). Here we present data on MALDI-TOF MS validation for fast and accurate identification of strains and clinical isolates of the species studied. MALDI-TOF MS allowed differentiation among all of the species in the C. haemulonii complex, producing secure species identification because all of the isolates could be identified to the species level with a reliability threshold of log scores of >1.7. The MALDI-TOF MS results were congruent with those obtained by AFLP typing.
Susceptibility tests of all of the strains included in this work established differences in their antifungal susceptibility patterns. In previous studies, all isolates of C. haemulonii were found to be resistant to both AMB and FLC (MIC range, 6.16 to 18.30 mg/liter). However, we found that our isolates from human, animal, and environmental sources demonstrated variable patterns of susceptibility to AMB (MIC range, 0.25 to >16 mg/liter) and FLC (MIC range, 8 to >64 mg/liter). Most of the FLC-resistant strains appeared to demonstrate azole cross-resistance (MIC of ITC, 4 to >8 mg/liter; MIC of VRC, >8 mg/liter; MIC of PSZ, >8 mg/liter), which was not described in previous reports, where most of the strains were reported to be susceptible to VRC and PSZ (16, 17, 34). Both C. haemulonii groups I and II have been reported to be susceptible to ANF and MCF (6, 17, 18, 34). In this study, the MICs of both MCF and ANF for all 30 isolates ranged from <0.03 to >16 mg/liter, thus showing that these two echinocandins are active against most of the 30 isolates in our collection. Some isolates, however, show high MICs of CAS, and some had a profile of cross-resistance to the three echinocandins, which has not been described before (MIC range, 16 to 18.34 mg/liter). When Candida spp. are grown in a medium containing a high concentration of an antifungal agent such as CAS, the result can be reduced activity of that agent against certain organisms. This phenomenon is called the Eagle effect or the paradoxical growth effect. The Eagle effect has previously been reported in many Candida species (12). In accordance with the results obtained with the C. haemulonii complex and the three echinocandins tested, more studies should be done to clarify the role of this paradoxical effect on the profile of susceptibility of these yeasts to these compounds.
With respect to biofilm formation, our data could not demonstrate significant differences between various species of the C. haemulonii complex. Recently, Oh et al. (28) reported that strains of C. haemulonii and C. pseudohaemulonii isolated from blood cultures showed good biofilm formation compared to C. auris isolates from ear specimens that did not form biofilms. The authors also correlated the extensive biofilm production with the origin of the isolates. Strains from central venous catheter-related fungemia recovered from patients receiving total parenteral nutrition did prominently form biofilms, whereas strains from ear specimens did not (28). In our study, the YNB medium was supplemented with 50 mM glucose (0.9%), which is the recommended medium for standard evaluation of biofilm formation (25, 28). In contrast, Shin et al. (37) used Sabouraud's dextrose broth supplemented with 8% glucose. These authors described differences in the ability to form biofilm among different Candida species cultivated in this high-glucose (8%) medium.
Here we propose a reclassification of the C. haemulonii complex and describe former C. haemulonii group II as a new species. We consider the differences in the DNA sequences of several genes among strains of C. haemulonii groups I and II to be too great to consider them conspecific, with similarity values found between the type strain of C. haemulonii group I (CBS 5149T) and the reference strain of C. haemulonii group II (CBS 7798) of 89.04% for ITS, 90.93% for D1/D2, 86.77% for RPB1, and 87.43% for RPB2. The different patterns of the two species in the MALDI-TOF MS and AFPL experiments corroborate the results of the molecular study. Differences in phenotypic and susceptibility features have also been observed. Different profiles of fermentation of raffinose and sucrose and assimilation of carbon compounds (for instance, l-sorbose, inulin, d-galactose, and succinate), different growth temperatures, and other discordant features were found. These different features could help to distinguish between the two species of the C. haemulonii species complex in the clinical setting when a molecular method or MALDI-TOF MS is not available. For this species, we propose the name C. duobushaemulonii sp. nov. (synonym: C. haemulonii group II).
In addition, a variety of C. haemulonii is described as C. haemulonii var. vulnera, which differs from other C. haemulonii isolates in the ITS sequence, some physiological and biochemical features, and its AMB susceptibility profile. Because we have only four isolates that probably accidentally were all obtained in Spain, we do not feel comfortable in commenting on this observation yet. If it turns out in the future that this variety is dominant in southern Europe or somewhere else, it may warrant some further mention.
The new taxa described here have a multiantifungal resistance profile that includes high MICs of AMB and cross-resistance to azole compounds and impairs the treatment of infections with these species with echinocandins and 5-FC. This fact is important in order to establish the correct treatment of patients with fungal infections due to these yeasts.
Fungemia due to C. haemulonii and its closely related new species is rare. Reliable identification is needed to start appropriate treatment and provide optimal management of infections due to these yeasts. Classical methods of identification are not able to identify these rare clinical isolates. Importantly, most of the uncommon yeasts that are incorrectly identified using conventional methods showed an antifungal resistance profile (i.e., C. haemulonii, Candida ciferri, Pichia anomala, Pichia membranifaciens, Pichia fermentans, Kodamaea ohmeri, and Candida rugosa) (4), and such incorrect identifications lead to inappropriate treatment and clinical management. C. haemulonii, C. haemulonii var. vulnera, C. duobushaemulonii, C. auris, and C. pseudohaemulonii are part of this group of rare clinical yeast that so far cannot be well differentiated by current commercial methods (17, 23, 30, 34). Identification of these species should be based on molecular methods such as PCR, sequencing analyses, and MALDI-TOF MS analyses. According to the results of sequencing and MALDI-TOF MS, we conclude that the best clinical identification at the species and variety levels can be done with the molecular barcode proposed for fungal identification, namely, the ITS region (36) and MALDI-TOF MS.
Supplementary Material
ACKNOWLEDGMENTS
In the past 5 years, M. Cuenca-Estrella has received grant support from Astellas Pharma, bioMérieux, Gilead Sciences, Merck Sharp & Dohme, Pfizer, Schering-Plough, and Soria Melguizo S.A. He has been an advisor/consultant to the Panamerican Health Organization, Gilead Sciences, Merck Sharp & Dohme, Pfizer, and Schering-Plough. He has been paid for talks on behalf of Gilead Sciences, Merck Sharp & Dohme, Pfizer, and Schering-Plough. M. Kostrzewa is an employee of Bruker Daltonik GmbH, the manufacturer of the MALDI Biotyper system, which was used for parts of this study.
This study was partially financed by The Spanish Society for Clinical Microbiology and Infectious Diseases (SEIMC, Spain) and by The European Consortium of Microbial Resource Centres (EMbaRC) training program. E. Cendejas-Bueno has a research contract from the Fondo de Investigaciones Sanitarias (grant CM08/0083, Spain). A. Alastruey-Izquierdo has a research contract from the Spanish Network for Research in Infectious Diseases (REIPI RD06/0008).
Footnotes
Published ahead of print 5 September 2012
Supplemental material for this article may be found at http://jcm.asm.org/.
REFERENCES
- 1.Alcoba-Flórez J, et al. 2005. Phenotypic and molecular characterization of Candida nivariensis sp. nov., a possible new opportunistic fungus. J. Clin. Microbiol. 43:4107–4111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bolano A, et al. 2001. Rapid methods to extract DNA and RNA from Cryptococcus neoformans. FEMS Yeast Res. 1:221–224 [DOI] [PubMed] [Google Scholar]
- 3.Borst A, et al. 2003. Use of amplified fragment length polymorphism analysis to identify medically important Candida spp., including C. dubliniensis. J. Clin. Microbiol. 41:1357–1362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cendejas-Bueno E, Gomez-Lopez A, Mellado E, Rodríguez-Tudela JL, Cuenca-Estrella M. 2010. Identification of pathogenic rare yeast species in clinical samples: comparison between phenotypical and molecular methods. J. Clin. Microbiol. 48:1895–1899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Correia A, Sampaio P, James S, Pais C. 2006. Candida bracarensis sp. nov., a novel anamorphic yeast species phenotypically similar to Candida glabrata. Int. J. Syst. Evol. Microbiol. 56:313–317 [DOI] [PubMed] [Google Scholar]
- 6.Crouzet J, Sotto A, Picard E, Lachaud L, Bourgeois N. 2011. A case of Candida haemulonii osteitis: clinical features, biochemical characteristics, and antifungal resistance profile. Clin. Microbiol. Infect. 17:1068–1070 [DOI] [PubMed] [Google Scholar]
- 7.Cuenca-Estrella M, et al. 2008. Update on the epidemiology and diagnosis of invasive fungal infection. Int. J. Antimicrob. Agents 32(Suppl 2):S143–S147 [DOI] [PubMed] [Google Scholar]
- 8.Cuenca-Estrella M, et al. 2006. Head-to-head comparison of the activities of currently available antifungal agents against 3,378 Spanish clinical isolates of yeasts and filamentous fungi. Antimicrob. Agents Chemother. 50:917–921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cuenca-Estrella M, et al. 2003. Multicenter evaluation of the reproducibility of the proposed antifungal susceptibility testing method for fermentative yeasts of the Antifungal Susceptibility Testing Subcommittee of the European Committee on Antimicrobial Susceptibility Testing (AFST-EUCAST). Clin. Microbiol. Infect. 9:467–474 [DOI] [PubMed] [Google Scholar]
- 10.Cuenca-Estrella M, et al. 2005. In vitro susceptibilities of bloodstream isolates of Candida species to six antifungal agents: results from a population-based active surveillance programme, Barcelona, Spain, 2002-2003. J. Antimicrob. Chemother. 55:194–199 [DOI] [PubMed] [Google Scholar]
- 11.de Hoog GS, Gerrits van den Ende AH. 1998. Molecular diagnostics of clinical strains of filamentous basidiomycetes. Mycoses 41:183–189 [DOI] [PubMed] [Google Scholar]
- 12.Fleischhacker M, Radecke C, Schulz B, Ruhnke M. 2008. Paradoxical growth effects of the echinocandins caspofungin and micafungin, but not of anidulafungin, on clinical isolates of Candida albicans and C. dubliniensis. Eur. J. Clin. Microbiol. Infect. Dis. 27:127–131 [DOI] [PubMed] [Google Scholar]
- 13.Gargeya IB, Pruitt WR, Meyer SA, Ahearn DG. 1991. Candida haemulonii from clinical specimens in the USA. J. Med. Vet. Mycol. 29:335–338 [PubMed] [Google Scholar]
- 14.Holá V, Ruzicka F, Horka M. 2010. Microbial diversity in biofilm infections of the urinary tract with the use of sonication techniques. FEMS Immunol. Med. Microbiol. 59:525–528 [DOI] [PubMed] [Google Scholar]
- 15.Jin Y, Yip HK, Samaranayake YH, Yau JY, Samaranayake LP. 2003. Biofilm-forming ability of Candida albicans is unlikely to contribute to high levels of oral yeast carriage in cases of human immunodeficiency virus infection. J. Clin. Microbiol. 41:2961–2967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Khan ZU, et al. 2007. Outbreak of fungemia among neonates caused by Candida haemulonii resistant to amphotericin B, itraconazole, and fluconazole. J. Clin. Microbiol. 45:2025–2027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim MN, et al. 2009. Candida haemulonii and closely related species at 5 university hospitals in Korea: identification, antifungal susceptibility, and clinical features. Clin. Infect. Dis. 48:e57–e61 [DOI] [PubMed] [Google Scholar]
- 18.Kim S, et al. 2011. Catheter-related candidemia caused by Candida haemulonii in a patient in long-term hospital care. J. Korean Med. Sci. 26:297–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kolecka A, Hernandez-Barbado R, Rupp S, Bujdakova H. 2011. Biofilm formation and adhesive/invasive properties of Candida dubliniensis in comparison with Candida albicans. Centr. Eur. J. Biol. 6:893–901 [Google Scholar]
- 20.Kurtzman CP, Fell JW, Boekhout T. 2011. The yeasts: a taxonomic study, 5th edition. Elsevier, Amsterdam, the Netherlands [Google Scholar]
- 21.Kurtzman CP, Robnett CJ. 1997. Identification of clinically important ascomycetous yeasts based on nucleotide divergence in the 5′ end of the large-subunit (26S) ribosomal DNA gene. J. Clin. Microbiol. 35:1216–1223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lavarde V, Daniel F, Saez H, Arnold M, Faguer B. 1984. Peritonite mycosique a Torulopsis haemulonii. Bull. Soc. Fr. Mycol. Med. 13:173–176 [Google Scholar]
- 23.Lee WG, et al. 2011. First three reported cases of nosocomial fungemia caused by Candida auris. J. Clin. Microbiol. 49:3139–3142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lehmann PF, Wu LC, Pruitt WR, Meyer SA, Ahearn DG. 1993. Unrelatedness of groups of yeasts within the Candida haemulonii complex. J. Clin. Microbiol. 31:1683–1687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li X, Yan Z, Xu J. 2003. Quantitative variation of biofilms among strains in natural populations of Candida albicans. Microbiology 149:353–362 [DOI] [PubMed] [Google Scholar]
- 26.Liu YJ, Whelen S, Hall BD. 1999. Phylogenetic relationships among ascomycetes: evidence from an RNA polymerase II subunit. Mol. Biol. Evol. 16:1799–1808 [DOI] [PubMed] [Google Scholar]
- 27.Marklein G, et al. 2009. Matrix-assisted laser desorption ionization–time of flight mass spectrometry for fast and reliable identification of clinical yeast isolates. J. Clin. Microbiol. 47:2912–2917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oh BJ, et al. 2011. Biofilm formation and genotyping of Candida haemulonii, Candida pseudohaemulonii, and a proposed new species (Candida auris) isolates from Korea. Med. Mycol. 49:98–102 [DOI] [PubMed] [Google Scholar]
- 29.Pinto A, et al. 2011. Matrix-assisted laser desorption ionization–time of flight mass spectrometry identification of yeasts is contingent on robust reference spectra. PLoS One 6:e25712 doi:10.1371/journal.pone.0025712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rodero L, et al. 2002. Transient fungemia caused by an amphotericin B-resistant isolate of Candida haemulonii. J. Clin. Microbiol. 40:2266–2269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rodríguez-Tudela JL, et al. 2007. Correlation of the MIC and dose/MIC ratio of fluconazole to the therapeutic response of patients with mucosal candidiasis and candidemia. Antimicrob. Agents Chemother. 51:3599–3604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rodriguez-Tudela JL, et al. 2008. EUCAST technical note on fluconazole. Clin. Microbiol. Infect. 14:193–195 [DOI] [PubMed] [Google Scholar]
- 33.Rodriguez-Tudela JL, et al. 2008. EUCAST technical note on voriconazole. Clin. Microbiol. Infect. 14:985–987 [DOI] [PubMed] [Google Scholar]
- 34.Ruan SY, Kuo YW, Huang CT, Hsiue HC, Hsueh PR. 2010. Infections due to Candida haemulonii: species identification, antifungal susceptibility and outcomes. Int. J. Antimicrob. Agents 35:85–88 [DOI] [PubMed] [Google Scholar]
- 35.Satoh K, et al. 2009. Candida auris sp. nov., a novel ascomycetous yeast isolated from the external ear canal of an inpatient in a Japanese hospital. Microbiol. Immunol. 53:41–44 [DOI] [PubMed] [Google Scholar]
- 36.Schoch CL, et al. 2012. Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for fungi. Proc. Natl. Acad. Sci. U. S. A. 109:6241–6246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shin JH, et al. 2002. Biofilm production by isolates of Candida species recovered from nonneutropenic patients: comparison of bloodstream isolates with isolates from other sources. J. Clin. Microbiol. 40:1244–1248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sugita T, Takashima M, Poonwan N, Mekha N. 2006. Candida pseudohaemulonii sp. nov., an amphotericin B- and azole-resistant yeast species, isolated from the blood of a patient from Thailand. Microbiol. Immunol. 50:469–473 [DOI] [PubMed] [Google Scholar]
- 39.Tavanti A, Davidson AD, Gow NA, Maiden MC, Odds FC. 2005. Candida orthopsilosis and Candida metapsilosis spp. nov. to replace Candida parapsilosis groups II and III. J. Clin. Microbiol. 43:284–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.van Uden N, Kolipinsky MC. 1962. Torulopsis haemulonii nov. spec., a yeast from the Atlantic Ocean. Antonie Van Leeuwenhoek 28:78–80 [DOI] [PubMed] [Google Scholar]
- 41.Yarrow D, Meyer SA. 1978. Proposal for amendment of the diagnosis of the genus Candida Berkhout nom. cons. Int. J. Syst. Bacteriol. 28:611–615 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.