Abstract
Campylobacter upsaliensis is a zoonotic, emerging pathogen that is not readily recovered in traditional stool culture. This case represents the first report of persistent bloody diarrhea with C. upsaliensis that was confirmed by filtration culture, PCR, and sequencing.
CASE REPORT
An 83-year-old male presented to the emergency department (ED) with an acute history of severe bloody diarrhea. His symptoms began with nausea, vomiting, and abdominal cramping, which he mistook for constipation. The patient took a single dose of laxative and shortly thereafter experienced numerous episodes of profuse bloody diarrhea that continued for several hours. He did not have fevers, chills, or sweats. The patient was found by his wife at home, collapsed in a chair, and was brought to the ED for evaluation. The patient's past medical history was significant for irritable bowel syndrome but no history of bloody diarrhea or rectal bleeding. His social history revealed contact with his sister-in-law and two canine pets, all with bloody diarrhea. On arrival in the ED, the patient's physical examination was unremarkable; however, out of concern for a lower gastrointestinal bleed, the patient was admitted for observation and further testing. Stool studies were negative for all gastrointestinal pathogens, including Salmonella, Shigella, Campylobacter, Aeromonas, Plesiomonas, and Vibrio by stool culture, Cryptosporidium, Giardia, and Shiga-toxigenic Escherichia coli (STEC) by enzyme immunoassay (EIA), and Clostridium difficile by PCR. In addition, ovum and parasite exams of the stool were performed and were remarkable only for numerous erythrocytes and leukocytes. These findings were consistent with the grossly bloody stool and a positive fecal lactoferrin EIA. The patient's symptoms gradually improved but did not resolve over the course of 48 h. He remained afebrile and hemodynamically stable and was discharged on hospital day 2 with no known etiologic cause of diarrhea. Five days later, he was treated with ciprofloxacin for continuing diarrhea out of concern for possible person-to-person spread of a still-unidentified gastrointestinal pathogen but only after STEC was ruled out by Shiga toxin EIA.
The clinical microbiology laboratory was consulted for additional testing for enteroinvasive or enteroaggregative E. coli, given the widely publicized enteroaggregative/Shiga-toxigenic E. coli O104:H4 outbreak that had recently concluded in Germany. No testing was available for these organisms; however, the stool was filtered through a 0.6-μm filter (Pall Life Sciences, Ann Arbor, MI) onto a brucella blood agar plate (Hardy Diagnostics, Santa Maria, CA) and cultured in an increased-hydrogen atmosphere of approximately 6.5 to 12.5% (7) (BioBag Type Cfj; BD, Franklin Lakes, NJ) at 37°C to enhance the detection of hydrogen-requiring Campylobacter spp. (21). For direct stool PCR, the specimen was treated with AL stool lysis buffer (Qiagen, Valencia, CA) and heated at 95°C for 10 min. The DNA was extracted with the Maxwell Cell LEV DNA purification kit on a Maxwell 16 automated extraction platform (Promega, Madison, WI). The PCR targeted a conserved region of the Campylobacter 16S rRNA gene and was positive (22). The amplified gene product was then sequenced, with a sequence 100% identical to that of Campylobacter upsaliensis. The subsequent filter culture grew small glistening colonies at 72 h of incubation that were weakly oxidase-positive, catalase-negative, Gram-negative “gull-shaped” rods (Fig. 1A and B). 16S rRNA sequencing was then performed on the colonies, and the result was also 100% identical to the sequence of C. upsaliensis (1,119-bp identity). The isolate had low MIC values for quinolones and macrolides (Table 1) (6). Retrospective stool specimens from the sister-in-law and both dogs (collected >3 weeks after the patient's admission) were tested by filtration culture and PCR. All specimens were culture negative. The sister-in-law was positive by PCR for the Campylobacter genus (22); however, the species could not be definitively determined by 16S rRNA sequencing.
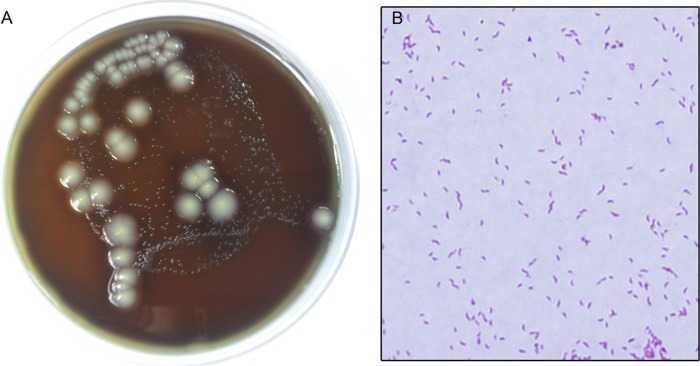
Fig 1.
Filter culture of the patient's stool sample. Small glistening colonies were identified as Campylobacter upsaliensis and grew in 72 h (A). The characteristic Gram stain morphology for C. upsaliensis is seen in panel B. Large white creamy colonies stained as Gram-positive rods, grew in less than 24 h, are regularly seen on Campylobacter-negative cultures, and were not consistent with Campylobacter (A).
Table 1.
Antimicrobial susceptibility profile of C. upsaliensis isolated from the patient's stool by filtration culturea
| Drug | MIC(s) (μg/ml) |
|---|---|
| Ampicillin-sulbactam | 1, 0.5 |
| Amoxacillin-clavulanate | 0.5, 0.25 |
| Cefoxitin | 16 |
| Trimethoprim-sulfamethoxasole | >2, 38 |
| Ceftriaxone | 16 |
| Meropenem | <0.25 |
| Clindamycin | 1 |
| Ampicillin | 2 |
| Moxifloxacin | <0.06 |
| Piperacillin-tazobactam | 32, 4 |
| Metronidazole | 8 |
| Vancomycin | >8 |
| Penicillin | >2 |
| Nitrofurantoin | >8 |
| Tetracycline | 0.5 |
| Azithromycin | <0.25 |
MICs were obtained by broth microdilution performed according to CLSI standards for C. jejuni/C. coli (6).
Campylobacter upsaliensis has been isolated from human blood, placental tissue, breast abscess, and stool (8, 13, 20, 25). There have also been rare/controversial reports of hemolytic-uremic syndrome and Guillain-Barré syndrome associated with C. upsaliensis (4, 9, 14). In both pediatric and immunocompromised hosts, C. upsaliensis is recognized as a clinically important emerging diarrheal pathogen (10, 17, 19). However, human cases of C. upsaliensis gastroenteritis with severe persistent bloody diarrhea similar to that observed in this patient have not been described. It is unclear what role treatment plays in recovery from these infections, and limited data are available for antibiotic resistance rates in C. upsaliensis. In several studies, erythromycin resistance has ranged from 10 to 15% for C. upsaliensis (11, 25, 28). Ciprofloxacin resistance in one study was approximately 5%, and resistance to quinolones in C. upsaliensis has been associated with prior patient exposure to quinolones in the setting of chronic unresolved diarrhea (17, 28). The rate of quinolone resistance for C. upsaliensis is low compared to those for Campylobacter jejuni and Campylobacter coli, which have surpassed 40% (23). This likely reflects the fact that C. coli and C. jejuni have been regularly exposed to sarafloxacin and enrofloxacin in the poultry industry, whereas domestic felines and canines (the reservoir for C. upsaliensis) would not have similar exposure to drive resistance (24).
The source of this infection could not be determined in this investigation, since only the patient was culture and PCR positive. C. upsaliensis is a well-documented cause of diarrhea in felines and canines. Therefore, one hypothesis is that the symptomatic dogs transmitted this infection to the patients (3, 26). Reports of serious infections attributed to pet-to-human transmission from both cats and dogs have been documented (12, 13). We were unable to detect C. upsaliensis in either dog's stool >3 weeks after symptoms; however, identification of C. upsaliensis in the dogs' stool would not have been definitive evidence of a point source, because up to 58% of nondiarrheic dogs are thought to be transiently colonized with C. upsaliensis (5). An alternative hypothesis is that the sister-in-law (who had molecular evidence of recent infection with a Campylobacter species and was treated empirically) may have acquired Campylobacter by drinking unfiltered spring water while recently hiking and subsequently transmitted the infection to the patient while visiting his home during her illness. Acquisition of C. jejuni from drinking contaminated groundwater has been previously described for hikers (27), and human-to-human transmission of both C. upsaliensis and C. jejuni have been described (10, 18). It is also possible that the patient acquired the infection from a source unrelated to the canines' or sister-in-law's illnesses.
The routine work-up for stool culture performed in most clinical labs in the United States does not include nonselective Campylobacter culture, such as the procedure that was performed in this study (15). While many Campylobacter species may “break through” the conditions optimized for C. jejuni and C. coli in a standard Campylobacter culture (i.e., 42°C incubation, cephalosporin-containing selective media, microaerobic environment, and culture discard at 72 h), many species of Campylobacter cannot tolerate one or more of these restrictive conditions. In particular, C. upsaliensis is typically sensitive to cephalosporins, thrives in an increased-hydrogen atmosphere, and typically grows in 96 h or more. Goossens et al. (11) found that of 99 C. upsaliensis isolates obtained by filtration culture, only 4 grew in a standard selective Campylobacter culture. This patient's routine Campylobacter culture was plated on cephalosporin-containing medium and incubated in a hydrogen-deficient atmosphere (Mitsubishi AnaeroPouch-MicroAero; Remel, Lenexa, KS), and the plates were discarded after 72 h, likely accounting for the lack of organism recovery. Use of similar conditions may account for an underappreciation of this species as a gastrointestinal pathogen. Studies in South Africa using filtration culture have found C. upsaliensis accounting for 23% of Campylobacter stool isolates (19), whereas Irish and Canadian studies using filtration culture and/or PCR detected C. upsaliensis in only 0.7 to 2.1% of stool specimens (1, 2, 16). These data suggest that the prevalence of this species may be geographically variable. Importantly, filtration culture and direct stool PCR/sequencing for the Campylobacter genus are not routinely available to most laboratories.
This report draws attention to the clinical importance of C. upsaliensis and reveals an emerging pathogen, capable of causing severe persistent bloody diarrhea. In this patient, the etiological agent of this infection would not have been identified were filtration culture and PCR not employed. The approach taken in this case also argues for the use of alternative methodologies amenable to the detection of other clinically relevant Campylobacter species that are intolerant to routine selective culture conditions.
Footnotes
Published ahead of print 22 August 2012
REFERENCES
- 1. Bullman S, et al. 2011. Emerging dynamics of human campylobacteriosis in Southern Ireland. FEMS Immunol. Med. Microbiol. 63:248– 253 [DOI] [PubMed] [Google Scholar]
- 2. Bullman S, O'Leary J, Corcoran D, Sleator RD, Lucey B. 2012. Molecular-based detection of non-culturable and emerging campylobacteria in patients presenting with gastroenteritis. Epidemiol. Infect. 140:684– 688 [DOI] [PubMed] [Google Scholar]
- 3. Burnens AP, Nicolet J. 1992. Detection of Campylobacter upsaliensis in diarrheic dogs and cats, using a selective medium with cefoperazone. Am. J. Vet. Res. 53:48– 51 [PubMed] [Google Scholar]
- 4. Carter JE, Cimolai N. 1996. Hemolytic-uremic syndrome associated with acute Campylobacter upsaliensis gastroenteritis. Nephron 74:489. [DOI] [PubMed] [Google Scholar]
- 5. Chaban B, Ngeleka M, Hill JE. 2010. Detection and quantification of 14 Campylobacter species in pet dogs reveals an increase in species richness in feces of diarrheic animals. BMC Microbiol. 10:73 doi:10.1186/1471-2180-10-73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. CLSI 2010. Methods for antimicrobial dilution and disk susceptibility testing of infrequently isolated or fastidious bacteria; approved guideline, 2nd ed CLSI document M45-A2. CLSI, Wayne, PA [Google Scholar]
- 7. Garcia LS. (ed). 2010. Clinical microbiology procedures handbook, 3rd ed, vol 1 ASM Press, Washington, DC [Google Scholar]
- 8. Gaudreau C, Lamothe F. 1992. Campylobacter upsaliensis isolated from a breast abscess. J. Clin. Microbiol. 30:1354– 1356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Goddard EA, Lastovica AJ, Argent AC. 1997. Campylobacter 0:41 isolation in Guillain-Barre syndrome. Arch. Dis. Child. 76:526– 528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Goossens H, et al. 1995. Investigation of an outbreak of Campylobacter upsaliensis in day care centers in Brussels: analysis of relationships among isolates by phenotypic and genotypic typing methods. J. Infect. Dis. 172:1298– 1305 [DOI] [PubMed] [Google Scholar]
- 11. Goossens H, et al. 1990. Characterization and description of “Campylobacter upsaliensis” isolated from human feces. J. Clin. Microbiol. 28:1039– 1046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Goossens H, et al. 1991. Campylobacter upsaliensis enteritis associated with canine infections. Lancet 337:1486– 1487 [DOI] [PubMed] [Google Scholar]
- 13. Gurgan T, Diker KS. 1994. Abortion associated with Campylobacter upsaliensis. J. Clin. Microbiol. 32:3093– 3094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ho TW, et al. 1997. Motor nerve terminal degeneration provides a potential mechanism for rapid recovery in acute motor axonal neuropathy after Campylobacter infection. Neurology 48:717– 724 [DOI] [PubMed] [Google Scholar]
- 15. Hurd S, et al. 2012. Clinical laboratory practices for the isolation and identification of Campylobacter in Foodborne Diseases Active Surveillance Network (FoodNet) sites: baseline information for understanding changes in surveillance data. Clin. Infect. Dis. 54(Suppl. 5):S440–S445 [DOI] [PubMed] [Google Scholar]
- 16. Inglis GD, Boras VF, Houde A. 2011. Enteric campylobacteria and RNA viruses associated with healthy and diarrheic humans in the Chinook health region of southwestern Alberta, Canada. J. Clin. Microbiol. 49:209– 219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jenkin GA, Tee W. 1998. Campylobacter upsaliensis-associated diarrhea in human immunodeficiency virus-infected patients. Clin. Infect. Dis. 27:816– 821 [DOI] [PubMed] [Google Scholar]
- 18. Jones A, Harrop C. 1981. A study of Campylobacter enteritis. J. Int. Med. Res. 9:40– 43 [DOI] [PubMed] [Google Scholar]
- 19. Lastovica AJ. 2006. Emerging Campylobacter spp.: the tip of the iceberg. Clin. Microbiol. Newsl. 28:49– 56 [Google Scholar]
- 20. Lastovica AJ, Le Roux E, Penner JL. 1989. “Campylobacter upsaliensis” isolated from blood cultures of pediatric patients. J. Clin. Microbiol. 27:657– 659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Le Roux E, Lastovica AJ. 1998. The Cape Town protocol: how to isolate the most campylobacters for your dollar, pound, franc, yen, etc., p 31–33 In Lastovica AJ, Newell DG, Lastovica EE. (ed), Proceedings of the 9th International Workshop on Campylobacter, Helicobacter and related organisms. Institute of Child Health, Cape Town, South Africa [Google Scholar]
- 22. Linton D, Owen RJ, Stanley J. 1996. Rapid identification by PCR of the genus Campylobacter and of five Campylobacter species enteropathogenic for man and animals. Res. Microbiol. 147:707– 718 [DOI] [PubMed] [Google Scholar]
- 23. Nachamkin I, Ung H, Li M. 2002. Increasing fluoroquinolone resistance in Campylobacter jejuni, Pennsylvania, USA,1982-2001. Emerg. Infect. Dis. 8:1501– 1503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Nelson JM, Chiller TM, Powers JH, Angulo FJ. 2007. Fluoroquinolone-resistant Campylobacter species and the withdrawal of fluoroquinolones from use in poultry: a public health success story. Clin. Infect. Dis. 44:977– 980 [DOI] [PubMed] [Google Scholar]
- 25. Patton CM, et al. 1989. Human disease associated with “Campylobacter upsaliensis” (catalase-negative or weakly positive Campylobacter species) in the United States. J. Clin. Microbiol. 27:66– 73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Steinhauserova I, Fojtikova K, Klimes J. 2000. The incidence and PCR detection of Campylobacter upsaliensis in dogs and cats. Lett. Appl. Microbiol. 31:209– 212 [DOI] [PubMed] [Google Scholar]
- 27. Taylor DN, McDermott KT, Little JR, Wells JG, Blaser MJ. 1983. Campylobacter enteritis from untreated water in the Rocky Mountains. Ann. Intern. Med. 99:38– 40 [DOI] [PubMed] [Google Scholar]
- 28. Vandenberg O, et al. 2006. Antimicrobial susceptibility of clinical isolates of non-jejuni/coli campylobacters and arcobacters from Belgium. J. Antimicrob. Chemother. 57:908– 913 [DOI] [PubMed] [Google Scholar]