Abstract
Candida parapsilosis has become a significant cause of invasive fungal infections in seriously ill patients. Nosocomial outbreaks through direct and indirect contact have been described. The aim of this study was the molecular characterization of what appeared to be an ongoing C. parapsilosis outbreak at the cardiothoracic intensive care unit of the University Hospital of Vienna between January 2007 and December 2008. Using two different molecular typing methods—automated repetitive sequence-based PCR (DiversiLab; bioMérieux) and microsatellite genotyping—we investigated the genetic relationship of 99 C. parapsilosis isolates. Eighty-three isolates originated from the cardiothoracic intensive care unit, while 16 isolates were random control isolates from other intensive care units and a different Austrian hospital. The 99 C. parapsilosis isolates analyzed by repetitive-element PCR all showed identical genotypes, suggesting an ongoing outbreak. In contrast, microsatellite genotyping showed a total of 56 different genotypes. Two major genotypes were observed in 10 and 15 isolates, respectively, whereas another 13 genotypes were observed in 2 to 4 isolates each. Forty-one genotypes were observed only once. Closely related genotypes that differed in only a single microsatellite marker were grouped into clonal complexes. When it comes to C. parapsilosis, microsatellite genotyping is a more discriminative method than repetitive-element PCR genotyping to investigate outbreaks.
INTRODUCTION
Invasive fungal infections are severe and life-threatening complications in immunocompromised or seriously ill patients. Candida species are the cause of nosocomial sepsis, septic shock, and lethal disseminated infection in critically ill patients admitted to intensive care units (ICUs), rising over the past decade to the fourth most common pathogens causing nosocomial bloodstream infections in the United States (3). The increasing frequency rate of serious Candida hospital infections in intensive care patients has multifactorial causes, including increasing usage of intravascular catheters, broad-spectrum antibiotics, extensive surgery, and immunosuppression for neoplastic disease or allograft preservation (2, 8). Despite the availability of potent antifungal agents, the mortality rate from invasive Candida infection is up to 65% (16).
With Candida albicans being still the most common pathogen isolated from immunosuppressed patients, the incidence of isolation of so-called non-albicans Candida, C. glabrata or C. parapsilosis, has been increasing in Europe (1, 19, 20). Candida parapsilosis strains, though, are more heterogeneous than other Candida species. Tavanti and colleagues suggested a division of the C. parapsilosis group (sensu lato) into three species named C. parapsilosis sensu stricto (or simply Candida parapsilosis), C. orthopsilosis, and C. metapsilosis (18). Although frequently associated with neonates and the use of parenteral nutrition, the epidemiology of C. parapsilosis is more complex, as demonstrated in a recent multicenter study by Cantón and coworkers. They found C. parapsilosis most frequently in adult intensive care unit, surgery, and internal medicine departments, and in their study, most C. parapsilosis candidemias were observed in adults (4). Moreover, the new class of echinocandin broad-spectrum antifungals—although very active against C. albicans or C. glabrata—shows increased MICs against most clinical isolates of the C. parapsilosis group (15), with differences at the species level (3).
Candida parapsilosis nosocomial outbreaks have been reported to occur not only through direct and indirect contact via the hands of health care workers but also through contaminated patient care equipment (5, 12, 13, 21). The close genetic relatedness of C. parapsilosis strains in clinical settings emphasizes the need for reliable identification methods to clarify potential transmission routes. As current methods for yeast identification are based on phenotypic features, they can neither differentiate among the three closely related C. parapsilosis sensu lato species nor reliably discriminate isolates below the species level within the C. parapsilosis sensu stricto group. To ensure optimal therapeutic options and study nosocomial cross-transmission, molecular identification of C. parapsilosis outbreak isolates at the species level is therefore of utmost importance (10).
During a 2-year observation period, a conspicuous colonization with C. parapsilosis was observed at one adult cardiothoracic intensive care unit at the University Hospital of Vienna. Genotyping with conventional methods indicated a pseudo-outbreak involving 50 patients with 83 colonizing isolates. Here we report the use of additional microsatellite genotyping of C. parapsilosis using a six-marker microsatellite panel.
MATERIALS AND METHODS
Study setting.
Our study population was derived from a larger prospective study (unpublished data) conducted at the University Hospital of Vienna—a 2,200-bed tertiary care medical university teaching hospital—between December 2006 and December 2008. During a 2-year period, all nonneutropenic cardiac surgery patients admitted to the ICU longer than 7 days were consecutively enrolled into the prospective study. To detect Candida colonization, surveillance culture samples were taken twice weekly from different superficial sites, open wounds, and anus and analyzed for the presence of Candida using routine methods for culture and identification. Over the 24-month period, a total of 199 patients were enrolled. Of 199 patients, 148 were colonized with Candida spp. at any site. Candida albicans was the predominant colonizing species, recovered from 142 patients, followed by C. parapsilosis, recovered from 50 patients. Molecular characterization followed for all colonizing isolates from that prospective study found to be C. parapsilosis. The study was approved by the Ethics Review Committee of the Medical University of Vienna (EC no. 510/2006).
Samples.
A total of 99 colonizing isolates were investigated using two different molecular typing methods. Eighty-three colonizing isolates from urine samples (n = 6) and from superficial sites, including anus (n = 4), axilla (n = 10), groin (n = 29), nose (n = 17), respiratory tract (n = 14), wound (n = 1), and other (n = 2), of 50 patients were collected during the period of the above-mentioned prospective study. Nine samples from patients of other intensive care units at our hospital as well as seven isolates from another Austrian hospital were also included in our analysis and used for comparison purposes.
Culture and identification.
All isolates were grown for 48 h on Sabouraud glucose agar medium (Heipha Diagnostika, Eppelheim, Germany) at 37°C. Identification was done using the API ID 32C (bioMérieux Austria GmbH) according to the manufacturer's instructions.
DNA extraction.
DNA from each of the 99 colonizing isolates was extracted using the UltraClean microbial DNA isolation kit (Mo Bio Laboratories) by following the manufacturer's instructions. We used 5 U/μl of the enzyme Zymolyase (Zymo Research Corporation, Irvine, CA). The DNA concentration was adjusted to approximately 25 ng/μl for each sample.
Molecular characterization by automated rep-PCR typing system (DiversiLab).
All DNA samples obtained from the 99 colonizing isolates were amplified using the DiversiLab Candida kit for DNA fingerprinting (bioMérieux Austria GmbH) by following the manufacturer's instructions. Briefly, 25 ng genomic DNA, 2.5 U AmpliTaq polymerase, 2.5 μl 10× PCR buffer (Applied Biosystems, CA), and 2 μl primer mix were added to the repetitive sequence-based PCR (rep-PCR) master mix in a total volume of 25 μl per reaction. Thermal cycling parameters were as follows: initial denaturation of 94°C for 2 min, followed by 35 cycles of denaturation at 92°C for 30 s, annealing at 50°C for 30 s, and extension at 70°C for 90 s, with a final extension at 70°C for 3 min. Detection of rep-PCR products was done by microfluidic chips and a model B2100 bioanalyzer (Agilent Technologies, Inc., Palo Alto, CA) of the DiversiLab system, and analysis was performed with DiversiLab software, version 3.4. The resulting DNA fingerprint patterns were viewed as electropherograms, and the results included a dendrogram with a similarity matrix and a virtual gel image of the fingerprint for each DNA sample (11).
Microsatellite typing.
A panel of six short tandem repeat (STR) markers was used for genotyping the C. parapsilosis isolates. Three trinucleotide repeat markers as described earlier by Sabino et al. (17) were amplified in a multiplex PCR. For these markers, we redesigned the amplification markers to generate shorter PCR products and to amplify all three in one multiplex PCR. Instead of the dinucleotide marker described before, we added a panel of three hexanucleotide markers. In each panel, one of the amplification primers (Table 1) was labeled with either 6-carboxyfluorescein (FAM), hexachlorofluorescein (HEX), or 6-carboxytetramethylrhodamine (TAMRA). Amplification reactions were performed in a final volume of 25 μl containing approximately 1 ng genomic DNA, 0.5 μM amplification primers, 0.2 mM deoxynucleoside triphosphates (dNTPs), 3 mM MgCl2, and 1 U FastStart Taq DNA polymerase (Roche Diagnostics, Almere, the Netherlands) in 1× reaction buffer (Roche Diagnostics). Thermocycling was performed in a T1 thermocycler (Biometra, Göttingen, Germany) using the following conditions: initial denaturation for 10 min at 95°C, followed by 35 cycles of 30 s of denaturation at 95°C, 30 s of annealing at 60°C, and 1 min of extension at 72°C. A final incubation of 10 min at 72°C was included before the reactions were cooled to room temperature. Following PCR, the amplification products were diluted 30-fold with distilled water and a 1-μl aliquot of diluted PCR product was added to 8.75 μl distilled water and 0.25 μl internal size marker ET-ROX 400 (GE Healthcare, Diegem, Belgium). After denaturation of the samples at 95°C for 1 min and rapid cooling to 4°C, they were injected and run onto a MegaBACE 500 platform (GE Healthcare) equipped with a 48-capillary array as recommended by the manufacturer. The number of repeats in each marker was determined by comparing the relative size of each allele to those obtained using the reference C. parapsilosis strain CDC317, whose genome has been fully sequenced. Diploid genotypes were converted to binary data by scoring the presence (“1”) or absence (“0”) of each possible allele. Similarities between genotypes were visualized by constructing a minimum spanning tree using BioNumerics, version 6.0 (Applied Maths, St.-Martens-Latem, Belgium), treating the data as categorical information.
Table 1.
Overview of amplification primers used in this study
| Marker | Labeled primer (5′–3′) | Unlabeled primer (5′–3′)a |
|---|---|---|
| 3A | FAM-CCTGGCTTGCAATTTCATTT | GCCTCATCGGTGGTGGAATTA |
| 3B | HEX-TTGGAGTAACAAGCGCAGAA | GTCGCTTGGACAACTGGTGTA |
| 3C | TAMRA-CAATAGCAGCAATGGAGCAG | GTGCTTTTGGTTTGTCCTTGG |
| 6A | FAM-CCAGGTTGGACTATCACTG | GGTTTCATTTTGTTGTGAAAA |
| 6B | HEX-CCCTTTCAAAAGAAACAGACA | GTTCTATAGATAAAACACACCCCATACA |
| 6C | TAMRA-TGGCGTTAGTATTGGCGTTA | GATTGTATCACGCGGGAACTC |
The underlined G nucleotide in the unlabeled primer sequence is not a match to the genomic DNA but was introduced to minimize the formation of minus-A peaks (7).
RESULTS
By using two different molecular typing methods—automated repetitive sequence-based PCR (rep-PCR) (DiversiLab) and microsatellite-based genotyping—we investigated the genetic relationship of 99 Candida parapsilosis isolates from 56 different patients. Eighty-three colonizing isolates from 50 patients were issued from that prospective study, while nine samples from other ICUs and seven samples from another Austrian hospital were included for comparison purposes.
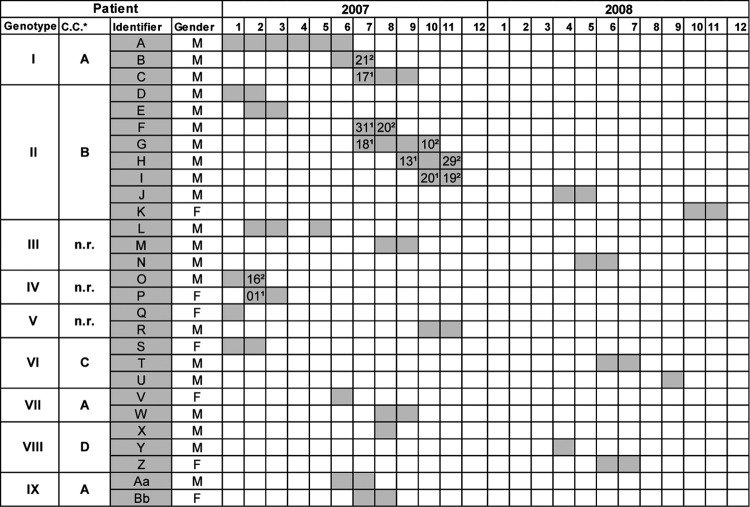
Genotyping of the 99 C. parapsilosis isolates using repetitive-element PCR showed identical rep-PCR genotype patterns (data not shown) irrespective of the patient's identity, the patient's location, and the time of sampling. In contrast, microsatellite genotyping using a panel of six STR markers identified 56 different genotypes, of which 41 were observed only once. Fifteen genotypes were found multiple times and were numbered I to XV. The two most prevalent genotypes (I and II) were found in 10 and 15 isolates and were retrieved from three and eight patients, respectively. Seven genotypes (III to IX) each contained two to four isolates from different patients (Table 2), while the remaining six genotypes (X to XV) involved only one patient each, with two isolates from different anatomic sites taken from this patient over time. A timeline of the nine genotypes involving more than one patient is given in Fig. 1. While with genotypes I, II, and IV there was an overlap in the times of stay of some of the patients, this was not the case for genotypes III, V to VIII, and IX (Fig. 1). The relationship between the obtained genotypes is illustrated in Fig. 2. From the minimum spanning tree it is clear that some genotypes may have evolved by spontaneous changes in one of the microsatellite markers. This can be recognized from the complexes of apparently closely related genotypes that differed in only one of the six microsatellite markers (indicated as A to F in Fig. 2A). Six of such complexes containing a total of 53 isolates were recognized. From 18 patients, multiple isolates were available. In nine patients, this involved only a single genotype, whereas in two patients, two different but closely related genotypes were found. In the remaining seven patients, two or more clearly different genotypes were found over time. In one patient, different genotypes were recovered on the same day but from different body sites.
Table 2.
Candida parapsilosis strain typing results using microsatellite genotyping: relationship between genotypes I to IX and clonal complexes of genotypes for 28 patients harboring genotypes I to IX at the cardiothoracic intensive care unit of the University Hospital of Vienna during the 24-month study period
| Genotype | No. of isolates (patients) per genotype | Concatenated diploid genotype | Clonal complexa |
|---|---|---|---|
| I | 10 (3) | 262799106404410128877 | A |
| II | 15 (8) | 26265151404010108877 | B |
| III | 4 (3) | 26275050474710108877 | NR |
| IV | 3 (2) | 26264545404110108877 | NR |
| V | 3 (2) | 26273636404619198877 | NR |
| VI | 3 (3) | 25273636374314148877 | C |
| VII | 2 (2) | 262799106444410128877 | A |
| VIII | 3 (3) | 15155959434414148877 | D |
| IX | 2 (2) | 2627106106404410128877 | A |
A to D indicate clonal complexes of genotypes differing in only one microsatellite marker. NR, not part of a clonal complex.
Fig 1.
Timeline showing length of stay and potential overlap of stay for 28 patients harboring genotypes I to IX at the cardiothoracic intensive care unit of the University Hospital of Vienna during the 24-month study period. The relationship between genotypes I to IX and clonal complexes of genotypes is also shown. * C.C., clonal complex of genotypes where A to D indicate clonal complexes of genotypes differing in only one of the six microsatellite markers. n.r., there is no relationship to a clonal complex. Numbers with a superscript “1” and a superscript “2” are dates of admission to and discharge from the cardiothoracic intensive care unit, respectively. Admission or discharge dates are given only if there is an overlap of stay of patients with the same genotype.
Fig 2.
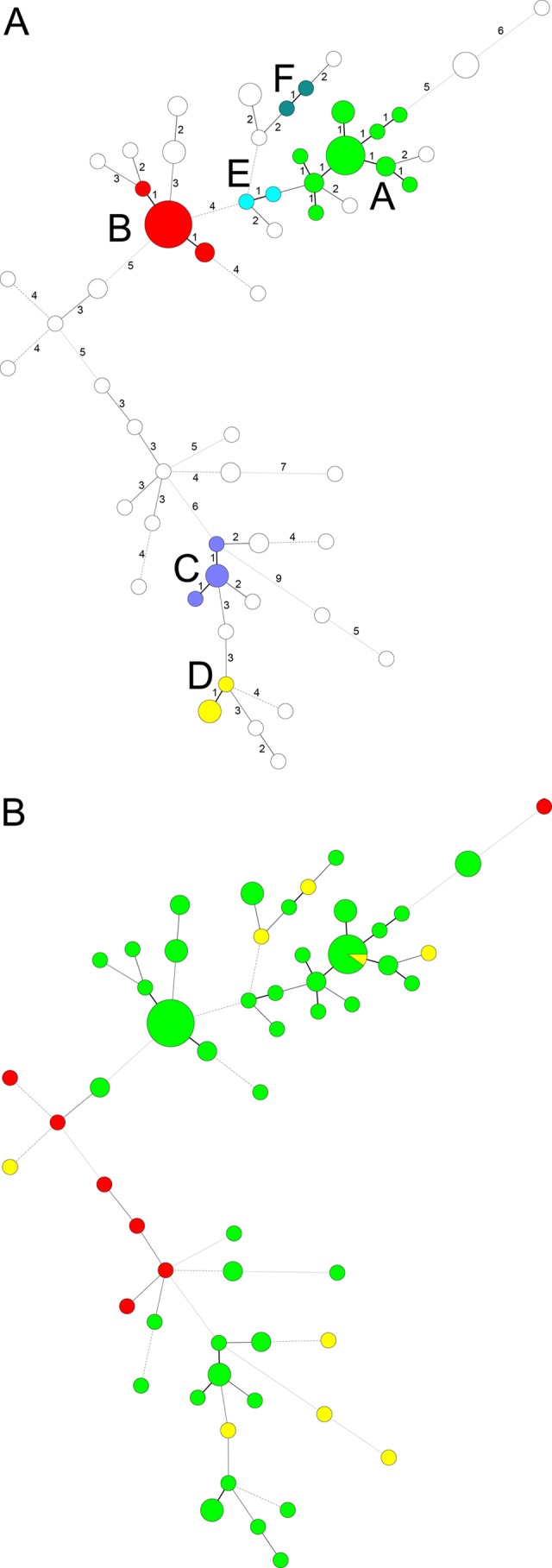
Minimum spanning tree showing the differences between the genotypes based on a categorical analysis. Each circle represents a unique genotype. The size of the circle corresponds to the number of isolates of that genotype. Numbers correspond to the number of differences between the genotypes. (A) Clonal complexes of genotypes differing in only one microsatellite marker are indicated with colors and lettered A to F. (B) Origin of the samples: green, study collection; yellow, isolates from different wards; red, isolates from another hospital.
To illustrate the discriminatory power of the microsatellite panel, we compared the genotypes to those found from different wards and from a different hospital. All nine control isolates from different wards yielded different genotypes. Only one of these genotypes was also found in our study population, and another isolate was of a different but related genotype. None of the seven isolates from a different hospital were found in our study collection (Fig. 2B).
DISCUSSION
Different molecular typing methods, such as random amplified polymorphic DNA typing, restriction fragment length polymorphism, and multilocus sequence typing to classify C. parapsilosis sensu lato into three distinct species according to Tavanti and coworkers, have been described previously (18, 22). In 2006, Lasker and colleagues reported a microsatellite method based on dinucleotide repeats able to distinguish isolates within the C. parapsilosis sensu stricto group (14). More recently, Sabino and coworkers also described a new DNA typing method with even higher discriminatory power involving microsatellite loci able to distinguish among C. parapsilosis sensu stricto isolates (17).
Surveillance culture samples were taken twice weekly from different superficial sites of all patients hospitalized on the cardiothoracic intensive care unit of the Vienna University Hospital between December 2006 and December 2008. Eighty-three isolates from 50 patients all from this cardiothoracic intensive care unit were found to be C. parapsilosis. Repetitive-element PCR genotyping results suggested that we were facing an ongoing outbreak of C. parapsilosis in this ICU. For comparison purposes, nine additional isolates were obtained from other ICUs at our hospital, and seven isolates were obtained from another Austrian hospital. All additional nonoutbreak isolates analyzed by automated repetitive-sequence-based PCR typing system did as well show the identical rep-PCR genotype. In order to clarify this potential outbreak, the isolates were then further analyzed by microsatellite genotyping. Microsatellite genotyping identified all 16 nonoutbreak isolates as being 16 clearly different genotypes. Within the 83 outbreak isolates, 26 genotypes were observed only once. The remaining 15 genotypes each represented a separate cluster. Our findings suggest that for 82% (41/50) of our patients, C. parapsilosis isolates were already present as part of the colonizing flora of the patients and were selected in the intensive care setting, where antimicrobial and antifungal treatment is widely administered. Because of the intrinsic instability of the microsatellite loci, isolates with only very minor changes in one of the microsatellite markers are being considered to be clonally related though not identical, as was the case for four of our patients. The isolation of patient-specific or clonally related genotypes of C. parapsilosis from multiple anatomical sites over time in 24% (12/50) of our patients further supports the evidence of an endogenous colonization with those Candida isolates (23). Nevertheless, nosocomial transmission cannot be excluded with isolates of genotypes I, II, and IV, since there was an overlap in the times of stay of some of the patients, as well as for those seven patients for whom two or three clearly distinct genotypes were found. Prevention strategies aimed at C. parapsilosis must therefore take into account appropriate control measures against both exogenous and endogenous infection reservoirs.
In our daily infection control, routine DNA typing methods have proven to be very valuable in studying the genetic relatedness among microorganisms involved in possible outbreak settings. DiversiLab typing is a rapid and easy-to-perform method for initial screening during outbreak investigation for different bacterial species, as described by Fluit and colleagues for Acinetobater spp., Stenotrophomonas maltophilia, and the Enterobacter cloacae complex (9) and as recently described by Deplano and coworkers for Enterococcus faecium and Klebsiella pneumoniae (6). Regarding Candida species, Wise and colleagues did show that DiversiLab was able to discriminate between the medically important members of the genus Candida at the species level (24). However, when it comes to C. parapsilosis strain differentiation, our study could demonstrate that DiversiLab typing has limited discriminatory power and therefore needs to be replaced by a more discriminative method such as microsatellite genotyping to investigate outbreaks where C. parapsilosis is involved.
ACKNOWLEDGMENTS
We thank Andrea Grisold from the Institute of Hygiene, Microbiology and Environmental Medicine of the Medical University of Graz, Graz, Austria, for the Candida parapsilosis isolates provided for comparison purposes from her hospital.
We declare that we have no conflict of interest.
Footnotes
Published ahead of print 8 August 2012
REFERENCES
- 1. Arendrup MC. 2010. Epidemiology of invasive candidiasis. Curr. Opin. Crit. Care 16: 445–452 [DOI] [PubMed] [Google Scholar]
- 2. Banerjee SN, et al. 1991. Secular trends in nosocomial primary bloodstream infections in the United States, 1980–1989. National Nosocomial Infections Surveillance System. Am. J. Med. 91: 86S–89S [DOI] [PubMed] [Google Scholar]
- 3. Cantón E, Espinel-Ingroff A, Peman J, del Castillo L. 2010. In vitro fungicidal activities of echinocandins against Candida metapsilosis, C. orthopsilosis, and C. parapsilosis evaluated by time-kill studies. Antimicrob. Agents Chemother. 54: 2194–2197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cantón E, et al. 2011. Prospective multicenter study of the epidemiology, molecular identification, and antifungal susceptibility of Candida parapsilosis, Candida orthopsilosis, and Candida metapsilosis isolated from patients with candidemia. Antimicrob. Agents Chemother. 55: 5590–5596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Clark TA, et al. 2004. Epidemiologic and molecular characterization of an outbreak of Candida parapsilosis bloodstream infection in a community hospital. J. Clin. Microbiol. 42: 4468–4472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Deplano A, et al. 2011. Controlled performance evaluation of the DiversiLab repetitive-sequence-based genotyping system for typing multidrug-resistant health care-associated bacterial pathogens. J. Clin. Microbiol. 49: 3616–3620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. de Valk HA, Meis JF, Klaassen CH. 2007. Microsatellite based typing of Aspergillus fumigatus: strengths, pitfalls and solutions. J. Microbiol. Methods 69: 268–272 [Epub ahead of print.] doi:10.1016/j.mimet.2007.01.009. [DOI] [PubMed] [Google Scholar]
- 8. Eggimann P, et al. 2005. Invasive candidiasis: comparison of management choices by infectious disease and critical care specialists. Intensive Care Med. 31: 1514–1521 [DOI] [PubMed] [Google Scholar]
- 9. Fluit AC, et al. 2010. Evaluation of the DiversiLab system for detection of hospital outbreaks of infections by different bacterial species. J. Clin. Microbiol. 48: 3979–3989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Garcia-Effron G, et al. 2011. Assessment of two new molecular methods for identification of Candida parapsilosis sensu lato species. J. Clin. Microbiol. 49: 3257–3261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Healy M, et al. 2005. Microbial DNA typing by automated repetitive-sequence-based PCR. J. Clin. Microbiol. 43: 199–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hernández-Castro R, et al. 2010. Outbreak of Candida parapsilosis in a neonatal intensive care unit: a health care workers source. Eur. J. Pediatr. 169: 783–787 [DOI] [PubMed] [Google Scholar]
- 13. Kuhn DM, et al. 2004. Candida parapsilosis characterization in an outbreak setting. Emerg. Infect. Dis. 10: 1074–1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lasker B, Butler G, Lott TJ. 2006. Molecular genotyping of Candida parapsilosis group I clinical isolates by analysis of polymorphic microsatellite markers. J. Clin. Microbiol. 44: 750–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Pfaller MA, et al. 2012. Frequency of decreased susceptibility and resistance to echinocandins among fluconazole-resistant bloodstream isolates of Candida glabrata: results from the SENTRY antimicrobial surveillance program (2006–2010) and the Centers for Disease Control and Prevention population-based surveillance (2008–2010). J. Clin. Microbiol. 50: 1199–1203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Presterl E, Daxbock F, Graninger W, Willinger B. 2007. Changing pattern of candidaemia 2001–2006 and use of antifungal therapy at the University Hospital of Vienna, Austria. Clin. Microbiol. Infect. 13: 1072–1076 [DOI] [PubMed] [Google Scholar]
- 17. Sabino R, et al. 2010. New polymorphic microsatellite markers able to distinguish among Candida parapsilosis sensu stricto isolates. J. Clin. Microbiol. 48: 1677–1682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tavanti A, Davidson AD, Gow NA, Maiden MC, Odds FC. 2005. Candida orthopsilosis and Candida metapsilosis spp. nov. to replace Candida parapsilosis groups II and III. J. Clin. Microbiol. 43: 284–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Van Asbeck EC, Clemons KV, Markham AN, Stevens DA. 2008. Candida parapsilosis Global Epidemiology Group. Molecular epidemiology of the global and temporal diversity of Candida parapsilosis. Scand. J. Infect. Dis. 40: 827–834 [DOI] [PubMed] [Google Scholar]
- 20. van Asbeck EC, Clemons KV, Stevens DA. 2009. Candida parapsilosis: a review of its epidemiology, pathogenesis, clinical aspects, typing and antimicrobial susceptibility. Crit. Rev. Microbiol. 35: 283–309 [DOI] [PubMed] [Google Scholar]
- 21. van Asbeck EC, Huang YC, Markham AN, Clemons KV, Stevens DA. 2007. Candida parapsilosis fungemia in neonates: genotyping results suggest healthcare workers hands as source, and review of published studies. Mycopathologia 164: 287–293 [DOI] [PubMed] [Google Scholar]
- 22. van Asbeck EC, Clemons KV, Markham AN, Stevens DA. 2009. Correlation of restriction fragment length polymorphism genotyping with internal transcribed spacer sequence, randomly amplified polymorphic DNA and multilocus sequence groupings for Candida parapsilosis. Mycoses 52: 493–498 [DOI] [PubMed] [Google Scholar]
- 23. Voss A, Hollis RJ, Pfaller MA, Wenzel RP, Doebbeling BN. 1994. Investigation of the sequence of colonization and candidemia in nonneutropenic patients. J. Clin. Microbiol. 32: 975–980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wise MG, et al. 2007. Species identification and strain differentiation of clinical Candida isolates using the DiversiLab system of automated repetitive sequence-based PCR. J. Med. Microbiol. 56: 778–787 [DOI] [PubMed] [Google Scholar]