Abstract
We compared the performance characteristics of culture and the Cepheid Xpert vanA assay for routine surveillance of vancomycin-resistant enterococci (VRE) from rectal swabs in patients at high risk for VRE carriage. The Cepheid Xpert vanA assay had a limit of detection of 100 CFU/ml and correctly detected 101 well-characterized clinical VRE isolates with no cross-reactivity in 27 non-VRE and related culture isolates. The clinical sensitivity, specificity, positive predictive value, and negative predictive value of the Xpert vanA PCR assay were 100%, 96.9%, 91.3%, and 100%, respectively, when tested on 300 consecutively collected rectal swabs. This assay provides excellent predictive values for prompt identification of VRE-colonized patients in hospitals with relatively high rates of VRE carriage.
INTRODUCTION
Vancomycin-resistant enterococci (VRE) are recognized as nosocomial pathogens alongside methicillin-resistant Staphylococcus aureus (MRSA) and Clostridium difficile. Vancomycin resistance in enterococci species is conferred mainly by the presence of the vanA or vanB gene, although the presence of other genes, including vanC, vanD, vanE, and vanG, can also result in a resistant phenotype (7, 25). In North America, the vanA gene is the most prevalent resistance marker in enterococci species, followed by the vanB gene, which can be found in bacteria other than enterococci (4, 10). Both the vanA and vanB genes are carried on transposable plasmids, and transfer of these plasmids to other enterococci and S. aureus has been shown both in vitro and in vivo (7).
Several reports have shown that in allogeneic hematopoietic stem cell transplant recipients, VRE colonization, prior to stem cell transplantation, is a significant risk factor for the development of VRE bacteremia, which is associated with poor clinical outcomes (3, 14, 22, 24). In order to decrease the spread of VRE in hospital settings, the Hospital Infection Control Practices Advisory Committee (HICPAC) recommends a multipronged approach that includes rapid identification and reporting of VRE-positive stools or rectal swabs by the microbiology laboratory in order to ensure prompt isolation of colonized patients (2).
Currently, VRE surveillance is performed at our institution using traditional culture. This procedure requires 48 to 96 h to obtain a final result and involves multiple media and incubation steps. Recently, the Food and Drug Administration (FDA) approved a rapid molecular assay, the Xpert vanA (Cepheid, Sunnyvale, CA), for the detection of VRE directly from rectal swab specimens only. The assay is a real-time, one-step PCR assay performed on the GeneXpert instrument and provides results in less than 1 h, compared to 48 to 96 h with culture. In addition to providing rapid results for timely isolation of colonized patients, rapid and more sensitive detection of VRE may also result in the timely identification of patients at risk for the development of VRE bacteremia. The objective of the present study was to evaluate the performance characteristics of this novel PCR assay compared to those of traditional culture for the detection of VRE from rectal swabs. To our knowledge, this is the first reported evaluation of the Xpert vanA assay in a patient population at high risk for VRE colonization.
(This study was presented in part at the 52nd Interscience Conference on Antimicrobial Agents and Chemotherapy.)
MATERIALS AND METHODS
Isolates and patient specimens.
One hundred and twenty-eight archived, previously characterized clinical isolates of enterococci (including both vancomycin-resistant [n = 101] and vancomycin-susceptible/intermediate [n = 7] isolates) and other nonenterococci isolates (n = 20) were tested to determine the analytical sensitivity and specificity of the Xpert vanA PCR assay (Table 1). Additionally, 300 consecutive rectal swabs (BBLCulturette; BD Diagnostics, Sparks, MD) from 162 patients that were submitted during a 4-week period to the laboratory for VRE surveillance culture were tested to determine the clinical sensitivity and specificity of the Xpert vanA PCR assay. The study was approved by the Memorial-Sloan Kettering Cancer Center (MSKCC) institutional review board.
Table 1.
Analytical sensitivity and specificity of the Xpert vanA PCR
| Organism | No. of strains/replicates tested | No. of Xpert vanA-positive isolates |
|---|---|---|
| E. faecium (vancomycin resistant) (101 CFU/ml) | 5 | 2 |
| E. faecium (vancomycin resistant) (102 CFU/ml)a | 5 | 5 |
| E. faecium (vancomycin resistant) (103 CFU/ml) | 2 | 2 |
| E. faecium (vancomycin resistant) (104 CFU/ml) | 2 | 2 |
| E. faecium (vancomycin resistant) (105 CFU/ml) | 2 | 2 |
| E. faecium (vancomycin resistant) (106 CFU/ml) | 2 | 2 |
| E. faecium (vancomycin resistant) (107 CFU/ml) | 2 | 2 |
| E. faecium (vancomycin resistant) | 81 | 81 |
| E. faecalis (vancomycin resistant) | 20 | 18b |
| E. faecium (vancomycin susceptible) | 2 | 0 |
| E. faecium (vancomycin intermediate) | 1 | 0 |
| E. faecalis (vancomycin susceptible) | 2 | 0 |
| Enterococcus raffinosus | 1 | 0 |
| Enterococcus gallinarum | 1 | 0 |
| S. aureus | 2 | 0 |
| Glycopeptide-intermediate S. aureus | 3 | 0 |
| Lactobacillus johnsonii | 1 | 0 |
| Gram-negative bacillic | 14 | 0 |
The lower limit of detection was 100 CFU/ml.
Two isolates were E. faecalis vanB positive.
Includes several Enterobacteriaceae isolates.
Surveillance culture.
VRE surveillance culture was performed by streaking a rectal swab onto a Campy agar plate containing cefoperazone, vancomycin, and amphotericin B (CVA) (BD Diagnostics, Sparks, MD), followed by incubation at 37°C in 5 to 10% CO2 for 24 to 48 h. Suspicious colonies were Gram stained and tested for the presence of pyrrolidonyl arylamidase activity and the lack of catalase activity. Any isolates consistent with Enterococcus species were tested for vancomycin susceptibility by the Kirby-Bauer method using a 30-μg vancomycin disk according to Clinical and Laboratory Standards Institute (CLSI) guidelines (6). The final species identification was generated using the MicroScan dried Gram-positive identification (ID) type 3 panel on the automated MicroScan instrument (Siemens, West Sacramento, CA).
Xpert vanA PCR.
A PCR assay was performed according to the manufacturer's instructions using rectal swabs collected for VRE surveillance culture. The limit of detection (LOD) of the assay was determined by testing a dilution series (0 CFU/ml to 107 CFU/ml) of a vancomycin-resistant Enterococcus faecium isolate (identification confirmed by culture) in 2 to 5 replicates.
Additional assays.
The vancomycin-teicoplanin Etest (AB Biodisk North America, Inc., Culver City, CA) was used to determine the phenotype of any VRE culture isolates that were negative by the Xpert vanA assay. VRE isolates with a vancomycin MIC of >32 μg/ml and a teicoplanin MIC of >32 μg/ml were considered vanA positive, and any VRE isolates with a vancomycin MIC of >32 μg/ml and a teicoplanin MIC of <32 μg/ml were considered vanB positive. Enriched broth culture was also used on discordant results and performed by inoculating the rectal swab in Trypticase soy broth for 5 days, followed by subculture and further testing as described above for surveillance culture.
vanA and vanB real-time PCRs.
Additional real-time PCR assays were developed to confirm the presence of the vanA or vanB gene in Xpert vanA PCR-positive, culture-negative specimens. Primers (vanA PCR forward primer, GGCTGTTTCGGGCTGTGA-3′; vanA PCR reverse primer, 5′-ACTAACGCGGCACTGTTTCC-3′; vanB PCR forward primer, 5′-GGGAACGAGGATGATTTGATTG-3′; vanB PCR reverse primer, 5′-CGTGGCTCAGCCGGATT-3′) were designed using the Applied Biosystems Primer Express software version 3.0 (Life Technology Corp., Carlsbad, CA). The analytical specificity was determined by performing a Basic Local Alignment Search Tool (BLAST) search of each primer and the entire amplicon sequence using the National Center for Biotechnology Information website (http://www.ncbi.nlm.nih.gov) and testing of isolates listed in Table 1. The analytical sensitivity of this laboratory-developed PCR assay was determined by performing serial dilution of vanA/vanB-positive VRE. Detection of the amplified product was performed using Fast SYBR green master mix (Life Technology Corp., Carlsbad, CA) on the 7500 real-time PCR system (Life Technology Corp., Carlsbad, CA) in a final volume of 20 μl with the following thermal cycler profile: 1 cycle of 95°C for 2 min, 40 cycles at 95°C for 5 s, 60°C for 10 s, and 75°C for 35 s, and a dissociation step of 95°C for 15 s, 60°C for 1 min, and 95°C for 15 s. The amplified sequences were run on a 2% gel (E-gel; Life Technology Corp., Carlsbad, CA) to confirm their correct size. Known positive and negative VRE isolates were included in each run and on each gel. To test culture isolates, 2 to 3 colonies of each isolate were diluted in 500 μl of nuclease-free water (Roche Applied Sciences, Indianapolis, IN), vortexed for 10 s at high speed, and boiled for 10 min at 95°C. Five microliters of the supernatant was used for amplification. To test rectal swab specimens, 5 μl of the remaining sample reagent buffer used for the Xpert vanA PCR was used for the real-time PCR.
Discordant result analysis.
The reference standard used to determine true-positive and false-negative results was a combination standard (i.e., a true-positive sample was a specimen that was positive by at least two methods). Any specimen with a discordant result was further analyzed by (i) a review of medical records to determine if the patient had a recent positive VRE culture (within 4 weeks) and/or (ii) additional testing of rectal swabs by enriched broth culture. Discordant test results were considered true positive only if the enriched broth culture and/or chart review confirmed the presence of VRE and true negative if neither the broth culture nor the chart review confirmed the presence of VRE.
Statistical analysis.
The sensitivity, specificity, positive predictive value (PPV), and negative predictive value (NPV) were calculated for both the Xpert vanA assay and direct culture using the reference standard described above. The significance of the observed difference was determined using Fisher's test for sensitivity and specificity and the 1-way analysis of variance (ANOVA) test to compare the median semiquantitative culture results to the corresponding median cycle threshold (CT) values of the Xpert vanA PCR. A P value of ≤0.05 was considered significant. Statistical analysis was performed using the GraphPad Prism software (GraphPad Software, Inc., La Jolla, CA).
RESULTS
Out of 128 well-characterized isolates, the Xpert vanA PCR correctly identified 101 clinical VRE isolates with no cross-reactivity with 27 non-VRE isolates (Table 1). Three VRE isolates originally tested negative by Xpert vanA PCR. Additional testing of these isolates with the vancomycin-teicoplanin Etest strips (AB Biodisk North America, Inc., Culver City, CA) identified two isolates as vancomycin-susceptible E. faecium (vancomycin MICs of 1.0 and 1.5 μg/ml and teicoplanin MICs of 1.5 and 2.0 μg/ml) and one isolate as vancomycin-resistant E. faecalis with a vanB phenotype (a vancomycin MIC of >32 μg/ml and a teicoplanin MIC of 2 μg/ml). Testing of this isolate using the vanB PCR described above confirmed the presence of the vanB gene (data not shown). Following resolution of discordant results, both the analytical sensitivity and specificity of the Xpert vanA assay were 100%.
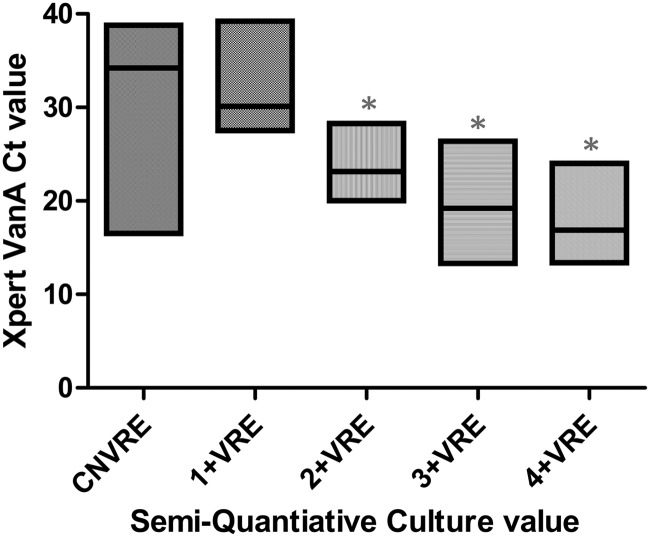
A total of 300 specimens from 162 patients were tested by both Xpert vanA PCR and direct culture. The vanA gene was detected in 81 specimens from 60 patients (37.0% of patients), while VRE isolates were recovered in 56 specimens from 46 patients (28.4% of patients). The lower limit of detection of the assay, as determined by 10-fold serial dilutions of VRE, was 100 CFU/ml (Table 1). The median CT value was compared to the corresponding semiquantitative result of the surveillance culture (Fig. 1) to further compare the sensitivity of the PCR assay to the results obtained by culture. The median CT value for PCR-positive, culture-negative specimens was 34.1, which was approximately one dilution away from the median CT value (30.1) for 1+ (1 to 9 colonies) positive cultures (Fig. 1). This suggests that the observed discrepancy might be due to a bacterial load below the sensitivity of direct culture, although the difference in CT values between these two groups was not statistically significant (P > 0.05).
Fig 1.
Xpert vanA CT values versus those of semiquantitative VRE culture. The horizontal line in each floating box represents the median CT value, and the length of each box reflects the range (minimum to maximum) of CT values for each semiquantitative result. *, P value of <0.0001 compared to culture-negative VRE (CNVRE). VRE, vancomycin-resistant enterococcus species. 1+, 1 to 9 colonies; 2+, 1 to 49 colonies; 3+, 5 to 300 colonies; 4+, >300 colonies.
Among the 25 PCR-positive, culture-negative swabs, 13 (52%) had a positive culture within 3 weeks (range, 1 day to 21 days; median, 7 days) of the PCR results and were considered true positive. The remaining 12 discordant swabs were incubated for 5 days in Trypticase soy broth, and VRE was detected in 5/12 swabs for a total of 18/25 (72%) true-positive PCR results. Although only 5/12 swabs became positive by enriched broth culture, 11/12 swabs tested by a second, laboratory-developed real-time PCR (LOD, 100 CFU/ml; specificity, 100%; data not shown) were positive for the vanA gene.
Following resolution of discordant results, the clinical sensitivity, specificity, positive predictive value (PPV), and negative predictive value (NPV) of the Xpert vanA PCR assay were 100%, 96.9%, 91.4%, and 100%, respectively, while those of direct culture were 75.7%, 100%, 100%, and 92.6%, respectively (Table 2). The difference between Xpert vanA and direct culture results was statistically significant (P < 0.001).
Table 2.
Comparison of VRE culture to Xpert vanA PCRa
| Test | No. of isolates with each set of resultsb |
Sensitivity (%) | Specificity (%) | PPV (%) | NPV (%) | |||
|---|---|---|---|---|---|---|---|---|
| Reference+, test+ | Reference+, test− | Reference−, test+ | Reference−, test− | |||||
| VRE direct culture | 56 | 18 | 0 | 226 | 75.7 (64.3–84.9) | 100 (98.4–100) | 100 (93.6–100) | 92.6 (88.6–95.6) |
| Xpert vanA PCR | 74 | 0 | 7 | 219 | 100 (95.1–100) | 96.9 (93.7–98.8) | 91.3 (83.0–96.5) | 100 (98.3–100) |
Values in parentheses are the 95% confidence interval.
Reference, direct culture and results of enriched broth culture and/or chart review for discordant specimens; reference+, positive result for the reference; test+, positive result for the indicated test.
DISCUSSION
We report our evaluation of the FDA-approved Cepheid Xpert vanA assay for the detection of VRE from rectal swabs compared to that by direct culture. Phenotypic identification of VRE isolates by culture is based on a MIC of ≥32 μg/ml (CLSI). Using the Campy agar plate, which contains 10 μg/ml of vancomycin, as our primary plating medium for VRE surveillance culture, both intermediate (8 to 16 μg/ml) and resistant (>32 μg/ml) enterococci can be isolated. Enterococci isolates growing on the Campy agar plate are then further tested using a 30-μg vancomycin disk to identify resistant strains. The Xpert vanA assay identifies VRE based solely on the presence of the vanA gene, which confers a high level of inducible resistance to both vancomycin and teicoplanin (7).
Similar studies have evaluated the performance of a Conformité Européenne (CE)-marked version of this assay that detects the presence of both the vanA and vanB genes (Xpert vanA/vanB assay) in rectal swabs, perianal swabs, and stool specimens (5, 9, 11, 16, 23) (Table 3). Bourdon et al. (5) tested 804 rectal swabs and detected 127 swabs positive for vanA or vanB by Xpert vanA/vanB assay, with 11 swabs positive by both Xpert PCR and culture. The high sensitivity (100%) and low positive predictive value (8.7%) of the assay in the Bourdon et al. study were attributed mainly to the high detection of the vanB gene (n = 115), which was considered a false-positive result due to the lack of enterococcus species growth in culture. The PPV of the assay for the vanA gene alone, although better, was also relatively low at 66.7%. Consequently, the authors recommended that all Xpert positive results be confirmed by culture, which negates, in part, the value of this rapid test. However, in a setting in which the prevalence of VRE is low, such an approach might be beneficial. Dekeyser et al. tested 565 rectal swabs during and following an outbreak of VRE in their hospital (9). VRE prevalences during and after the outbreak were 5.9% and 1%, respectively. However, the PPV of the assay remained low (15% during the outbreak versus 2.8% after the outbreak), primarily due to detection of the vanB gene. The poor PPV of PCR for vanB VRE has also been reported for other PCR assays, including the BD GeneOhmVanR assay (BD GeneOhm, San Diego, CA) and the Roche LightCycler analyte-specific reagents (ASRs) (15, 17, 21). Different from Bourdon et al., our findings showed a higher PPV (91%, versus 66.7% in the study by Bourdon et al.) after resolution of discrepant results for the vanA gene.
Table 3.
Comparison of published Xpert vanA/vanB test characteristicsa
| Study | No. of specimens tested | VRE prevalence | VRE positivity rate | Gene target | Sensitivity (%) | Specificity (%) | PPV (%) | NPV (%) |
|---|---|---|---|---|---|---|---|---|
| Marner et al. (16)b,c | 184 | 46.7 | NA | vanA/vanB | 96.4 (89.6–99.2) | 93.0 (86.0–96.8) | 92.0 (84.2–96.3) | 96.9 (86.5–99.4) |
| This study | 300 | 30.0 | NA | vanA | 100 (95.1–100) | 96 (93.7–98.8) | 91.4 (83.0–96.5) | 100 (98.3–100) |
| Bourdon et al. (5) | 804 | NA | 1.4 | vanA | 100 (62.8–100) | 99.5 (98.7–99.9) | 66.7 (38.8–86.5) | 100 (99.4–100) |
| vanB | 100 (38.2–100) | 85.6 (82.9–87.8) | 2.6 (0.6–7.7) | 100 (99.3–100) | ||||
| Dekeyser et al. (9) | 447 | 1.0 | NA | vanA/vanB | 100 (ND) | 76.8 (ND) | 2.8 (ND) | 100 (ND) |
| 118 | 5.9 | NA | vanA/vanB | 100 (ND) | 69.3 (ND) | 15 (ND) | 100 (ND) | |
| Gazin et al. (11)b,d | 50 | NA | 44 | vanA | 73.9 (ND) | 92.3 (ND) | 89.5 (ND) | 80.6 (ND) |
| vanB | 87.5 (ND) | 14.7 (ND) | 32.6 (ND) | 71.4 (ND) | ||||
| Zabicka et al. (23)d | 37 | 35.1 | NA | vanA/vanB | 61.5 (32.3–84.8) | 79.2 (57.3–92.1) | 61.5 (32.3–84.9) | 79.2 (57.3–92.1) |
PPV, positive predictive value; NPV, negative predictive value; ND, not determined; NA, not available. Values in parentheses are the 95% confidence interval.
Study not done on consecutive specimens. When prevalence was not available, the positivity rate obtained with the culture method is listed.
Perianal swab specimens.
Stool specimens.
The prevalence of VRE for patients screened in our hospital was calculated for the year 2011 at 30.0%, which is close to the prevalence during our study period. The higher prevalence of VRE-colonized patients in our population may explain the marked difference in the PPV between the two studies (Table 3). Screening of our VRE isolate library revealed a low incidence of vanB VRE isolates; only one culture isolate negative by Xpert vanA was determined to be positive for the vanB gene by a vancomycin-teicoplanin Etest and a vanB PCR assay. Similar to our data, Stamper et al. detected vanB-positive enterococcus species by the BD GeneOhm VanR assay (BD GeneOhm, San Diego, CA) in only 3/147 specimens positive by culture (21). In their evaluation of the Xpert vanA/vanB assay, Marner and colleagues also detected a low number (5/88) of perianal swabs positive only for the vanB gene, with only 1 confirmed by culture (16). These results confirm the lower prevalence of the vanB gene in Enterococcus species in North America compared to the prevalence of the vanB gene in Enterococcus species in Europe, as previously reported by the SENTRY antimicrobial surveillance program (10). Unlike Marner et al., our evaluation was performed (i) on consecutive rectal swabs rather than on a selected set of perianal swabs, (ii) using a different reference method as the gold standard, and (iii) targeting only the vanA gene. These differences may explain the variations in the observed sensitivity, specificity, PPV, and NPV between the two studies. Two additional studies evaluating the performance of the Xpert vanA showed remarkably lower sensitivity by the assay than by culture (11, 23). The limited number of specimens tested (≤50), as well as the use of stool specimens rather than rectal swabs, might explain the suboptimal performance of the Xpert vanA assay in those studies.
Although culture is the most common method used for surveillance screening, a higher bacterial burden is necessary to obtain a positive result. D'Agata et al. (8) showed that the sensitivity of a rectal swab culture varied from 0% when VRE density was ≤4.5 log10 CFU/g of stool to 100% when the VRE density was ≥7.5 log10 CFU/g of stool (average of 58% sensitive). In our study, the sensitivity of the surveillance culture was 75.7% (Table 2). Additionally, there are variations in the sensitivity and specificity of different culture methods, including chromogenic agars, which, as previously reported, result in a range of sensitivities (1, 13, 20). The lower sensitivity of a culture, therefore, should be taken into consideration when following the HICPAC recommendation to terminate isolation following three negative cultures (2). Multiple studies have also shown that spontaneous decolonization occurs only in a limited number of patients; however, reappearance of the VRE within a few weeks of decolonization is common (12, 18, 19). All Xpert vanA-positive discordant results were tested by a second independent laboratory-developed real-time PCR to confirm that the false-positive results were truly due to the presence of the vanA gene. Additional VRE isolates were also detected when broth-enriched culture, which we do not perform routinely, was used to analyze discrepant results. Further review of our Xpert vanA-positive, culture-negative results suggested that 68% of our discrepant results can be attributed to low bacterial load because the PCR results preceded or followed a recent positive culture result. The significance of a low bacterial VRE load detected by PCR only and its impact on nosocomial transmission of VRE are unknown and will have to be studied further, especially as it applies to discontinuation of contact precautions for PCR-positive patients. Since a VRE PCR-positive result with a CT value of >34 (range, 16.5 to 38.8) often corresponded to a negative culture, a quantitative or semiquantitative PCR assay rather than a qualitative assay might be more relevant for infection control purposes, although this remains to be determined.
Our study has some limitations. First, the Xpert vanA PCR was performed using the same swab used to set up the culture. Although this algorithm did not affect the sensitivity of the assay, more specimens might have been positive if the swabs were tested directly as opposed to following culture inoculation. Second, it is possible that false-negative results occurred due to the presence of vanB VRE, which are not detected by this assay. We did not have any rectal swabs that were culture positive and Xpert vanA negative, although, as described earlier, the sensitivity of the culture is not optimal.
At MSKCC, active surveillance for VRE is performed in units with high-risk patients, including those in intensive care and bone marrow transplant units. Implementation of the Xpert vanA PCR would provide several advantages, including rapid identification and prompt reporting of VRE-colonized patients for immediate isolation, identification of patients at high-risk for developing VRE bacteremia, and decreased labor and turnaround time associated with traditional culture. In theory, identification and isolation of VRE-colonized patients should result in a decreased rate of VRE infections as well as a decreased rate of nosocomial cases. However, rapid identification and isolation of colonized patients is only part of the equation; other factors, such as prudent use of vancomycin and hospital staff knowledge and adherence to isolation precautions, also contribute to the overall VRE hospital rate (2). Practical issues associated with the use of PCR for VRE identification include the inability to save VRE culture isolates (necessary for epidemiological studies in case of outbreak) and the inability to differentiate between E. faecium and Enterococcus faecalis. Furthermore, similar to other commercial PCR assays, the cost of the Xpert vanA PCR is significantly higher than that of culture. Therefore, depending on the rate of VRE colonization and VRE infection in a hospital, the implementation of a more sensitive, albeit more costly, test might be justified for efficient infection control and rapid identification of patients at increased risk for VRE infections.
In conclusion, the excellent sensitivity and specificity and rapid turnaround time of the Cepheid Xpert vanA assay make it an attractive option for routine surveillance of VRE from rectal swabs. This assay will significantly reduce the labor and time associated with the traditional surveillance culture method.
ACKNOWLEDGMENTS
We thank Steve Tulumba and Darlene Reid in the Bacteriology Laboratory for assistance with testing of rectal swabs by Xpert vanA PCR.
Footnotes
Published ahead of print 12 September 2012
REFERENCES
- 1.Adler H, Oezcan S, Frei R. 2010. Vancomycin-resistant enterococci of vanB genotype may pose problems for screening with highly selective media. J. Clin. Microbiol. 48:2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Anonymous. 1995. Recommendations for preventing the spread of vancomycin resistance. Recommendations of the Hospital Infection Control Practices Advisory Committee (HICPAC). MMWR Recommend. Rep. 44(RR-12):1–13 [PubMed] [Google Scholar]
- 3.Avery R, et al. 2005. Early vancomycin-resistant enterococcus (VRE) bacteremia after allogeneic bone marrow transplantation is associated with a rapidly deteriorating clinical course. Bone Marrow Transplant. 35:497–499 [DOI] [PubMed] [Google Scholar]
- 4.Ballard SA, Grabsch EA, Johnson PD, Grayson ML. 2005. Comparison of three PCR primer sets for identification of vanB gene carriage in feces and correlation with carriage of vancomycin-resistant enterococci: interference by vanB-containing anaerobic bacilli. Antimicrob. Agents Chemother. 49:77–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bourdon N, et al. 2010. Rapid detection of vancomycin-resistant enterococci from rectal swabs by the Cepheid Xpert vanA/vanB assay. Diagn. Microbiol. Infect. Dis. 67:291–293 [DOI] [PubMed] [Google Scholar]
- 6.CLSI 2010. Performance standards for antimicrobial susceptibility testing; 20th informational supplement. CLSI M100-S20. Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 7.Courvalin P. 2006. Vancomycin resistance in gram-positive cocci. Clin. Infect. Dis. 42(Suppl 1:S25–S34 [DOI] [PubMed] [Google Scholar]
- 8.D'Agata EM, Gautam S, Green WK, Tang YW. 2002. High rate of false-negative results of the rectal swab culture method in detection of gastrointestinal colonization with vancomycin-resistant enterococci. Clin. Infect. Dis. 34:167–172 [DOI] [PubMed] [Google Scholar]
- 9.Dekeyser S, Beclin E, Descamps D. 2011. Implementation of vanA and vanB genes by PCR technique research interest in system (Xpert vanA/vanB CepheidR) closed in a laboratory of microbiology in managing an outbreak to Enterococcus faecium resistant glycopeptide (EfRG). Pathol. Biol. 59:73–78 (In French.) [DOI] [PubMed] [Google Scholar]
- 10.Deshpande LM, Fritsche TR, Moet GJ, Biedenbach DJ, Jones RN. 2007. Antimicrobial resistance and molecular epidemiology of vancomycin-resistant enterococci from North America and Europe: a report from the SENTRY antimicrobial surveillance program. Diagn. Microbiol. Infect. Dis. 58:163–170 [DOI] [PubMed] [Google Scholar]
- 11.Gazin M, Lammens C, Goossens H, Malhotra-Kumar S. 2012. Evaluation of GeneOhm VanR and Xpert vanA/vanB molecular assays for the rapid detection of vancomycin-resistant enterococci. Eur. J. Clin. Microbiol. Infect. Dis. 31:273–276 [DOI] [PubMed] [Google Scholar]
- 12.Huckabee CM, Huskins WC, Murray PR. 2009. Predicting clearance of colonization with vancomycin-resistant enterococci and methicillin-resistant Staphylococcus aureus by use of weekly surveillance cultures. J. Clin. Microbiol. 47:1229–1230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jenkins SG, Raskoshina L, Schuetz AN. 2011. Comparison of performance of the novel chromogenic Spectra VRE agar to that of bile esculin azide and Campylobacter agars for detection of vancomycin-resistant enterococci in fecal samples. J. Clin. Microbiol. 49:3947–3949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kamboj M, et al. 2010. The changing epidemiology of vancomycin-resistant enterococcus (VRE) bacteremia in allogeneic hematopoietic stem cell transplant (HSCT) recipients. Biol. Blood Marrow Transplant. 16:1576–1581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mak A, Miller MA, Chong G, Monczak Y. 2009. Comparison of PCR and culture for screening of vancomycin-resistant enterococci: highly disparate results for vanA and vanB. J. Clin. Microbiol. 47:4136–4137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marner ES, et al. 2011. Diagnostic accuracy of the Cepheid GeneXpert vanA/vanB assay ver. 1.0 to detect the vanA and vanB vancomycin resistance genes in Enterococcus from perianal specimens. Diagn. Microbiol. Infect. Dis. 69:382–389 [DOI] [PubMed] [Google Scholar]
- 17.Mehta MS, et al. 2008. Optimization of a laboratory-developed test utilizing Roche analyte-specific reagents for detection of Staphylococcus aureus, methicillin-resistant S. aureus, and vancomycin-resistant Enterococcus species. J. Clin. Microbiol. 46:2377–2380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Park I, et al. 2011. Rectal culture screening for vancomycin-resistant enterococcus in chronic haemodialysis patients: false-negative rates and duration of colonisation. J. Hosp. Infect. 79:147–150 [DOI] [PubMed] [Google Scholar]
- 19.Patel R, et al. 2001. Natural history of vancomycin-resistant enterococcal colonization in liver and kidney transplant recipients. Liver Transpl. 7:27–31 [DOI] [PubMed] [Google Scholar]
- 20.Sloan LM, et al. 2004. Comparison of the Roche LightCycler vanA/vanB detection assay and culture for detection of vancomycin-resistant enterococci from perianal swabs. J. Clin. Microbiol. 42:2636–2643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stamper PD, Cai M, Lema C, Eskey K, Carroll KC. 2007. Comparison of the BD GeneOhm VanR assay to culture for identification of vancomycin-resistant enterococci in rectal and stool specimens. J. Clin. Microbiol. 45:3360–3365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Weinstock DM, et al. 2007. Colonization, bloodstream infection, and mortality caused by vancomycin-resistant enterococcus early after allogeneic hematopoietic stem cell transplant. Biol. Blood Marrow Transplant. 13:615–621 [DOI] [PubMed] [Google Scholar]
- 23.Zabicka D, et al. 2012. Efficiency of the Cepheid Xpert vanA/vanB assay for screening of colonization with vancomycin-resistant enterococci during hospital outbreak. Antonie Van Leeuwenhoek 101:671–675 [DOI] [PubMed] [Google Scholar]
- 24.Zirakzadeh A, et al. 2008. Vancomycin-resistant enterococcal colonization appears associated with increased mortality among allogeneic hematopoietic stem cell transplant recipients. Bone Marrow Transplant. 41:385–392 [DOI] [PubMed] [Google Scholar]
- 25.Zirakzadeh A, Patel R. 2006. Vancomycin-resistant enterococci: colonization, infection, detection, and treatment. Mayo Clin. Proc. 81:529–536 [DOI] [PubMed] [Google Scholar]