Abstract
We describe a multiplex PCR assay to detect the Mycobacterium tuberculosis Beijing genotype variant B0/W148, which is considered a “successful” clone of M. tuberculosis, widespread in Russia. Specificity and sensitivity of the assay were 100% based on the analysis of a collection of 516 M. tuberculosis isolates of different genotypes and origins. This assay may be used for accurate and simple detection and surveillance of this clinically and epidemiologically important variant of M. tuberculosis.
TEXT
Tuberculosis caused by Mycobacterium tuberculosis is one of the most devastating infectious diseases, but some of this pathogen's lineages and strains are more prone to disseminate and cause active disease than others. A clonal group designated B0 (8, 9) or W148 (1), comprising one-fourth of the Beijing genotype isolates in different parts of the former Soviet Union as well as isolates recovered from Russian immigrants, has been identified (1, 2). The Beijing B0/W148 variant was initially identified using IS6110 restriction fragment length polymorphism (RFLP) typing (1, 8). B0/W148 isolates and isolates with similar profiles sharing the same characteristic double band in the upper part of the profile were more recently defined as the B0/W148 cluster (6).
Relative to other Beijing genotype isolates, B0/W148 isolates demonstrate an increased virulence in the macrophage model (3), a stronger association with multidrug resistance (9), and an increased transmissibility (10). It has been suggested that the Beijing B0/W148 cluster represents a “successful” clone of M. tuberculosis in Russia (6). We sought to develop and evaluate a method for the simple and rapid detection of Beijing B0/W148 cluster isolates.
Retrospective collections of M. tuberculosis DNA used in this study were partly described previously (3, 4, 7, 12) and have been characterized by IS6110 RFLP and spoligotyping (see Table S1 in the supplemental material).
An in silico genome analysis and identification of the B0/W148-specific IS6110 insertion in contig 1.39 of strain W-148 (GenBank no. ACSX01000039) are described in detail below. A multiplex PCR targeting the B0/W148-specific IS6110 insertion was performed using the primers INS1 (5′-CGTGAGGGCATCGAGGTGGC) (11), Rv2665R (5′-CTCGGCCGTACGGACGACGATC), and W139F2 (5′-GCGTTCCAACGGTTCGGGCC) in a C1000 thermal cycler (Bio-Rad) in 25 μl (1 unit of Taq polymerase [Sileks, Russia], 2 mM MgCl2, 20 pmol of Rv2665R and W139F2 each, and 25 pmol of INS1) under the following conditions: 95°C for 5 min; 6 cycles of 94.5°C for 45 s, 66°C for 45 s, and 72°C for 1 min 20 s; 34 cycles of 94.5°C for 45 s, 67°C for 45 s, and 72°C for 1 min 20 s; and 72°C for 8 min. The amplified fragments were electrophoresed in 1.2% agarose gels.
The digoxigenin (DIG)-labeled PCR fragments amplified with either primers INS1 and INS2 (5′-GCGTAGGCGTCGGTGACAAA) (11) or primers W139F2 and W139R (5′-TCAGGAGACGGCGGGTCGC) were hybridized separately to the same blots with immobilized PvuII-digested M. tuberculosis DNA. The hybridization signals were revealed using anti-DIG-antibodies conjugated to alkaline phosphatase, nitroblue tetrazolium, and 5-bromo-4-chloro-3-indolylphosphate. The resulting fragments were compared to the IS6110 RFLP profiles of the same isolates.
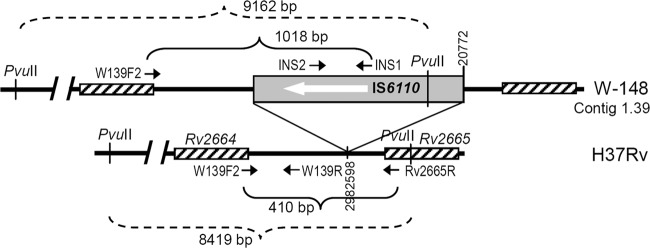
An in silico analysis of the complete genome sequence of M. tuberculosis strain W148 (GenBank no. ACSX00000000.1) identified an inverted IS6110 in contig 1.39 (positions 19418 to 20772) located within the 9,162-bp PvuII digest (Fig. 1). Theoretically, it corresponded to the largest (∼9.2-kb) band in the IS6110 RFLP profile of the B0/W148 strain (see Fig. S1 in the supplemental material). This insertion corresponds to positions 2982598/9 in the complete genome sequence of strain H37Rv (GenBank NC_000962.2) located in a short intergenic region between genes Rv2664 and Rv2665.
Fig 1.
Schematic view of the genome region containing the IS6110 insertion specific of strain W148 (not to scale). Short arrows indicate the primers. PvuII site-flanked fragments are shown by dashed lines.
The specificity of this particular IS6110 insertion for clinical isolates with the Beijing B0/W148 profile was experimentally confirmed by Southern hybridization of the PvuII-digested DNA of the same isolates with probes: (i) the IS6110 fragment flanked by primers INS1 and INS2 and (ii) a fragment of the genome region flanked by primers W139F2 and W139R and located upstream of the aforementioned IS6110 in strain W148 contig 1.39 (Fig. 1). A comparison of the hybridization profiles confirmed that the 9.2-kb fragment in the IS6110 RFLP profile of the Beijing B0/W148 isolates did indeed span the above-mentioned IS6110 insertion in the Rv2664/Rv2665 intergenic region. Strain H37Rv presented a 8.4-kb band, thus confirming the absence of IS6110 in its Rv2664-Rv2665 intergenic region (Fig. 2b).
Fig 2.
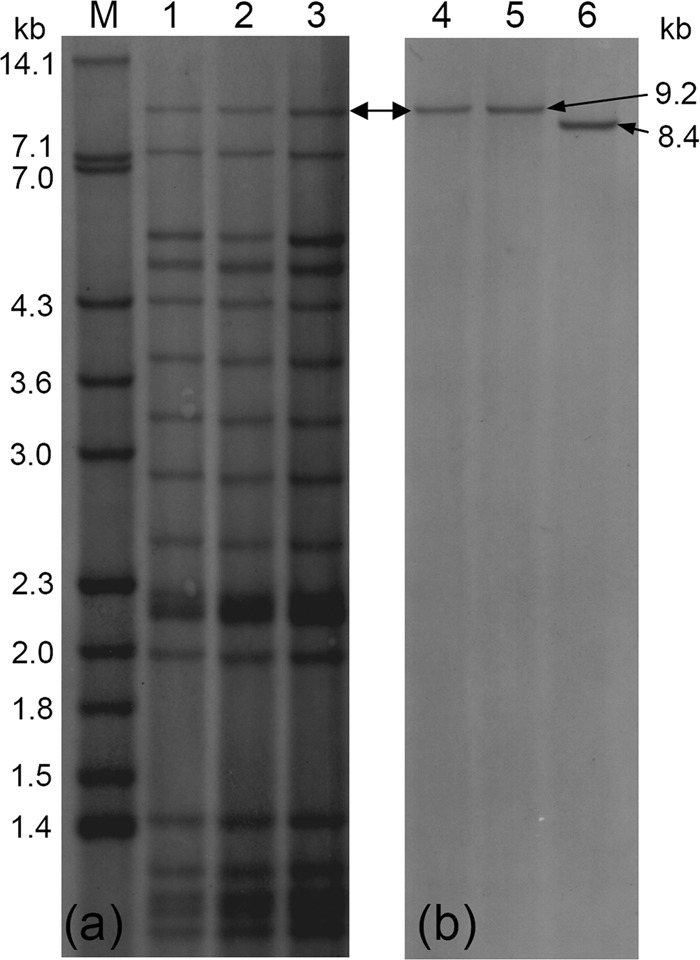
Southern hybridization of PvuII-digested DNA of M. tuberculosis isolates with probes. (a) Internal IS6110 fragment (primers INS1 and INS2); (b) Rv2664-Rv2665 fragment (primers W139F2 and W139R). Lanes: 1 to 5, Beijing B0/W148 isolates; 6, strain H37Rv; M, strain Mt14323 used as a molecular mass marker. The double-headed arrow indicates fragments corresponding to IS6110 RFLP profiles and the PvuII digest spanning the Rv2664-Rv2665 region in the Beijing B0/W148 isolates.
The Rv2664-Rv2665 intergenic region is both intact and identical in all other published complete genomes of M. tuberculosis complex strains of different genotypes available in GenBank (as of 31 July 2012), except for Mycobacterium bovis and M. bovis BCG strains with an A-to-G substitution (position 2935950; GenBank no. NC_008769.1) and Mycobacterium cannetti (no similarity found).
Accordingly, a multiplex PCR procedure was designed for detection of the Beijing B0/W148 cluster isolates (Fig. 1 and 3). M. tuberculosis isolates with an intact Rv2664-Rv2665 region are characterized by a 410-bp band amplified with primers W139F2 and Rv2665R. M. tuberculosis isolates with the Beijing B0/W148-specific IS6110 insertion in this genome region are characterized by a 1,018-bp band amplified with primers W139F2 and INS1 (Fig. 1). The assay was optimized with reference strains H37Rv, BCG, and Mt14323 and a limited number of the Beijing genotype isolates previously characterized by IS6110 RFLP typing (by definition, the “gold standard” method for detecting a B0/W148 strain) and spoligotyping (Fig. 3).
Fig 3.
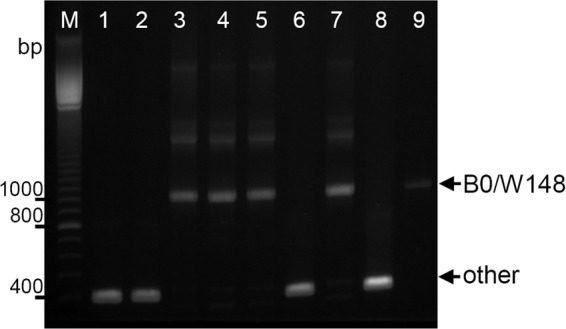
Agarose gel electrophoresis of the multiplex PCR products of M. tuberculosis isolates. Lanes: 3 to 5, 7, and 9, Beijing B0/W148 cluster isolates; 1 and 2, isolates of other Beijing variants; 6, H37Rv; 7, BCG; M, 100-bp DNA ladder (GE Healthcare). Arrows show specific bands for Beijing B0/W148 (1,018 bp) and other genotypes (410 bp).
The specificity and sensitivity of the method were evaluated with an enlarged collection of 227 Beijing and 164 non-Beijing isolates from different locations, characterized by spoligotyping (all isolates) and IS6110 RFLP typing (Beijing genotype isolates) (see Table S1 in the supplemental material). All 62 isolates of the B0/W148 cluster, i.e., the B0/W148 profile and similar profiles (see Fig. S1 in the supplemental material), were correctly identified by the multiplex PCR assay. All non-Beijing isolates (n = 164) and all non-B0/W148 Beijing isolates (n = 165) were characterized by amplification of the 410-bp band, correctly indicating their non-B0/W148 status. Thus, the specificity and sensitivity of the multiplex PCR assay in detecting the Beijing B0/W148 variant were 100%.
The method was additionally applied to the available DNA collections previously characterized by spoligotyping only. This allowed us to detect the B0/W148 genotype in 18 of 49 Beijing isolates from St. Petersburg, Russia, in 7 of 39 Beijing isolates from Pskov, Russia, and in none of the isolates from Vietnam (see Table S1 in the supplemental material).
In total, the B0/W148 cluster in this study was found only in former Soviet Union settings, and its rate (of all Beijing isolates) varied from 18.0% in Pskov and 23.3% in Karelia to a higher rate of 28.8% in St. Petersburg and even 52.5% in Belorussia. However, the latter collection included only drug-resistant isolates and was thus biased. In contrast, no B0/W148 cluster strain was identified among isolates from China, Vietnam, and Brazil (see Table S1 in the supplemental material). Since none of these countries received a significant influx of immigrants from the former Soviet Union (5), this finding is not unexpected. Indeed, the major direction of the Russian (ex-Soviet Union) emigration has been toward regions in western Europe and North America (5) where B0/W148 isolates have been detected (1, 2).
In conclusion, we have developed a multiplex PCR-based procedure for fast, simple, and reliable detection of the epidemiologically and clinically important genetic variant of M. tuberculosis known as the Beijing B0/W148 cluster. We propose this assay for screening and surveillance of this genotype in the areas of its epidemic circulation, such as the countries of the former Soviet Union, and in areas receiving immigrants from these countries.
Supplementary Material
ACKNOWLEDGMENTS
This work was partly supported by the European Commission (ORCHID project, grant agreement no. 261378), the Russian Foundation for Basic Research (grant no. 11-04-91172 GFEN_a), and the National Natural Science Foundation of China (grant no. 81071315).
Footnotes
Published ahead of print 29 August 2012
Supplemental material for this article may be found at http://jcm.asm.org/.
REFERENCES
- 1.Bifani PJ, Mathema B, Kurepina NE, Kreiswirth BN. 2002. Global dissemination of the Mycobacterium tuberculosis W-Beijing family strains. Trends Microbiol. 10:45–52 [DOI] [PubMed] [Google Scholar]
- 2.Ghebremichael S, et al. 2010. Drug resistant Mycobacterium tuberculosis of the Beijing genotype does not spread in Sweden. PLoS One 5:e10893 doi:10.1371/journal.pone.0010893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lasunskaia E, et al. 2010. Emerging multi-drug resistant Mycobacterium tuberculosis strains of the Beijing genotype circulating in Russia express a pattern of biological properties associated with enhanced virulence. Microbes Infect. 12:467–475 [DOI] [PubMed] [Google Scholar]
- 4.Markelov Y, Narvskaya O. 2010. Circulation of multidrug-resistant tuberculosis pathogen strains in the Republic of Karelia. Tuberk. Bolezn. Legk. 2:54–56. (In Russian.) [PubMed] [Google Scholar]
- 5.Ministry of Foreign Affairs of Russian Federation. Russian language in the world. 2003. www.ln.mid.ru/Brp_4.nsf/arh/B6BE784B3E2ABD1343256DF8003AC21C?OpenDocument.
- 6.Mokrousov I, et al. 2008. Mycobacterium tuberculosis Beijing genotype in Russia: in search of informative VNTR loci. J. Clin. Microbiol. 46:3576–3584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mokrousov I, et al. 2006. Rapid detection of the Mycobacterium tuberculosis Beijing genotype and its ancient and modern sub-lineages by IS6110-based inverse PCR. J. Clin. Microbiol. 44:2851–2856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Narvskaya O, Mokrousov I, Otten T, Vishnevsky B. 1999. Genetic marking of polyresistant Mycobacterium tuberculosis strains isolated in the north-west of Russia. Probl. Tuberk. 3:39–41 (In Russian) [PubMed] [Google Scholar]
- 9.Narvskaya O, Mokrousov I, Otten T, Vishnevsky B. 2005. Molecular markers: application for studies of Mycobacterium tuberculosis population in Russia, p 111–125. In Read MM. (ed), Trends in DNA fingerprinting research. Nova Science Publishers, New York, NY [Google Scholar]
- 10.Pardini M, et al. 2009. Characteristics of drug-resistant tuberculosis in Abkhazia (Georgia), a high-prevalence area in Eastern Europe. Tuberculosis (Edinburgh) 89:317–324 [DOI] [PubMed] [Google Scholar]
- 11.van Embden JDA, et al. 1993. Strain identification of Mycobacterium tuberculosis by DNA fingerprinting: recommendations for a standardized methodology. J. Clin. Microbiol. 31:406–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vasilenko N, et al. 2006. Spacer oligonucleotide typing of drug-resistant Mycobacterium tuberculosis circulating on the territory of Belarus. Immunopathol. Allergol. Infektol. 4:70–74 (In Russian.) [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.