Abstract
Herpes infections are among the most common sexually transmitted infections (STI), but diagnostic methods for genital herpes have not kept pace with the movement toward molecular testing. Here, we describe an FDA-approved molecular assay that identifies and types herpes simplex virus (HSV) infections for use in routine clinical settings. Paired samples from anogenital lesions were tested using the BD ProbeTec HSV Qx (HSVQx) system, HSV culture and, a laboratory-developed PCR assay. Family planning, obstetrics/gynecology (OB/GYN), or sexually transmitted disease (STD) clinics in the United States served as recruitment sites. Sensitivity and specificity estimates, head-to-head comparisons, measures of agreement, and latent-class analyses were performed to provide robust estimates of performance. A total of 508 participants (174 men and 334 women) with anogenital lesions were included; 260 HSV-2 and 73 HSV-1 infections were identified. No differences in test performance based on gender, clinic type, location of the lesion, or type of lesion were observed. The sensitivity of HSV-2 detection ranged from 98.4 to 100% depending on the analytical approach, while the specificity ranged from 80.6%, compared to the less sensitive culture method, to 97.0%, compared to PCR. For HSV-1, the sensitivity and specificity ranges were 96.7 to 100% and 95.1 to 99.4%, respectively. This assay may improve our ability to accurately diagnose anogenital lesions due to herpes infection.
INTRODUCTION
Genital herpes is a widespread sexually transmitted infection directly and indirectly linked to a spectrum of morbidities that include adverse outcomes of pregnancy and increased risk for HIV infection. In 2008, herpes simplex virus 2 (HSV-2), the predominant cause of genital herpes, was estimated to affect approximately 17% (over 45 million) of Americans, most of whom are unaware that they are infected (9, 17). Serological testing has helped to define the prevalence of infections and is a useful tool for evaluation of selected patient groups; however, it is not recommended for routine screening of the general population (5). In contrast, microbiologic demonstration of virus provides information critical to clinical management of infection. Laboratory testing for herpes simplex virus (HSV) is critical for accurate diagnosis of genital herpes infections and may have a profound impact on management decisions for persons with and at risk for genital herpes.
Multiple factors currently contribute to underdiagnosis of herpes infections. Important contributors include the varied clinical presentations of genital herpes lesions, failure to test lesions, and the lack of sensitivity of culture when testing is performed. Further, even experienced clinicians have been shown to be incorrect (e.g., categorizing a lesion as herpes when it is not) about 20% of the time when a clinical diagnosis of genital herpes is made strictly on the basis of clinical presentation (12). Testing for the presence of HSV is currently most often performed using herpesvirus culture methods despite the fact that nucleic acid amplification tests have been repeatedly demonstrated to be 3 to 5 times more sensitive for virus detection than carefully performed culture (5).
Type-specific testing of herpesviruses also plays an important role in management decisions for persons with genital herpes, since an increasing proportion of genital herpes is now due to HSV-1. The natural history of genital HSV-1 markedly differs from that of genital HSV-2. Compared to genital HSV-2, HSV-1 genital outbreaks recur less frequently and are less likely to cause asymptomatic virus shedding (8). These factors may influence the risk for transmission of infection to others and thus management decisions, including what schedule of herpes treatment, if any, should be offered to a patient.
Here, we present the results of a multicenter study evaluating the performance of a recently FDA-approved, commercially available, type-specific nucleic acid amplification test for HSV: the BD ProbeTec HSV Qx (HSVQx) system.
(This work was presented in part at the 19th biennial meeting of the International Society for STD Research, Quebec City, Canada, July 2011.)
MATERIALS AND METHODS
Study population and specimens.
Participant enrollment took place at nine clinical centers comprised of 5 sexually transmitted disease (STD) and 4 family planning clinics. Two laboratory sites provided viral-culture results, three performed HSVQx testing, and one provided PCR results only. The study protocol was approved by institutional review boards for human research (IRBs) at each participating site. All participants provided written informed consent for specimen collection. Inclusion criteria included the presence of a lesion (either vesicle or ulcer) in the anogenital (“boxer shorts”) region, i.e., the perineum, perianal, groin, buttocks, thigh, vulvar, penis, and scrotal areas. Exclusion criteria included lesions in nongenital areas, crusted lesions, or use of antiherpesvirus medications (e.g., acyclovir) within the 3 weeks prior to sample collection. HIV serostatus was not assessed as part of this study.
Lesions were sampled using a polyester swab, which was placed directly into universal viral transport medium (UVT) (Becton, Dickinson [BD], Sparks, MD), and then using a second, foam-tipped swab, which was placed directly into the BD Qx liquid wet-swab transport medium (Qx WS). UVT samples were collected first in order to optimize the integrity of culture specimens and were stored at 2 to 8°C for no more than 36 h prior to aliquoting for use in multiple assays (Fig. 1). Specimens collected in UVT were divided into three aliquots: one was tested using the ElvisHSV ID/Typing Test System (Diagnostic Hybrids Inc., San Diego, CA), the second aliquot (500 μl) was placed into BD Qx transport medium for testing using the new assay (designated Qx UVT) on the Viper instrument, and the third 500-μl aliquot was stored at −70°C for subsequent testing by PCR at the University of Washington (16). The second swab, the Qx WS specimen, was immediately placed in the BD Qx transport medium and stored at 2 to 30°C for up to 7 days prior to testing on the Viper system. This dual-swab specimen collection and aliquoting design allowed head-to-head testing of a single sample using culture, a highly regarded research PCR (10, 14, 16), and the new assay, as well as comparison with a second swab collected directly into the Qx WS transport medium. All participants had samples tested by all three methods: viral culture, PCR, and HSVQx. Specimens from all participants were sent to one of three laboratories (located at Indiana University, Louisiana State University, and University of Alabama—Birmingham) for testing with the BD Viper system, one of two laboratories (located at LabCorp and Louisiana State University) performed all viral cultures, and all PCR testing was performed at the University of Washington. All testing sites participated in pretrial training and performance validation prior to testing any participant samples.
Fig 1.
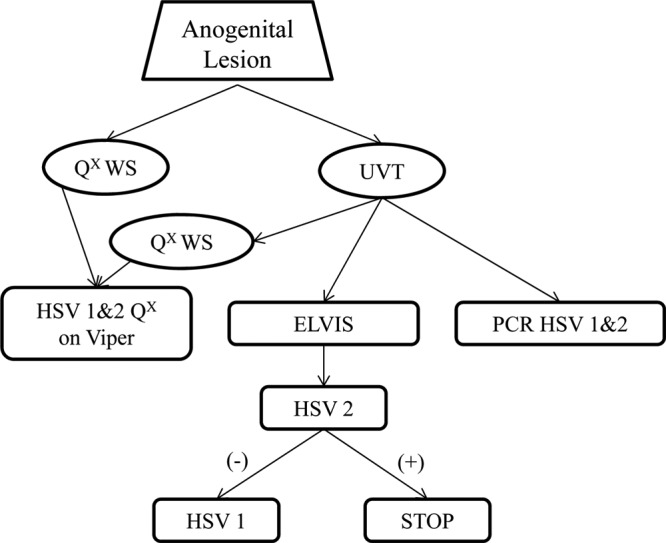
Sample collection and testing scheme. UVT samples were aliquoted for use in all three HSV assays (Elvis culture, PCR, and HSVQx). HSV-1 testing in the Elvis system could be performed only on HSV-2-negative samples. The sample collected using the assay collection kit (Qx WS) and the UVT aliquoted into the collection kit were both tested on the HSVQx system. +, positive; −, negative.
HSV-1 and -2 assays. (i) HSVQx using Viper instrumentation.
The procedural and processing details of the BD HSVQx assays are identical to those of the BD Chlamydia trachomatis Qx assay on the Viper instrument, which has been described elsewhere (14). Briefly, the steps of the assay are as follows. Samples in transport medium were warmed at 114°C to dissolve mucus and homogenize the specimen matrix. After cooling at room temperature for 15 min, the specimens were loaded onto the Viper instrument, which then performed all the steps necessary for extraction and amplification of target DNA without further user intervention. The HSVQx amplified-DNA assays are based on the simultaneous amplification and detection of target DNA using amplification primers and a fluorescently labeled detector probe. The primers for each assay target a type-specific DNA sequence (HSV-1, 126 bp; HSV-2, 124 bp) within the herpes simplex virus glycoprotein G gene. The presence or absence of HSV DNA is determined by calculating the peak fluorescence, which is called the maximum relative fluorescence units, over the course of the amplification process and by comparing this measurement to a predetermined threshold value.
(ii) Elvis culture.
The Elvis culture system uses a genetically engineered cell line for culture that expresses β-galactosidase in the presence of HSV-1 or -2 replication (7). Accumulation of this reporter product can be detected within 17 to 24 h following inoculation with samples. Cultures demonstrating β-galactosidase reactivity are then sequentially stained with fluorescence-tagged monoclonal antibodies for virus typing. Type-specific fluorescent staining is first performed for HSV-2. If negative for HSV-2, the cultures are restained with an HSV-1 type-specific reagent. As a result, for those cultures giving a positive result for HSV-2, no further staining is possible, since the fluorescent tag is fluorescein isothiocyanate (FITC) in both cases. Thus, dual infections with both HSV-1 and HSV-2 cannot be identified by this method.
(iii) Real-time PCR.
While there are many laboratory-developed PCR assays described in the literature, a lack of thorough evaluation and standardization prevents cross-study comparison of the performances of these tests. The University of Washington PCR assay for detection and typing of HSV has been well described and is recognized as the gold standard assay for use in FDA drug treatment and vaccine clinical trials (6, 10, 11, 16). Thus, this assay was chose as the comparator assay for the evaluation of the HSVQx assay. Briefly, UVT samples were used for DNA extraction, and the resulting DNA was amplified in two stages. In the first amplification, genus-specific primers are used in real-time PCR to amplify either HSV-1 or -2. The assay has a linear output that also provides quantification of HSV DNA present in the specimen. If positive, a second, multiplex PCR assay containing primers and probes specific for both HSV-1 and -2 is performed. Type specificity is determined based on melting curves. Although the assay is not commercially available, it is considered to be a highly sensitive detection method and has been utilized to describe the pathogenesis of HSV and the association of HSV with HIV-1 infection and to measure clinical outcomes for an HSV vaccine trial (1, 4, 15). The assay can detect mixed infections with up to a 3-log-unit difference in DNA copy numbers and is estimated to be 3 to 5 times more sensitive than culture performed in the same laboratory (6, 16).
Analysis.
The results of the HSVQx assay, using either the Qx WS or Qx UVT sample, were compared head to head with Elvis and with PCR results. The performances of the Qx WS and Qx UVT were also compared to one another and to a patient infected standard (PIS). The patient infected standard was defined as a positive result by either Elvis or the PCR assay. The intent of this analysis was to identify all potential infections and to allow the best estimation of the sensitivity of the HSVQx assay. For all head-to-head comparisons, percent agreement and κ scores were estimated. κ scores are used for assessing the agreement between paired binary outcomes with a rule of thumb interpretation of 0.60 to 0.80 as good agreement and >0.80 as very good agreement. Sensitivity and specificity were calculated compared to the PIS. The analysis for HSV-1 compared to culture is based only on those samples that were negative for HSV-2, since Elvis staining for HSV-1 could not be performed for HSV-2-positive cultures. Differences in performance based on gender, age, and location of the lesion were assessed. Finally, latent-class analysis was used in an attempt to provide an unbiased estimate of sensitivity and specificity based on the latent class of infection status (2). Using results from all of the assays, latent-class analysis determines the number of classes into which results can be categorized; for diagnostic testing, this is ideally two classes (infected and uninfected). After classes are identified based on the total of all testing results, an estimation of the accuracy of each assay is made based on those categorizations. An alpha of 0.05 was used for all analyses.
RESULTS
Samples were obtained from 564 participants, 56 of whom were subsequently excluded based on predetermined study inclusion/exclusion criteria. For the final analyses, 498 Qx WS results and 501 Qx UVT results were compared to culture and PCR for HSV-2 (Table 1). The 189 (37.2%) HSV-2-positive culture specimens were not able to be tested by subsequent staining with the HSV-1 type-specific antibody, and one HSV-1 Qx wet-swab result was unable to be determined due to an extraction control failure. Thus, while all compliant Qx results were compared to PCR for HSV-1, there were 308 (62%) Qx WS and 312 (62%) Qx UVT results that could be analyzed using HSV-1 culture results. Exclusions were based on improper sample collection, improper specimen handling, or improper storage of specimens; 2 samples were not available for PCR testing, 7 were missing Elvis results, and 3 were lacking Qx WS samples.
Table 1.
Description of participants and samples collected
| Parameter | Valuea |
||
|---|---|---|---|
| Female | Male | Total | |
| Sample size | 334 | 174 | 508 |
| Median age (yr) | 25 | 25 | 25 |
| Clinic type | |||
| Family planning | 221 | 35 | 256 |
| STD | 113 | 139 | 252 |
| Lesion type | |||
| First episode | 233 | 122 | 355 |
| Recurrent | 101 | 52 | 153 |
| Lesion location | |||
| Glans | NA | 28 | 28 |
| Penis | NA | 106 | 106 |
| Skin | 16 | 37 | 53 |
| Perianal | 17 | 3 | 20 |
| Perineum | 55 | NA | 55 |
| Vulva | 243 | NA | 243 |
Number, except for median age. NA, not applicable.
The study participants consisted of 334 women and 174 men ranging in age from 17 to 71 years with an overall median age of 25 years (Table 1). Approximately half (49.6%) were enrolled from STD clinics. The lesions tested were sampled from the following anogenital sites: glans or penis (77.0% of men), vulva (74.5% of women), perineum (15.9% of women), perianal area (3.8% of all participants), and unspecified skin locations (10.2% of all participants). Based on estimates derived from the PCR assay, the positivity rates of HSV-2 in these symptomatic patients were 55.0 and 41.2% in STD and family planning clinics, respectively. The positivity rates of HSV-1 were lower but remained substantial, with 8.0 and 20.4% identified from patients attending STD and family planning clinics, respectively.
HSV-2.
The UVT specimens were tested by both the Elvis culture system and the HSVQx assay. This sample type run on the HSVQx assay detected 186 of the 189 (98.4%) samples that were positive in the Elvis culture system (Table 2) and yielded an additional 51 positive results from specimens with negative Elvis culture results. The Qx WS specimen type, which was run only on HSVQx and not culture, detected 186 of the culture-positive participants while yielding positive results for an additional 60 patients with negative cultures. The κ scores for the Qx UVT samples were 0.782, 0.956, and 0.945 when calculated for culture, PCR, or the patient infected standard, respectively. For the Qx WS samples, the κ scores were 0.746, 0.944, and 0.932. The estimated sensitivities of the HSVQx assay, based on the patient infected standard, were 96.4% and 97.6% for Qx UVT and Qx WS, respectively. There was no difference in assay performance estimates by age, gender, or lesion location (all P values, >0.3). The latent-class analysis model was saturated with two latent classes, suggesting that the results could be divided into infected and uninfected participants. For HSV-2 infections, the sensitivities and specificities estimated using latent-class analysis were as follows: 79.7% ± 1.9% and 98.9% ± 0.5% for culture, 100% and 98.3% ± 0.6% for PCR, and 100% and 97.0% ± 0.8% for HSVQx. The ability to detect infections based on the PCR-determined organism load is shown in Fig. 2 for the HSVQx assay and Elvis cultures. The data for HSVQx were generated with pooled data from both sample types. Thus, the commercially available HSVQx assay performs substantially better than culture and at least as well as the laboratory-developed PCR assay for detection of HSV-2.
Table 2.
HSV-2 performance compared to culture and PCR
| Assaya | Resultb | No. (%) with result: |
|||||
|---|---|---|---|---|---|---|---|
| Elvisc |
PCRd |
PIS |
|||||
| + | − | + | − | + | − | ||
| Qx UVT | + | 186 (98.4) | 51 | 237 (97.5) | 5 | 238 (96.4) | 5 |
| − | 3 | 261 (83.7) | 6 | 258 (98.1) | 9 | 254 (98.1) | |
| Total | 189 | 312 | 243 | 263 | 247 | 259 | |
| Qx WS | + | 186 (98.4) | 60 | 240 (98.8) | 11 | 241 (97.6) | 11 |
| − | 3 | 249 (80.6) | 3 | 249 (95.8) | 6 | 245 (95.7) | |
| Total | 189 | 309 | 243 | 260 | 247 | 256 | |
Qx UVT, HSVQx assay performed using samples collected in universal transport medium; Qx WS, HSVQx assay performed using the wet-swab transport system provided with the assay.
+, positive; −, negative.
Elvis culture system results.
PCR performed by the University of Washington.
Fig 2.
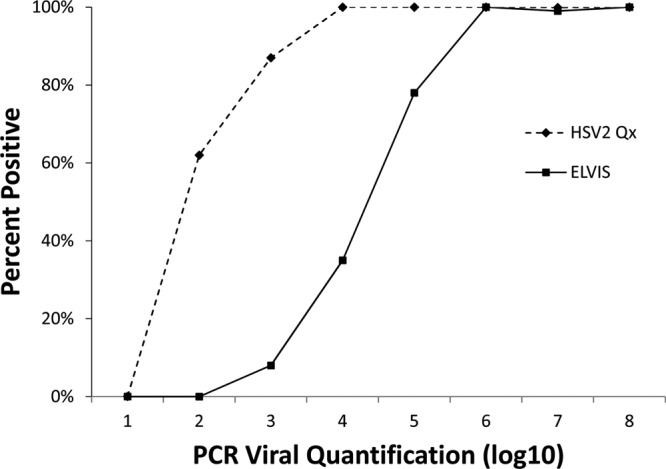
Assay sensitivity by organism load for HSV-2. PCR-positive samples generated a viral-load value. The percent positivity of the other two assays (Elvis culture and HSVQx) for HSV-2 is shown based on the PCR-determined organism load.
HSV-1.
As described in Materials and Methods, the Elvis culture system does not allow testing of specimens for HSV-1 if the culture is positive for HSV-2. As a result, 189 specimens culture positive for HSV-2 were not available to provide HSV-1 culture results. The HSVQx assay using the same UVT (Qx UVT) samples that were used in the culture system detected 60 (96.7%) of the 62 HSV-1 culture-positive samples. This sample type also resulted in detection of 6 additional positive samples that were negative by culture (Table 3). The HSVQx assay identified 59 (96.7%) HSV DNAs in 61 culture-positive specimens and had an additional 12 positive results when using the Qx WS sample type. The κ scores for the Qx UVT samples were 0.921, 0.992, and 0.973 compared to culture, PCR, or the patient infected standard, respectively. For the Qx WS samples, the κ scores were 0.865, 0.960, and 0.938. The estimated sensitivities of the HSVQx assay, based on the patient infected standard, were 95.9% and 97.3% for Qx UVT and Qx WS, respectively. There was no difference in assay performance estimates by age, gender, or lesion location (all P values, >0.3). For HSV-1 infections, the latent-class analysis-estimated sensitivities and specificities were as follows: 90.2% ± 2.6% and 99.2% ± 0.4% for culture, 100% and 99.9% ± 0.1% for PCR, and 100% and 99.4% ± 0.3% for HSVQx. The detection of infections based on the organism load is shown in Fig. 3 for the HSVQx assay and Elvis cultures. These data demonstrate that the HSVQx assay performs as well as the other assays on a fully automated system that provides results within a few hours rather than many days.
Table 3.
HSV-1 performance compared to culture and PCR
| Assaya | Resultb | No. (%) with result: |
|||||
|---|---|---|---|---|---|---|---|
| Elvisc |
PCRd |
PIS |
|||||
| + | − | + | − | + | − | ||
| Qx UVT | + | 60 (96.8) | 6 | 71 (98.6) | 0 | 71 (95.9) | 0 |
| − | 2 | 244 (97.6) | 1 | 434 (100) | 3 | 242 (100) | |
| Total | 62 | 250 | 72 | 434 | 74 | 242 | |
| Qx WS | + | 59 (96.7) | 12 | 71 (100) | 5 | 71 (97.3) | 5 |
| − | 2 | 235 (95.1) | 0 | 426 (98.8) | 2 | 234 (97.9) | |
| Total | 61 | 247 | 71 | 431 | 73 | 239 | |
Qx UVT, HSVQx assay performed using samples collected in universal transport medium; Qx WS, HSVQx assay performed using the wet-swab transport system provided with the assay.
+, positive; −, negative.
Elvis culture system results. The assay could be performed only on HSV-2 culture-negative samples, while PCR was performed on all specimens. Thus, Elvis and PIS comparisons have lower total sample sizes.
PCR performed by the University of Washington.
Fig 3.
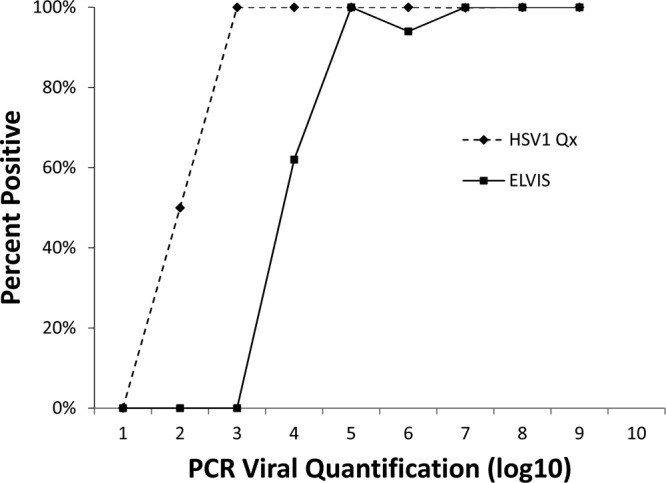
Assay sensitivity by organism load for HSV-1. PCR-positive samples generated a viral-load value. The percent positivity of the other two assays (Elvis culture and HSVQx) for HSV-1 is shown based on the PCR-determined organism load.
DISCUSSION
The data from this large, multisite clinical trial demonstrate the excellent performance of the HSVQx for detection of both HSV-1 and HSV-2 from anogenital lesions. The assay provides a potential mechanism for increased availability of highly sensitive detection of HSV in persons with lesions. Like well-characterized laboratory-developed research PCR assays (16), the system detected HSV from far more patients than the FDA-approved commercially available culture-based assay used for comparison in the study. Further, when the results were compared to those of PCR testing performed in a respected research laboratory, the results of the two assays were comparable. Indeed, the DNA-based assays had the capacity to detect dual HSV-1 and HSV-2 infections, which cannot be accomplished using the culture-based system. Although the impact of not having an HSV-1 culture result was negligible in this study (only 2/504 [<0.4%] participants had dual infections [data not shown]), it is useful to have any assay that provides results accurately and quickly for those populations in which dual infections might be more common. In addition to increased sensitivity, the HSVQx assay provides a convenient method for collecting specimens in transport medium, which can be stocked at room temperature and easily transported to laboratories for testing. As a commercially available assay, the BD Qx assay for HSV detection also has the potential advantage of being added to a laboratory platform already in wide use for detection of gonococcal and chlamydial infections.
Our analyses were rigorous, employing multiple methods. Confirmation of the sensitivity and specificity estimates derived by comparison with the patient infected standard was obtained using latent-class analysis. In fact, the latent-class analysis-estimated sensitivity was even higher than the estimate obtained in comparison with the patient infected standard. This is likely due to the fact that the latent-class analysis does not assume that culture-negative results represent uninfected individuals. The improved limit of detection of the HSVQx assay (Fig. 2 and 3), and thus the ability to correctly identify infections with fewer organisms in the specimen, results in improved clinical sensitivity over culture.
This study is not without limitations. The study was limited to evaluation of lesions occurring in the anogenital region, and thus, further study will be needed to evaluate the utility of the test for detection of HSV from oral lesions and from other locations. The study design did not allow assessment of asymptomatic genital shedding, an important cause of HSV transmission that can be detected by PCR from about 10% of specimens in persons with genital HSV-2 infections.
Highly sensitive and specific tools for HSV diagnosis and typing are needed. Serological testing has proven helpful in some settings but is not currently recommended for routine screening (5). Antibody responses may take months to develop following infection, delaying diagnosis and potentially resulting in either spread of infection to others by persons unaware that they have HSV or troublesome anxiety in persons awaiting test results. Finally, in persons with HSV infections, a positive serological test does not identify the location of infection, a fact that may hamper adoption of measures to reduce transmission to others.
HSV is the most common cause of genital lesions, irrespective of appearance, worldwide. Globally, it is the most common cause of genital ulceration and many lesions that are not “classical.” In contrast, the most common cause of a “typical” chancre in multiple studies is still HSV (13). Conversely, about 20 to 25% of lesions identified as HSV by experts are not actually attributable to HSV and are due to other causes. As a result, with the wide availability of more sensitive tests, there is now an opportunity to operationalize the recommendation that all genital lesions that are not known to be a recurring problem with a known diagnosis (i.e., HSV or other dermatological process) should be tested for HSV.
The changing epidemiology of genital herpes also warrants increased testing with emphasis on determining, not only the presence or absence of infection, but, when herpes is diagnosed, the type of virus present. The proportion of genital HSV caused by HSV-1 is increasing, and in some studies, the proportion of genital herpes caused by HSV-1 equals or exceeds the proportion caused by HSV-2. While the presentations of initial genital herpes due to HSV-1 and HSV-2 are clinically indistinguishable (3), compared to genital HSV-1 infection, genital HSV-2 infection recurs more often and is associated with higher rates of asymptomatic genital shedding, differences that may impact management decisions. While there are data to encourage the provision of chronic suppressive antiviral therapy to persons with HSV-2 infection, there are not similar recommendations for HSV-1. Addition of the HSVQx to the laboratory diagnostic menu will positively impact our ability to identify, and therefore manage, this highly prevalent infection.
ACKNOWLEDGMENTS
We thank Hanne Harbison, Paula Dixon, and Connie Lenderman at UAB for their assistance in this trial.
This study was sponsored by BD Diagnostics, Sparks, MD.
Barbara Van Der Pol has consulted or performed research projects for Abbott Molecular Diagnostics, Beckman Coulter, BD Diagnostics, Cepheid and Roche Diagnostics. Terri Warren has been awarded grant funding from BD Diagnostics for research. Stephanie N. Taylor has been awarded grant funding from Louisiana State University Health Sciences Center. Mark Martens has been awarded grant funding from PPAEO to collect samples. Keith R. Jerome has been awarded grant funding from BD Diagnostics; he is a board member of Eragen Biosciences, Idaho Technologies, and Gentura Dx; and he has been a speaker for BD Diagnostics and has been paid royalties by Elsevier. Leandro Mena has been awarded grant funding from BD Diagnostics. Joel Lebed and Savita Ginde have no conflict of interest to disclose. Paul Fine has been awarded grant funding from BD Diagnostics. Edward W. Hook III has been awarded funding from Becton, Dickinson, Cepheid, Roche Molecular, Gen-Probe, Inc., and Cempra; he has consulted for Cempra; and he has been a speaker for BD Diagnostics and has been paid royalties by McGraw-Hill.
Footnotes
Published ahead of print 8 August 2012
REFERENCES
- 1. Baeten JM, et al. 2008. Herpes simplex virus (HSV)-suppressive therapy decreases plasma and genital HIV-1 levels in HSV-2/HIV-1 coinfected women: a randomized, placebo-controlled, cross-over trial. J. Infect. Dis. 198: 1804–1808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Baughman AL, et al. 2008. Utility of composite reference standards and latent class analysis in evaluating the clinical accuracy of diagnostic tests for pertussis. Clin. Vaccine Immunol. 15: 106–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Benedetti J, Corey L, Ashley R. 1994. Recurrence rates in genital herpes after symptomatic first-episode infection. Ann. Intern. Med. 121: 847–854 [DOI] [PubMed] [Google Scholar]
- 4. Cattamanchi A, et al. 2008. Phase I study of a herpes simplex virus type 2 (HSV-2) DNA vaccine administered to healthy, HSV-2-seronegative adults by a needle-free injection system. Clin. Vaccine Immunol. 15: 1638–1643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Centers for Disease Control and Prevention 2010. Sexually transmitted diseases treatment guidelines, 2010. MMWR Morb. Mortal. Weekly Rep. 59: 1–112 [Google Scholar]
- 6. Corey L, Huang M, Selke S, Wald A. 2005. Differentiation of herpes simplex virus types 1 and 2 in clinical samples by a real-time Taqman PCR assay. J. Med. Virol. 76: 350–355 [DOI] [PubMed] [Google Scholar]
- 7. Crist GA, Langer JM, Woods GL, Procter M, Hillyard DR. 2004. Evaluation of the ELVIS plate method for the detection and typing of herpes simplex virus in clinical specimens. Diagn. Microbiol. Infect. Dis. 49: 173–177 [DOI] [PubMed] [Google Scholar]
- 8. Engelberg P, Carrell D, Krantz E, Corey L, Wald A. 2003. Natural history of genital herpes simplex virus type 1 infection. Sex. Transm. Dis. 30: 174–177 [DOI] [PubMed] [Google Scholar]
- 9. Fleming DT, et al. 1997. Herpes simplex virus type 2 in the United States, 1976 to 1994. N. Engl. J. Med. 337: 1105–1111 [DOI] [PubMed] [Google Scholar]
- 10. Hobbs MM, et al. 2008. From the NIH: proceedings of a workshop on the importance of self-obtained vaginal specimens for detection of sexually transmitted infections. Sex. Transm. Dis. 35: 8–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Jerome K, Huang M-L, Wald A, Selke S, Corey L. 2002. Quantitative stability of DNA after extended storage of clinical specimens as determined by real-time polymerase chain reaction. J. Clin. Microbiol. 40: 2609–2611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Langenberg AGM, et al. 1999. A prospective study of infections with herpes simplex virus type 1 and 2. N. Engl. J. Med. 341: 1432–1438 [DOI] [PubMed] [Google Scholar]
- 13. Mertz KJ, et al. 1998. Etiology of genital ulcers and prevalence of human immunodeficiency virus coinfection in 10 US cities. J. Infect. Dis. 178: 1795–1798 [DOI] [PubMed] [Google Scholar]
- 14. Taylor SN, et al. 2011. Clinical evaluation of the BD ProbeTec(TM) Chlamydia trachomatis Qx Amplified DNA Assay on the BD Viper System with XTR technology. Sex. Transm. Dis. 38: 603–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Tronstein E, et al. 2011. Genital shedding of herpes simplex virus among symptomatic and asymptomatic persons with HSV-2 infection. JAMA 305: 1441–1449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wald A, Huang M, Selke S. 2003. Polymerase chain reaction for detection of herpes simplex virus (HSV) on mucosal surfaces: comparison with HSV isolation in cell culture. J. Infect. Dis. 188: 1345–1351 [DOI] [PubMed] [Google Scholar]
- 17. Xu F, et al. 2006. Trends in herpes simplex virus type 1 and type 2 seroprevalence in the United States. JAMA 296: 964–973 [DOI] [PubMed] [Google Scholar]


