Abstract
Although coronaviruses are known to infect various animals by adapting to new hosts, interspecies transmission events are still poorly understood. During a surveillance study from 2005 to 2010, a novel alphacoronavirus, BatCoV HKU10, was detected in two very different bat species, Ro-BatCoV HKU10 in Leschenault's rousettes (Rousettus leschenaulti) (fruit bats in the suborder Megachiroptera) in Guangdong and Hi-BatCoV HKU10 in Pomona leaf-nosed bats (Hipposideros pomona) (insectivorous bats in the suborder Microchiroptera) in Hong Kong. Although infected bats appeared to be healthy, Pomona leaf-nosed bats carrying Hi-BatCoV HKU10 had lower body weights than uninfected bats. To investigate possible interspecies transmission between the two bat species, the complete genomes of two Ro-BatCoV HKU10 and six Hi-BatCoV HKU10 strains were sequenced. Genome and phylogenetic analyses showed that Ro-BatCoV HKU10 and Hi-BatCoV HKU10 represented a novel alphacoronavirus species, sharing highly similar genomes except in the genes encoding spike proteins, which had only 60.5% amino acid identities. Evolution of the spike protein was also rapid in Hi-BatCoV HKU10 strains from 2005 to 2006 but stabilized thereafter. Molecular-clock analysis dated the most recent common ancestor of all BatCoV HKU10 strains to 1959 (highest posterior density regions at 95% [HPDs], 1886 to 2002) and that of Hi-BatCoV HKU10 to 1986 (HPDs, 1956 to 2004). The data suggested recent interspecies transmission from Leschenault's rousettes to Pomona leaf-nosed bats in southern China. Notably, the rapid adaptive genetic change in BatCoV HKU10 spike protein by ∼40% amino acid divergence after recent interspecies transmission was even greater than the ∼20% amino acid divergence between spike proteins of severe acute respiratory syndrome-related Rhinolophus bat coronavirus (SARSr-CoV) in bats and civets. This study provided the first evidence for interspecies transmission of coronavirus between bats of different suborders.
INTRODUCTION
Coronaviruses (CoVs) infect a wide variety of animals, causing respiratory, enteric, hepatic, and neurological diseases of varying severity. Traditionally, CoVs have been classified into groups 1, 2, and 3, based on genotypic and serological characteristics (29, 79). Recently, the nomenclature and taxonomy of CoVs were revised by the Coronavirus Study Group of the International Committee for Taxonomy of Viruses (ICTV). CoVs are now classified into three genera, Alphacoronavirus, Betacoronavirus, and Gammacoronavirus, which replace the three traditional groups (5). Novel CoVs, which represented a novel genus, Deltacoronavirus, have also been identified (72, 73). While CoVs from all four genera can be found in mammals, bat CoVs are likely the gene source of Alphacoronavirus and Betacoronavirus, and avian CoVs are the gene source of Gammacoronavirus and Deltacoronavirus (9, 41, 73). CoVs are known to possess high frequency of recombination and mutation rates, which may allow them to adapt to new hosts and ecological niches (21, 29, 35, 68, 71, 78).
The severe acute respiratory syndrome (SARS) epidemic, caused by SARS CoV (SARS-CoV) (17, 27, 43), has boosted interest in the discovery of novel CoVs in both humans and animals (12, 20, 36, 41, 63, 65, 66, 72). In particular, a previously unknown variety of CoVs have been identified in bats from China and other countries, including SARS-related Rhinolophus bat CoVs (SARSr-Rh-BatCoVs) in horseshoe bats, suggesting that bats are important reservoirs of CoVs (8, 13, 30, 31, 33, 40, 49, 59, 67, 70). However, our understanding of the diversity, evolution, and interspecies transmission of CoVs in animals is still limited. For example, it remains unknown if bats are the direct origin of SARS-CoV in civets and humans, as the spike (S) protein of SARSr-Rh-BatCoV possesses only ∼80% amino acid identity to that of civet SARSr-CoV, with significant differences in the receptor binding domain (30, 32, 40, 51).
During a continuous surveillance study, in an attempt to better understand the role of bats in the evolution of CoVs and search for other bat species which may have served as intermediate hosts for interspecies transmission of SARSr-CoVs, a potentially novel alphacoronavirus, BatCoV HKU10, was detected in two very different bat species. After its first detection in a Leschenault's rousette in Guangdong (70), the virus was also found in Pomona leaf-nosed bats in Hong Kong. In the present study, the epidemiology of BatCoV HKU10 in different bat species was determined. To investigate possible interspecies transmission events, complete genome sequencing and analysis of eight BatCoV HKU10 strains from the two bat species was performed. The results revealed that viruses from the two bat species were highly similar, except for their S proteins, which shared only ∼60% amino acid identities. Positive selection and molecular-clock analysis showed that interspecies transmission of BatCoV HKU10 from Leschenault's rousettes in Guangdong to Pomona leaf-nosed bats in Hong Kong is likely to have occurred recently, with rapid evolution of the S protein in the latter bat species.
MATERIALS AND METHODS
Collection of bat samples.
Bats of various species were captured from different locations in Hong Kong and in the Guangdong province of southern China over a 5-year period (September 2005 to August 2010). Respiratory and alimentary specimens were collected using procedures described previously (30, 77). To prevent cross contamination, specimens were collected using disposable swabs with protective gloves, which were changed between samples. All specimens were immediately placed in viral transport medium before transportation to the laboratory for RNA extraction.
RNA extraction.
Viral RNA was extracted from the respiratory and alimentary specimens using a QIAamp viral RNA minikit (Qiagen, Hilden, Germany). The RNA was eluted in 50 μl of AVE buffer and was used as the template for reverse transcription-PCR (RT-PCR).
RT-PCR for CoVs and DNA sequencing.
CoV detection was performed by amplifying a 440-bp fragment of the RNA-dependent RNA polymerase (RdRp) gene of CoVs using conserved primers (5′-GGTTGGGACTATCCTAAGTGTGA-3′ and 5′-CCATCATCAGATAGAATCATCATA-3′) designed by multiple alignments of the nucleotide sequences of available RdRp genes of known CoVs as described previously (32, 66). Reverse transcription was performed using a SuperScript III kit (Invitrogen, San Diego, CA). The PCR mixture (25 μl) contained cDNA, PCR buffer (10 mM Tris-HCl [pH 8.3], 50 mM KCl, 3 mM MgCl2, and 0.01% gelatin), a 200 μM concentration of each deoxynucleoside triphosphate (dNTP), and 1.0 U Taq polymerase (Applied Biosystems, Foster City, CA). The mixtures were amplified with 60 cycles of 94°C for 1 min, 48°C for 1 min, and 72°C for 1 min and a final extension at 72°C for 10 min in an automated thermal cycler (Applied Biosystems, Foster City, CA). Standard precautions were taken to avoid PCR contamination, and no false positives were observed in negative controls.
The PCR products were gel purified using a QIAquick gel extraction kit (Qiagen, Hilden, Germany). Both strands of the PCR products were sequenced twice with an ABI Prism 3700 DNA analyzer (Applied Biosystems, Foster City, CA), using the two PCR primers. The sequences of the PCR products were compared with known sequences of the RdRp genes of CoVs in the GenBank database. Phylogenetic tree construction was performed using neighbor-joining method with ClustalX 1.83.
Statistical analysis.
Comparison of body weights of bats between different groups was performed using Student's t test and covariate analysis (SPSS version 11.5). A P value of <0.05 was regarded as statistically significant.
Viral culture.
Four samples positive for BatCoV HKU10 were cultured in FRhK-4 (rhesus monkey kidney; ATCC CRL-1688), Vero E6 (African green monkey kidney; ATCC CRL-1586), and HRT-18G (human colorectal adenocarcinoma; ATCC CRL-11663) cell lines and primary bat kidney and lung fibroblast cells derived from a Chinese horseshoe bat.
Complete genome sequencing of Ro-BatCoV HKU10 and Hi-BatCoV HKU10.
Six complete genomes of Hipposideros bat CoV HKU10 (Hi-BatCoV HKU10) and two complete genomes of Rousettus bat CoV HKU10 (Ro-BatCoV HKU10) detected in the present study were amplified and sequenced using the RNA directly extracted from the alimentary specimens as templates according to previously described strategies (31, 32). The RNA was converted to cDNA by a combined random-priming and oligo(dT) priming strategy. As the initial results revealed that they belong to Alphacoronavirus, the cDNA was amplified by degenerate primers designed by a multiple alignment of the genomes of human CoV 229E (HCoV 229E) (GenBank accession no. NC_002645), porcine epidemic diarrhea virus (PEDV) (GenBank accession no. NC_003436), porcine transmissible gastroenteritis virus (TGEV) (GenBank accession no. NC_002306), feline infectious peritonitis virus (FIPV) (GenBank accession no. AY994055), HCoV NL63 (GenBank accession no. NC_005831), and Rhinolophus bat CoV HKU2 (Rh-BatCoV HKU2) (GenBank accession no. EF203067), and additional primers covering the original degenerate primer sites were designed from the results of the first and subsequent rounds of sequencing. These primer sequences are shown in Tables S1 and S2 in the supplemental material. The 5′ ends of the viral genomes were confirmed by rapid amplification of cDNA ends (RACE) using the 5′-3′ RACE kit (Roche, Germany). Sequences were assembled and manually edited to produce final sequences of the viral genomes. For the other positive samples not included in complete genome sequencing, additional PCR targeted to other genome sites, including partial fragments of the helicase (Hel) and S genes, was also performed using the genome sequencing primers to exclude false positives due to PCR contamination.
Genome analysis.
The nucleotide sequences of the genomes and the deduced amino acid sequences of the open reading frames (ORFs) were compared to those of other CoVs. Phylogenetic trees were constructed using the maximum-likelihood method (18), with bootstrap values calculated from 100 trees. Protein family analysis was performed using PFAM and InterProScan (1, 2). Prediction of transmembrane domains was performed using TMHMM (55).
Sequencing of the complete RdRp and S genes of Ro-BatCoV HKU10 and Hi-BatCoV HKU10 strains.
To allow more accurate analysis of positive selection and divergence time, the complete RdRp genes of 25 Hi-BatCoV HKU10 strains and the complete S genes of one Ro-BatCoV HKU10 and four Hi-BatCoV HKU10 strains, in addition to the eight strains with complete genome sequences, were amplified and sequenced using primers available from genome sequencing as described above. The sequences of the PCR products were assembled manually to produce the complete RdRp and S gene sequences.
Estimation of synonymous- and nonsynonymous-substitution rates.
The number of synonymous substitutions per synonymous site, Ks, and the number of nonsynonymous substitutions per nonsynonymous site, Ka, for each coding region were calculated using the Nei-Gojobori method (Jukes-Cantor) in MEGA version 5 (57).
Recombination analysis and detection of positive selection.
Recombination detection was performed among genomes of BatCoV HKU10 strains using bootscan analysis and genetic algorithm recombination detection (GARD) with the Kishino-Hasegawa (KH) test as described previously (25, 32, 36, 47). While bootscan analysis is a widely used tool for detection of recombination with the window size having strong influence on recombination inference, GARD is often used to accurately locate the recombination breakpoints and determine the level of statistical significance (25). For estimation of positive selection, BatCoV HKU10 strains were grouped based on the year of sampling and the host species after removal of duplicates: Ro-BatCoV HKU10, Hi-BatCoV HKU10 from 2005–2006, Hi-BatCoV HKU10 from 2007–2008, Hi-BatCoV HKU10 from 2005–2008, and Hi-BatCoV HKU10 from 2005–2010. Sites under positive selection in the S gene were inferred using single-likelihood ancestor counting (SLAC), fixed-effects likelihood (FEL), and random-effects likelihood (REL) methods as implemented in the DataMonkey server (http://www.datamonkey.org) (48). Positive selection for a site was considered to be statistically significant if the P value was <0.1 for the SLAC and FEL methods or posterior probability was ≥90% for the REL method. An unrestricted random-effects branch site model, branch site REL, was implemented for detecting lineage-specific selection (26). This method is usually used to identify branches in a tree with evidence of episodic diversifying selection and is known to be more robust to errors because it does not enforce uniform selective pressure on all background branches (26).
Estimation of divergence time.
Divergence time was calculated using RdRp gene sequence data of Hi-BatCoV HKU10 and Ro-BatCoV HKU10 strains and the Bayesian Markov chain Monte Carlo (MCMC) approach as implemented in BEAST (version 1.6.2) as described previously (10, 32, 35, 36). One parametric model (constant size) and one nonparametric model (Bayesian Skyline with five groups) for tree priors were used for the inference. Analyses were performed with the SRD06 substitution model using both strict and relaxed [uncorrelated lognormal (Ucld) and uncorrelated exponential (Uced)] molecular clocks. The MCMC run was 1 × 108 steps long, with sampling every 1,000 steps. Convergence was assessed on the basis of the effective sampling size after a 10% burn-in using Tracer software version 1.5 (10). The mean time of the most recent common ancestor (tMRCA) and the highest posterior density regions at 95% (HPD) were calculated, and the best-fitting model was selected by a Bayes factor, using marginal likelihoods implemented in Tracer (56). Bayesian Skyline under a relaxed-clock model with Uced was adopted for making inferences, as Bayes factor analysis indicated that this model fitted the data better than other models tested (data not shown). The trees were summarized in a target tree by the Tree Annotator program included in the BEAST package by choosing the tree with the maximum sum of posterior probabilities (maximum clade credibility) after a 10% burn-in.
Nucleotide sequence accession numbers.
The nucleotide sequences of the eight genomes of BatCoV HKU10 have been deposited in the GenBank sequence database under accession no. JQ989266 to JQ989273.
RESULTS
Detection of a novel alphacoronavirus in Leschenault's rousettes and Pomona leaf-nosed bats.
A total of 9,443 respiratory and alimentary specimens from 4,796 bats of 22 species were obtained in Hong Kong and Guangdong Province in southern China. RT-PCRs for a 440-bp fragment in the RdRp genes of CoVs were positive for the potentially novel alphacoronavirus BatCoV HKU10 in the alimentary samples from three (0.7%) of 416 Leschenault's rousettes (Rousettus leschenaulti) and in the alimentary and respiratory samples from 36 (7.2%) and 3 (0.6%) of 524 Pomona leaf-nosed bats (Hipposideros pomona), respectively (Table 1). Sequencing of the PCR products showed that these viral sequences formed a separate cluster distinct from known CoVs upon phylogenetic analysis, with <82% nucleotide identities to the corresponding sequences of Rh-BatCoV A977 (GenBank accession no. DQ648855). All positive samples were confirmed by RT-PCR of multiple genome sites using primers targeted to Hel or S genes. All Leschenault's rousettes positive for Ro-BatCoV HKU10 were from Guangdong Province, and all Pomona leaf-nosed bats positive for Hi-BatCoV HKU10 were from 6 of 15 sampling locations in Hong Kong (Table 1).
Table 1.
Detection of Ro-BatCoV HKU 10 and Hi-BatCoV HKU10 in bats by RT-PCR
| Bat |
No. of bats tested | No. (%) of bats positive for BatCoV HKU10 in: |
||
|---|---|---|---|---|
| Suborder, family, and specific name | Common name | Respiratory samples | Alimentary samples | |
| Megachiroptera | ||||
| Pteropodidae | ||||
| Cynopterus sphinx | Short-nosed fruit bat | 24 | 0 (0) | 0 (0) |
| Rousettus leschenaulti | Leschenault's rousette | 416 | 0 (0) | 3 (0.7)a |
| Microchiroptera | ||||
| Hipposideridae | ||||
| Hipposideros armiger | Himalayan leaf-nosed bat | 207 | 0 (0) | 0 (0) |
| Hipposideros larvatus | Intermediate leaf-nosed bat | 2 | 0 (0) | 0 (0) |
| Hipposideros pomona | Pomona leaf-nosed bat | 524 | 3 (0.6)b | 36 (7.2)b |
| Rhinolophidae | ||||
| Rhinolophus affinus | Intermediate horseshoe bat | 339 | 0 (0) | 0 (0) |
| Rhinolophus osgoodi | Osgood's horseshoe bat | 1 | 0 (0) | 0 (0) |
| Rhinolophus pusillus | Least horseshoe bat | 83 | 0 (0) | 0 (0) |
| Rhinolophus sinicus | Chinese horseshoe bat | 1,671 | 0 (0) | 0 (0) |
| Vespertilionidae | ||||
| Hypsugo pulveratus | Chinese pipistrelle | 1 | 0 (0) | 0 (0) |
| Miniopterus magnater | Greater bent-winged bat | 14 | 0 (0) | 0 (0) |
| Miniopterus pusillus | Lesser bent-winged bat | 380 | 0 (0) | 0 (0) |
| Miniopterus schreibersii | Common bent-winged bat | 525 | 0 (0) | 0 (0) |
| Myotis chinensis | Chinese myotis | 86 | 0 (0) | 0 (0) |
| Myotis horsfieldii | Horsfield's bat | 7 | 0 (0) | 0 (0) |
| Myotis muricola | Whiskered myotis | 3 | 0 (0) | 0 (0) |
| Myotis ricketti | Rickett's big-footed bat | 175 | 0 (0) | 0 (0) |
| Nyctalus noctula | Brown noctule | 38 | 0 (0) | 0 (0) |
| Pipistrellus abramus | Japanese pipistrelle | 198 | 0 (0) | 0 (0) |
| Pipistrellus tenuis | Least pipistrelle | 11 | 0 (0) | 0 (0) |
| Scotophilus kuhlii | Lesser yellow bat | 16 | 0 (0) | 0 (0) |
| Tylonycteris pachypus | Lesser bamboo bat | 75 | 0 (0) | 0 (0) |
Ro-BatCoV HKU10 was detected in three (0.8%) of 350 Leschenault's rousette bats in Guangdong but none of 66 Leschenault's rousette bats in Hong Kong.
Hi-BatCoV HKU10 was detected in 37 (7%) of 523 Pomona leaf-nosed bats in Hong Kong but not in one Pomona leaf-nosed bat in Guangdong.
No obvious disease was observed in bats positive for Ro-BatCoV HKU10 or Hi-BatCoV HKU10. However, lower body weights were observed in Pomona leaf-nosed bats positive for Hi-BatCoV HKU10 (body weight [mean ± standard deviation], 6.67 ± 0.4 g) than those negative for CoVs (6.95 ± 0.8 g) (P = 0.038 by Student's t test). Since all 37 infected Pomona leaf-nosed bats were adults (juvenile and adult bats are differentiated by their fur color and finger joints), comparison was also performed using only adult Pomona leaf-nosed bats negative for CoVs (body weight, 7.00 ± 0.8 g) (P = 0.016 by Student's t test). To control for the confounding effect of variation in body weights in different seasons, e.g., after hibernation, covariate analysis was performed using only data from the months with positive detection (March, August, October, November, and December). Results showed that Hi-BatCoV HKU10 carriage was an independent factor in association with lower body weights (P = 0.016). Attempts to stably passage BatCoV HKU10 in cell cultures were unsuccessful, with no cytopathic effect or viral replication being detected.
Complete genome characterization of Ro-BatCoV HKU10 and Hi-BatCoV HKU10.
Since the partial RdRp sequences suggested the presence of closely related viruses belonging to a potentially novel alphacoronavirus in two bat species, the complete genome sequences of two strains of Ro-BatCoV HKU10, 175A and 183A (from alimentary samples of two Leschenault's rousettes), and six strains of Hi-BatCoV HKU10, TLC1310A, TLC1347A, TLC1343A, TT3A, SL12A, and LSH5A (from alimentary samples of six Pomona leaf-nosed bats), were determined to look for genomic differences between viruses from the two bat species and evidence of interspecies transmission. The eight genomes possessed genome sizes of 28,483 to 28,494 nucleotides, with a G+C content of 38% to 39%. The two genomes of Ro-BatCoV HKU10 from Leschenault's rousettes had 99% overall nucleotide identity, while the six genomes of Hi-BatCoV HKU10 from Pomona leaf-nosed bats had 99% overall nucleotide identity. On the other hand, comparison between Ro-BatCoV HKU10 and Hi-BatCoV HKU10 genomes showed only 93 to 97% nucleotide identity. Their genome organization was similar to that of other alphacoronaviruses (Table 2; Fig. 1). In both Ro-BatCoV HKU10 and Hi-BatCoV HKU10 genomes, a putative transcription regulatory sequence (TRS) motif, 5′-CUAAAC-3′, similar to that in other alphacoronaviruses was identified at the 3′ end of the leader sequence and precedes each ORF except the NS3 and envelope (E) genes (Table 2) (11, 22). Preceding the E gene, an alternative TRS motif, 5′-CUAAAU-3′, was also identified in both the Ro-BatCoV HKU10 and Hi-BatCoV HKU10 genomes (Table 2).
Table 2.
Coding potential and putative transcription regulatory sequences of Ro-BatCoV HKU10 and Hi-BatCoV HKU10
| Coronavirus | ORF | Nucleotide position | No. of nucleotides | No. of amino acids | Frame | Putative TRS |
|
|---|---|---|---|---|---|---|---|
| Nucleotide position in genome | TRS sequencea | ||||||
| Ro-BatCoV HKU10 183A | 1ab | 303–20644 | 20,342 | 6,780 | +3, +2 | 74 | CUAAAC(220)AUG |
| S | 20641–24690 | 4,050 | 1,349 | +1 | 20628 | CUAAAC(4)AUG | |
| NS3 | 24690–25346 | 657 | 218 | +3 | 24655 | ||
| E | 25375–25602 | 228 | 75 | +1 | 25353 | CUAAAU(13)AUG | |
| M | 25608–26297 | 690 | 229 | +3 | 25596 | CUAAAC(3)AUG | |
| N | 26308–27516 | 1,209 | 402 | +2 | 26296 | CUAAAC(4)AUG | |
| NS7a | 27532–27777 | 246 | 81 | +1 | 27518 | CUAAAC(5)AUG | |
| NS7b | 27787–28248 | 462 | 153 | +1 | |||
| NS7c | 27986–28216 | 231 | 76 | +2 | |||
| Hi-BatCoV HKU10 TLC1310A | 1ab | 303–20647 | 20,345 | 6,781 | +3, +2 | 74 | CUAAAC(220)AUG |
| S | 20644–24699 | 4,056 | 1,351 | +1 | 20631 | CUAAAC(4)AUG | |
| NS3 | 24699–25355 | 657 | 218 | +3 | 24664 | ||
| E | 25384–25611 | 228 | 75 | +1 | 25362 | CUAAAU(13)AUG | |
| M | 25617–26297 | 681 | 226 | +3 | 25605 | CUAAAC(3)AUG | |
| N | 26309–27508 | 1,200 | 399 | +2 | 26296 | CUAAAC(4)AUG | |
| NS7a | 27524–27766 | 243 | 80 | +2 | 27510 | CUAAAC(5)AUG | |
| NS7b | 27776–28237 | 462 | 153 | +2 | |||
| NS7c | 27975–28205 | 231 | 76 | +3 | |||
The number in parentheses is the number of nucleotides between the TRS and start codon.
Fig 1.
Genome organizations of Ro-BatCoV HKU10, Hi-BatCoV HKU10, and representative CoVs from each group. Genes for papain-like proteases (PL1pro, PL2pro, and PLpro), 3C-like protease (3CLpro), and RNA-dependent RNA polymerase (RdRp) are represented by orange boxes. Genes for hemagglutinin esterase (HE), spike protein (S), envelope protein (E), membrane protein (M), and nucleocapsid protein (N) are represented by green boxes. Genes for putative accessory proteins are represented by blue boxes. BatCoV HKU10 strains detected in this study are shown in bold.
The characteristics of putative nonstructural proteins (NSPs) of ORF1 of Ro-BatCoV HKU10 and Hi-BatCoV HKU10 and their predicted cleavage sites are summarized in Table 3. A unique putative cleavage site at NSP10/11 or NSP10/12 was observed in both Ro-BatCoV HKU10 and Hi-BatCoV HKU10, where the P1′ position was occupied by alanine instead of serine or glycine as in other alphacoronaviruses. This amino acid substitution was due to mutations from TC(A/T), AG(T/C), or GGC to GCT in Ro-BatCoV HKU10 and Hi-BatCoV HKU10.
Table 3.
Characteristics of putative nonstructural proteins of ORF1ab in Ro-BatCoV HKU10 and Hi-BatCoV HKU10
| NSP | Putative function or domaina | Amino acidsb |
|
|---|---|---|---|
| Ro-BatCoV HKU10 (183A) | Hi-BatCoV HKU10 (LSH5A) | ||
| NSP1 | Unknown | M1-A195 | M1-A195 |
| NSP2 | Unknown | K196-G888 | K196-G888 |
| NSP3 | ADRP, Putative PLpro domain PL1pro, PL2pro | S889-G2518 | S889-G2519 |
| NSP4 | Hydrophobic domain | S2519-Q2996 | S2520-Q2997 |
| NSP5 | 3CLpro | S2997-Q3298 | S2998-Q3299 |
| NSP6 | Hydrophobic domain | S3299-Q3574 | S3300-Q3575 |
| NSP7 | Unknown | S3575-Q3657 | S3576-Q3658 |
| NSP8 | Unknown | S3658-Q3852 | S3659-Q3853 |
| NSP9 | Unknown | N3853-Q3960 | N3854-Q3961 |
| NSP10 | Unknown | A3961-Q4097 | A3962-Q4098 |
| NSP11 | Unknown (short peptide at the end of ORF1a) | A4098-N4115 | A4099-N4116 |
| NSP12 | RdRp | A4098-Q5024 | A4099-Q5025 |
| NSP13 | Hel | S5025-Q5621 | S5026-Q5622 |
| NSP14 | ExoN, N7-MTase | A5622-Q6139 | A5623-Q6140 |
| NSP15 | NendoU | S6140-Q6478 | S6141-Q6479 |
| NSP16 | 2′-O-MT | S6479-R6780 | S6480-R6781 |
ADRP, ADP-ribose-1″-phosphatase; PLpro, papain-like protease; 3CLpro, 3C-like protease; RdRp, RNA-dependent RNA polymerase; Hel, helicase; ExoN, 3′-to-5′ exonuclease; N7-MTase, (guanine-N7)-methyltransferase; NendoU, nidoviral uridylate-specific endoribonuclease; and 2′-O-MT, 2′-O-ribose methyltransferase.
Given in the format first residueposition-last residueposition. The alanine at the P1′ position of the unique putative cleavage site at NSP10/11 or NSP10/12 is shown in bold.
One ORF, which encodes a putative 218-aa nonstructural protein, NS3, was observed between the S and E genes of Ro-BatCoV HKU10 and Hi-BatCoV HKU10. This NS3, which is highly conserved among different strains of Ro-BatCoV HKU10 and Hi-BatCoV HKU10 with ≥98.2% amino acid identities, possessed only ≤47% amino acid identities to NS3 of Mi-BatCoV HKU8 and other alphacoronaviruses. TMHMM analysis showed three putative transmembrane domains in NS3 of Ro-BatCoV HKU10 strain 175A and all six Hi-BatCoV HKU10 strains (at residues 33 to 53, 62 to 82, and 88 to 106, respectively), while only two putative transmembrane domains were observed in NS3 of Ro-BatCoV HKU10 strain 183A (at residues 33 to 53 and 74 to 96, respectively).
The most striking difference between Ro-BatCoV HKU10 and Hi-BatCoV HKU10 genomes was observed in their S proteins, which consisted of 1,349 to 1,351 aa. In contrast to products of other regions of the genome, such as 3C-like protease (3CLpro), RdRp, Hel, E, membrane (M), and nucleocapsid (N) proteins, where they possessed high sequence similarity (>96% amino acid identities), the S proteins of Ro-BatCoV HKU10 and Hi-BatCoV HKU10 shared only about 60.5% amino acid identities, as a result of frequent amino acid substitutions observed throughout their S-protein sequences (Table 4; also, see Fig. S1 in the supplemental material). The S protein of Ro-BatCoV HKU10 and Hi-BatCoV HKU10 shared <52% amino acid identity to the S proteins of other alphacoronaviruses (Table 4). As in other alphacoronaviruses (6), no cleavage site was identified between S1 and S2. InterProScan analysis predicted them as type I membrane glycoproteins, with most of the protein (residues 23/24/28 to 1292/1294) exposed on the outside of the virus and with a transmembrane domain (residues 1293/1295 to 1327/1329) at the C terminus, followed by a cytoplasmic tail rich in cysteine residues. Two heptad repeats (HR), important for membrane fusion and viral entry (4), were located at residues 959 to 1085/1086 (HR1) and 1234 to 1284 (HR2) for Ro-BatCoV HKU10 and at residues 952/955/959 to 1048/1051/1053 (HR1) and 1235/1237 to 1284/1286 (HR2) for Hi-BatCoV HKU10. Aminopeptidase N (CD13) has been shown to be the receptor for various alphacoronaviruses, including HCoV 229E, canine CoV (CCoV), FIPV, PEDV, and TGEV (7, 75). On the other hand, human angiotensin-converting enzyme 2 (hACE2) has been found to be the receptor for both HCoV NL63, an alphacoronavirus, and SARS-CoV, a betacoronavirus, although they utilize different receptor-binding sites (23, 38). The S proteins of Ro-BatCoV HKU10 and Hi-BatCoV HKU10 did not exhibit significant homology to the known receptor-binding domains of other CoVs, including HCoV 229E (3, 24, 28, 44, 74).
Table 4.
Comparison of genome sizes and amino acid identities between predicted proteins of Ro-BatCoV HKU10, Hi-BatCoV HKU10, and other CoVs
| Coronavirusa | Genome size (bases) | Pairwise amino acid identity (%) |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ro-BatCoV HKU10 183A |
Hi-BatCoV HKU10 LSH5A |
||||||||||||||
| 3CLpro | RdRp | Hel | S | E | M | N | 3CLpro | RdRp | Hel | S | E | M | N | ||
| Alphacoronavirus | |||||||||||||||
| TGEV | 28,586 | 63.6 | 75 | 77 | 42.8 | 32.9 | 47 | 36.7 | 63.2 | 74.7 | 76.8 | 42.5 | 32.9 | 44.9 | 36.7 |
| FIPV | 29,355 | 62.6 | 75.2 | 77 | 42.8 | 32.9 | 44.9 | 33.3 | 62.3 | 74.9 | 76.8 | 42.8 | 32.9 | 42.6 | 35.3 |
| PRCV | 27,550 | 63.6 | 74.8 | 76.6 | 41.1 | 36.6 | 47.1 | 36.1 | 63.2 | 74.5 | 76.5 | 41.1 | 36.6 | 45.4 | 36.3 |
| CCoV | 29,363 | 61.3 | 74.9 | 77.5 | 42.6 | 35.4 | 44.8 | 36.5 | 60.9 | 74.6 | 77.3 | 42.1 | 35.4 | 42.9 | 36.9 |
| MCoV | 28,894 | 58.9 | 74.0 | 76.5 | 43.3 | 31.7 | 45.4 | 34.7 | 59.3 | 73.8 | 76.4 | 42.1 | 31.7 | 44.6 | 34.4 |
| HCoV 229E | 27,317 | 67.9 | 79.3 | 82.9 | 42.8 | 46.8 | 60.7 | 42.4 | 67.5 | 79.2 | 82.7 | 43.2 | 46.8 | 59.7 | 42.3 |
| HCoV NL63 | 27,553 | 65.7 | 82.1 | 84.8 | 46.3 | 49.4 | 65.5 | 43.1 | 65.3 | 81.9 | 84.6 | 45.3 | 49.4 | 66.8 | 43.5 |
| PEDV | 28,033 | 74.5 | 82.5 | 86.3 | 44.9 | 51.9 | 72.9 | 43.8 | 74.2 | 82.2 | 86.1 | 46.8 | 51.9 | 73 | 43.2 |
| Rh-BatCoV HKU2 | 27,165 | 62.3 | 79.9 | 80.1 | 25.6 | 60 | 68.1 | 47 | 61.9 | 79.8 | 79.9 | 25.2 | 60 | 69 | 46.8 |
| Mi-BatCoV 1A | 28,326 | 70.5 | 83.4 | 84.3 | 47.4 | 52 | 65.9 | 47.8 | 70.2 | 83.3 | 84.1 | 47.5 | 52 | 64.7 | 47.8 |
| Mi-BatCoV 1B | 28,476 | 70.2 | 82.6 | 84.3 | 45.7 | 52 | 64.3 | 48.1 | 69.9 | 82.5 | 84.1 | 45.1 | 52 | 64.7 | 48.3 |
| Mi-BatCoV HKU8 | 28,773 | 74.5 | 84.1 | 86.1 | 51.4 | 50.7 | 67.7 | 46.3 | 74.5 | 83.8 | 85.9 | 51.2 | 50.7 | 66.9 | 45.8 |
| Sc-BatCoV 512 | 28,203 | 70.9 | 80.7 | 82.6 | 46.3 | 53.2 | 69.9 | 46.7 | 71.2 | 80.4 | 82.4 | 45.5 | 53.2 | 69.6 | 46.9 |
| Ro-BatCoV HKU10 183A | 28,494 | 99.7 | 99.5 | 99.8 | 60.5 | 100 | 96.1 | 97 | |||||||
| Hi-BatCoV HKU10 LSH5A | 28,492 | 99.7 | 99.5 | 99.8 | 60.5 | 100 | 96.1 | 97 | |||||||
| Betacoronavirus subgroup A | |||||||||||||||
| HCoV OC43 | 30,738 | 44.2 | 57 | 56.9 | 27 | 30.2 | 35.3 | 25.9 | 44.2 | 56.9 | 56.8 | 27 | 30.2 | 35.5 | 25.7 |
| BCoV | 31,028 | 43.9 | 56.9 | 57.1 | 27.4 | 30.2 | 36.1 | 24.9 | 43.9 | 56.7 | 56.9 | 25.4 | 30.2 | 36.1 | 25.1 |
| PHEV | 30,480 | 43.6 | 56.9 | 57.1 | 27 | 30.2 | 34.2 | 26.5 | 43.6 | 56.7 | 56.9 | 25.9 | 30.2 | 34.3 | 25.7 |
| GiCoV | 30,979 | 43.9 | 56.9 | 57.1 | 27.4 | 30.2 | 36.1 | 25.5 | 43.9 | 56.7 | 56.9 | 25.5 | 30.2 | 36.1 | 25.3 |
| MHV | 31,357 | 45.2 | 56.4 | 56.9 | 26.6 | 32.5 | 36.8 | 25 | 45.2 | 56.5 | 56.7 | 25.7 | 32.5 | 37.2 | 24.7 |
| HCoV HKU1 | 29,926 | 44.6 | 56.5 | 55.6 | 26.9 | 31 | 36.7 | 26.7 | 44.2 | 56.1 | 55.4 | 26.3 | 31 | 38 | 27.1 |
| Betacoronavirus subgroup B | |||||||||||||||
| SARS-CoV | 29,751 | 44.8 | 59.1 | 62 | 26.5 | 21.1 | 32.8 | 26.9 | 45.1 | 59 | 61.8 | 25.6 | 21.1 | 31.7 | 26.4 |
| SARSr-Rh-BatCoV HKU3 | 29,728 | 44.1 | 59.1 | 61.6 | 26 | 21.1 | 32.3 | 27.1 | 44.4 | 59 | 61.5 | 25.1 | 21.1 | 31.3 | 27.3 |
| Betacoronavirus subgroup C | |||||||||||||||
| Ty-BatCoV HKU4 | 30,286 | 45.3 | 59.9 | 62.5 | 27.8 | 24.4 | 35.7 | 26.5 | 45 | 59.8 | 62.3 | 26.7 | 24.4 | 36 | 26.9 |
| Pi-BatCoV HKU5 | 30,488 | 45.9 | 59.4 | 63.5 | 25.8 | 23.2 | 33 | 26.7 | 45.6 | 59.3 | 63.3 | 26.4 | 23.2 | 33.2 | 26.9 |
| Betacoronavirus subgroup D | |||||||||||||||
| Ro-BatCoV HKU9 | 29,114 | 43.8 | 59.3 | 61.8 | 26.5 | 15.2 | 33 | 23.4 | 44.2 | 59.1 | 61.6 | 27 | 15.2 | 32.2 | 23.7 |
| Gammacoronavirus | |||||||||||||||
| IBV | 27,608 | 41.4 | 59.5 | 57.8 | 26.5 | 15.7 | 25 | 24 | 41.1 | 59.4 | 57.6 | 26.7 | 15.7 | 25.4 | 22.7 |
| BWCoV SW1 | 31,686 | 40.8 | 57.2 | 58.5 | 27.5 | 22.7 | 25.1 | 27.3 | 40.5 | 57 | 58.4 | 26.8 | 22.7 | 24.6 | 26.9 |
| Deltacoroanvirus | |||||||||||||||
| BuCoV HKU11 | 26,476 | 34.8 | 49.1 | 50.8 | 38 | 21.7 | 26.9 | 21.2 | 34.8 | 49 | 50.7 | 37.6 | 21.7 | 25.7 | 21.7 |
| ThCoV HKU12 | 26,396 | 34.8 | 48.6 | 50.7 | 37.6 | 22.9 | 28 | 21.3 | 34.8 | 48.5 | 50.5 | 37.9 | 22.9 | 27.8 | 21.8 |
| MunCoV HKU13 | 26,552 | 34.2 | 49.4 | 50.9 | 38.3 | 24.1 | 25.1 | 19.8 | 34.2 | 49.3 | 50.7 | 36.3 | 24.1 | 25.7 | 20.2 |
| PorCoV HKU15 | 25,421 | 34.8 | 48.8 | 51.6 | 37.4 | 21.4 | 25.1 | 19.7 | 34.8 | 48.6 | 51.4 | 37.4 | 21.4 | 24.5 | 20.7 |
TGEV, porcine transmissible gastroenteritis virus; FIPV, feline infectious peritonitis virus; PRCV, porcine respiratory coronavirus; HCoV 229E, human coronavirus 229E; HCoV NL63, human coronavirus NL63; PEDV, porcine epidemic diarrhea virus; CCoV, canine coronavirus; MCoV, mink coronavirus; Rh-BatCoV HKU2, Rhinolophus bat coronavirus HKU2; Mi-BatCoV 1A, Miniopterus bat coronavirus 1A; Mi-BatCoV 1B, Miniopterus bat coronavirus 1B; Mi-BatCoV HKU8, Miniopterus bat coronavirus HKU8; Sc-BatCoV 512, Scotophilus bat coronavirus 512; Ro-BatCoV HKU10, Rousettus bat coronavirus HKU10; Hi-BatCoV HKU10, Hipposideros bat coronavirus HKU10; HCoV HKU1, human coronavirus HKU1; HCoV OC43, human coronavirus OC43; MHV, murine hepatitis virus; BCoV, bovine coronavirus; PHEV, porcine hemagglutinating encephalomyelitis virus; GiCoV, giraffe coronavirus; SARS-CoV, SARS coronavirus; SARSr-Rh-BatCoV HKU3, SARS-related Rhinolophus bat coronavirus HKU3; Ty-BatCoV HKU4, Tylonycteris bat coronavirus HKU4; Pi-BatCoV HKU5, Pipistrellus bat coronavirus HKU5; Ro-BatCoV HKU9, Rousettus bat coronavirus HKU9; IBV, infectious bronchitis virus; BWCoV SW1, beluga whale coronavirus SW1; BuCoV HKU11, bulbul coronavirus HKU11; ThCoV HKU12, thrush coronavirus HKU12; MunCoV HKU13, munia coronavirus HKU13; PorCoV HKU15, porcine coronavirus HKU15.
Downstream of the N gene, both Ro-BatCoV HKU10 and Hi-BatCoV HKU10 genomes (except those of strains TT3A and SL12A) possess three ORFs encoding nonstructural proteins NS7a, NS7b, and NS7c, of 80 or 81, 153, and 76 aa, respectively. Strains TT3A and SL12A possess NS7b and NS7c but not NS7a, as a result of a nucleotide substitution in the start codon of NS7a (ATG to ATT). And since Ro-BatCoV HKU10 and Hi-BatCoV HKU10 share only 60% amino acid identity in NS7a to other strains, this gene may be nonfunctional. In contrast, NS7b and NS7c were highly similar between the two viruses, sharing 92 to 95% and 88 to 90% amino acid identities, respectively. However, a BLAST search revealed no significant amino acid similarities between these putative nonstructural proteins and other known proteins. TMHMM analysis showed two putative transmembrane domains in NS7a (at residues 5 to 23 and 42 to 62/76) but none in NS7b. For NS7c, one putative transmembrane domain was observed (at residues 31 to 51) in all strains except Ro-BatCoV HKU10 strain 175A, which possessed no putative transmembrane domain in its NS7c. Some alphacoronaviruses, such as FIPV, TGEV, porcine respiratory CoV (PRCV), Rh-BatCoV HKU2, and Sc-BatCoV 512, are also known to possess genes downstream of that for N (Fig. 1). In FIPV, the two genes downstream of the N gene may be important for virulence, while in TGEV, the gene downstream of the N gene may play a role in membrane association of replication complexes or virus assembly (19, 42, 62). Further experiments will delineate the function of such ORFs in bat CoVs.
Phylogenetic analyses.
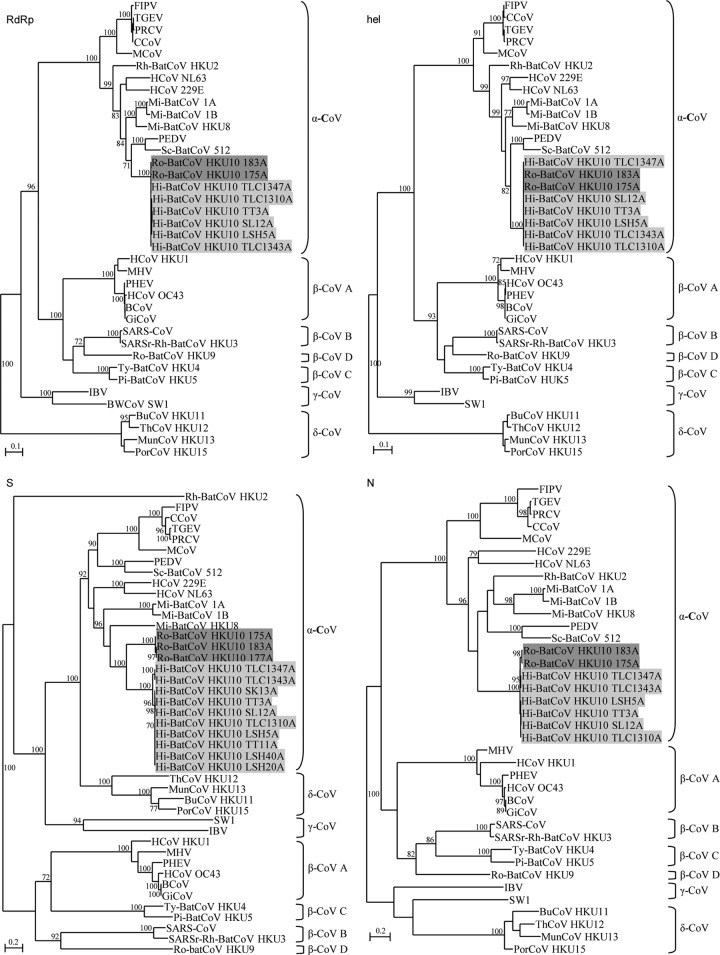
The phylogenetic trees constructed using the amino acid sequences of the RdRp, Hel, S, and N proteins of Ro-BatCoV HKU10, Hi-BatCoV HKU10, and other CoVs are shown in Fig. 2, and the corresponding pairwise amino acid identities are shown in Table 4. For Hel, RdRp, and N genes, the two strains of Ro-BatCoV HKU10 and six strains of Hi-BatCoV HKU10 clustered together with very short branch lengths, reflecting their high sequence similarities (Fig. 2). Moreover, comparison of the amino acid sequences of the seven conserved replicase domains or NSPs {ADP-ribose-1″-phosphatase, ADRP, NSP5 (3CLpro), NSP12 (RdRp), NSP13 (Hel), NSP14 [3′-to-5′ exonuclease, ExoN; (guanine-N7)-methyltransferase, N7-MTase], NSP15 (nidoviral uridylate-specific endoribonuclease, NendoU) and NSP16 (2′-O-ribose methyltransferase, 2′-O-MT)} for CoV species demarcation (5) showed that Ro-BatCoV HKU10 and Hi-BatCoV HKU10 possess <90% amino acid identities to those of other alphacoronaviruses but >90% amino acid identities to each other, indicating that they represented the same novel species of Alphacoronavirus.
Fig 2.
Phylogenetic analysis of RdRp, Hel, S, and N of Hi-BatCoV HKU10 and Ro-BatCoV HKU10. The trees were constructed by the maximum-likelihood method with bootstrap values calculated from 100 trees. A total of 951, 609, 1,899 and 572 amino acid positions in RdRp, Hel, S, and N, respectively, were included in the analysis. The scale bars indicate the estimated number of substitutions per 5 or 10 aa. HCoV 229E, human coronavirus 229E; PEDV, porcine epidemic diarrhea virus; TGEV, porcine transmissible gastroenteritis virus; FIPV, feline infectious peritonitis virus; PRCV, porcine respiratory coronavirus; HCoV NL63, human coronavirus NL63; Rh-BatCoV HKU2, Rhinolophus bat coronavirus HKU2; Mi-BatCoV 1A, Miniopterus bat coronavirus 1A; Mi-BatCoV 1B, Miniopterus bat coronavirus 1B; Mi-BatCoV HKU8, Miniopterus bat coronavirus HKU8; Sc-BatCoV 512, Scotophilus bat coronavirus 512; HCoV HKU1, human coronavirus HKU1, HCoV OC43, human coronavirus OC43; MHV, murine hepatitis virus; BCoV, bovine coronavirus; PHEV, porcine hemagglutinating encephalomyelitis virus; GiCoV, giraffe coronavirus; SARS-CoV, SARS coronavirus; SARSr-Rh-BatCoV HKU3, SARS-related Rhinolophus bat coronavirus HKU3; Ty-BatCoV HKU4, Tylonycteris bat coronavirus HKU4; Pi-BatCoV HKU5, Pipistrellus bat coronavirus HKU5; Ro-BatCoV HKU9, Rousettus bat coronavirus HKU9; IBV, infectious bronchitis virus; BWCoV SW1, beluga whale coronavirus SW1; BuCoV HKU11, bulbul coronavirus HKU11; ThCoV HKU12, thrush coronavirus HKU12; MunCoV HKU13, munia coronavirus HKU13; PorCoV HKU15, porcine coronavirus HKU15.
In contrast, marked sequence divergence was observed between the S proteins of Ro-BatCoV HKU10 and Hi-BatCoV HKU10 strains, forming two distinct clusters upon phylogenetic analysis, which was confirmed by further sequencing of the S genes of one additional Ro-BatCoV HKU10 and four additional Hi-BatCoV HKU10 strains (Table 4; Fig. 2). Moreover, among the 10 Hi-BatCoV HKU10 strains with available S-gene sequences, two strains, TLC43A and TLC47A, both detected in 2010, appeared to form a distinct cluster, sharing ∼95% amino acid identities to the other eight strains, with most of the substitutions being nonsynonymous substitutions localized within the S1 region.
Estimation of synonymous and nonsynonymous substitution rates.
As demonstrated in studies on the evolution and cross-species transmission of SARS-CoV-like viruses, high Ka/Ks ratios and substantial changes in the spike proteins of coronaviruses may reflect rapid viral evolution soon after introduction into a new animal host (54). Since results from genome analysis suggested that Ro-BatCoV HKU10 and Hi-BatCoV HKU10 possess highly similar genome sequences except in the S genes, we hypothesize that interspecies transmission between the two bat species occurred recently, with subsequent viral adaptation in the new host species. To test this hypothesis, the Ka/Ks ratios for the various coding regions in different strains of Ro-BatCoV HKU10 and Hi-BatCoV HKU10 were determined (Table 5). Compared to Ro-BatCoV HKU10 strains, higher Ka/Ks ratios were observed in S (0.277 versus 0.078) and N (0.235 versus 0.077) genes of Hi-BatCoV HKU10 strains. Interestingly, when the Ka/Ks ratios of S genes of additional strains from different sampling times were compared, a dramatic reduction of Ka/Ks ratios, from 2.000 among four Hi-BatCoV HKU10 strains from 2005-2006 to 0.333 among four Hi-BatCoV HKU10 strains from 2007-2008 and to 0 among two Hi-BatCoV HKU10 strains from 2010, was observed, compared to 0.070 among three Ro-BatCoV HKU10 strains from 2005. This suggests that the S gene of Hi-BatCoV HKU10 from 2005-2006 likely underwent rapid evolution under positive selection in Pomona leaf-nosed bats and that this evolution stabilized thereafter.
Table 5.
Estimation of nonsynonymous substitution and synonymous rates in the genomes of Ro-BatCoV HKU10 and Hi-BatCoV HKU10
| Gene |
Ka/Ks ratio |
|
|---|---|---|
| Hi-BatCoV HKU10 (6 strains) | Ro-BatCoV HKU10 (2 strains) | |
| NSP1 | 0.121 | 0.285 |
| NSP2 | 0.173 | 0.071 |
| NSP3 | 0.100 | 0.313 |
| NSP4 | 0.050 | 0.063 |
| NSP5 | 0.022 | 0.029 |
| NSP6 | 0.051 | Ka = 0, Ks = 0.037 |
| NSP7 | Ka = 0, Ks = 0.021 | 0.128 |
| NSP8 | 0.047 | Ka = 0, Ks = 0.023 |
| NSP9 | Ka = 0, Ks = 0.022 | Ka = 0, Ks = 0.027 |
| NSP10 | 0.022 | Ka = 0, Ks = 0.031 |
| NSP11 | Ka = 0, Ks = 0 | Ka = 0, Ks = 0 |
| NSP12 | 0.063 | 0.061 |
| NSP13 | 0.012 | Ka = 0, Ks = 0.043 |
| NSP14 | 0.028 | 0.021 |
| NSP15 | 0.053 | 0.012 |
| NSP16 | 0.087 | 0.043 |
| S | 0.277a | 0.078b |
| NS3 | 0.077 | 0.108 |
| E | Ka = 0, Ks = 0 | Ka = 0, Ks = 0 |
| M | 0.333 | 0.290 |
| N | 0.235 | 0.077 |
| NS7a | Ka = 0, Ks = 0 | 0.151 |
| NS7b | 1.000 | 0.740 |
| NS7c | 0.435 | 0.575 |
The Ka/Ks of S sequences of four Hi-BatCoV HKU10 strains from 2005–2006 was 2.000, that of four Hi-BatCoV HKU10 strains from 2007–2008 was 0.333, and that of two Hi-BatCoV HKU10 strains from 2010 was 0 (Ka = 0, Ks = 0.001).
The Ka/Ks of S sequences of three Ro-BatCoV HKU10 strains from 2005 was 0.070.
Recombination analysis and detection of positive selection.
No significant recombination breakpoint among BatCoV HKU10 genomes was detected by bootscan or GARD analysis. Significant positive selection was predicted by the REL method but not the SLAC and FEL methods, as the REL method is more powerful, since it pools signals from multiple sites to detect selection. Branch site REL analysis of S-gene sequences showed that only the branch of two Hi-BatCoV HKU10 strains from 2010 was under significant positive selection (P = 0.003), with the strength of positive selection (ω+) and the proportion of total branch length affected by positive selection (q+) being 4,378.93 and 0.03, respectively (Fig. 3A). This suggested that the S gene of Hi-BatCoV HKU10 evolved under positive selection along the year 2010 lineage on short segments of the branch. REL analysis found that 66 of the 1,351 codons in the S proteins of Hi-BatCoV HKU10 strains from 2005 to 2010 were under positive selection. Most of these sites were distributed within the S1 domain, indicating that this domain may have been under functional constraints (Fig. 3B). However, since detection of specific amino acid sites under positive selection using REL is unstable in the presence of heterotachy, only the trends of spatial localization were indicated.
Fig 3.
Selection pressure analysis of the S genes of BatCoV HKU10. (A) Detection of lineage-specific selection pressure. The branch with a P value of <0.01 is highlighted. ω+, strength of positive selection; q+, proportion of the total branch length influenced by the selective pressure. The scale bar indicates the estimated number of substitutions per 20 nucleotides. (B) Distribution of positively selected sites in S protein genes identified using REL among Hi-BatCoV HKU10 strains from 2005–2010. The receptor-binding domains (RBD) of the S proteins of TGEV, HCoV NL63, and HCoV 229E were mapped previously (3, 14, 74). Homology modeling of the RBD in the S proteins of Ro-BatCoV HKU10 and Hi-BatCoV HKU10 was performed using SwissModel in automated mode (52). The heptad repeat (HR) regions were predicted by using the coiled-coil prediction program MultiCoil2 (61).
Estimation of divergence dates.
Using the relaxed clock model with Uced on RdRp gene sequences, tMRCA of all BatCoV HKU10 strains was estimated at 1959.34 (HPDs, 1886.23 to 2002.77), approximately 53 years ago. The tMRCA of Hi-BatCoV HKU10 was estimated at 1986.88 (HPDs, 1956.17 to 2004.76) and that of Ro-BatCoV HKU10 at 1991.58 (HPDs, 1968.62 to 2004.41) (Fig. 4). The estimated mean substitution rate of the RdRp data set was 3.705 × 10−4 substitution per site per year, which is comparable to previous estimations for other CoVs (32, 35, 50, 64).
Fig 4.
Estimation of the tMRCA of BatCoV HKU10. The time-scaled phylogeny was summarized from all MCMC phylogenies of the RdRp gene data set analyzed under the relaxed-clock model with an exponential distribution (Uced) in BEAST version 1.6.2. Viruses characterized in this study are in bold.
DISCUSSION
In this study, we detected and characterized a novel alphacoronavirus, BatCoV HKU10, from two very different bat species in China. Ro-BatCoV HKU10 was detected in three Leschenault's rousettes in Guangdong Province, whereas Hi-BatCoV HKU10 was detected in 37 Pomona leaf-nosed bats in Hong Kong. The genomes of Ro-BatCoV HKU10 and Hi-BatCoV HKU10 were highly similar except in the S region, where the two viruses shared only 60.5% amino acid identities. Nevertheless, they formed a distinct cluster within Alphacoronavirus upon phylogenetic analysis, supporting the idea that BatCoV HKU10 represents a novel species. Since Ro-BatCoV HKU10 and Hi-BatCoV HKU10 have >90% amino acid identity in the seven conserved replicase domains for CoV species demarcation by ICTV (5), these two CoVs should be recognized as the same species infecting two different bat species.
The marked difference between the S proteins of Ro-BatCoV HKU10 and Hi-BatCoV HKU10 despite the high similarity between their genomes in other regions strongly suggested that they shared a recent common ancestor. Moreover, positive selection and molecular-clock analysis suggested that BatCoV HKU10 may have been transmitted to the new host, Pomona leaf-nosed bats, relatively recently. First the Ka/Ks ratio of the S gene of Hi-BatCoV HKU10 was higher than that of Ro-BatCoV HKU10, although the latter was detected only in Leschenault's rousettes sampled in 2005. Moreover, the drop in Ka/Ks ratio for S genes of Hi-BatCoV HKU10 from 2.000 among strains from 2005-2006 to 0 among strains from 2010 suggested that the S gene of Hi-BatCoV HKU10 was under strong positive selection during 2005-2006, which was probably due to recent interspecies transmission and adaptation in the new host species, Pomona leaf-nosed bats. Second, significant positive selection was observed at the branch of two Hi-BatCoV HKU10 strains from 2010, with most of the codons under selection being distributed within the S1 domain. This suggested that these most recent strains have undergone further rapid evolution in their S1 domains, which may have favored the emergence of a novel subtype to adapt to new host and/or environmental factors. Third, molecular-clock analysis of the RdRp genes dated the tMRCA of all BatCoV HKU10 strains at around 1959 (HPDs, 1886 to 2002) and that of Hi-BatCoV HKU10 at around 1986 (HPDs, 1956 to 2004), which supported the recent emergence of BatCoV HKU10 and recent interspecies transmission to Pomona leaf-nosed bats. Based on the above evidence, it is likely that BatCoV HKU10 was transmitted to Pomona leaf-nosed bats not long before 2005, most probably from Leschenault's rousettes, and the virus has been rapidly adapting in the new host by changing its S protein. However, as the number of bat samples, especially from Pomona leaf-nosed bats in Guangdong, was limited in this study, further studies on more samples and virus isolation in cell cultures derived from the two bat species may allow a more accurate determination of the directionality of interspecies transmission and exclude other possible explanations of the observed difference in S proteins, such as host selection driving rapid evolution. Moreover, detection of more strains of Ro-BatCoV HKU10 from Leschenault's rousettes in the near future for evolutionary studies may help further confirm that these bats are the primary reservoir of BatCoV HKU10. The S proteins of CoVs are responsible for receptor binding and host species adaptation, and their genes therefore constitute one of the most variable regions within CoV genomes (30, 31, 40). Previous studies on SARS-CoV have also provided clues on how changes in the CoV S protein, both within and outside the receptor-binding domain, may govern CoV cross-species transmission and emergence in new host populations (16, 45). The present results also suggested that the CoV S protein is able to evolve rapidly within a short time after viral transmission to a new host, analogous to the situation of SARSr-CoV evolution (39, 54, 76). In fact, the sequence divergence between the S proteins of Ro-BatCoV HKU10 and Hi-BatCoV HKU10 is even higher than that between SARSr-Rh-BatCoV and civet SARSr-CoV (30, 40), which in turn supported the idea that horseshoe bats could well be the reservoir for the direct ancestor of SARSr-CoVs in civets, with recent bat-to-civet transmission. Unfortunately, bat CoVs discovered so far cannot be cultivated in traditional in vitro cell lines, which has hampered studies on their receptor binding and host adaptation.
The interspecies transmission of BatCoV HKU10 could well be explained by the biological characteristics of its host species. Bats (order Chiroptera), which account for about 20% of all mammalian species, are classified into two suborders, Microchiroptera and Megachiroptera. Pomona leaf-nosed bats are small, insectivorous bats belonging to the suborder Microchiroptera, family Hipposideridae, with a body weight ranging from 6 to 8 g. In Hong Kong, they are very common and widespread throughout countryside areas and roost in colonies with up to several hundred individuals, mainly in water tunnels and abandoned mines or other enclosures with limited airflow. Interestingly, leaf-nosed bats belonging to Hipposideridae have also been found to harbor coronaviruses, including alphacoronaviruses closely related to HCoV 229E, with the most recent common ancestor of these alphacoronaviruses and HCoV 229E being dated to approximately 1686-1800 (15, 46). In contrast, Leschenault's rousettes are fruit bats belonging to the suborder Megachiroptera, family Pteropodidae, with large body size, weighing 54 to 155 g and with a forearm length up to 88 mm (53). This bat species is widely distributed in Asia and roosts in extremely densely packed colonies of up to several thousand individuals. It is also well known for a very long flying distance, >11 km, and the ability to tolerate diverse and harsh habitats. These special biological features probably explain the ability of Leschenault's rousettes to acquire various viruses as well as to transmit them to other bat species. Transmission of BatCoV HKU10 from Leschenault's rousettes residing in Guangdong to Pomona leaf-nosed bats in Hong Kong is possible, given that the two places are only about 140 km apart. According to survey records in Hong Kong, these two species have also been found to share roosting sites, which would allow indirect contact. Besides Ro-BatCoV HKU10, Leschenault's rousettes from China have also been found to carry other viruses, including diverse genotypes of Ro-BatCoV HKU9, a subgroup D betacoronavirus, arising from recombination, as well as Tuhokovirus 1, 2, and 3, which are rubulaviruses belonging to the family Paramyxoviridae (33, 34). Although no evidence for recombination was observed among the present BatCoV HKU10 strains, coinfection of different CoVs in the same bat species may potentially create opportunities for recombination and emergence of new viruses.
Although bats infected with BatCoV HKU10 appeared to be healthy, lower body weights were observed in Pomona leaf-nosed bats positive for Hi-BatCoV HKU10 than those negative for CoVs. This is similar to our previous findings that Chinese horseshoe bats infected with SARSr-Rh-BatCoV had lower body weights than those that were uninfected or infected with another CoV, Rh-BatCoV HKU2 (32). This supports the idea that certain bat CoVs may cause acute infection associated with weight loss in their host species. The fact that BatCoV HKU10 was detected mainly in alimentary samples also suggests an enteric tropism. However, further studies are required to understand the pathogenicity of BatCoV HKU10 in its host species.
The present study not only provides the first evidence for interspecies transmission of a CoV between two very different bats belonging to different suborders but also illustrates the power of genome sequencing and analysis in understanding the evolution and ecology of CoVs. The present Hi-BatCoV HKU10 genomes also represented the first genome data available for CoVs in bats belonging to the genus Hipposideros. While the existence of CoVs in bats was unknown until after the SARS epidemic, different bat populations from various countries are now known to harbor diverse CoVs, likely as a result of their species diversity, roosting behaviors, and migrating abilities (30, 40, 49, 58, 67, 70). The present data also support the idea that these warm-blooded flying vertebrates are ideal hosts for the gene source for Alphacoronavirus and Betacoronavirus to fuel coronavirus evolution and dissemination (73). Should recombination occur among these bat CoVs when bats are in proximity to other animals, such as in markets and restaurants in Guangdong (69), these animals could well be the source of new epidemics, like SARS. Bats are increasingly recognized as reservoirs for various zoonotic viruses, including lyssavirus, rabies virus, Hendra virus, Nipah Ebola virus, and influenza virus (37, 60). Continuous studies of viruses from different bat species and their genome analysis would help us better understand the role of bats in the ecology and evolution of CoVs and other zoonotic viruses.
Supplementary Material
ACKNOWLEDGMENTS
We thank Alan Chi-Kong Wong, Siu-Fai Leung, Chik-Chuen Lay, and Ping-Man So [HKSAR Department of Agriculture, Fisheries, and Conservation (AFCD)] and the Hong Kong Police Force for facilitation and support; Cynthia S. M. Chan and Joseph W. K. So from AFCD; and King-Shun Lo (Laboratory Animal Unit, The University of Hong Kong) and Cassius Chan for their excellent technical assistance and collection of animal specimens. We are grateful for the generous support of Carol Yu, Richard Yu, Hui Hoy, and Hui Ming in the genomic sequencing platform.
This work was partly supported by the Research Grant Council Grant, University Grant Council; a Committee for Research and Conference Grant, the Strategic Research Theme Fund, and the University Development Fund, The University of Hong Kong; the HKSAR Research Fund for the Control of Infectious Diseases of the Food and Health Bureau; the Shaw Foundation; Providence Foundation Limited in memory of the late Lui Hac Minh; a donation from Eunice Lam; and Consultancy Service for Enhancing Laboratory Surveillance of Emerging Infectious Disease for the HKSAR Department of Health.
Footnotes
Published ahead of print 29 August 2012
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1. Apweiler R, et al. 2001. The InterPro database, an integrated documentation resource for protein families, domains and functional sites. Nucleic Acids Res. 29:37–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bateman A, et al. 2002. The Pfam protein families database. Nucleic Acids Res. 30:276–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bonavia A, Zelus BD, Wentworth DE, Talbot PJ, Holmes KV. 2003. Identification of a receptor-binding domain of the spike glycoprotein of human coronavirus HCoV-229E. J. Virol. 77:2530–2538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chan WE, Chuang CK, Yeh SH, Chang MS, Chen SS. 2006. Functional characterization of heptad repeat 1 and 2 mutants of the spike protein of severe acute respiratory syndrome coronavirus. J. Virol. 80:3225–3237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. de Groot RJ, et al. 2011. Coronaviridae, p 806–828 In King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ. (ed), Virus taxonomy, classification and nomenclature of viruses. Ninth report of the International Committee on Taxonomy of Viruses, International Union of Microbiological Societies, Virology Division Elsevier Academic Press, London, United Kingdom [Google Scholar]
- 6. de Haan CA, Haijema BJ, Schellen P, Wichgers Schreur P, te Lintelo E, Vennema H, Rottier PJ. 2008. Cleavage of group 1 coronavirus spike proteins: how furin cleavage is traded off against heparan sulfate binding upon cell culture adaptation. J. Virol. 82:6078–6083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Delmas B, et al. 1992. Aminopeptidase N is a major receptor for the entero-pathogenic coronavirus TGEV. Nature 357:417–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dominguez SR, O'Shea TJ, Oko LM, Holmes KV. 2007. Detection of group 1 coronaviruses in bats in North America. Emerg. Infect. Dis. 13:1295–1300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dong BQ, et al. 2007. Detection of a novel and highly divergent coronavirus from Asian leopard cats and Chinese ferret badgers in southern China. J. Virol. 81:6920–6926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Drummond AJ, Rambaut A. 2007. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 7:214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dye C, Siddell SG. 2005. Genomic RNA sequence of feline coronavirus strain FIPV WSU-79/1146. J. Gen. Virol. 86:2249–2253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Fouchier RA, et al. 2004. A previously undescribed coronavirus associated with respiratory disease in humans. Proc. Natl. Acad. Sci. U. S. A. 101:6212–6216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gloza-Rausch F, et al. 2008. Detection and prevalence patterns of group I coronaviruses in bats, northern Germany. Emerg. Infect. Dis. 14:626–631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Godet M, Grosclaude J, Delmas B, Laude H. 1994. Major receptor-binding and neutralization determinants are located within the same domain of the transmissible gastroenteritis virus (coronavirus) spike protein. J. Virol. 68:8008–8016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gouilh MA, et al. 2011. SARS-coronavirus ancestor's foot-prints in South-East Asian bat colonies and the refuge theory. Infect. Genet. Evol. 11:1690–1702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Graham RL, Baric RS. 2010. Recombination, reservoirs, and the modular spike: mechanisms of coronavirus cross-species transmission. J. Virol. 84:3134–3146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Guan Y, et al. 2003. Isolation and characterization of viruses related to the SARS coronavirus from animals in southern China. Science 302:276–278 [DOI] [PubMed] [Google Scholar]
- 18. Guindon S, et al. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 59:307–321 [DOI] [PubMed] [Google Scholar]
- 19. Haijema BJ, Volders H, Rottier PJ. 2004. Live, attenuated coronavirus vaccines through the directed deletion of group-specific genes provide protection against feline infectious peritonitis. J. Virol. 78:3863–3871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hasoksuz M, et al. 2007. Biologic, antigenic, and full-length genomic characterization of a bovine-like coronavirus isolated from a giraffe. J. Virol. 81:4981–4990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Herrewegh AA, Smeenk I, Horzinek MC, Rottier PJ, de Groot RJ. 1998. Feline coronavirus type II strains 79-1683 and 79-1146 originate from a double recombination between feline coronavirus type I and canine coronavirus. J. Virol. 72:4508–4514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hiscox JA, Mawditt KL, Cavanagh D, Britton P. 1995. Investigation of the control of coronavirus subgenomic mRNA transcription by using T7-generated negative-sense RNA transcripts. J. Virol. 69:6219–6227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hofmann H, et al. 2005. Human coronavirus NL63 employs the severe acute respiratory syndrome coronavirus receptor for cellular entry. Proc. Natl. Acad. Sci. U. S. A. 102:7988–7993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hofmann H, et al. 2006. Highly conserved regions within the spike proteins of human coronaviruses 229E and NL63 determine recognition of their respective cellular receptors. J. Virol. 80:8639–8652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kosakovsky Pond SL, Posada D, Gravenor MB, Woelk CH, Frost SD. 2006. GARD: a genetic algorithm for recombination detection. Bioinformatics 22:3096–3098 [DOI] [PubMed] [Google Scholar]
- 26. Kosakovsky Pond SL, et al. 2011. A random effects branch-site model for detecting episodic diversifying selection. Mol. Biol. Evol. 28:3033–3043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ksiazek TG, et al. 2003. A novel coronavirus associated with severe acute respiratory syndrome. N. Engl. J. Med. 348:1953–1966 [DOI] [PubMed] [Google Scholar]
- 28. Kubo H, Yamada YK, Taguchi F. 1994. Localization of neutralizing epitopes and the receptor-binding site within the amino-terminal 330 amino acids of the murine coronavirus spike protein. J. Virol. 68:5403–5410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lai MM, Cavanagh D. 1997. The molecular biology of coronaviruses. Adv. Virus Res. 48:1–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lau SK, et al. 2005. Severe acute respiratory syndrome coronavirus-like virus in Chinese horseshoe bats. Proc. Natl. Acad. Sci. U. S. A. 102:14040–14045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lau SKP, et al. 2007. Complete genome sequence of bat coronavirus HKU2 from Chinese horseshoe bats revealed a much smaller spike gene with a different evolutionary lineage from the rest of the genome. Virology 367:428–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lau SKP, et al. 2010. Ecoepidemiology and complete genome comparison of different strains of severe acute respiratory syndrome-related Rhinolophus bat coronavirus in China reveal bats as a reservoir for acute, self-limiting infection that allows recombination events. J. Virol. 84:2808–2819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lau SKP, et al. 2010. Coexistence of different genotypes in the same bat and serological characterization of Rousettus bat coronavirus HKU9 belonging to a novel Betacoronavirus subgroup. J. Virol. 84:11385–11394 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 34. Lau SKP, et al. 2010. Identification and complete genome analysis of three novel paramyxoviruses, Tuhoko virus 1, 2 and 3, in fruit bats from China. Virology 404:106–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lau SKP, et al. 2011. Molecular epidemiology of human coronavirus OC43 reveals evolution of different genotypes over time and recent emergence of a novel genotype due to natural recombination. J. Virol. 85:11325–11337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lau SKP, et al. 2012. Isolation and characterization of a novel Betacoronavirus subgroup A coronavirus, rabbit coronavirus HKU14, from domestic rabbits. J. Virol. 86:5481–5496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Leroy EM, et al. 2005. Fruit bats as reservoirs of Ebola virus. Nature 438:575–576 [DOI] [PubMed] [Google Scholar]
- 38. Li W, et al. 2003. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature 426:450–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Li W, et al. 2005. Receptor and viral determinants of SARS-coronavirus adaptation to human ACE2. EMBO J. 24:1634–1643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Li W, et al. 2005. Bats are natural reservoirs of SARS-like coronaviruses. Science 310:676–679 [DOI] [PubMed] [Google Scholar]
- 41. Mihindukulasuriya KA, Wu G, St Leger J, Nordhausen RW, Wang D. 2008. Identification of a novel coronavirus from a beluga whale by using a panviral microarray. J. Virol. 82:5084–5088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Olsen CW. 1993. A review of feline infectious peritonitis virus: molecular biology, immunopathogenesis, clinical aspects, and vaccination. Vet. Microbiol. 36:1–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Peiris JS, et al. 2003. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet 361:1319–1325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Peng G, et al. 2011. Crystal structure of mouse coronavirus receptor-binding domain complexed with its murine receptor. Proc. Natl. Acad. Sci. U. S. A. 108:10696–10701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Perlman S, Netland J. 2009. Coronaviruses post-SARS: update on replication and pathogenesis. Nat. Rev. Microbiol. 7:439–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Pfefferle S, et al. 2009. Distant relatives of severe acute respiratory syndrome coronavirus and close relatives of human coronavirus 229E in bats, Ghana. Emerg. Infect. Dis. 15:1377–1384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Pond SL, Frost SD, Muse SV. 2005. HyPhy: hypothesis testing using phylogenies. Bioinformatics 21:676–679 [DOI] [PubMed] [Google Scholar]
- 48. Pond SL, Frost SD. 2005. Datamonkey: rapid detection of selective pressure on individual sites of codon alignments. Bioinformatics 21:2531–2533 [DOI] [PubMed] [Google Scholar]
- 49. Poon LL, et al. 2005. Identification of a novel coronavirus in bats. J. Virol. 79:2001–2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Pyrc K, et al. 2006. Mosaic structure of human coronavirus NL63, one thousand years of evolution. J. Mol. Biol. 364:964–973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Ren W, et al. 2006. Full-length genome sequences of two SARS-like coronaviruses in horseshoe bats and genetic variation analysis. J. Gen. Virol. 87:3355–3359 [DOI] [PubMed] [Google Scholar]
- 52. Schwede T, Kopp J, Guex N, Peitsch MC. 2003. SWISS-MODEL: an automated protein homology-modeling server. Nucleic Acids Res. 31:3381–3385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Shek CT. 2006. Leschenault's rousette, p 108–112 In A field guide to the terrestrial mammals of Hong Kong. Friends of Country Park and Cosmos Book Limited, Hong Kong [Google Scholar]
- 54. Song HD, et al. 2005. Cross-host evolution of severe acute respiratory syndrome coronavirus in palm civet and human. Proc. Natl. Acad. Sci. U. S. A. 102:2430–2435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Sonnhammer EL, von Heijne G, Krogh A. 1998. A hidden Markov model for predicting transmembrane helices in protein sequences. Proc. Int. Conf. Intell. Syst. Mol. Biol. 6:175–182 [PubMed] [Google Scholar]
- 56. Suchard MA, Weiss RE, Sinsheimer JS. 2001. Bayesian selection of continuous-time Markov chain evolutionary models. Mol. Biol. Evol. 18:1001–1013 [DOI] [PubMed] [Google Scholar]
- 57. Tamura K, et al. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 10:2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Tang XC, et al. 2006. Prevalence and genetic diversity of coronaviruses in bats from China. J. Virol. 80:7481–7490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Tong S, et al. 2009. Detection of novel SARS-like and other coronaviruses in bats from Kenya. Emerg. Infect. Dis. 15:482–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Tong S, et al. 2012. A distinct lineage of influenza A virus from bats. Proc. Natl. Acad. Sci. U. S. A. 109:4269–4274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Trigg J, Gutwin K, Keating AE, Berger B. 2011. Multicoil2: predicting coiled coils and their oligomerization states from sequence in the twilight zone. PLoS One 6:e23519 doi:10.1371/journal.pone.0023519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Tung FY, et al. 1992. The 9-kDa hydrophobic protein encoded at the 3′ end of the porcine transmissible gastroenteritis coronavirus genome is membrane-associated. Virology 186:676–683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. van der Hoek L, et al. 2004. Identification of a new human coronavirus. Nat. Med. 10:368–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Vijgen L, et al. 2005. Complete genomic sequence of human coronavirus OC43: molecular clock analysis suggests a relatively recent zoonotic coronavirus transmission event. J. Virol. 79:1595–1604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Vlasova AN, et al. 2011. Molecular characterization of a new species in the genus Alphacoronavirus associated with mink epizootic catarrhal gastroenteritis. J. Gen. Virol. 92:1369–1379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Woo PC, et al. 2005. Characterization and complete genome sequence of a novel coronavirus, coronavirus HKU1, from patients with pneumonia. J. Virol. 79:884–895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Woo PC, et al. 2006. Molecular diversity of coronaviruses in bats. Virology 351:180–187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Woo PC, et al. 2006. Comparative analysis of 22 coronavirus HKU1 genomes reveals a novel genotype and evidence of natural recombination in coronavirus HKU1. J. Virol. 80:7136–7145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Woo PC, Lau SK, Yuen KY. 2006. Infectious diseases emerging from Chinese wet-markets: zoonotic origins of severe respiratory viral infections. Curr. Opin. Infect. Dis. 19:401–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Woo PC, et al. 2007. Comparative analysis of 12 genomes of three novel group 2c and group 2d coronaviruses reveals unique group and subgroup features. J. Virol. 81:1574–1585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Woo PC, Lau SK, Huang Y, Yuen KY. 2009. Coronavirus diversity, phylogeny and interspecies jumping. Exp. Biol. Med. (Maywood) 234:1117–1127 [DOI] [PubMed] [Google Scholar]
- 72. Woo PC, et al. 2009. Comparative analysis of complete genome sequences of three avian coronaviruses reveals a novel group 3c coronavirus. J. Virol. 83:908–917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Woo PC, et al. 2012. Discovery of seven novel mammalian and avian coronaviruses in the genus Deltacoronavirus supports bat coronaviruses as the gene source of Alphacoronavirus and Betacoronavirus and avian coronaviruses as the gene source of Gammacoronavirus and Deltacoronavirus. J. Virol. 86:3995–4008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Wu K, Li W, Peng G, Li F. 2009. Crystal structure of NL63 respiratory coronavirus receptor-binding domain complexed with its human receptor. Proc. Natl. Acad. Sci. U. S. A. 106:19970–19974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Yeager CL, et al. 1992. Human aminopeptidase N is a receptor for human coronavirus 229E. Nature 357:420–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Yeh SH, et al. 2004. Characterization of severe acute respiratory syndrome coronavirus genomes in Taiwan: molecular epidemiology and genome evolution. Proc. Natl. Acad. Sci. U. S. A. 101:2542–2547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Yob JM, et al. 2001. Nipah virus infection in bats (order Chiroptera) in peninsular Malaysia. Emerg. Infect. Dis. 7:439–441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Zeng Q, Langereis MA, van Vliet AL, Huizinga EG, de Groot RJ. 2008. Structure of coronavirus hemagglutinin-esterase offers insight into corona and influenza virus evolution. Proc. Natl. Acad. Sci. U. S. A. 105:9065–9069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Ziebuhr J. 2004. Molecular biology of severe acute respiratory syndrome coronavirus. Curr. Opin. Microbiol. 7:412–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.