Abstract
Kaposi's sarcoma (KS)-associated herpesvirus (KSHV) is the causative agent of KS, an important AIDS-associated malignancy. KSHV expresses at least 18 different mature microRNAs (miRNAs). We identified interleukin-1 receptor (IL-1R)-associated kinase 1 (IRAK1) as a potential target of miR-K12-9 (miR-K9) in an array data set examining changes in cellular gene expression levels in the presence of KSHV miRNAs. Using 3′-untranslated region (3′UTR) luciferase reporter assays, we confirmed that miR-K9 and other miRNAs inhibit IRAK1 expression. In addition, IRAK1 expression is downregulated in cells transfected with miR-K9 and during de novo KSHV infection. IRAK1 is an important component of the Toll-like receptor (TLR)/IL-1R signaling cascade. The downregulation of IRAK1 by miR-K9 resulted in the decreased stimulation of NF-κB activity in endothelial cells treated with IL-1α and in B cells treated with a TLR7/8 agonist. Interestingly, miR-K9 had a greater effect on NF-κB activity than did a small interfering RNA (siRNA) targeting IRAK1 despite the more efficient downregulation of IRAK1 expression with the siRNA. We hypothesized that KSHV miRNAs may also be regulating a second component of the TLR/IL-1R signaling cascade, resulting in a stronger phenotype. Reanalysis of the array data set identified myeloid differentiation primary response protein 88 (MYD88) as an additional potential target. 3′UTR luciferase reporter assays and Western blot analysis confirmed the targeting of MYD88 by miR-K5. The presence of miR-K9 and miR-K5 inhibited the production of IL-6 and IL-8 upon the IL-1α stimulation of endothelial cells. These results demonstrate KSHV-encoded miRNAs regulating the TLR/IL-1R signaling cascade at two distinct points and suggest the importance of these pathways during viral infection.
INTRODUCTION
Kaposi's sarcoma (KS)-associated herpesvirus (KSHV) (human herpesvirus 8 [HHV-8]) is a member of the gammaherpesvirus family and is the causative agent of KS, primary effusion lymphoma (PEL), and multicentric Castleman's disease (MCD), all of which predominantly affect patients with HIV/AIDS (8, 31, 32). Like other herpesviruses, KSHV undergoes two stages of infection: lytic infection and latency. Most of the infected cells within KS and PEL are latently infected with KSHV, while a substantial percentage of infected MCD cells express lytic genes (35). Only a few viral proteins are expressed during latency, with functions associated primarily with the maintenance of the viral genome, cellular proliferation, and the activation of NF-κB and p38 mitogen-activated protein (MAP) kinase signaling cascades. In addition, 12 viral pre-microRNAs (pre-miRNAs) are expressed at various levels during latency, resulting in at least 18 mature miRNAs, as follows: five pre-miRNAs donate both strands of the duplex to the RNA-induced silencing complex (RISC) (miR-K12-3, -4, -6, -9, and -12), and one (miR-K12-10a) undergoes RNA editing, resulting in a single-base change in the seed sequence (miR-K12-10b) (37, 50, 54). The KSHV-encoded miRNAs were reported previously to regulate the expressions of some viral genes (4, 29) and a number of cellular targets (14), including BCLAF (55), TWEAKR (1), thrombospondin (39), BACH1 (16, 41), MAF (19), IκBα (27), Rbl2 (30), and p21 (15). Previously, Ziegelbauer et al. reported a collection of data from a series of microarrays examining changes in cellular gene expression in the presence of KSHV miRNAs under the following conditions: the transient transfection of B cells with KSHV pre-miRNAs, the stable transduction of B cells with clusters of pre-miRNAs, the de novo infection of endothelial cells, and the transient transfection of latently infected B cells with inhibitors of KSHV miRNAs (55). This array data set was used to identify BCLAF and TWEAKR as cellular targets of KSHV miRNAs (1, 55). Here, we again utilize this microarray data set to predict, validate, and provide functional data on two new cellular targets of KSHV miRNAs, interleukin-1 receptor (IL-1R)-associated kinase 1 (IRAK1) and myeloid differentiation primary response protein 88 (MYD88). These proteins are both components of the Toll-like receptor (TLR)/IL-1R signaling cascade.
IRAK1 is an important component of the TLR/IL-1R signaling cascade (reviewed in reference 13). There are four members of the IRAK family: IRAK1 and IRAK4 provide scaffolding and kinase activities important to TLR/IL-1R signaling, IRAK2 lacks kinase activity but has adapter functions redundant to those of IRAK1 in the context of IL-1R and most TLR-mediated signaling (excluding TLR7/9-mediated alpha interferon [IFN-α] production), and IRAK3 (IRAKM) is an inhibitor of signaling detected only in macrophages. The functions of IRAK1 and the TLR/IL-1R signaling cascades have been extensively reviewed (3, 13). The stimulation of the TLR/IL-1R signaling cascade leads to the recruitment of MYD88, the major adapter protein, and the subsequent recruitment of IRAK1 and IRAK4 via death domain interactions. The phosphorylation and activation of IRAK1 lead to the activation of MAP kinases and NF-κB, resulting primarily in the expression of proinflammatory cytokines, including IL-6, IL-8, tumor necrosis factor alpha (TNF-α), and IL-10. The knockdown or knockout of IRAK1 has been shown to reduce, but not eliminate, the production of proinflammatory cytokines upon stimulation with IL-1 and some TLR ligands (22, 42, 45, 48). However, upon TLR7/9 stimulation, IRAK1 was shown to be essential for the production of IFN-α in plasmacytoid dendritic cells (pDCs), while inflammatory-cytokine production is unaffected (49). In contrast, both MYD88 and IRAK4 knockouts result in the complete ablation of the TLR/IL-1R signaling cascade and subsequent cytokine production (2, 43, 44, 47). The regulation of IRAK1 and MYD88 results in similar but not identical effects on the TLR/IL-1R signaling cascade.
IL-1 is a proinflammatory-cytokine family with 11 members, including the prototype members IL-1α and IL-1β (40). While very similar in functional outcomes, IL-1α and IL-1β differ in their expression characteristics. IL-1β must be cleaved to be functional, is secreted, and is produced primarily by monocytes and macrophages. IL-1α does not require cleavage for function, remains associated with the membrane of the producing cell, and acts locally, and its expression is fairly ubiquitous, with endothelial cells, lymphocytes, and dying cells being the major sources of IL-1α. KS lesions have high mRNA levels of IL-1β, and cultured KS spindle cells express high levels of IL-1α, IL-1β, IL-6, and IL-8 (10), making this signaling pathway relevant for KSHV infections.
In this report, we validated IRAK1 as a target for KSHV miR-K12-9 (miR-K9), using 3′-untranslated region (3′UTR) luciferase reporter assays and Western blot analysis of cells transfected with miR-K9 mimics and cells undergoing de novo infection. We hypothesized that the main function of IRAK1 downregulation by KSHV miRNAs is the inhibition of NF-κB activation that is stimulated through TLR/IL-1R signaling and the subsequent reduction in proinflammatory-cytokine expression levels. We observed an inhibition of IL-1α- and TLR7/8-stimulated NF-κB activity mediated by miR-K9. A reanalysis of the array data set predicted that miR-K9 and miR-K5 also target MYD88. In the presence of miR-K9 and miR-K5, IL-1α-stimulated cells produced lower levels of IL-6 and IL-8. These results suggest that KSHV miRNAs provide a control mechanism for cytokine production upon stimulation with TLR agonists and IL-1α. This report demonstrates KSHV-encoded miRNAs regulating multiple components of the same signaling cascade, suggesting the importance of this pathway to viral infection.
MATERIALS AND METHODS
Cells and reagents.
293 cells and the KS-derived human endothelial cell line SLK (20) were maintained in Dulbecco's modified Eagle's medium (DMEM) containing 10% fetal bovine serum (FBS) and 1× penicillin and streptomycin (Pen Strep) glutamine solution (Gibco). BJAB (EBV-negative, KSHV-negative) and BCBL-1 (KSHV latently infected) human B cell lines were maintained in RPMI 1640 containing 10% FBS, 1× Pen Strep glutamine solution, and 55 μM β-mercaptoethanol. Primary human umbilical vein endothelial cells (HUVECs; Lonza) were maintained in EGM-2 (Lonza) for up to five passages. Synthetic KSHV miRNA mimics were obtained from Ambion. ON-TARGETplus SMARTpool small interfering RNAs (siRNAs) targeting IRAK1 and an ON-TARGETplus nontargeting pool were obtained from Dharmacon. IL-1α was purchased from Peprotech, and the R-848 TLR7/8 agonist was purchased from Santa Cruz Biotechnology. For the miRNA Western blot panel, HUVECs were seeded at 2 × 105 cells/well in a 6-well plate, transfected by using 1.5 μl/well DharmaFECT 1 reagent (Dharmacon) and 10 nM KSHV miRNA, and harvested at 48 h posttransfection (hpt).
3′UTR luciferase reporter assay.
293 cells were reverse transfected in 96-well plates by using 0.5 μl Lipofectamine 2000 (Invitrogen), 13 nM each KSHV miRNA (or a negative-control miRNA, neg2), and 0.5 ng/μl of the luciferase reporter plasmid, which expresses firefly luciferase driven by the herpes simplex virus (HSV) thymidine kinase (TK) promoter as an internal control and Renilla luciferase fused to the entire 3′UTR of IRAK1 or MYD88 driven by a simian virus 40 (SV40) promoter as the reporter (Protein Expression Laboratory, SAIC, Frederick, MD). A reporter plasmid lacking any 3′UTR adjacent to the Renilla luciferase served as a control for nonspecific responses to the KSHV miRNAs. Every miRNA was transfected with this empty vector control or the same vector containing a 3′UTR of interest. Site-directed mutagenesis was performed on the IRAK1 3′UTR reporter plasmid by using the QuikChange II kit (Stratagene) according to the manufacturer's instructions. The following primers and their reverse complements were used to introduce mutations (underlined) into the 3′UTR reporters: 5′-GTGGCAGAGGGGCCACTATTTAAGGTCTAGCTAGGCCC-3′ for IRAK1 mutant 1 (mut1), 5′-CATGCAGGGCGTCTGCAGTTTAGGCCTCTGGCAGCAG-3′ for IRAK1 mut2, and 5′-CGATATTTGCCATTCTTCCTATATCCTGGAATATATCTTGCTAAATGAGTTTATAATAATAAATAATATTCTACCTT-3′ for MYD88 mut1 (underlining indicates mutated bases compared to the wild type). Assays were performed by using the Dual-Luciferase reporter system (Promega) at 24 and 48 hpt. Each transfection was performed at least three independent times and was assayed in triplicate.
Inhibition of endogenous KSHV miRNAs.
Power locked nucleic acids (LNAs) against miR-K12-K5 or -K9 (Exiqon), at a total of 50 pmol (for miR-K5) or 100 pmol (for miR-K9), were electroporated into 2 × 106 BCBL-1 cells in Nucleofection solution V using program T-01, according to the manufacturer's instructions (Amaxa Inc.). Cells were harvested at 24 h postelectroporation, and total protein was extracted in radioimmunoprecipitation assay (RIPA) lysis buffer. Expression levels of the IRAK1 or MYD88 protein were quantitated relative to the actin expression level, as described below. Changes in protein levels upon LNA electroporation were compared to levels in BCBL-1 cells electroporated with the negative-control LNA (NegA).
NF-κB luciferase reporter assay.
SLK cells (1 × 104 cells/well) were reverse transfected in 96-well plates using 0.5 μl Lipofectamine 2000, 13 nM the indicated KSHV miRNAs (or a negative-control miRNA, neg2), 67.5 ng of the pGL4.32 NF-κB reporter plasmid (contains 5 copies of an NF-κB response element driving the transcription of firefly luciferase; Promega), and 7.5 ng of a pRL-null internal control plasmid (Renilla luciferase reporter containing no promoter or enhancer elements; Promega). At 64 hpt, cells were treated with 10 ng/ml IL-1α for 8 h and then harvested for analysis. Assays were performed by using the Dual-Luciferase reporter system; each transfection was performed at least three independent times and was assayed in triplicate. Three additional wells (untreated) were harvested in RIPA lysis buffer for analysis by Western blotting at 72 hpt.
BJAB cells were maintained for 3 days at a density of 2 × 105 cells/ml prior to electroporation. For each reaction, 3.5 × 106 cells were spun down, washed in serum-free RPMI 1640, and resuspended in 400 μl serum-free RPMI 1640. Cells were electroporated in 0.4-cm cuvettes with 100 nM miRNA or siRNA targeting IRAK1, 2 μg pGL4.32 NF-κB reporter plasmid, and 250 ng of the phRG-B internal control plasmid (Renilla luciferase reporter containing no promoter or enhancer elements; Promega) at 220 V, at 975 μF, and with the exponential wave function. Cells were then transferred into a 6-well plate containing 3 ml/well complete growth medium prewarmed to 37°C. At 24 hpt, cells were replated into a 24-well plate (3 × 105 cells/well) in RPMI 1640 containing 2% FBS and incubated for 1 h at 37°C. Cells were mock treated or treated with 0.5 μg/ml R-848 for 24 h and harvested for analysis by a luciferase assay. Assays were performed by using the Dual-Luciferase reporter system; each transfection was performed at least three independent times and was assayed in duplicate.
De novo KSHV infection.
KSHV was harvested from BCBL-1 cultures 6 days after induction with valproic acid and purified by centrifugation. SLK cells (at 50% confluence in DMEM containing 2% FBS) or HUVECs were infected with KSHV in medium containing 8 μg/ml Polybrene for 6 h at 37°C. Cells were washed twice with Dulbecco's phosphate-buffered saline (DPBS) and maintained in normal growth medium, and whole-cell lysates were prepared at 1, 2, and 3 days postinfection (dpi) (SLK cells) or at 7 dpi (HUVECs) for analysis by Western blotting.
Human tissue samples.
Six tissue samples were obtained from five patients with a confirmed history of HIV and KSHV coinfection who had consented to and were enrolled in an Institutional Review Board-approved protocol at the National Cancer Institute (study NCT00006518; principal investigator, R.Y.). These biopsy samples included three surgical lymph node biopsy specimens (one from a patient with MCD, one from a patient with PEL, and one from a patient with KS), one skin biopsy specimen of a KS skin lesion, and two lymph node samples (mediastinal and inguinal lymph nodes) obtained from a patient with PEL and advanced KS at autopsy. The two autopsy samples were evaluated for KSHV by an assessment of latency-associated nuclear antigen (LANA) by Western blotting, while the four surgical samples were analyzed for KSHV by LANA immunohistochemical (IHC) staining. Four of the samples showed evidence of LANA: one lymph node and one skin biopsy specimen showed LANA-positive spindle cell proliferations typical of Kaposi's sarcoma, and the two lymph nodes obtained at autopsy showed evidence of LANA by Western blotting. The other two lymph node biopsy specimens (one from a patient with MCD and one from a patient with PEL) had follicular hyperplasia without evidence of LANA by immunohistochemistry and were used as KSHV-negative controls. Other KSHV-negative controls studied included HUVEC primary cells (Lonza) and KSHV-uninfected commercial normal lymph node and skin samples (Abcam).
All research samples from the same specimens were homogenized and lysed in RIPA lysis buffer (Sigma). Soluble protein was analyzed by Western blotting, and MYD88 levels were normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) levels. Expression levels were normalized to levels in the normal lymph node sample (Abcam).
Western blot analysis.
Total cell protein was harvested from cell pellets by using RIPA lysis buffer (Sigma) supplemented with 1× Halt protease and phosphatase inhibitor cocktail (Thermo Scientific). Cells were lysed on ice for 10 min, and cell debris was removed by centrifugation at 13,000 rpm for 10 min. The Li-Cor Odyssey system was used for the detection and quantitation of protein bands. The following primary antibodies were used: mouse anti-IRAK1 (F-4; Santa Cruz Biotechnology), rabbit anti-MYD88 (catalog number D80F5; Cell Signaling), mouse anti-GAPDH (7B; Santa Cruz Biotechnology), and mouse anti-actin (AC-74, catalog number A5316; Sigma) antibodies. The following secondary antibodies conjugated to infrared (IR) fluorescing dyes were obtained from Li-Cor: goat anti-rabbit antibody IR800CW, goat anti-mouse antibody IR680, and goat anti-mouse antibody IR800CW. Results are presented as IRAK1 or MYD88 expression levels normalized to actin levels, relative to levels in mock-infected or negative-control miRNA-transfected cells.
RT-qPCR for IFN-α transcript levels and ELISA for IL-6 and IL-8 expression.
HUVECs were plated at 7 × 104 cells/well in a 12-well plate 1 day prior to transfection. Cells were transfected with 0.5 μl DharmaFECT1 and 2 nM KSHV miRNA or 10 nM siRNA targeting IRAK1. At 24 hpt, cells were mock treated or treated with 5 ng/ml IL-1α for 4 h. Supernatants were harvested and analyzed by using IL-6 and IL-8 enzyme-linked immunosorbent assays (ELISAs) (eBiosciences), according to manufacturer's instructions. Results are presented as the percent cytokine expression relative to that of the appropriate negative control. Cells were collected and split into two samples: one for the harvesting of total cell protein for Western blot analysis of IRAK1 and MYD88 expression and the other for the harvesting of total cell RNA for analysis of IFN-α transcript levels by quantitative reverse transcription-PCR (RT-qPCR). Total RNA was isolated from cell pellets by using the RNeasy microkit (Qiagen). cDNA was generated from 250 ng RNA in reverse transcription reactions using the SuperScript III first-strand synthesis kit (Invitrogen). PCRs were performed with a total volume of 25 μl using SYBR green PCR 2× master mix (Applied Biosystems), 500 nM each primer, and 5 ng cDNA template. The ABI Prism 7000 sequence detection system was used for amplification with the following PCR program: 2 min at 50°C, 10 min at 95°C, 40 cycles of denaturation at 95°C for 15 s, and annealing and extension at 60°C for 1 min. Results are presented as the change in IFN-α transcript levels normalized to β-actin transcript levels using the comparative threshold cycle (ΔΔCT) method, relative to the appropriate negative control (which was set to 1). The following primer sets were used: 5′-CTTCAACCTCTTTACCACAAAAGATTC-3′ and 5′-TGCTGGTAGAGTTCGGTGCA-3′ for IFN-α1 and 5′-GTGACATTAAGGAGAAGCTGTGCTA-3′ and 5′-CTTCATGATGGAGTTGAAGGTAGTT-3′ for β-actin.
Microarray data and rank sum rank analysis.
MIAME (minimum information about a microarray experiment)-compliant HUVEC array data can be found in the Princeton University MicroArray database (http://puma.princeton.edu/). Gene expression changes were ranked by comparing changes in the expression levels of all genes on one array. The most downregulated gene in the presence of an miRNA received a rank of 1. Calculated ranks for each gene from the appropriate array were added to compute the rank sum. The rank sum values were then ranked to generate the rank sum rank. The gene with the lowest rank sum rank (1) had the largest gene expression changes across all microarrays analyzed. This reflects the biological meaning that this was the most repressed gene in the presence a particular miRNA. The highest rank sum rank for this analysis was 10,870.
Statistical analysis.
Student's t test (unpaired and two tailed) was used to determine the statistical significance of data wherever indicated, and P values of less than 0.05 (denoted with asterisks) and less than 0.01 (denoted with double asterisks) were considered significant.
Microarray data accession numbers.
BJAB and BCBL-1 array data are available under Gene Expression Omnibus accession numbers GSE12967 and GSE40093, respectively.
RESULTS
We previously described an analysis of a microarray data set examining changes in cellular gene expression in response to KSHV miRNAs to make unbiased predictions of the cellular targets of KSHV miRNAs (1, 55). Using a rank sum rank method, we ranked genes based on their expression changes in the presence of KSHV miRNAs or when KSHV miRNAs were inhibited. The rank sum rank integrates microarray data from transient miRNA expression, stable miRNA expression, de novo KSHV infection, and the inhibition of miRNAs in infected cells. A low rank sum rank reflects suppression in the presence of a specific miRNA (see Materials and Methods). IRAK1 was selected as a predicted target of multiple miRNAs due to the low rank sum rank observed from the microarray data (Table 1) and potential miRNA target sites predicted by TargetScan (18) (Fig. 1A).
Table 1.
Summary of IRAK1 rank sum rank microarray data and 3′UTR luciferase reporter and Western blot results
| miRNA | Rank sum ranka | Luciferase at 48 hb | Western blot at 48 hb |
|---|---|---|---|
| Negative control | NA | 1.00 | 1.00 |
| K1 | 72 | 0.55 | 0.83 |
| K2 | 96 | 0.94 | 1.00 |
| K3 | 121 | 0.74 | 0.91 |
| K3* | 2,296 | 0.62 | 0.96 |
| K4-3 | 229 | 1.06 | 0.89 |
| K4-5 | 201 | 0.89 | 0.84 |
| K5 | 2,609 | 0.84 | 0.99 |
| K6-3 | 302 | 0.69 | 0.66 |
| K6-5 | 1,058 | 0.95 | 0.69 |
| K7 | 2,437 | 0.82 | 0.89 |
| K8 | 1,338 | 0.87 | 0.85 |
| K9 | 307 | 0.58 | 0.59 |
| K9* | 655 | 0.83 | 0.73 |
| K10a | 3,852 | 1.03 | 0.99 |
| K10b | 2,576 | 0.93 | 0.85 |
| K11 | 12 | 0.92 | 0.98 |
NA, not applicable.
Relative change.
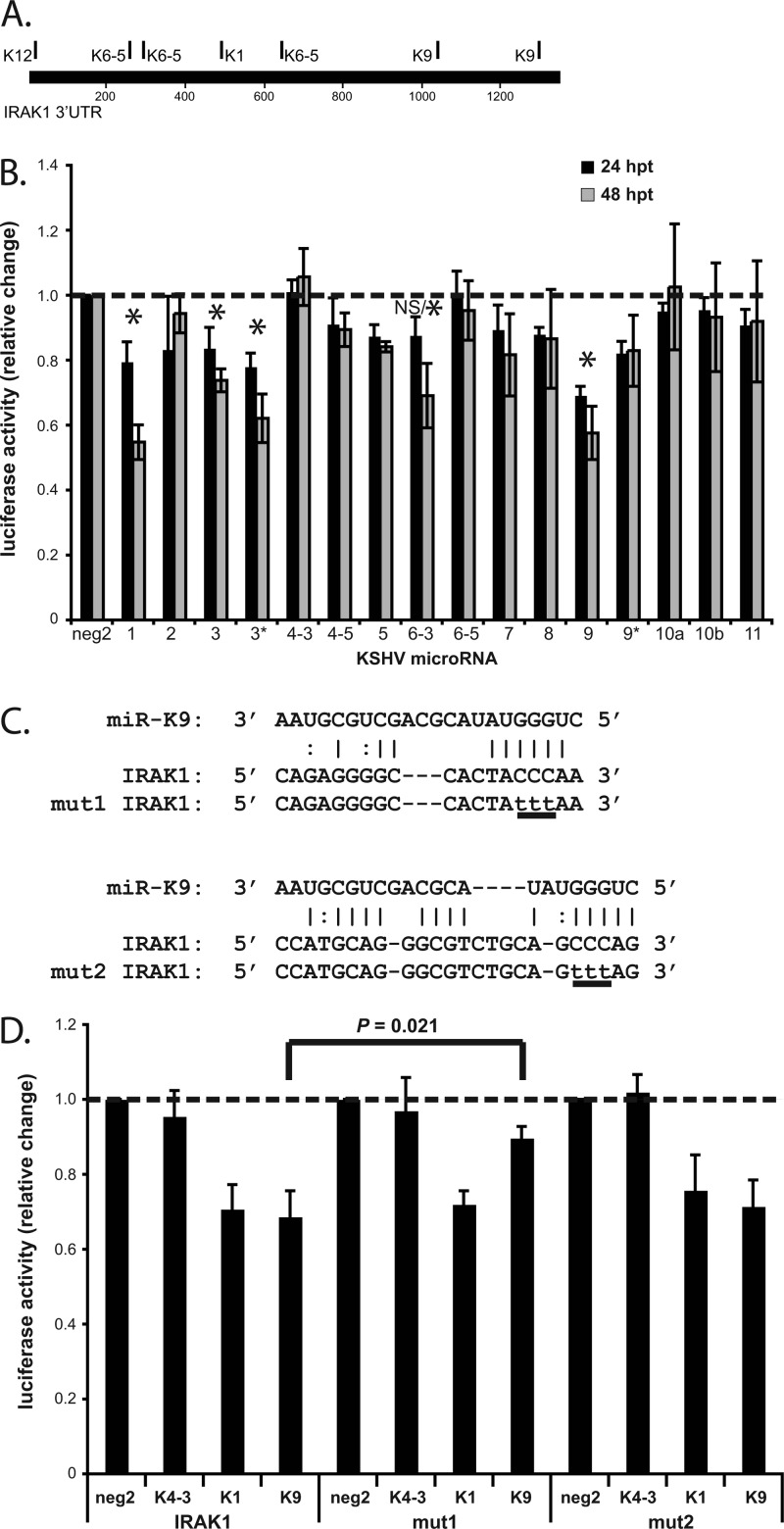
Fig 1.
miR-K9 targets the 3′UTR of IRAK1. (A) Map of the IRAK1 3′UTR with TargetScan-predicted hits for KSHV miRNAs. (B) 293 cells were transfected with each of the KSHV miRNAs and the reporter plasmid expressing Renilla luciferase fused to the 3′UTR of IRAK1. Lysates were analyzed by luciferase assays at 24 and 48 h posttransfection (hpt), and results are presented as the change in normalized relative light units relative to negative-control miRNA (neg2). Averages and standard deviations were calculated from three independent experiments. NS/* indicates that the change in activity was not significant at 24 hpt but was significant at 48 hpt. (C) The sequence of miR-K9 and its putative binding sites within the 3′UTR of IRAK1 are shown, based on predictions from miRanda and TargetScan. The same 3-bp mutation (underlined lowercase letters) was made at two different sites in the reporter plasmid containing the 3′UTR of IRAK1 (mut1 and mut2), to disrupt the binding of miR-K9. (D) Luciferase assays were performed at 24 hpt as described above for panel B, using only neg2, miR-K4-3, miR-K1, and miR-K9 and the reporter plasmid containing the wild-type 3′UTR (IRAK1) or the 3′UTR mutated as shown in panel C (mut1 and mut2). Averages and standard deviations were calculated from three independent experiments.
miR-K9 targets the 3′UTR of IRAK1.
To determine if virus-encoded miRNAs directly target IRAK1, we transfected cells with each of the KSHV miRNAs and a luciferase reporter plasmid containing the 3′UTR of IRAK1 (Fig. 1B). Luciferase activity was significantly downregulated in the presence of miR-K9 (42% downregulated at 48 hpt), miR-K1 (45% downregulated at 48 hpt), miR-K3 (26% downregulated at 48 hpt), and miR-K3* (38% downregulated at 48 hpt) relative to the activity in the presence of a negative-control miRNA. The effect with miR-K9 was the strongest and most consistent at both time points. We scanned the 3′UTR of IRAK1 for potential binding sites of miR-K9 using two different miRNA binding-site prediction programs, TargetScan (18) and miRanda (9). Using site-directed mutagenesis, we made mutations in the top site predicted by each program to disrupt miR-K9 binding in the context of the luciferase reporter plasmid (mut1, site predicted by TargetScan; mut2, site predicted by miRanda) (Fig. 1C). The reporter plasmid containing mut1 showed a significant restoration of luciferase activity in the presence of miR-K9, indicating that a valid, functional miR-K9 binding site had been disrupted (Fig. 1D). The reporter plasmid containing mut2 showed no difference in miR-K9-mediated inhibition from the wild-type reporter plasmid (IRAK1), suggesting that this site is not targeted by miR-K9. The mutations designed to disrupt miR-K9 binding did not affect the regulation of luciferase activity in the presence of miR-K4-3 or miR-K1. These data demonstrate the binding of miR-K9 to a specific site within the IRAK1 3′UTR.
miR-K9 downregulates the expression of IRAK1.
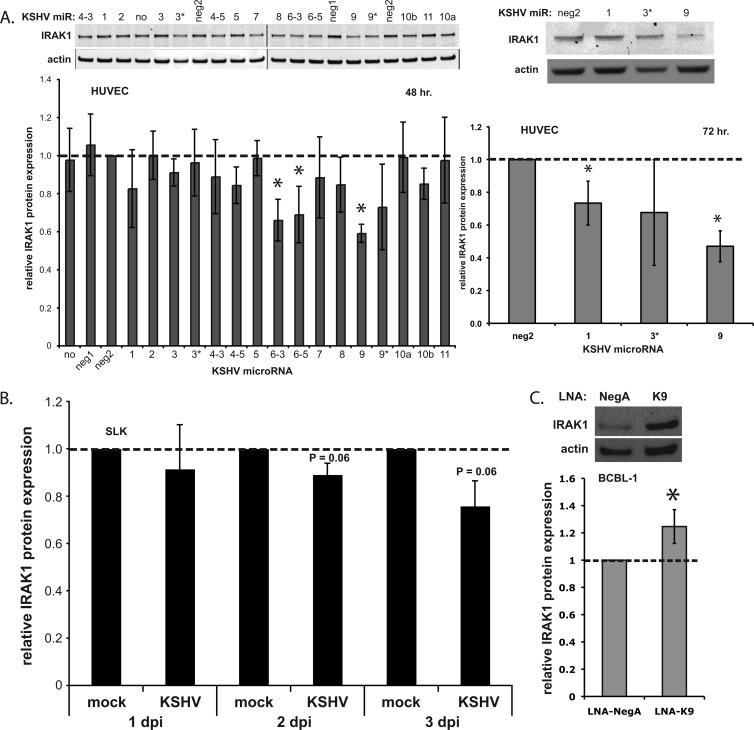
To examine the effect of KSHV miRNAs on IRAK1 protein expression, HUVECs were transfected with each of the miRNAs, and total cell lysates were analyzed by quantitative Western blotting. We observed that miR-K9, -K6-3, and -K6-5 significantly downregulated IRAK1 expression at 48 hpt (Fig. 2A). miR-K6-3 and miR-K6-5 failed to inhibit luciferase activity in the 3′UTR reporter assay (Fig. 1B), suggesting either that the binding sites for these miRNAs are located outside the 3′UTR included in the reporter or that the regulatory effects result indirectly from the downregulation of another cellular protein. miR-K9 demonstrated the most robust downregulation of IRAK1, with a stronger effect at 72 hpt (53% downregulated in HUVECs [Fig. 2A] and 63% downregulated in SLK cells [data not shown]). After the de novo infection of SLK cells, we observed a modest but consistent downregulation of IRAK1 (Fig. 2B). Similarly, we observed a significant repression of IRAK1 7 days after the de novo infection of HUVECs compared to mock-infected cells (see Fig. 5A). This result is important for showing the inhibition of IRAK1 under physiologically relevant conditions and correlates well with the level of downregulation at the mRNA level observed for the array data set (see Fig. S1 in the supplemental material). Finally, the inhibition of miR-K9 in infected BCBL-1 cells using LNAs correlated with an increase in the IRAK1 protein expression level (Fig. 2C). Together, these data validate IRAK1 as a target of KSHV miR-K9.
Fig 2.
miR-K9 downregulates the expression of IRAK1. (A) HUVECs were transfected with each of the KSHV miRNAs. Total cell lysates were harvested at 48 h (left) or 72 h (right) after transfection and analyzed by Western blotting. The top panels show representative images of IRAK1 and actin expression; the bottom panels show the average change in IRAK1/actin ratios relative to levels in negative-control miRNA-transfected cells (neg2). Averages and standard deviations were calculated from four independent experiments. “no” denotes no miRNA; “neg1” and “neg2” denote two different negative-control miRNAs. (B) SLK cells were infected with KSHV, and total cell lysates were harvested at 1, 2, and 3 dpi for analysis by Western blotting. Results are presented as the average changes in normalized IRAK1 expression levels relative to levels in mock-infected cells. (C) BCBL-1 cells were transfected with control LNAs (LNA-NegA) or LNAs targeting miR-K9 (LNA-K9) and analyzed by Western blotting. Results are presented as the average changes in normalized IRAK1 expression levels relative to levels in the control. Averages and standard deviations were calculated from three independent experiments. The dashed line indicates IRAK1 expression in the neg2 control.
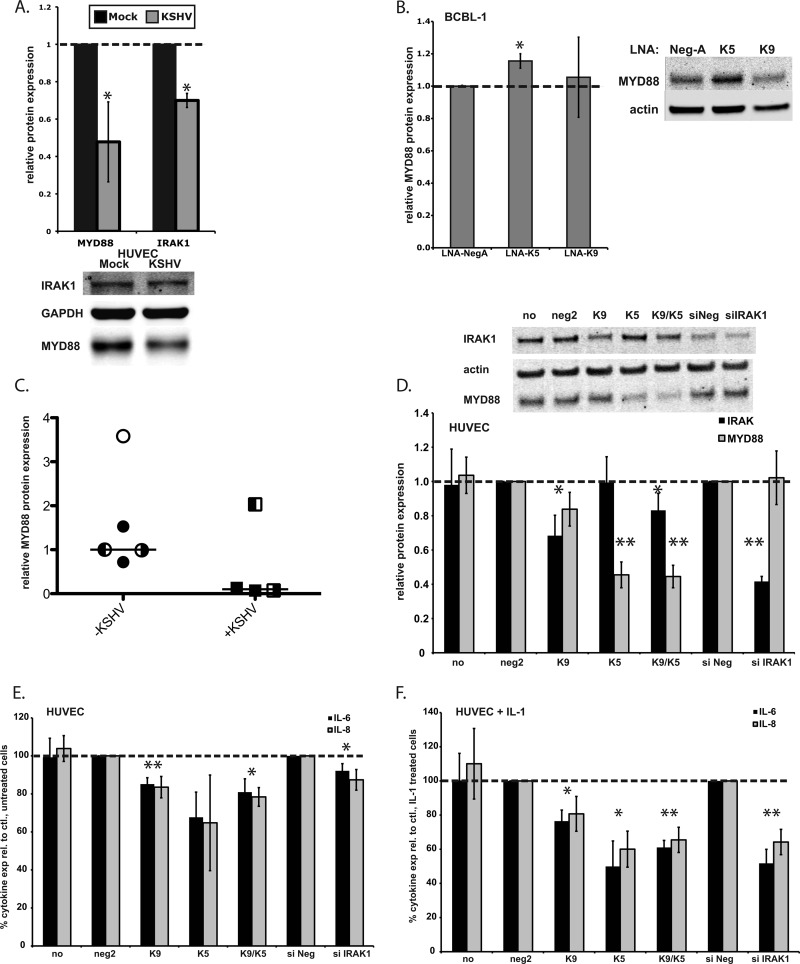
Fig 5.
KSHV infection represses MYD88 and IRAK1 expressions. miR-K9 and miR-K5 dampen the production of proinflammatory cytokines in response to IL-1α stimulation. (A) HUVECs were infected with KSHV and harvested after 7 days. The relative changes in IRAK1 and MYD88 protein expression levels normalized to GAPDH levels are shown compared to mock-infected cells from three independent experiments. The bottom panel shows a representative Western blot. (B) KSHV-infected BCBL-1 cells were transfected with control LNA miRNA inhibitors (LNA-NegA) or inhibitors of miR-K5 or miR-K9. The relative change in the protein expression level normalized to the actin level was calculated from four independent experiments. (C) Relative levels of the MYD88 protein normalized to GAPDH levels were calculated for KSHV-negative samples (open circle, HUVECs; filled circles, follicular hyperplasia of lymph node; left-filled circle, normal lymph node; right-filled circle, normal skin) or KSHV-positive samples (filled squares, PEL/KS of lymph node; left-filled square, KS of lymph node; right-filled square, KS of skin). Horizontal lines indicate median values. (D) HUVECs were transfected with miR-K9, miR-K5, or siRNA targeting IRAK1, and total cell lysates were harvested at 28 hpt and analyzed by Western blotting. Shown are the average changes in normalized IRAK1 or MYD88 expression levels relative to levels in neg2-transfected cells. Averages and standard deviations were calculated from at least three independent experiments. “no” denotes no miRNA; “neg2” denotes a negative-control miRNA. (E and F) HUVECs were transfected for 24 h, as described above for panel D. Cells were treated with IL-1α for 4 h, and culture supernatants were harvested 28 h after transfection and analyzed for IL-6 and IL-8 expression levels in unstimulated (E) or IL-1α-stimulated (F) cells. Results are presented as percent cytokine expression relative to the appropriate negative control. Averages and standard deviations were calculated from at least three independent experiments.
miR-K9-mediated downregulation of IRAK1 inhibits IL-1α- and TLR7/8 agonist-induced NF-κB activation.
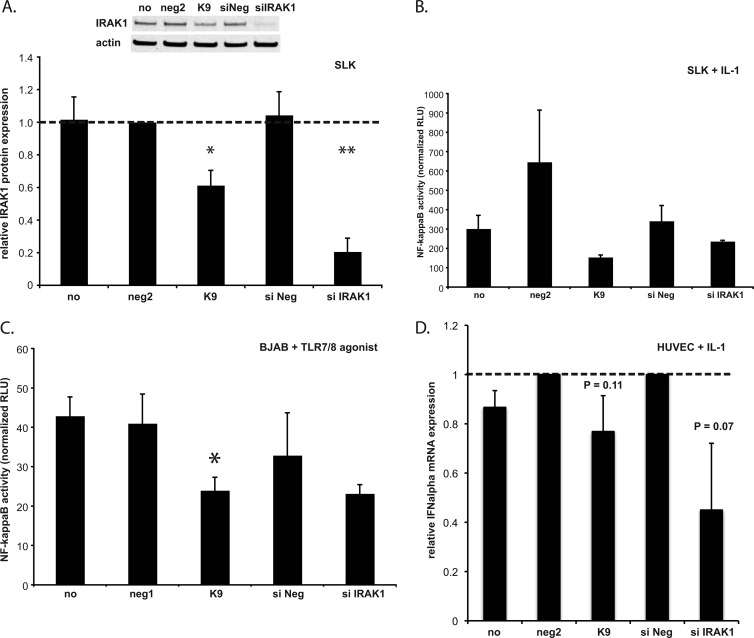
IRAK1 is an important component of the TLR/IL-1R signaling cascade, leading to NF-κB activation and the production of proinflammatory cytokines. We hypothesized that KSHV might benefit from the miRNA-mediated inhibition of IRAK1 by blocking this signaling cascade and dampening cytokine production and immune activation induced by external stimuli to the infected cell. SLK cells were used to study NF-κB activation in the context of IL-1α stimulation because they express both IL-1α and IL-1R and also because endothelial cells are relevant for KSHV infection in the context of KS. Transfection with miR-K9 or siRNA targeting IRAK1 resulted in nearly 40% and 80% reductions in IRAK1 expression levels, respectively (Fig. 3A). We then determined NF-κB activity following IL-1α treatment. The treatment of cells with IL-1α stimulated a 25- to 60-fold increase in NF-κB activation, as measured by luciferase activity (data not shown). In the presence of miR-K9, however, NF-κB activation was approximately 4-fold lower than in cells transfected with negative-control miRNA (Fig. 3B). We also saw a 50% reduction in basal NF-κB activation in the presence of miR-K9 in unstimulated cells (data not shown). siRNA targeting IRAK1 resulted in less than a 2-fold inhibition of NF-κB activation, which was surprising considering the dramatic reduction in the IRAK1 expression level (Fig. 3A). This observation suggested that miR-K9 might be regulating the expression of a second component of the TLR/IL-1R signaling cascade, resulting in a more dramatic phenotype than that with siRNA, which targets only IRAK1.
Fig 3.
miR-K9-mediated downregulation of IRAK1 inhibits IL-1α- and TLR agonist-induced NF-κB activation and IFN-α expression. (A) SLK cells were transfected with either miR-K9 or siRNAs targeting IRAK1, and total cell lysates were harvested at 72 hpt and analyzed by Western blotting. Results are presented as the average changes in normalized IRAK1 expression levels relative to levels in neg2-transfected SLK cells. Averages and standard deviations were calculated from three independent experiments. “no” denotes no miRNA; “neg2” denotes a negative-control miRNA; “siNeg” denotes a negative-control siRNA. (B) SLK cells were cotransfected with the NF-κB luciferase reporter plasmid and the indicated small RNA, treated with IL-1α for 8 h, and assayed for luciferase activity as a measurement of NF-κB activity at 72 hpt. Results are presented as the relative level of NF-κB activity in IL-1α-treated cells (in relative light units [RLU] normalized to the internal control). Averages and standard deviations were calculated from three independent experiments. (C) BJAB cells were transfected as described in Materials and Methods, treated with a TLR7/8 agonist for 24 h, and assayed for luciferase activity as a measurement of NF-κB activation. Results are presented as the relative activity in agonist-treated cells (in relative light units normalized to the internal control). Averages and standard deviations were calculated from three independent experiments. “neg1” denotes an additional negative-control miRNA (similar to neg2). (D) HUVECs were transfected with miR-K9 and treated with IL-1α as described in the legend of Fig. 5. RNA was harvested, and RT-qPCR was performed to measure transcript levels. Results show expression changes relative to negative-control miRNA (neg2) or control siRNA (siNeg) levels. Averages and standard deviations were calculated from three independent experiments.
In the context of viral infections, TLR3, -7, -8, and -9 are established as the predominant TLRs for viral recognition (23) and have been shown to play a role in KSHV recognition and reactivation (17, 52). However, the TLR3 signaling cascade does not involve IRAK1 as a major component, and TLR7, -8, and -9 are not expressed on endothelial cells. Therefore, we used KSHV-negative, Epstein-Barr virus (EBV)-negative B cells (BJAB cells) transfected with miR-K9 or siRNA targeting IRAK1 to demonstrate an effect on TLR-stimulated signaling in the NF-κB luciferase reporter assay. The transfection efficiency using the electroporation of BJAB cells was only 30 to 40%, as determined by using a green fluorescent protein (GFP) expression plasmid (data not shown). In the luciferase assays, only transfected cells will contribute to the results, making the phenotype driven by miR-K9 readily observable. Cells treated with a TLR7/8 agonist showed a significant reduction in NF-κB activity in the presence of miR-K9, similar to that mediated by siRNA targeting IRAK1 (Fig. 3C). These data demonstrate that the KSHV miR-K9-mediated downregulation of IRAK1 results in a functional inhibition of NF-κB activity stimulated by two different ligands (IL-1α and TLR7/8 agonist) in two different, relevant cell types for KSHV infection (endothelial cells and B cells). Furthermore, the IL-1α stimulation of alpha interferon mRNA was modestly decreased by miR-K9 and siRNAs targeting IRAK1 (Fig. 3D). Taken together, the repression of IRAK1 by miR-K9 inhibits the expression of known downstream components of TLR and IL-1α signaling.
miR-K9, miR-K5, and components of the TLR/IL-1R signaling cascade.
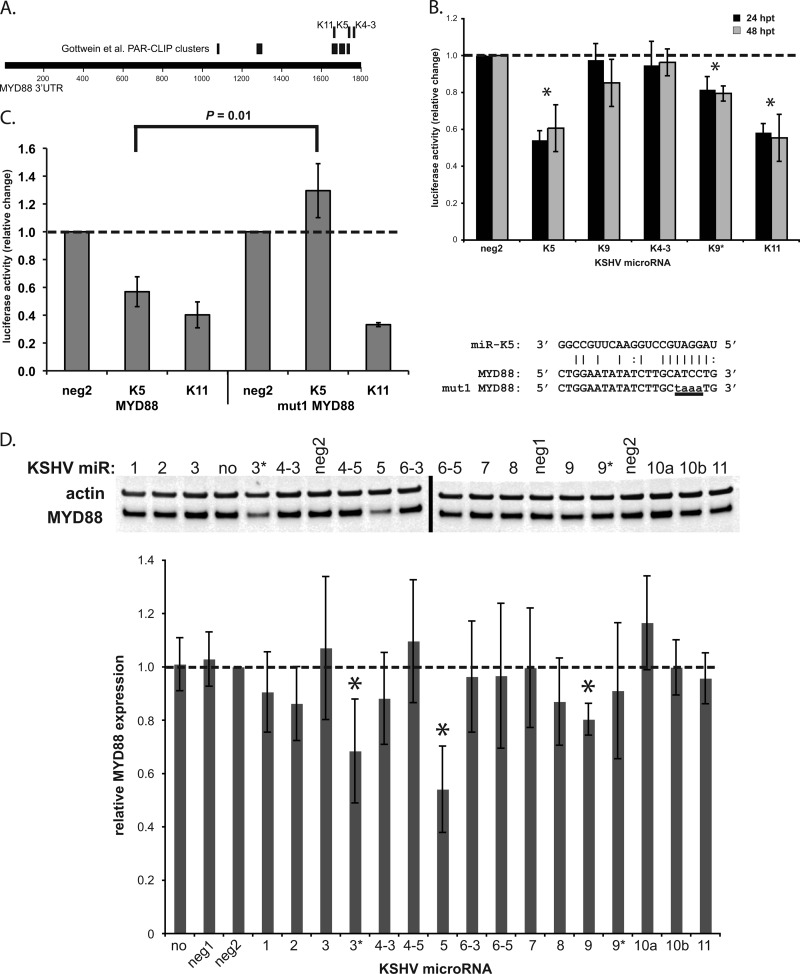
In Fig. 3B, we observed that miR-K9 inhibited IL-1α-stimulated NF-κB activity more strongly than did siRNA targeting IRAK1, which more robustly downregulated IRAK1 expression. This finding suggests that miR-K9 may target more than one component of the TLR/IL-1R signaling cascade. To explore this possibility, we reanalyzed the microarray data set for other high-probability targets of miR-K9 that are involved in this pathway. Interestingly, MYD88, a well-studied adapter protein critical for TLR/IL-1R signaling, displayed a low rank sum rank for miR-K9 (relative to those of other miRNAs) (Table 2), contained previously described argonaute-associated sequences (PAR-CLIP clusters) (14), and contained several TargetScan-predicted miRNA binding sites, suggesting that MYD88 may also be targeted by miR-K9 and other KSHV miRNAs (Fig. 4A). Several KSHV miRNAs were examined for their ability to target MYD88 by use of the 3′UTR luciferase reporter assay (Fig. 4B). Luciferase activity was significantly downregulated in the presence of miR-K5 (39% downregulated at 48 hpt), miR-K9* (20% downregulated at 48 hpt), and miR-K11 (44% downregulated at 48 hpt) relative to that in the presence of negative-control miRNA. The mutation of the predicted miR-K5 site (within the PAR-CLIP cluster region) completely abolished the repression of the MYD88 3′UTR by miR-K5, while the mutant was still repressed by miR-K11 (Fig. 4C). Interestingly, miR-K9 did not appear to target the 3′UTR of MYD88 (Fig. 4B). In transiently transfected HUVECs, however, we found that miR-K9 mediated a modest downregulation of MYD88 at 48 hpt (20%) (Fig. 4D). In addition, miR-K5 and miR-K3* had a robust inhibitory effect on MYD88 protein expression, resulting in 45% and 31% downregulation, respectively (Fig. 4D). After de novo infection of HUVECs, we observed a repression of MYD88 protein expression (47% downregulated) (Fig. 5A) compared to the expression level in mock-infected cells. We also observed an increase in MYD88 protein expression levels when BCBL-1 cells were treated with LNA miRNA inhibitors targeting miR-K5 (Fig. 5B). LNAs targeting miR-K9 did not show a significant change in MYD88 protein expression. Finally, we observed a decrease in MYD88 protein expression levels in three out of four KSHV-infected clinical samples (two lymph node samples from a patient with KS and PEL and one skin biopsy specimen from a KS lesion), whereas none of the five KSHV-negative samples tested (primary endothelial cells, normal skin, normal lymph node, and lymph node with follicular hyperplasia) had substantially decreased levels (Fig. 5C). Together, these data confirm that miR-K5 and miR-K9 downregulate the expressions of two components of the TLR/IL-1R signaling cascade. However, the modest repression of the MYD88 protein by miR-K9 is unlikely to explain our observations in the NF-κB activity assay (Fig. 3B). The redundancy of KSHV miRNAs targeting IRAK1 and MYD88 suggests the importance of regulating these components during viral infection.
Table 2.
Summary of MYD88 rank sum rank microarray data and 3′UTR luciferase reporter and Western blot resultsa
| miRNA | Rank sum rank | Luciferase at 48 hb | Western blot at 48 hb |
|---|---|---|---|
| Negative control | NA | 1.00 | 1.00 |
| K1 | 9,550 | ND | 0.91 |
| K2 | 8,612 | ND | 0.86 |
| K3 | 9,710 | ND | 1.07 |
| K3* | 3,958 | ND | 0.69 |
| K4-3 | 6,881 | 0.96 | 0.88 |
| K4-5 | 7,076 | ND | 1.10 |
| K5 | 9,926 | 0.61 | 0.54 |
| K6-3 | 7,296 | ND | 0.96 |
| K6-5 | 8,933 | ND | 0.97 |
| K7 | 6,460 | ND | 1.00 |
| K8 | 2,851 | ND | 0.87 |
| K9 | 3,077 | 0.85 | 0.80 |
| K9* | 7,100 | 0.79 | 0.91 |
| K10a | 7,296 | ND | 1.17 |
| K10b | 6,096 | ND | 1.00 |
| K11 | 5,168 | 0.55 | 0.96 |
NA, not applicable; ND, not determined.
Relative change.
Fig 4.
MYD88, a second component of the TLR/IL-1R signaling cascade, is targeted by KSHV miRNAs. (A) Map of the MYD88 3′UTR with TargetScan-predicted hits for KSHV miRNAs (top) and PAR-CLIP clusters (miRNA/argonaute-associated sequences) described previously (14) (middle). (B) 293 cells were transfected with the indicated KSHV miRNAs and the reporter plasmid expressing Renilla luciferase fused to the 3′UTR of MYD88. Lysates were analyzed by a luciferase assay at 24 and 48 h posttransfection (hpt), and results are presented as the changes in normalized relative light units relative to the negative-control miRNA (neg2). Averages and standard deviations were calculated from three independent experiments. (C) Luciferase assays were performed at 24 hpt as in panel B with the reporter plasmid containing the wild-type 3′UTR (MYD88) or the mutated 3′UTR (mut1, as shown). Averages and standard deviations were calculated from three independent experiments. (D) HUVECs were transfected with each of the KSHV miRNAs. Total cell lysates were harvested at 48 hpt and analyzed by Western blotting. Top panels show representative images of MYD88 and actin expression levels; the bottom panel shows the average change in the normalized MYD88/actin expression level relative to levels in negative-control miRNA-transfected cells (neg2). Averages and standard deviations were calculated from four independent experiments. The dashed line indicates MYD88 expression in the neg2 control. “no” denotes no miRNA; “neg1” and “neg2” denote two different negative-control miRNAs.
miR-K9 and miR-K5 dampen production of proinflammatory cytokines in response to IL-1α stimulation.
The primary result of signaling through IL-1R is the production of proinflammatory cytokines, especially IL-6 and IL-8 in endothelial cells (40). We hypothesized that the miR-K9- and miR-K5-mediated knockdown of IRAK1 and MYD88 would inhibit the IL-1α-driven expression of these cytokines, blocking the proinflammatory effects of IL-1R signaling. HUVECs transfected with miR-K9, miR-K5, or a mix of both miRNAs showed a consistent reduction in IRAK1 and MYD88 expression levels at 28 h posttransfection (hpt) (Fig. 5D). At 24 hpt, miRNA-transfected HUVECs were treated with IL-1α for 4 h, and total cell RNA and culture supernatants were harvested for analyses of IL-6 and IL-8 mRNA and protein expression levels. RT-qPCR analysis revealed significant decreases in the levels of both IL-6 and IL-8 mRNAs in cells transfected with the combination of miR-K9 and miR-K5 (miR-K9/K5) upon stimulation with IL-1α (data not shown). The effect on cells transfected with miR-K5 alone was similar for IL-6 mRNA and less robust for IL-8 mRNA. miR-K9 alone had no effect on cytokine mRNA levels, possibly due to the relatively weak downregulation of both target proteins at this early time point. siRNA targeting IRAK1 had a stronger effect than the viral miRNAs on IL-6 mRNA and a similar effect on IL-8 mRNA. In the absence of IL-1α stimulation, IL-8 mRNA levels were reduced by 60% in the presence of miR-K5 or miR-K9/K5, showing that MYD88 regulation also affects the basal expression of IL-8 (data not shown). We observed a similar but more variable reduction in IL-6 and IL-8 mRNA levels in SLK cells transfected with miR-K9 for 68 h followed by IL-1α stimulation for 4 h (data not shown). These results confirm the findings for a second cell type and suggest that a longer exposure to miR-K9 could produce a more prominent phenotype on cytokine mRNA expression.
Culture supernatants were analyzed for the presence of secreted IL-6 and IL-8 by ELISAs. In unstimulated cells, the presence of miR-K9, miR-K5, miR-K9/K5, or siRNA targeting IRAK1 resulted in a similar 10 to 20% reduction of IL-6 and IL-8 levels (Fig. 5E). These data indicate that signaling occurs at low levels in the absence of stimuli and that the downregulation of IRAK1 and MYD88 using KSHV miRNAs can blunt the resulting cytokine expression. Upon stimulation with IL-1α, the presence of miR-K9/K5 resulted in a 40% reduction in IL-6 and IL-8 expression levels, similar to that observed in the presence of siRNA targeting IRAK1 (Fig. 5F). This correlates well with both the cytokine mRNA levels and the relative inhibition of IRAK1 and MYD88 expressions. These data demonstrate that a functional outcome of the miR-K9- and miR-K5-mediated downregulation of IRAK1 and MYD88 is a reduction of IL-1α-stimulated proinflammatory-cytokine production.
DISCUSSION
In this report, we have characterized two new cellular targets of KSHV-encoded miRNAs, IRAK1 and MYD88, both of which are involved in the TLR/IL-1R signaling cascade. It is important to note that each target prediction method (microarrays, seed matching, and others) and validation method (luciferase assays and quantitative Western blotting) have various limitations. For example, the genes repressed in the microarray analysis could be direct miRNA targets or indirect targets. Since each assay measures gene expression differently, we cannot expect a perfect correlation between various assays (Tables 1 and 2). Our analysis sought to use a variety of methods and focus on miRNAs and targets which showed the best correlation across multiple target prediction and validation assays. We showed the targeting of IRAK1 by miR-K9 and MYD88 by miR-K5 in 3′UTR luciferase reporter assays, by assessments of protein expression after the transfection of miRNAs or de novo KSHV infections, and by assessments of the alleviation of regulation in latently infected B cells in the presence of LNA miRNA inhibitors. MYD88 was also shown to be downregulated in three out of four tested KSHV-infected clinical samples, highlighting the relevance of this target. Using an NF-κB reporter assay, we observed that miR-K9 inhibits NF-κB activation in endothelial cells stimulated with IL-1α and in B cells stimulated with a TLR7/8 agonist. The transfection of endothelial cells with miR-K9 and miR-K5 resulted in the downregulation of IL-6 and IL-8 mRNAs and protein expression stimulated by IL-1α. Overall, we have validated IRAK1 and MYD88 as two new targets of KSHV miRNAs and demonstrated an inhibition of proinflammatory-cytokine expression as a functional consequence of this regulation. We do not propose that these KSHV miRNAs function to repress NF-κB, IL-6, and IL-8 expression in all contexts, since NF-κB activity can be beneficial to the virus in KSHV-infected lymphomas (25). Instead, we suggest that they function to suppress activation stimulated by a host immune response and external stimuli like IL-1α.
IRAK1 expression is regulated by a cellular miRNA, miR-146a, which also targets TRAF6 and IRAK2 (21, 46). The expression of miR-146a is induced by NF-κB activation mediated by IRAK1, leading to a negative-feedback loop during TLR/IL-1R stimulation. This mechanism likely prevents the overstimulation and overproduction of proinflammatory cytokines (36). KSHV and EBV are both members of the gammaherpesvirus family yet share no homologous miRNAs (51). EBV LMP1 induces the expression of miR-146a by the activation of NF-κB in infected cells, resulting in the downregulation of IRAK1 and the inhibition of genes involved in the interferon response (6, 33). miR-146a expression was not detected in B cells latently infected with KSHV but was highly expressed in B cells latently infected with EBV; the same has been shown for the cellular microRNA miR-155, for which KSHV encodes a homolog, miR-K12-11 (16, 33, 41). Thus, while EBV modulates the expression of NF-κB-induced cytokines by acting on cellular miRNAs (miR-146a and miR-155), KSHV encodes its own miRNAs that regulate the same cellular targets. miR-146a was, however, upregulated during the early stages of KSHV infection in lymphatic endothelial cells (26) and during infection of HUVECs (38), suggesting differences in this signaling cascade between different cell types. Overall, these reports show that the regulation of IRAK1 and the TLR/IL-1R signaling cascade is important for gammaherpesvirus infections.
In this report, we have demonstrated the KSHV miRNA-mediated regulation of IL-1α- and TLR7/8-mediated signaling (Fig. 3 and 5). As mentioned previously, IRAK1 is essential for TLR7/9-stimulated IFN-α production, an antiviral response which occurs primarily in plasmacytoid dendritic cells (pDCs) and also requires MYD88 (24, 49). KSHV was recently shown to infect pDCs, resulting in early activation and IFN-α secretion, with IFN-α levels reaching a plateau or tapering off at around 72 h postinfection (53). Thus, KSHV infection of pDCs is relevant, and the expression of miR-K9 and miR-K5 at later stages of infection may play a role in turning off the antiviral interferon response.
The role of the miR-K9- and miR-K5-mediated targeting of IRAK1 and MYD88 during KSHV infection may extend beyond the inhibition of the immune response. Several previous reports suggested that the reactivation of KSHV from latency to lytic infection is controlled by TLR signaling. Agonists of TLR7/8 induced the reactivation of latent KSHV in B cells, leading to viral lytic gene transcription and replication, while other TLR agonists had no effect (17). However, previously reported experiments in an epithelial cell line with a KSHV miRNA deletion mutant suggested that increased NF-κB activity stimulated by miRNAs suppresses lytic reactivation (27). This is the opposite effect of NF-κB activity on lytic reactivation compared to the effects of TLR7/8 agonists (which activate NF-κB). Similar to KSHV infection in B cells, murine gammaherpesvirus 68 (MHV68) can be reactivated by signaling through TLR3, -4, -5, and -9, and the establishment of MHV68 latency requires MYD88 (11, 12). Thus, the KSHV-mediated downregulation of IRAK1 and MYD88 may blunt TLR7/8 signaling to prevent KSHV reactivation, providing an additional means of controlling the latent-lytic switch. KSHV has been shown to upregulate and activate TLR3 signaling, leading to the expression of CXCL10 and IFN-β during infections of monocytes by KSHV (52). Interestingly, TLR3 signals primarily in a MYD88-independent manner (3) and therefore would not be regulated by the mechanisms described in this report. Finally, IL-1R-mediated signaling plays a role in establishing and maintaining senescence through the senescence-associated secretory phenotype (SASP) (7). The SASP involves primarily the expression of IL-6 and IL-8 stimulated by IL-1α and is negatively regulated by miR-146a by inhibiting IRAK1 expression (5, 34). A recent paper showed that the KSHV latent protein v-cyclin promotes senescence in cells and that this pathway is blocked by viral FLICE inhibitory protein (v-FLIP) (28). v-FLIP was previously shown to upregulate miR-146a upon ectopic expression and during KSHV infection of HUVECs (38). miR-K9 and miR-K5 may also participate in this coordinated regulation of senescence during latency by reducing levels of cytokines involved in the SASP.
Our current study demonstrates that KSHV-encoded miRNAs regulate the TLR/IL-1R signaling cascade by targeting two components of this pathway. Similarly, EBV upregulates a cellular miRNA that also regulates multiple components of the TLR/IL-1R pathway, suggesting the importance of this signaling cascade for herpesvirus infections. The downregulation of proinflammatory-cytokine expression is demonstrated here as a functional consequence of miRNA targeting; however, future reports may reveal additional functions in controlling viral reactivation and cellular senescence. These studies will lead to a greater understanding of KSHV biology and pathogenesis.
Supplementary Material
ACKNOWLEDGMENTS
We thank members of the Ziegelbauer laboratory for help, support, and critical review of the manuscript. We also thank Mark Polizzotto, Kathleen Wyvill, Karen Aleman, Stefanie Pittaluga, and other members of the HIV and AIDS Malignancy Branch and Medical Oncology Branch for clinical services.
This work was supported by the Intramural Research Program of the Center for Cancer Research, National Cancer Institute, National Institutes of Health. J.R.A. was supported by an intramural AIDS research fellowship.
Footnotes
Published ahead of print 15 August 2012
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1. Abend JR, Uldrick T, Ziegelbauer JM. 2010. Regulation of tumor necrosis factor-like weak inducer of apoptosis receptor protein (TWEAKR) expression by Kaposi's sarcoma-associated herpesvirus microRNA prevents TWEAK-induced apoptosis and inflammatory cytokine expression. J. Virol. 84:12139–12151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Adachi O, et al. 1998. Targeted disruption of the MyD88 gene results in loss of IL-1- and IL-18-mediated function. Immunity 9:143–150 [DOI] [PubMed] [Google Scholar]
- 3. Akira S, Takeda K. 2004. Toll-like receptor signalling. Nat. Rev. Immunol. 4:499–511 [DOI] [PubMed] [Google Scholar]
- 4. Bellare P, Ganem D. 2009. Regulation of KSHV lytic switch protein expression by a virus-encoded microRNA: an evolutionary adaptation that fine-tunes lytic reactivation. Cell Host Microbe 6:570–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bhaumik D, et al. 2009. MicroRNAs miR-146a/b negatively modulate the senescence-associated inflammatory mediators IL-6 and IL-8. Aging (Albany NY) 1:402–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cameron JE, et al. 2008. Epstein-Barr virus latent membrane protein 1 induces cellular microRNA miR-146a, a modulator of lymphocyte signaling pathways. J. Virol. 82:1946–1958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Coppe JP, Desprez PY, Krtolica A, Campisi J. 2010. The senescence-associated secretory phenotype: the dark side of tumor suppression. Annu. Rev. Pathol. 5:99–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Engels EA, et al. 2008. Cancer risk in people infected with human immunodeficiency virus in the United States. Int. J. Cancer 123:187–194 [DOI] [PubMed] [Google Scholar]
- 9. Enright AJ, et al. 2003. MicroRNA targets in Drosophila. Genome Biol. 5:R1 doi:10.1186/gb-2003-5-1-r1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ensoli B, Sturzl M. 1998. Kaposi's sarcoma: a result of the interplay among inflammatory cytokines, angiogenic factors and viral agents. Cytokine Growth Factor Rev. 9:63–83 [DOI] [PubMed] [Google Scholar]
- 11. Gargano LM, Forrest JC, Speck SH. 2009. Signaling through Toll-like receptors induces murine gammaherpesvirus 68 reactivation in vivo. J. Virol. 83:1474–1482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gargano LM, Moser JM, Speck SH. 2008. Role for MyD88 signaling in murine gammaherpesvirus 68 latency. J. Virol. 82:3853–3863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gottipati S, Rao NL, Fung-Leung WP. 2008. IRAK1: a critical signaling mediator of innate immunity. Cell. Signal. 20:269–276 [DOI] [PubMed] [Google Scholar]
- 14. Gottwein E, et al. 2011. Viral microRNA targetome of KSHV-infected primary effusion lymphoma cell lines. Cell Host Microbe 10:515–526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gottwein E, Cullen BR. 2010. A human herpesvirus microRNA inhibits p21 expression and attenuates p21-mediated cell cycle arrest. J. Virol. 84:5229–5237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gottwein E, et al. 2007. A viral microRNA functions as an orthologue of cellular miR-155. Nature 450:1096–1099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gregory SM, et al. 2009. Toll-like receptor signaling controls reactivation of KSHV from latency. Proc. Natl. Acad. Sci. U. S. A. 106:11725–11730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Grimson A, et al. 2007. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol. Cell 27:91–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hansen A, et al. 2010. KSHV-encoded miRNAs target MAF to induce endothelial cell reprogramming. Genes Dev. 24:195–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Herndier BG, et al. 1994. Characterization of a human Kaposi's sarcoma cell line that induces angiogenic tumors in animals. AIDS 8:575–581 [DOI] [PubMed] [Google Scholar]
- 21. Hou J, et al. 2009. MicroRNA-146a feedback inhibits RIG-I-dependent type I IFN production in macrophages by targeting TRAF6, IRAK1, and IRAK2. J. Immunol. 183:2150–2158 [DOI] [PubMed] [Google Scholar]
- 22. Kanakaraj P, et al. 1998. Interleukin (IL)-1 receptor-associated kinase (IRAK) requirement for optimal induction of multiple IL-1 signaling pathways and IL-6 production. J. Exp. Med. 187:2073–2079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kawai T, Akira S. 2010. The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nat. Immunol. 11:373–384 [DOI] [PubMed] [Google Scholar]
- 24. Kawai T, et al. 2004. Interferon-alpha induction through Toll-like receptors involves a direct interaction of IRF7 with MyD88 and TRAF6. Nat. Immunol. 5:1061–1068 [DOI] [PubMed] [Google Scholar]
- 25. Keller SA, et al. 2006. NF-kappaB is essential for the progression of KSHV- and EBV-infected lymphomas in vivo. Blood 107:3295–3302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lagos D, et al. 2010. miR-132 regulates antiviral innate immunity through suppression of the p300 transcriptional co-activator. Nat. Cell Biol. 12:513–519 [DOI] [PubMed] [Google Scholar]
- 27. Lei X, et al. 2010. Regulation of NF-kappaB inhibitor IkappaBalpha and viral replication by a KSHV microRNA. Nat. Cell Biol. 12:193–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Leidal AM, Cyr DP, Hill RJ, Lee PW, McCormick C. 2012. Subversion of autophagy by Kaposi's sarcoma-associated herpesvirus impairs oncogene-induced senescence. Cell Host Microbe 11:167–180 [DOI] [PubMed] [Google Scholar]
- 29. Lin X, et al. 2011. miR-K12-7-5p encoded by Kaposi's sarcoma-associated herpesvirus stabilizes the latent state by targeting viral ORF50/RTA. PLoS One 6:e16224 doi:10.1371/journal.pone.0016224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lu F, Stedman W, Yousef M, Renne R, Lieberman PM. 2010. Epigenetic regulation of Kaposi's sarcoma-associated herpesvirus latency by virus-encoded microRNAs that target Rta and the cellular Rbl2-DNMT pathway. J. Virol. 84:2697–2706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Mbulaiteye SM, et al. 2006. Spectrum of cancers among HIV-infected persons in Africa: the Uganda AIDS-Cancer Registry Match Study. Int. J. Cancer 118:985–990 [DOI] [PubMed] [Google Scholar]
- 32. Mosam A, et al. 2009. Increasing incidence of Kaposi's sarcoma in black South Africans in KwaZulu-Natal, South Africa (1983–2006). Int. J. STD AIDS 20:553–556 [DOI] [PubMed] [Google Scholar]
- 33. Motsch N, Pfuhl T, Mrazek J, Barth S, Grasser FA. 2007. Epstein-Barr virus-encoded latent membrane protein 1 (LMP1) induces the expression of the cellular microRNA miR-146a. RNA Biol. 4:131–137 [DOI] [PubMed] [Google Scholar]
- 34. Orjalo AV, Bhaumik D, Gengler BK, Scott GK, Campisi J. 2009. Cell surface-bound IL-1alpha is an upstream regulator of the senescence-associated IL-6/IL-8 cytokine network. Proc. Natl. Acad. Sci. U. S. A. 106:17031–17036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Parravicini C, et al. 2000. Differential viral protein expression in Kaposi's sarcoma-associated herpesvirus-infected diseases: Kaposi's sarcoma, primary effusion lymphoma, and multicentric Castleman's disease. Am. J. Pathol. 156:743–749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Perry MM, et al. 2008. Rapid changes in microRNA-146a expression negatively regulate the IL-1beta-induced inflammatory response in human lung alveolar epithelial cells. J. Immunol. 180:5689–5698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Pfeffer S, et al. 2005. Identification of microRNAs of the herpesvirus family. Nat. Methods 2:269–276 [DOI] [PubMed] [Google Scholar]
- 38. Punj V, et al. 2010. Kaposi's sarcoma-associated herpesvirus-encoded viral FLICE inhibitory protein (vFLIP) K13 suppresses CXCR4 expression by upregulating miR-146a. Oncogene 29:1835–1844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Samols MA, et al. 2007. Identification of cellular genes targeted by KSHV-encoded microRNAs. PLoS Pathog. 3:e65 doi:10.1371/journal.ppat.0030065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sims JE, Smith DE. 2010. The IL-1 family: regulators of immunity. Nat. Rev. Immunol. 10:89–102 [DOI] [PubMed] [Google Scholar]
- 41. Skalsky RL, et al. 2007. Kaposi's sarcoma-associated herpesvirus encodes an ortholog of miR-155. J. Virol. 81:12836–12845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Song KW, et al. 2009. The kinase activities of interleukin-1 receptor associated kinase (IRAK)-1 and 4 are redundant in the control of inflammatory cytokine expression in human cells. Mol. Immunol. 46:1458–1466 [DOI] [PubMed] [Google Scholar]
- 43. Suzuki N, et al. 2002. Severe impairment of interleukin-1 and Toll-like receptor signalling in mice lacking IRAK-4. Nature 416:750–756 [DOI] [PubMed] [Google Scholar]
- 44. Suzuki N, et al. 2003. IL-1R-associated kinase 4 is required for lipopolysaccharide-induced activation of APC. J. Immunol. 171:6065–6071 [DOI] [PubMed] [Google Scholar]
- 45. Swantek JL, Tsen MF, Cobb MH, Thomas JA. 2000. IL-1 receptor-associated kinase modulates host responsiveness to endotoxin. J. Immunol. 164:4301–4306 [DOI] [PubMed] [Google Scholar]
- 46. Taganov KD, Boldin MP, Chang KJ, Baltimore D. 2006. NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc. Natl. Acad. Sci. U. S. A. 103:12481–12486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Takeda K, Kaisho T, Akira S. 2003. Toll-like receptors. Annu. Rev. Immunol. 21:335–376 [DOI] [PubMed] [Google Scholar]
- 48. Thomas JA, et al. 1999. Impaired cytokine signaling in mice lacking the IL-1 receptor-associated kinase. J. Immunol. 163:978–984 [PubMed] [Google Scholar]
- 49. Uematsu S, et al. 2005. Interleukin-1 receptor-associated kinase-1 plays an essential role for Toll-like receptor (TLR)7- and TLR9-mediated interferon-alpha induction. J. Exp. Med. 201:915–923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Umbach JL, Cullen BR. 2010. In-depth analysis of Kaposi's sarcoma-associated herpesvirus microRNA expression provides insights into the mammalian microRNA-processing machinery. J. Virol. 84:695–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Walz N, Christalla T, Tessmer U, Grundhoff A. 2010. A global analysis of evolutionary conservation among known and predicted gammaherpesvirus microRNAs. J. Virol. 84:716–728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. West J, Damania B. 2008. Upregulation of the TLR3 pathway by Kaposi's sarcoma-associated herpesvirus during primary infection. J. Virol. 82:5440–5449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. West JA, Gregory SM, Sivaraman V, Su L, Damania B. 2011. Activation of plasmacytoid dendritic cells by Kaposi's sarcoma-associated herpesvirus. J. Virol. 85:895–904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Ziegelbauer JM. 2010. Functions of Kaposi's sarcoma-associated herpesvirus microRNAs. Biochim. Biophys. Acta 1809:623–630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Ziegelbauer JM, Sullivan CS, Ganem D. 2009. Tandem array-based expression screens identify host mRNA targets of virus-encoded microRNAs. Nat. Genet. 41:130–134 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.