Abstract
Although clinical trials with human subjects are essential for determination of safety, infectivity, and immunogenicity, it is desirable to know in advance the infectiousness of potential candidate live attenuated influenza vaccine strains for human use. We compared the replication kinetics of wild-type and live attenuated influenza viruses, including H1N1, H3N2, H9N2, and B strains, in Madin-Darby canine kidney (MDCK) cells, primary epithelial cells derived from human adenoids, and human bronchial epithelium (NHBE cells). Our data showed that despite the fact that all tissue culture models lack a functional adaptive immune system, differentiated cultures of human epithelium exhibited the greatest restriction for all H1N1, H3N2, and B vaccine viruses studied among three cell types tested and the best correlation with their levels of attenuation seen in clinical trials with humans. In contrast, the data obtained with MDCK cells were the least predictive of restricted viral replication of live attenuated vaccine viruses in humans. We were able to detect a statistically significant difference between the replication abilities of the U.S. (A/Ann Arbor/6/60) and Russian (A/Leningrad/134/17/57) cold-adapted vaccine donor strains in NHBE cultures. Since live attenuated pandemic influenza vaccines may potentially express a hemagglutinin and neuraminidase from a non-human influenza virus, we assessed which of the three cell cultures could be used to optimally evaluate the infectivity and cellular tropism of viruses derived from different hosts. Among the three cell types tested, NHBE cultures most adequately reflected the infectivity and cellular tropism of influenza virus strains with different receptor specificities. NHBE cultures could be considered for use as a screening step for evaluating the restricted replication of influenza vaccine candidates.
INTRODUCTION
Influenza A and B viruses infect 5 to 15% of the global population annually and cause an estimated 250,000 to 500,000 deaths (35, 54). Outbreaks and epidemics of influenza virus regularly cause excess mortality among the elderly and considerable morbidity in all ages during the influenza season (32, 35). Vaccination is the most effective way of preventing disease caused by influenza viruses. Since influenza A and B viruses undergo continuous antigenic change, the influenza vaccine components often need to be updated annually to antigenically match the circulating strains. The two influenza vaccines currently licensed in the United States are the inactivated trivalent influenza vaccine, given by intramuscular injection, and the live attenuated influenza vaccine, administered intranasally (30, 35). It is recognized that live attenuated influenza virus vaccines are more efficacious than inactivated vaccines in young children (1–3, 5, 8, 38) and that both vaccines could afford protection with differing efficacy against drifted strains in adults (4, 8, 27, 33, 36).
Live attenuated influenza virus vaccine contains hemagglutinin (HA) and neuraminidase (NA) gene segments from the three currently circulating influenza strains (H1N1, H3N2, and B) and the six internal protein gene segments (PB1, PB2, PA, NP, M, and NS) from master donor A and B viruses (21, 30). Donor strains were independently developed by sequential passages at lower temperature (25°C) in the United States and the former Soviet Union from virulent H2N2 and B isolates (A/Ann Arbor/6/60 and B/Ann Arbor/1/66, respectively, in the United States and A/Leningrad/134/57 and B/USSR/60/69, respectively, in the former Soviet Union) (20, 51). Two influenza A virus donor strains were prepared in Russia: A/Leningrad/134/17/57 (H2N2), the “17× passaged” variant of the master strain, for use in adults and A/Leningrad/134/47/57 (H2N2), the “47× passaged” variant of the same parent (which received an extra 30 passages at low temperatures), for use in children (15, 20). Both influenza A and B donor viruses are cold adapted (ca) (replicate efficiently at 25°C and 33°C), temperature sensitive (ts) (do not replicate at temperatures above 39°C), and attenuated (att) (do not produce classic influenza-like illness and are restricted in replication in the lower respiratory tracts of ferrets) (22, 30). These specific phenotypes, mediated by mutations in the internal gene segments (15, 17, 20, 30), lead to limited replication in the respiratory tract of the infected host and stimulate both systemic and cellular immune responses (30, 42, 54). The ca U.S. and Russian master donor strains have not been directly compared for infectivity, immunogenicity, and safety in clinical trials with humans.
As live attenuated influenza vaccines replicate in the nasopharynx of the recipient, infectious vaccine virus can be cultured from upper respiratory tract secretions after vaccination, a phenomenon termed “virus shedding.” Previous studies have estimated the median human infectious dose required for infection with live attenuated seasonal influenza vaccine to be 2.5 to 4.5 log10 50% tissue culture infective doses (TCID50) in seronegative children and 5.0 to 6.4 log10 TCID50 in seronegative adults (12, 31, 42, 49). There is a direct correlation between the magnitude of shedding of influenza virus and the illness experienced by the host (30). Therefore, for reasons of safety, infectivity, and immunogenicity, it is desirable to know in advance the levels of replication of potential candidate live attenuated vaccine strains for human use.
In addition to yearly outbreaks and epidemics, influenza A viruses cause periodic pandemics, in which viruses containing novel HA and/or NA are introduced into susceptible human populations (54). In preparation for the next influenza pandemic, a number of strategies to develop pandemic vaccines are under way, including the use of live attenuated vaccines. Unfortunately, it is hard to predict the levels of replication in humans of candidate vaccines bearing HA influenza virus subtypes with pandemic potential (H2, H5, H7, and H9 HA subtypes) before performing human clinical trials (45). The replication of such attenuated vaccine strains in mice and ferrets is not predictive of replication of these viruses in humans. For example, H5N1 and H9N2 ca vaccine strains replicated minimally in humans but were readily recovered by culture in small-animal models (9, 18, 19, 47). The reasons for this discrepancy are not completely understood, but it may be related to (i) preexisting antibodies to HAs and/or NAs in human serum that cross-react with the avian HAs and/or NAs and decrease virus vaccine replication, (ii) cellular immunity, or (iii) decreased affinity of the avian HAs for sialic acid (SA) receptors in the human upper airways (44). Human influenza virus HAs preferentially bind to cell surface receptors terminating in SA α2,6-galactose (SAα2,6Gal), whereas avian influenza viruses preferentially bind to receptors terminating in SAα2,3Gal (39, 44). Thus, new screening tools or models that predict the infectivity of ca influenza viruses in the human host need to be developed.
In the present study, we compared the replication kinetics of wild-type (wt) and ca influenza viruses, including H1N1, H3N2, H9N2, and B strains, in Madin-Darby canine kidney (MDCK) cells and human epithelial cells derived from adenoids (HAEC cells) and bronchial epithelium (NHBE cells). We also compared the replication abilities of the U.S. (ca A/Ann Arbor/6/60 [H2N2]) and three Russian (ca A/Leningrad/134/17/57 [H2N2], ca A/Leningrad/134/47/57 [H2N2], and ca A/Leningrad/134/80/57 [H2N2]) vaccine donor strains side by side in MDCK and NHBE cells. Since live attenuated influenza vaccines could potentially bear HA and NA genes of different origin (human, avian, swine, or equine), we also assessed which of the three cell cultures could optimally evaluate the infectivity and cellular tropism of influenza viruses from different hosts and with different receptor specificities.
MATERIALS AND METHODS
Cells.
MDCK cells were obtained from the American Type Culture Collection (Manassas, VA) and were maintained as described elsewhere (16). Primary NHBE cells from human tracheal/bronchial tissues were obtained from Lonza (Walkersville, MD). Cells of passage 2 were grown on membrane supports (6.5-mm Transwell; Corning Inc., Corning, NY) at the air-liquid interface in serum-free and hormone- and growth factor-supplemented medium as described previously (16, 25). Fully differentiated 4- to 8-week-old cultures were used for all experiments.
Adenoids were obtained at the time of adenoidectomy performed for independently defined clinical indications under a protocol approved by the Vanderbilt Institutional Review Board (Nashville, TN). The isolation and growth of primary epithelial cells from adenoidal tissue (HAEC cells) were previously described (13, 53). Briefly, optimal recovery of viable epithelial cells was obtained by placing the whole tissue in minimal essential medium with 0.1% pronase type 14 (Sigma Chemicals, St. Louis, MO) and antibiotics and rocking overnight at 4°C. The superficial layers of cells were further dispersed by pipetting, and cells were placed in medium containing 10% fetal calf serum to inactivate the pronase. The cells were then centrifuged, resuspended in 50% Ham's F-12 medium (Mediatech Inc., Manassas, VA)–50% Eagle's minimal essential medium with supplements (insulin, 5 μg/ml; transferrin, 5 μg/ml; epidermal growth factor, 10 ng/ml; cholera toxin, 10 ng/ml; hydrocortisone, 10−6 M; bovine hypothalamic extract, 15 μg/ml; HEPES buffer, 0.015 M; retinol, 10−7 M; gentamicin, 50 μg/ml; penicillin G, 15 U/ml; streptomycin, 15 U/ml; and fetal calf serum, 0.5%) and seeded on 24-well tissue culture plates coated with a collagen matrix of Vitrogen 100 (Cohesion, Palo Alto, CA). The cells were incubated at 37°C under 5% CO2 until they reached 90% confluence.
Virus isolates.
The wt and ca A/California/10/78 (H1N1), A/Alaska/6/77 (H3N2), A/Washington/897/80 (H3N2), and ca A/Ann Arbor/6/60 (H2N2) influenza viruses were kindly provided by Kanta Subbarao at the National Institute of Allergy and Infectious Diseases, Bethesda, MD. The wt and ca A/New Caledonia/20/99 (H1N1), A/Panama/2007/99 (H3N2), A/Wyoming/03/03 (H3N2), B/Hong Kong/330/01, and ca A/chicken/HK/G9/97 (H9N2) influenza viruses were obtained from the Influenza Division of the Centers for Disease Control and Prevention, Atlanta, GA. The wt A/Leningrad/134/57 (H2N2), ca A/Leningrad/134/17/57 (H2N2), ca A/Leningrad/134/47/57 (H2N2), and ca A/Leningrad/134/80/57 (H2N2) viruses were obtained from the Institute for Experimental Medicine, Russian Academy of Medical Science, St. Petersburg, Russia. Human (A/Tottori/849/94 AL3 [H3N2], A/Tottori/849/94 K4 [H3N2], A/Tottori/872/94 AL3 [H3N2]), avian (A/duck/Ukraine/1/63 [H3N8], A/duck/Hokkaido/8/80 [H3N8]), equine (A/equine/TN/5/86 [H3N8]), and swine (A/swine/Italy/635/87 [H3N2]) strains were kindly provided by Yoshihiro Kawaoka at the University of Wisconsin, Madison, WI. Stock viruses were prepared by one passage in the allantoic cavities of 10-day-old embryonated chicken eggs for 48 h at 37°C (or at 33°C for ca and B viruses), and aliquots were stored at −70°C until used. All experimental work was performed in a biosafety level 2 laboratory approved for use with these strains by the U.S. Department of Agriculture and the U.S. Centers for Disease Control and Prevention.
Infectivities of influenza viruses.
The infectivities of H1N1, H2N2, H3N2, and H3N8 influenza A and influenza B viruses were determined as PFU/ml in MDCK cells. All the viruses were titrated in MDCK cells due to inability to plaque in other cell culture models despite the fact that the use of MDCK cells for determining infectious titers could be regarded as a potential confounding factor of this study. Briefly, confluent MDCK cells were incubated at 37°C (or at 33°C for wt and ca reassortant viruses) for 1 h with 10-fold serial dilutions of virus. The cells were then washed and overlaid with minimal essential medium containing 1 μg/ml l-(tosylamido-2-phenyl)ethylchloromethylketone (TPCK)-treated trypsin, 0.3% bovine serum albumin (BSA), and 0.9% Bacto agar. After 3 days of incubation at 37°C (or 33°C), cells were stained with 0.1% crystal violet in 10% formaldehyde solution, and the PFU per milliliter were determined.
The infectivity of ca A/chicken/HK/G9/97 (H9N2) virus was defined as log10 50% tissue culture infective dose (TCID50) as described previously (16), because this H9N2 virus did not produce plaques in MDCK cells. Briefly, confluent monolayers of MDCK cultures growing in 96-well plates were inoculated with serial virus dilutions (each dilution was added to five wells) in the presence of 1 μg/ml TPCK-treated trypsin. After 3 days, virus was titrated by hemagglutination assay, and virus titers were expressed as log10TCID50/ml by the endpoint method of Reed and Muench (40).
Replication kinetics.
To determine multistep growth curves, HAEC and MDCK cells were infected with viruses at an identical multiplicity of infection (MOI) of 0.01 PFU/cell at 33°C. After 1 h of incubation, the cells were washed and overlaid with infection medium (minimal essential medium with 0.3% BSA); 1 μg/ml TPCK-treated trypsin was added only in MDCK cells, because HAEC cells support the growth of influenza viruses without exogenous trypsin (13). Supernatants were collected at 1, 24, 48, and 72 h postinfection and stored at −70°C for titration by plaque assay.
To determine multistep growth curves in NHBE cells, triplicate cell cultures growing in 6.5-mm-diameter inserts were washed extensively with sterile phosphate-buffered saline (PBS) to remove mucus secretions on the apical surface prior to infection and then were inoculated via the apical side with each influenza virus at an MOI of 0.01 at 33°C. After 1 h of incubation, the inoculum was removed. Viruses released into the apical compartment of NHBE cells were harvested at the indicated time points by the apical addition and collection of 300 μl of medium allowed to equilibrate for 30 min at 33°C. The virus titers were determined as log10 PFU/ml in MDCK cells.
The area under the curve (AUC) viral load was defined as the area under the multistep growth curve and calculated by the trapezoidal rule, using exact viral titers at 24, 48, and 72 h postinfection as determined by plaque assay in MDCK cells.
Immunostaining and light microscopy.
MDCK and NHBE cells were infected with H3 human, avian, swine, and equine viruses at an MOI of 0.01 and fixed for 30 min in 4% formaldehyde at 8 and 24 h postinfection. Fixed cultures were permeabilized with 0.5% Triton X-100, blocked with 3% BSA, and stained with mouse anti-NP IgG diluted in 3% BSA in PBS. After a 1-h incubation, the cells were then incubated with goat horseradish peroxidase (HRP)-labeled anti-mouse IgG (Sigma-Aldrich, St. Louis, MO). For localization of ciliated cells, fixed NHBE cells were costained with anti-β-tubulin IV antibody and HRP-labeled secondary antibody for detection. The cultures were mounted using Crystal Mount (Biomeda, Foster City, CA).
For cell counting, the cultures were observed en face by using a Nikon microscope at 40× and 100× objectives. In microscopic fields, the percentage of infected cells with respect to the total number of cells was calculated. In NHBE cells, the percentage of ciliated infected cells with respect to the total number of infected cells was also calculated. For each sample, 20 fields were analyzed and the results were averaged.
Receptor-binding assay.
The binding of H3 influenza viruses to fetuin (containing α2,3- and α2,6-linked sialyl receptors) was measured in a direct solid-phase assay using the immobilized virus and horseradish peroxidase-conjugated fetuin, as described previously (14). The affinity of viruses for synthetic 3′- and 6′-sialylglycopolymers obtained by conjugation of a 1-N-glycyl derivative of 3′- or 6′-sialyllactose (3′SL or 6′SL) or a 3-aminopropylglycoside of 3′- or 6′-sialyllactosamine (3′SLN or 6′SLN) with poly(4-phenylacrylate) (7) was measured in a competitive assay based on the inhibition of binding to peroxidase-labeled fetuin (24). 3′SLN and 6′SLN-acrylic polymers contain an additional amino group compared to 3′SL or 6′SL and more closely approximate “avian-type” or “human-type” sialyl receptors, respectively (26). Association constant (Ka) values were determined as the sialic acid (Neu5Ac) concentration at the point Amax/2 on Scatchard plots.
Statistical analysis.
The virus yields, AUCs, mean peak viral titers, and binding to sialyl receptors of influenza A and B viruses were compared either by analysis of variance (ANOVA) or by use of an unpaired t test. A probability value of 0.05 was prospectively chosen to indicate that the findings were not the result of chance alone.
RESULTS
Replication kinetics of wt and ca A/California/10/78 (H1N1), A/Alaska/6/77 (H3N2), and A/Washington/897/80 (H3N2) influenza viruses in HAEC, MDCK, and NHBE cells.
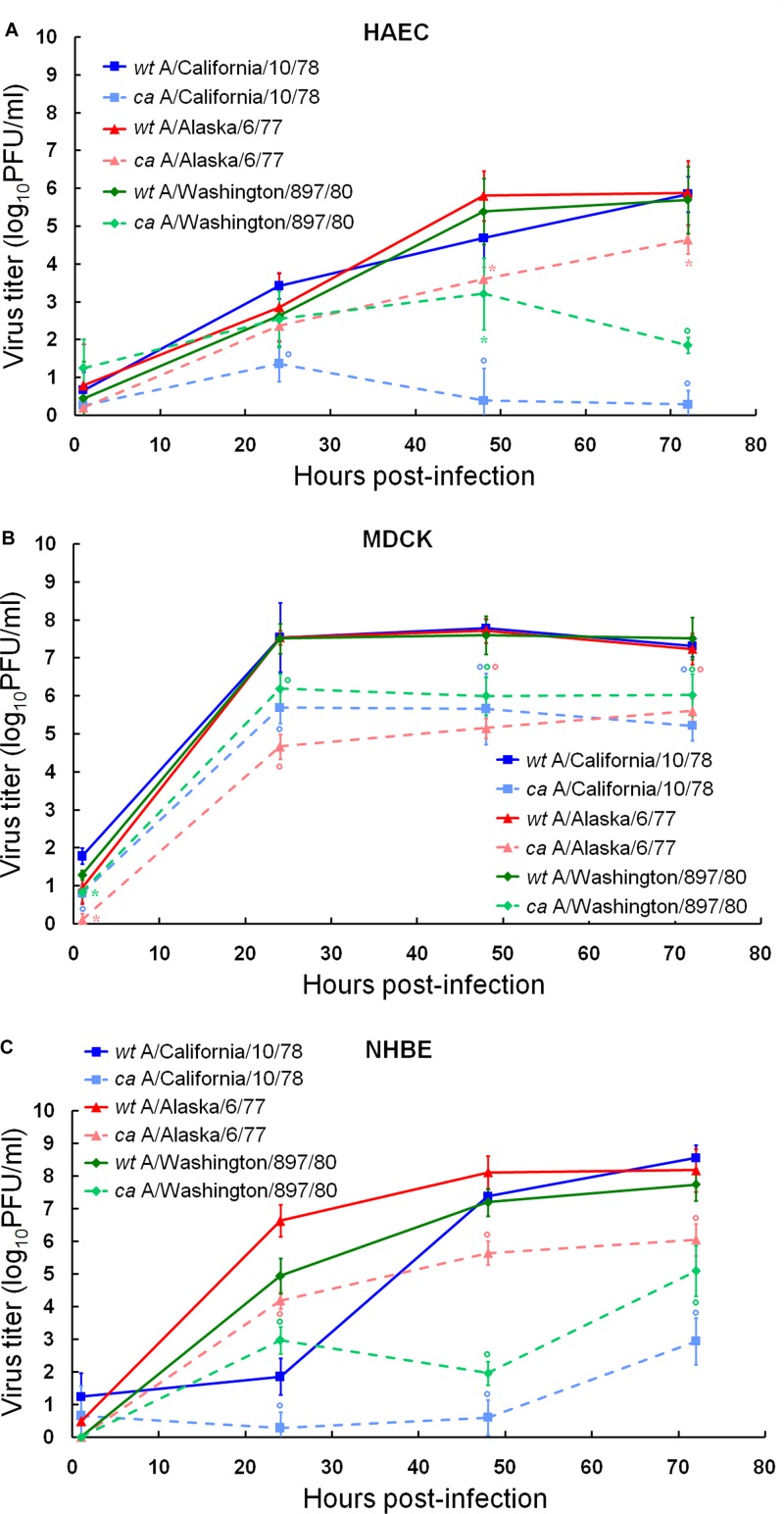
To assess which cell culture system could be used to adequately evaluate the viral growth of attenuated vaccine candidates, we first determined the levels of replication of early ca live influenza A virus vaccine strains, i.e., ca A/California/10/78 (H1N1), ca A/Alaska/6/77 (H3N2), and ca A/Washington/897/80 (H3N2) reassortants, in comparison with their respective wt viruses in HAEC, MDCK, and NHBE cells (Fig. 1; Table 1). Viral replication was compared by inoculating all three cultures with the wt and ca viruses at an MOI of 0.01 at 33°C and determining yields of the viral progeny at 1, 24, 48, and 72 h postinfection by plaque titration in MDCK cells. The growth of influenza viruses was supplemented by the addition of trypsin in MDCK cells, whereas no proteolytic enzymes were added to the epithelial cell systems.
Fig 1.
Replication kinetics of wt and ca A/California/10/78 (H1N1), A/Alaska/6/77 (H3N2), and A/Washington/897/80 (H3N2) influenza viruses in HAEC (A), MDCK (B), and NHBE (C) cell cultures. Cultures were infected with viruses at an MOI of 0.01 PFU/cell at 33°C. Supernatants were collected at 1, 24, 48, and 72 h postinfection, and the virus titers were determined as log10 PFU/ml in MDCK cells. *, P < 0.05; °, P < 0.01 (compared with the value for the respective wt virus, determined with an unpaired t test).
Table 1.
Areas under the curve and mean peak viral titers for wt and ca A/California/10/78, A/Alaska/6/77, and A/Washington/897/80 influenza viruses in HAEC, MDCK, and NHBE cell cultures
| Virus | Subtype | Mean AUCa (mean peak titer, log10 PFU/ml) |
||
|---|---|---|---|---|
| HAEC | MDCK | NHBE | ||
| A/California/10/78 | ||||
| wt | H1N1 | 223.6 (5.9) | 364.8 (7.8) | 301.8 (8.6) |
| ca | H1N1 | 28.8 (1.4) | 266.7 (5.7) | 53.0 (2.9) |
| Degree of restrictionb | 194.8 (4.5) | 98.1 (2.1) | 248.8 (5.7) | |
| A/Alaska/6/77 | ||||
| wt | H3N2 | 244.0 (5.9) | 362.5 (7.7) | 372.1 (8.2) |
| ca | H3N2 | 170.3 (4.6) | 246.7 (5.6) | 258.2 (6.0) |
| Degree of restriction | 73.7 (1.3) | 115.8 (2.1) | 113.9 (2.2) | |
| A/Washington/897/80 | ||||
| wt | H3N2 | 229.1 (5.7) | 362.5 (7.6) | 324.6 (7.7) |
| ca | H3N2 | 129.7 (3.2) | 290.5 (6.2) | 143.6 (5.1) |
| Degree of restriction | 99.4 (2.5) | 72.0 (1.4) | 181.0 (2.6) | |
The area under the curve (AUC) represents the total viral load at 24, 48, and 72 h postinfection.
The degree of restriction of viral replication is expressed as the value for wt virus minus the value for the corresponding ca virus.
Our data showed that wt A/California/10/78 (H1N1), wt A/Alaska/6/77 (H3N2), and wt A/Washington/897/80 (H3N2) replicated to the same extent in each cell line tested, as seen by similar total amounts of viral load (AUC) and almost equal peak viral titers (Table 1). All wt strains grew to significantly higher titers than the respective ca viruses at 48 and 72 h after infection in HAEC, MDCK, and NHBE cells (1.2 to 6.8 log units; P < 0.01) (Fig. 1). The replication abilities of the ca reassortant viruses (i.e., AUCs, peak viral titers, and virus yields at each time point) differed significantly (P < 0.05) from each other in HAEC and NHBE cells but not in MDCK cells (Fig. 1; Table 1).
We further performed pairwise comparisons of the cumulative amounts of each virus and peak viral titers in the three cell types (see Table S1 in the supplemental material). The levels of replication of the wt and ca viruses together with the levels of decrease of replication of the ca viruses were almost always significantly different in the three cell culture systems. The most limited growth of both wt and ca viruses was observed in HAEC cells (P < 0.05). Overall, our parallel experiments demonstrated that all three ca reassortants, i.e., ca A/California/10/78 (H1N1), ca A/Alaska/6/77 (H3N2), and ca A/Washington/897/80 (H3N2), exhibited the most attenuated growth in NHBE cells (i.e., decreases of viral peak titers were 5.7, 2.2, and 2.6 log units in comparison to the respective wt viruses, respectively) (Table 1).
Replication kinetics of wt and ca A/New Caledonia/20/99 (H1N1), A/Panama/2007/99 (H3N2), and A/Wyoming/03/03 (H3N2) influenza viruses in MDCK and NHBE cells.
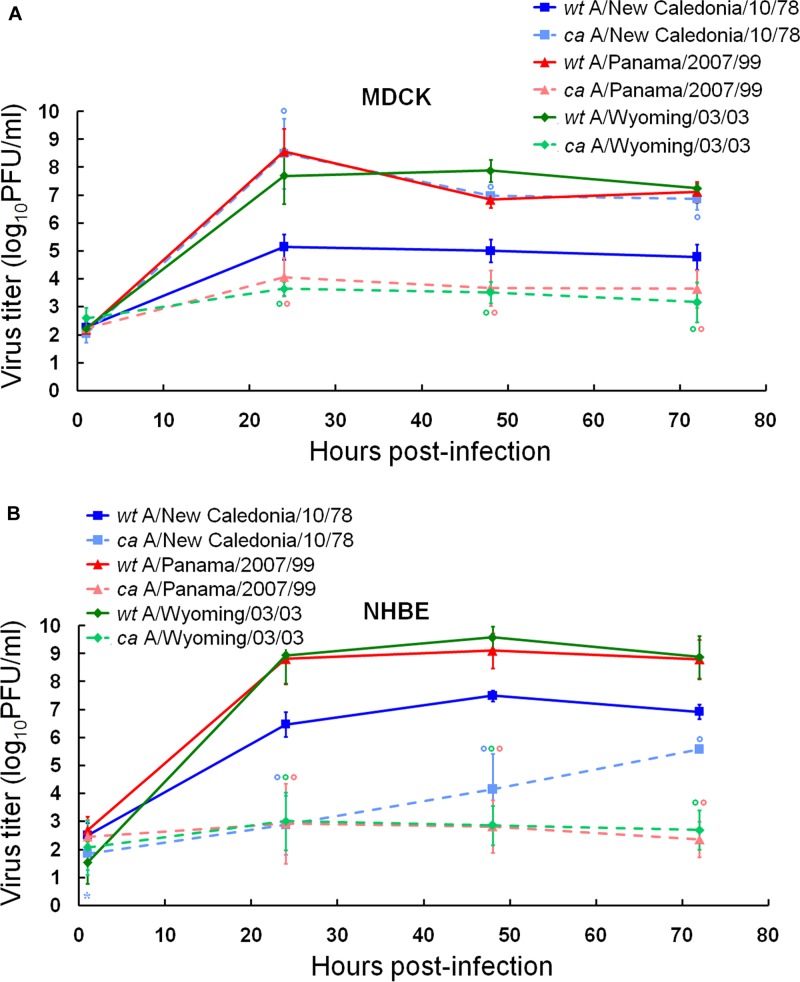
We evaluated the replication abilities of more contemporary ca influenza A virus vaccine strains, i.e., ca A/New Caledonia/20/99 (H1N1), ca A/Panama/2007/99 (H3N2), and ca A/Wyoming/03/03 (H3N2), in comparison with their respective wt viruses in MDCK and NHBE cells (Fig. 2; Table 2). We observed that two wt H3N2 viruses, wt A/Panama/2007/99 and wt A/Wyoming/03/03, replicated similarly (i.e., no significantly different AUCs, peak viral titers, or virus yields were observed) and to significantly higher titers than wt A/New Caledonia/20/99 (H1N1) at 24, 48, and 72 h after infection in both cell types (1.6 to 3.4 log units; P < 0.01). The replication kinetics of two ca H3N2 reassortant viruses, ca A/Panama/2007/99 and ca A/Wyoming/03/03, did not differ significantly from each other (Fig. 2) or between the two cell systems (see Table S1 in the supplemental material). They replicated to significantly lower titers than the respective wt viruses at 24, 48, and 72 h after infection in MDCK and NHBE cells (3.2 to 6.7 log units; P < 0.01) (Fig. 2). Notably, ca A/New Caledonia/20/99 (H1N1) virus replicated to a significantly higher extent than the respective wt virus and the two other ca H3N2 reassortants in MDCK cells, as seen by its significantly higher AUCs, peak viral titers, and virus yields in this cell line (P < 0.01) (Table 2). In contrast, ca A/New Caledonia/20/99 (H1N1) virus exhibited a viral load similar to those of ca H3N2 viruses ca A/Panama/2007/99 and ca A/Wyoming/03/03 and significantly limited growth with a decrease of peak viral titer of 1.9 log units (P < 0.01) in comparison to the respective wt virus in NHBE cells (Fig. 2; Table 2). Taken together, our experiments clearly showed that differentiated NHBE cultures exhibited the greatest restriction for all studied ca H1N1 and H3N2 vaccine viruses among three cell types tested.
Fig 2.
Replication kinetics of wt and ca A/New Caledonia/20/99 (H1N1), A/Panama/2007/99 (H3N2), and A/Wyoming/03/03 (H3N2) influenza viruses in MDCK (A) and NHBE (B) cell cultures. Cultures were infected with viruses at an MOI of 0.01 PFU/cell at 33°C. Supernatants were collected at 1, 24, 48, and 72 h postinfection, and the virus titers were determined as log10 PFU/ml in MDCK cells. *, P < 0.05; °, P < 0.01 (compared with the value for the respective wt virus, determined with an unpaired t test).
Table 2.
Areas under the curve and mean peak viral titers for wt and ca A/New Caledonia/20/99, A/Panama/2007/99, and A/Wyoming/03/03 influenza viruses in MDCK and NHBE cell cultures
| Virus | Subtype | Mean AUC (mean peak titer, log10 PFU/ml)a |
|
|---|---|---|---|
| MDCK | NHBE | ||
| A/New Caledonia/20/99 | |||
| wt | H1N1 | 239.5 (5.2) | 340.8 (7.5) |
| ca | H1N1 | 351.8 (8.5) | 201.2 (5.6) |
| Degree of restrictionb | 112.3 (3.1) | 139.6 (1.9) | |
| A/Panama/2007/99 | |||
| wt | H3N2 | 352.2 (8.6) | 429.9 (9.1) |
| ca | H3N2 | 180.7 (4.1) | 130.9 (2.9) |
| Degree of restriction | 171.5 (4.5) | 299.0 (6.2) | |
| A/Wyoming/03/03 | |||
| wt | H3N2 | 368.6 (7.9) | 443.8 (9.6) |
| ca | H3N2 | 166.2 (3.7) | 137.1 (3.0) |
| Degree of restriction | 202.4 (4.3) | 306.7 (6.6) | |
The area under the curve (AUC) represents the total viral load at 24, 48, and 72 h postinfection. Bold indicates an increase in titer and lack of restricted replication of ca influenza vaccine virus.
The degree of restriction of viral replication is expressed as the value for wt virus minus the value for the corresponding ca virus.
Comparison of replication kinetics of U.S. and Russian ca influenza vaccine donor strains in MDCK and NHBE cells.
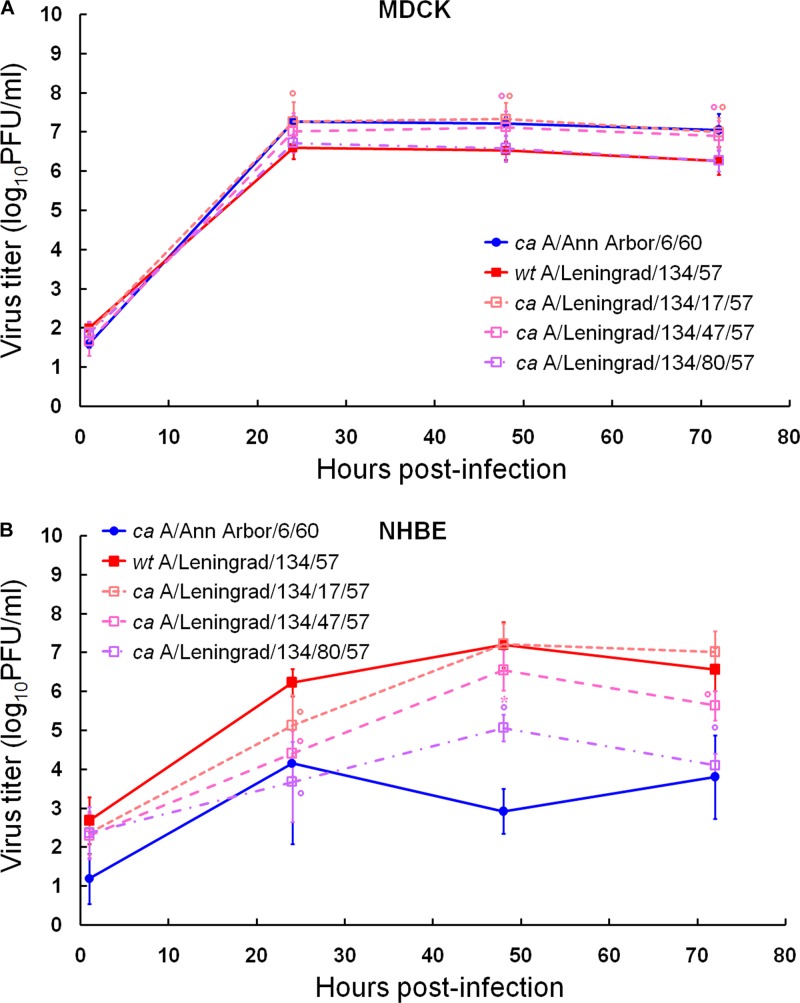
Since there is a correlation between the level of replication of influenza virus and its capacity to induce immunity (30), we compared side by side the replication abilities of different ca H2N2 vaccine donor strains, i.e., ca A/Ann Arbor/6/60, ca A/Leningrad/134/17/57, and ca A/Leningrad/134/47/57, in MDCK and NHBE cells (Fig. 3; Table 3). wt A/Leningrad/134/57 (H2N2) and its “80× passaged” variant ca A/Leningrad/134/80/57 (H2N2) were also included in the comparison. In MDCK cells, we observed that although ca A/Leningrad/134/17/57 and ca A/Leningrad/134/47/57 exhibited significantly higher peak viral titers and virus yields than wt A/Leningrad/134/57 and ca A/Leningrad/134/80/57 at 24, 48, and 72 h after infection (∼0.6 log units; P < 0.01) (Fig. 3A), all four strains shed similar amount of virus (Table 3). In NHBE cells, wt A/Leningrad/134/57 and ca A/Leningrad/134/17/57 replicated similarly, whereas ca A/Leningrad/134/47/57 and ca A/Leningrad/134/80/57 showed significantly lower peak viral titers and virus yields at 24, 48, and 72 h after infection (0.7 to 2.9 log units; P < 0.05) (Fig. 3B). ca A/Leningrad/134/80/57 (H2N2) virus replicated to a significantly lower extent, as seen by AUC, than the respective wt strain (P < 0.01) (Table 3), which was consistent with its higher in vitro passage history.
Fig 3.
Replication kinetics of ca A/Ann Arbor/6/60, wt A/Leningrad/134/57, ca A/Leningrad/134/17/57, ca A/Leningrad/134/47/57, and ca A/Leningrad/134/80/57 influenza viruses in MDCK (A) and NHBE (B) cell cultures. Cultures were infected with viruses at an MOI of 0.01 PFU/cell at 33°C. Supernatants were collected at 1, 24, 48, and 72 h postinfection, and the virus titers were determined as log10 PFU/ml in MDCK cells. *, P < 0.05; °, P < 0.01 (compared with the value for wt A/Leningrad/134/57 virus, determined by one-way ANOVA for Russian strains only).
Table 3.
Areas under the curve and mean peak viral titers for ca A/Ann Arbor/6/60, wt A/Leningrad/134/57, ca A/Leningrad/134/17/57, ca A/Leningrad/134/47/57, and ca A/Leningrad/134/80/57 influenza viruses in MDCK and NHBE cell cultures
| Virus | Subtype | Mean AUC (mean peak titer, log10 PFU/ml)a |
|
|---|---|---|---|
| MDCK | NHBE | ||
| ca A/Ann Arbor/6/60 | H2N2 | 344.7 (7.3) | 165.5 (4.2) |
| wt A/Leningrad/134/57 | H2N2 | 310.8 (6.6c) | 326.2c (7.2c) |
| ca A/Leningrad/134/17/57 | H2N2 | 346.8 (7.3) | 318.9c (7.2c) |
| Degree of restrictionb | 36.0 (0.7) | 7.3 (0) | |
| ca A/Leningrad/134/47/57 | H2N2 | 337.7 (7.1) | 277.6c (6.6c) |
| Degree of restriction | 26.9 (0.5) | 48.6 (0.6) | |
| ca A/Leningrad/134/80/57 | H2N2 | 313.8 (6.7c) | 214.7 (5.1) |
| Degree of restriction | 3.0 (0.1) | 111.5 (2.1) | |
The area under the curve (AUC) represents the total viral load at 24, 48, and 72 h postinfection. Bold indicates an increase in titer and lack of restricted replication of ca influenza vaccine virus.
The degree of restriction of viral replication is expressed as the value for wt virus minus the value for the corresponding ca virus.
P < 0.01 compared with the value for ca A/Ann Arbor/6/60 (H2N2) virus, by one-way ANOVA.
We further evaluated the replication abilities of the Russian influenza A vaccine donor strains in comparison with the U.S. donor strain, ca A/Ann Arbor/6/60 (H2N2), in MDCK and NHBE cells (Fig. 3; Table 3). We did not observe significantly different AUCs for all the viruses in MDCK cells; however, wt A/Leningrad/134/57 (H2N2) and its “80× passaged” ca A/Leningrad/134/80/57 (H2N2) variant exhibited significantly lower peak viral titers and virus yields at 24, 48, and 72 h after infection than ca A/Ann Arbor/6/60 in this cell line (∼0.7 log units; P < 0.05). In NHBE cells, the replication kinetics of ca A/Ann Arbor/6/60 and ca A/Leningrad/134/80/57 did not differ from each other (as seen by similar AUCs and peak viral titers) (Table 3). However, wt A/Leningrad/134/57, ca A/Leningrad/134/17/57, and ca A/Leningrad/134/47/57 grew to significantly higher titers at 48 and 72 h after infection than ca A/Ann Arbor/6/60 (1.8 to 4.3 log units; P < 0.01) (Fig. 3B) and shed significantly larger amounts of virus (P < 0.01) (Table 3). Taken together, our data showed statistically significant differences between the replication abilities of the U.S. and Russian ca vaccine donor strains in NHBE cells.
Replication kinetics of influenza viruses isolated from different species in HAEC, MDCK, and NHBE cells.
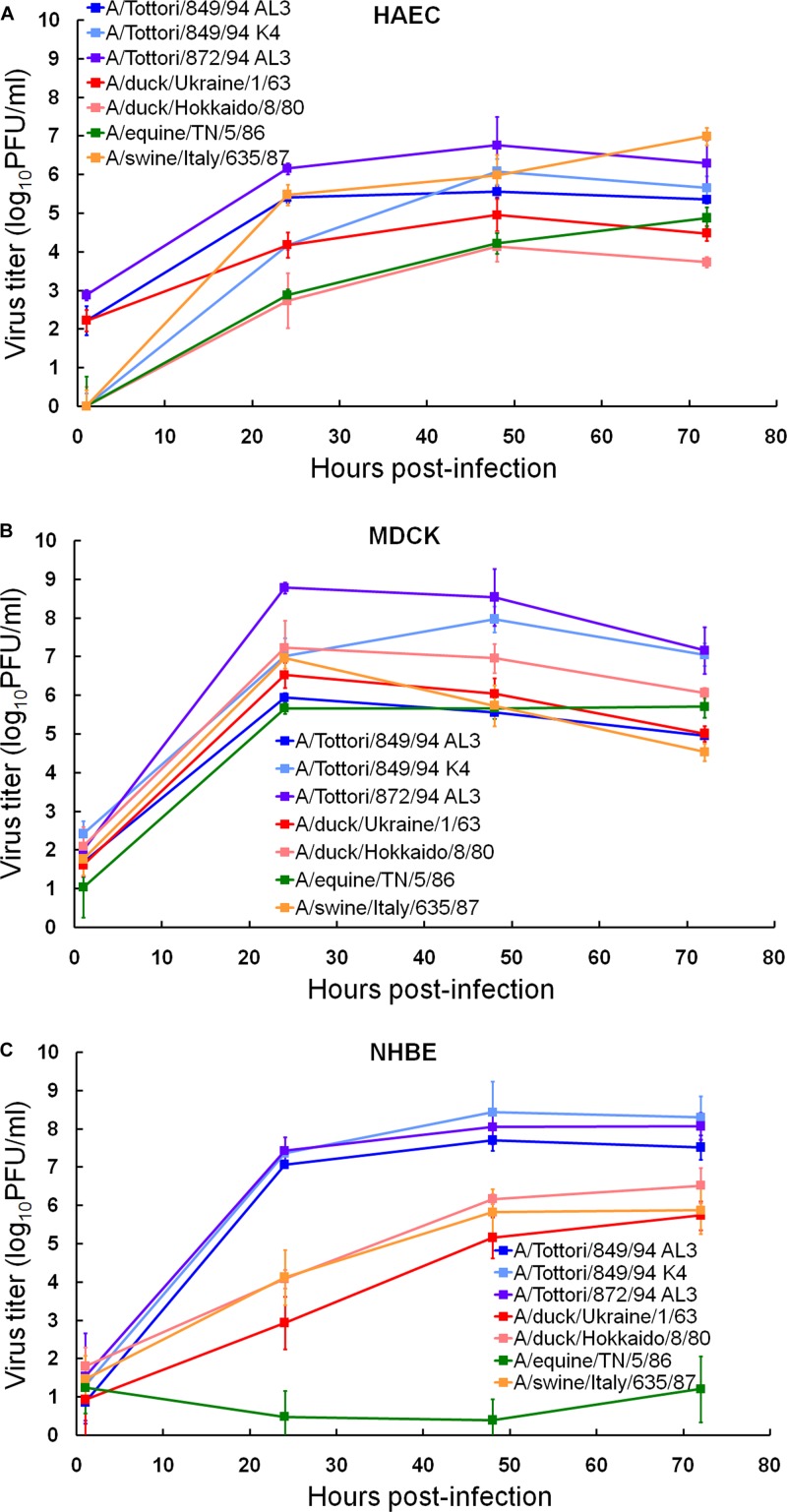
In this study, we assessed which of the three cell culture models could reflect the load of human infection based on the source (host) of the virus. For this purpose, we chose seven viruses of the H3 HA subtype that were isolated from different animal species, i.e., humans, birds, pigs, and horses, and assayed their yields after multiple replication cycles in HAEC, MDCK, and NHBE cultures (Fig. 4). Our results demonstrated no statistically significant difference between the replication kinetics of all the strains in HAEC and MDCK cells (Fig. 4A and B). Human, avian, swine, and equine isolates replicated to titers of 104.1 to 8.8 PFU/ml, suggesting that these two cell lines were totally susceptible to influenza viruses of different origin and, therefore, could not predict the risk of human infection (Table 4). In contrast, we observed three different patterns of replication corresponding to the origin of influenza virus in NHBE cells (Fig. 4C). (i)Three human isolates, A/Tottori/849/94 (H3N2) AL3, A/Tottori/849/94 (H3N2) K4, and A/Tottori/872/94 (H3N2) AL3, grew to significantly higher titers (1.0 to 8.0 log units higher; P < 0.05) than the rest of the viruses. (ii) The replication abilities of the avian viruses, A/duck/Ukraine/1/63 (H3N8) and A/duck/Hokkaido/8/80 (H3N8), did not differ from that of the swine A/swine/Italy/635/87 (H3N2) virus, and the yields of avian and swine isolates were approximately 66% of those of the human viruses at 24, 48, and 72 h after infection. (iii) The equine A/equine/TN/5/86 (H3N8) isolate exhibited the least replication in NHBE cells (mean peak titer of 1.2 log10 PFU/ml) (Fig. 4C; Table 4), indicating that this virus possessed minimal infectivity compared to the other viruses tested. Statistically significant differences between cumulative amounts of viral load and mean peak viral titers of human versus equine versus avian and swine isolates were observed in NHBE but not in HAEC or MDCK cells (P < 0.01; Table 4).
Fig 4.
Replication kinetics of H3 human, avian, swine, and equine influenza A viruses in HAEC (A), MDCK (B), and NHBE (C) cell cultures. Cultures were infected with viruses at an MOI of 0.01 PFU/cell. Supernatants were collected at the indicated time points and titrated in MDCK cells by plaque assay. Representative results expressed as log10 PFU/ml from 3 independent experiments are shown.
Table 4.
Areas under the curve, mean peak viral titers, and cell tropism of H3 influenza viruses isolated from different hosts in HAEC, MDCK, and NHBE cell cultures
| Virus | Subtype | AUC,a mean ± SD (mean peak viral titer, log10 PFU/ml) |
% of infected cells at 8 h (24 h) postinfection, mean ± SDb |
|||
|---|---|---|---|---|---|---|
| HAEC | MDCK | NHBE | MDCK | NHBE | ||
| A/Tottori/849/94 AL3 | H3N2 | 262.4 ± 6.2 (5.6) | 263.9 ± 6.2 (5.9) | 359.8 ± 11.0 (7.7) | 9.5 ± 2.3 (100) | 0.9 ± 0.4 (8.1 ± 2.6) |
| A/Tottori/849/94 K4 | H3N2 | 263.9 ± 13.0 (6.1) | 360.0 ± 17.2 (8.0) | 390.2 ± 27.2 (8.4) | 3.5 ± 0.1 (100) | 0.5 ± 0.2 (9.3 ± 3.4) |
| A/Tottori/872/94 AL3 | H3N2 | 311.5 ± 26.6 (6.8) | 396.2 ± 26.6 (8.8) | 379.1 ± 19.4 (8.1) | 3.9 ± 1.3 (100) | 0.6 ± 0.3 (7.3 ± 5.5) |
| A/duck/Ukraine/1/63 | H3N8 | 222.7 ± 16.1 (5.0) | 283.2 ± 16.1 (6.5) | 227.7 ± 25.7 (5.7) | 2.3 ± 0.4 (100) | ≤0.01 (1.3 ± 0.7) |
| A/duck/Hokkaido/8/80 | H3N8 | 176.7 ± 19.2 (4.1) | 326.4 ± 19.2 (7.2) | 275.2 ± 11.4 (6.5) | 6.0 ± 0.7 (100) | ≤0.01 (2.6 ± 0.8) |
| A/equine/TN/5/86 | H3N8 | 194.2 ± 11.6 (4.9) | 272.3 ± 11.6 (5.7) | 29.5 ± 31.6 (1.2) | 1.1 ± 0.4 (53.0 ± 19.9) | ≤0.01 (∼0.07) |
| A/swine/Italy/635/87 | H3N2 | 293.2 ± 18.6 (7.0) | 275.5 ± 18.6 (7.0) | 259.5 ± 30.7 (5.9) | 3.7 ± 0.9 (100) | ≤0.01 (0.7 ± 0.5) |
The area under the curve (AUC) represents the total viral load at 24, 48, and 72 h postinfection.
MDCK, HAEC, and NHBE cells were infected with H3 influenza viruses of different origins at an MOI of 0.01. Due to similar results being observed in MDCK and HAEC cells, data for HAEC cells are not shown.
Cellular tropism and virus spread of influenza viruses isolated from different species in HAEC, MDCK, and NHBE cells.
To determine why NHBE cultures are capable of reflecting the capacity of influenza strains to infect humans, we further assessed cell-specific tropism of influenza viruses of different origin in NHBE and MDCK cells. We infected two cell lines with either virus at an MOI of 0.01, fixed the cells at 8 h postinfection (i.e., after the first cycle of viral replication), and then immunostained the cultures for viral antigen (Table 4). The patterns of infection with H3 influenza virus strains of different origin were strikingly different between cell lines. All viruses were able to infect MDCK cells in the presence of trypsin, with significantly different capacities (P < 0.05). These data suggested that human, avian, swine, and equine viruses possessed no host range limitation in this cell line. In contrast, in NHBE cells at 8 h postinfection, only three human isolates, A/Tottori/849/94 (H3N2) AL3, A/Tottori/849/94 (H3N2) K4, and A/Tottori/872/94 (H3N2) AL3, showed replication (Table 4), suggesting that these viruses from the human host possess better tropism for NHBE cells than avian, swine, and equine isolates.
We further compared patterns of virus spread by H3 influenza viruses of different origin at 24 h after viral inoculation. Human, avian, and swine strains infected all cells in MDCK monolayers, and equine isolate A/equine/TN/5/86 (H3N8) infected ∼53% of all the cells (Table 4). In contrast, limited growth and the focal nature of the influenza infection were observed in NHBE cultures. After infection (24 h), human viruses A/Tottori/849/94 (H3N2) AL3, A/Tottori/849/94 (H3N2) K4, and A/Tottori/872/94 (H3N2) AL3 infected ∼7-fold more cells than avian and swine strains (Table 4). In addition, human isolates produced continuous foci of infected cells, which included both nonciliated and ciliated (∼46%) (data not shown) cells. Avian viruses A/duck/Ukraine/1/63 (H3N8) and A/duck/Hokkaido/8/80 (H3N8) infected the same amount of cells as the swine A/swine/Italy/635/87 (H3N2) virus (∼1.5%) (Table 3), and most of the infected cells were ciliated (65%) (data not shown). The equine A/equine/TN/5/86 (H3N8) isolate exhibited negligible infection in NHBE cells (Table 4). Taken together, our results suggested that NHBE cell cultures could approximate the cellular tropism of influenza viruses isolated from different species to the human respiratory tract.
Receptor specificity of H3 influenza A viruses isolated from different species.
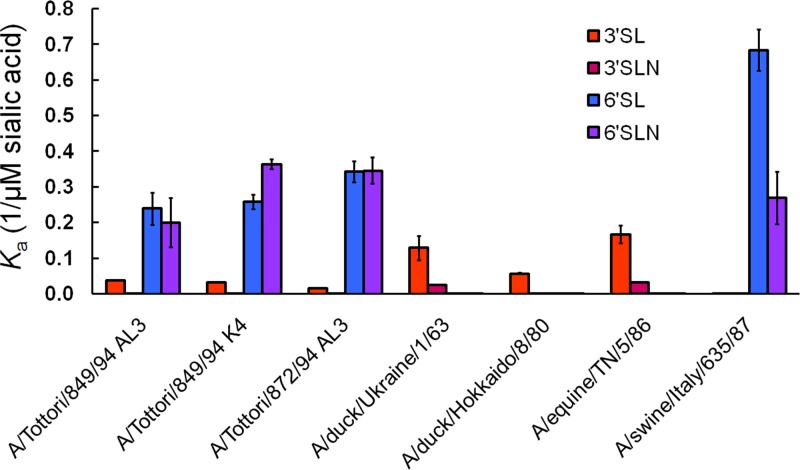
To examine whether the observed cellular tropism and virus spread of influenza viruses of different origin in NHBE cells were mediated by the receptor specificity of the viral HA, we measured the receptor specificities of the H3 influenza virus isolates to synthetic sialic substrates (3′SL/N and 6′SL/N) (Fig. 5). As shown by the Ka values, human viruses A/Tottori/849/94 (H3N2) AL3, A/Tottori/849/94 K4, and A/Tottori/872/94 (H3N2) AL3 exhibited increased affinity for 6′SL/N (synthetic sialosaccharides with the “human-type” SAα2,6Gal linkage), whereas the binding to the “avian-type” 3′ substrates 3′SL/N was negligible. A similar pattern was observed for the swine A/swine/Italy/635/87 (H3N2) virus. Two avian H3 viruses, A/duck/Ukraine/1/63 (H3N8) and A/duck/Hokkaido/8/80 (H3N8), and one equine isolate, A/equine/TN/5/86 (H3N8), bound strongly to 3′SL only (Fig. 5). Therefore, our experiments showed that differential cell tropism and virus spread of H3 influenza viruses isolated from different species were dependent, but only partially, on HA receptor specificity in NHBE cells.
Fig 5.
Receptor specificities of H3 human, avian, swine, and equine influenza A viruses. Association constants (Ka) of virus complexes with synthetic sialylglycopolymers conjugated to 3′SL(N) and 6′SL(N) are shown. Higher Ka values indicate stronger binding. Values are the means ± standard deviations (SD) from 4 independent experiments.
Replication kinetics of ca A/chicken/HK/G9/97 (H9N2) and wt and ca B/Hong Kong/330/01 influenza viruses in MDCK and NHBE cells.
Live attenuated A/Ann Arbor/6/60 ca influenza vaccines have been made with HA influenza subtypes with pandemic potential (H2, H5, H7, and H9 HA subtypes). The replication of such vaccine strains in mice and ferrets has not been predictive of replication of these viruses in humans (18, 19, 45, 46). Therefore, we assessed the growth capacity of the ca H9N2 pandemic vaccine strain, ca A/chicken/HK/G9/97, in MDCK and NHBE cells. We observed that the level of replication of ca A/chicken/HK/G9/97 (H9N2) was significantly different in the two cell culture systems (Table 5; see Table S1 in the supplemental material). In MDCK cells, ca H9N2 virus replicated to a significantly higher extent than other ca H1N1 and H3N2 reassortants studied, as seen by its significantly higher AUC (P < 0.01) (Tables 1, 2, and 5). In contrast, ca A/chicken/HK/G9/97 (H9N2) exhibited a viral load similar to those of other ca viruses, except ca A/California/10/78 (H1N1) (P < 0.01) (Table 1) and ca A/Panama/2007/99 (H3N2) (P < 0.05) (Table 2), in NHBE cultures. We observed significantly different peak viral titers of all studied ca vaccine strains in MDCK cells (P < 0.05). Conversely, no statistically significant difference was found between peak viral titers of ca reassortants, except ca A/Alaska/6/77 (H3N2), in NHBE cells (P < 0.05) (Tables 1, 2, and 5).
Table 5.
Areas under the curve and mean peak viral titers of ca A/Chicken/HK/G9/97 and wt and ca B/Hong Kong/330/01 influenza viruses in MDCK and NHBE cell cultures
| Virus | Subtype | Mean AUC (Mean peak titer, log10 PFU/ml)a |
|
|---|---|---|---|
| MDCK | NHBE | ||
| ca A/chicken/HK/G9/97 | H9N2 | 443.2 (7.1) | 216.6 (3.8) |
| B/Hong Kong/330/01 | |||
| wt | 218.2 (3.3) | 342.8 (5.4) | |
| ca | 235.0 (3.7) | 217.1c (3.5d) | |
| Degree of restrictionb | 16.8 (0.4) | 125.7 (1.9) | |
The area under the curve (AUC) represents total viral load at 24, 48, and 72 h postinfection. ca A/chicken/HK/G9/97 (H9N2) influenza virus did not produce plaques in MDCK cells; these values are defined as log10 TCID50/ml ± SD determined in MDCK cells after 3 days of incubation at 33°C with 10-fold serially diluted virus. Bold indicates an increase in titer and lack of restricted replication of ca influenza vaccine virus.
The degree of restriction of viral replication was expressed as the value for wt virus minus the value for the corresponding ca virus.
P < 0.05 compared with the value for wt B/Hong Kong/330/01 virus, determined with an unpaired t test.
P < 0.01 compared with the value for wt B/Hong Kong/330/01 virus, determined with an unpaired t test.
Finally, we assessed whether the NHBE culture model could adequately reflect the restriction of replication of the ca live influenza B virus vaccine strain, ca B/Hong Kong/330/01, in comparison with the respective wt B/Hong Kong/330/01 virus. Both isolates replicated to titers of ∼103.5 PFU/ml in MDCK cells, showing no statistically significant difference between their replication kinetics in this cell line (Table 5). In contrast, wt and ca B viruses replicated to significantly different titers at 24, 48, and 72 h after infection in NHBE cells (2.0-log-unit difference; P < 0.01) (data not shown). The ca B/Hong Kong/330/01 strain exhibited significantly limited growth, with a decrease of peak viral titer of 1.9 log unit (P < 0.01) compared to that of the wt virus in the epithelial cells (Table 5). Therefore, our experiments showed that differentiated NHBE cultures could reflect the restricted replication of ca influenza B viruses in humans.
DISCUSSION
We compared the replication kinetics of ca live attenuated vaccine candidates, including H1N1, H3N2, and B strains, in comparison with their respective wt viruses in MDCK cells, human adenoid epithelial cells, and bronchial airway epithelium to assess which cell culture model could more consistently and accurately reflect their infectivities in humans. Since the levels of replication of several wt and ca influenza A virus strains used in this study have been previously evaluated in clinical trials with adult and seronegative pediatric volunteers (10, 11, 28, 29), we were able to compare their viral growth in humans and three tissue culture systems. Previous studies demonstrated that three ca reassortant viruses, ca A/California/10/78 (H1N1), ca A/Alaska/6/77 (H3N2), and ca A/Washington/897/80 (H3N2), exhibited different degrees of attenuation (>3.9, >4.5, and ∼2.8 log units, respectively), as measured by comparison of mean peak viral titers (log10 TCID50/ml of nasopharyngeal wash sample) between the wt and ca viruses administered at similar doses to humans (10, 28, 29). Unfortunately, we were unable to determine any significant correlation by Spearman's rank correlation analysis (P > 0.05) between degrees of restriction of ca A/California/10/78 (H1N1), ca A/Alaska/6/77 (H3N2), and ca A/Washington/897/80 (H3N2) strains in humans and those in any of the cell culture systems tested. However, our data demonstrated that the levels of attenuation of these three ca viruses in human subjects were consistently higher (∼2.8 logs) (10, 28–30) than those in MDCK cells. Furthermore, the ca A/New Caledonia/20/99 (H1N1) reassortant replicated to significantly higher viral titers than the respective wt H1N1 virus (P < 0.01), and no statistically significant difference was observed between the replication abilities of wt and ca B/Hong Kong/330/01 viruses in MDCK cells. Taking these findings together, we can speculate that data obtained in MDCK cells could be the least predictive of restricted viral replication of ca live attenuated vaccine viruses in humans.
In this study, we observed that limited growth of ca vaccine viruses was reflected in HAEC and NHBE cultures. However, HAEC cells remain very difficult to grow and could vary between different donors, whereas the two other cell types studied are commercially available and therefore could be used from the same source/donor for different experiments. Our experiments showed that differentiated NHBE cultures exhibited a consistently greater restriction for all studied ca H1N1, H3N2, and B vaccine viruses among the three cell lines tested. A similar pattern was observed for ca A/California/10/78 (H1N1) virus in NHBE cells isolated from a different human donor (data not shown). Despite the fact that all tissue culture models lack a functional adaptive immune system, which would obviously play a role in virus replication in the host, NHBE cells exhibited the best correlation between the degrees of restriction of viral replication of ca A/California/10/78 (H1N1), ca A/Alaska/6/77 (H3N2), and ca A/Washington/897/80 (H3N2) and those seen in clinical trials with humans (10, 28–30). ca A/New Caledonia/20/99 (H1N1), ca A/Panama/2007/99 (H3N2), and ca A/Wyoming/03/03 (H3N2) showed the lowest virus yields in NHBE cells, which were in good correlation with recently published data (6, 23). Therefore, NHBE cells may be considered for use as the screening step for evaluating the restricted replication phenotype of potential influenza virus vaccine candidates in humans.
There has been a long-standing parallel development in Russia of potential live attenuated vaccines (20, 51). To date, there are no reports of side-by-side comparisons of the U.S. and Russian ca H2N2 vaccine donor strains, ca A/Ann Arbor/6/60, ca A/Leningrad/134/17/57, and ca A/Leningrad/134/47/57, in cell culture models or in humans. So far, in only one study were immune responses to these donor strains in the lungs and mediastinal lymph nodes of mice compared (52). Overall, combined data for viral clearance, antibody-secreting cells, and cytokine responses suggested that ca A/Leningrad/134/17/57 is a superior immunogen to ca A/Leningrad/134/47/57, which in turn is superior to ca A/Ann Arbor/6/60 (52). In addition, two reassortant vaccines prepared from the ca A/Ann Arbor/6/60 and ca A/Leningrad/134/17/57 donor strains with the surface antigens of A/Korea/1/82 (H3N2) were compared in rats, ferrets, and humans (34). A reassortant prepared from the Russian strain induced slightly better rates of seroconversion, but conclusions as to their relative immunogenicities could not be made because of differences in the number of internal genes present in each reassortant. Here, for the first time, we compared side by side the replication abilities of the U.S. and Russian ca H2N2 vaccine donor strains in two cell lines, MDCK and NHBE cells. We did not observe statistically significant differences in replication kinetics of these viruses (as seen by similar AUCs, viral peak titers, and virus yields at all time points) in MDCK cells. In contrast, a statistically significant difference between the replication abilities of the U.S. and Russian ca vaccine donor strains was detected in NHBE cultures (P < 0.05). ca A/Leningrad/134/17/57, which has become the main vaccine donor strain in Russia, replicated to significantly higher titers (∼1 log unit; P < 0.05) than ca A/Leningrad/134/47/57, which in turn replicated to significantly higher titers (∼2.7 log units; P < 0.01) than ca A/Ann Arbor/6/60 at 48 and 72 h after infection in human airway epithelium. Therefore, we observed that NHBE cultures were able to rank the levels of replication of various ca live attenuated influenza vaccines compared to each other. This property can assist in choosing the ca vaccine donor strain with the appropriate balance between replicative capacity and lack of reactogenicity.
For use in pandemic preparedness, live attenuated influenza vaccines will likely need to represent novel HA and NA genes from different species (45, 46). In this study, we assessed whether NHBE cultures could reflect the ability of an animal influenza virus to infect humans. Taking into account that previous studies demonstrated transmission of avian and swine viruses to humans (41) but that horse-to-human transmission remains to be reported (48), we can speculate that the pattern of virus infectivity of the studied H3 influenza viruses from different species of origin (i.e., human, avian, swine, and equine) in NHBE cells could parallel the pattern of infectivity of these strains in humans.
Our results also showed that NHBE cell cultures most adequately approximated the cellular tropism of influenza viruses isolated from different species to the human respiratory tract. Previous studies have suggested that airway epithelial cell cultures contain ciliated, nonciliated, and goblet cells. Although both types of receptors (SAα2,6 and SAα2,3) are present on the cell surface in NHBE cultures (as they are in HAEC cultures [43]), NHBE cultures express abundantly more SAα2,6, while SAα2,3 is expressed at a lower level (25, 37). It was shown that SAα2,6 receptor moieties were abundant on the apical surface of nonciliated cells and were particularly concentrated on the microvilli. However, a lower level of SAα2,6 was also observed on the apical surface of some ciliated and goblet cells (37, 50), suggesting that α2,6-linked SA is distributed across all cell types in NHBE cultures. “Avian-type” SAα2,3 receptors were shown to be predominant on the apical surface of most, but not all ciliated cells at the base of the cilial shaft in the region of microvilli and were also found to a much lesser degree on some nonciliated cells (25, 50). Taken together, our experiments showed that differential cell tropism and virus spread of human and avian influenza isolates were dependent, but only partially, on HA receptor specificity. However, our data also demonstrated that despite the fact that both human and swine viruses exhibited human-like virus receptor specificity, only human viruses replicated to a higher extent in NHBE cells. Among H3 viruses of different origin with avian-like receptor specificity, only avian and not equine strains were able to establish some limited infection in human airway epithelium. Overall, possible explanations for the more limited growth of the avian, swine, and equine strains compared to the human viruses could be innate immunity and some unknown host range mechanisms that are present in NHBE cells but not in MDCK or HAEC cells.
Finally, in this study we assessed the replication ability of the ca A/chicken/HK/G9/97 (H9N2) pandemic vaccine strain, which contained avian surface glycoproteins and was tested in a clinical trial in humans (18), in MDCK and NHBE cells. We compared its replication with those of human ca H1N1 and H3N2 vaccine strains in an attempt to explain the robust difference seen in replication between seasonal and avian ca influenza vaccines in healthy adult volunteers (9, 18, 19, 45, 46). The ca H9N2 virus replicated similarly to other classical ca reassortants and exhibited similar viral titers in NHBE cultures. However, the degree of restriction differed from that observed in the clinical trial in humans (18). We can speculate that although NHBE cultures are able to reflect the restricted phenotype of ca influenza vaccine viruses, the lack of replication of the avian ca H9N2 vaccine virus seen in adults (18), but not in the NHBE cell model, suggests the presence of host immune factors in humans that induce innate and heterosubtypic protection against infection with avian ca vaccines. Such heterosubtypic immunity could be conferred by previous infection with influenza viruses belonging to another HA subtypes and appears to be enough to restrict to some extent the replication of human ca vaccines as well (10, 28, 29).
In conclusion, in the present study we compared three tissue culture systems, i.e., HAEC, MDCK, and NHBE cells, for their ability to model the levels of restriction of ca influenza vaccine viruses and the risk of infection and cellular tropism of influenza strains of different origin with different receptor specificities. Our data showed that differentiated cultures of human airway epithelium exhibited the best approximation to the events in humans. Although clinical trials with human subjects are essential, NHBE cultures could be considered for use as a screening step for evaluating and possibly ranking the restricted replication of influenza vaccine candidates having HA proteins of different origin.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services, under contract no. HHSN272200900026C and ARRA funding and by the Intramural Research Program of NIAID, NIH.
We thank MedImmune (Mountain View, CA) for manufacturing ca influenza vaccine strains. We gratefully acknowledge the gift of sialic polymer substrates from Nicolai Bovin (Moscow, Russia).
Footnotes
Published ahead of print 22 August 2012
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1. Ashkenazi S, et al. 2006. Superior relative efficacy of live attenuated influenza vaccine compared with inactivated influenza vaccine in young children with recurrent respiratory tract infections. Pediatr. Infect. Dis. J. 25:870–879 [DOI] [PubMed] [Google Scholar]
- 2. Belshe RB, et al. 1992. Immunization of infants and young children with live attenuated trivalent cold recombinant influenza A H1N1 and H3N2 and B vaccine. J. Infect. Dis. 165:727–732 [DOI] [PubMed] [Google Scholar]
- 3. Belshe RB, et al. 2007. Live attenuated versus inactivated influenza vaccine in infants and young children. N. Engl. J. Med. 356:685–696 [DOI] [PubMed] [Google Scholar]
- 4. Belshe RB, Lee MS, Walker RE, Stoddard J, Mendelman PM. 2004. Safety, immunogenicity and efficacy of intranasal, live attenuated influenza vaccine. Expert Rev. Vaccines 3:643–654 [DOI] [PubMed] [Google Scholar]
- 5. Belshe RB, et al. 1998. The efficacy of live attenuated, cold-adapted, trivalent, intranasal influenzavirus vaccine in children. N. Engl. J. Med. 338:1405–1412 [DOI] [PubMed] [Google Scholar]
- 6. Block SL, et al. 2008. Shedding and immunogenicity of live attenuated influenza vaccine virus in subjects 5-49 years of age. Vaccine 26:4940–4946 [DOI] [PubMed] [Google Scholar]
- 7. Bovin NV, et al. 1993. Synthesis of polymeric neoglycoconjugates based on N-substituted polyacrylamides. Glycoconj. J. 10:142–151 [DOI] [PubMed] [Google Scholar]
- 8. Carter NJ, Curran MP. 2011. Live attenuated influenza vaccine (FluMist®; FluenzTM). Drugs 71:1591–1622 [DOI] [PubMed] [Google Scholar]
- 9. Chen H, et al. 2003. Generation and characterization of a cold-adapted influenza A H9N2 reassortant as a live pandemic influenza virus vaccine candidate. Vaccine 21:4430–4436 [DOI] [PubMed] [Google Scholar]
- 10. Clements ML, Betts RF, Maassab HF, Murphy BR. 1984. Dose response of influenza A/Washington/897/80 (H3N2) cold-adapted reassortant virus in adult volunteers. J. Infect. Dis. 149:814–815 [DOI] [PubMed] [Google Scholar]
- 11. Clements ML, Betts RF, Murphy BR. 1984. Advantage of live attenuated cold-adapted influenza A virus over inactivated vaccine for A/Washington/80 (H3N2) wild-type virus infection. Lancet 31:705–708 [DOI] [PubMed] [Google Scholar]
- 12. Edwards KM, et al. 1991. Safety and immunogenicity of live attenuated cold-adapted influenza B/Ann Arbor/1/86 reassortant virus vaccine in infants and children. J. Infect. Dis. 163:740–745 [DOI] [PubMed] [Google Scholar]
- 13. Endo Y, Carroll KN, Ikizler MR, Wright PF. 1996. Growth of influenza A virus in primary, differentiated epithelial cells derived from adenoids. J. Virol. 70:2055–2058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gambaryan AS, Matrosovich MN. 1992. A solid-phase enzyme-linked assay for influenza virus receptor-binding activity. J. Virol. Methods 39:111–123 [DOI] [PubMed] [Google Scholar]
- 15. Ghendon Y, et al. 1984. Recombinants of cold-adapted attenuated influenza A vaccines for use in chidren: molecular and genetic analysis of the cold-adapted donor and recombinants. Infect. Immun. 44:730–733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ilyushina NA, Govorkova EA, Gray TE, Bovin NV, Webster RG. 2008. Human-like receptor specificity does not affect the neuraminidase-inhibitor susceptibility of H5N1 influenza viruses. PLoS Pathog. 4:e1000043 doi:10.1371/journal.ppat.1000043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jin H, et al. 2003. Multiple amino acid residues confer temperature sensitivity to human influenza virus vaccine strains (FluMist) derived from cold-adapted A/Ann Arbor/6/60. Virology 306:18–24 [DOI] [PubMed] [Google Scholar]
- 18. Karron RA, et al. 2009. A live attenuated H9N2 influenza vaccine is well tolerated and immunogenic in healthy adults. J. Infect. Dis. 199:711–716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Karron RA, et al. 2009. Evaluation of two live attenuated cold-adapted H5N1 influenza virus vaccines in healthy adults. Vaccine 27:4953–4960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kendal AP, Maassab HF, Alexandrova GI, Ghendon YZ. 1981. Development of cold-adapted recombinant live, attenuated influenza A vaccines in the USA and USSR. Antiviral Res. 1:339–365 [Google Scholar]
- 21. Maassab HF, Bryant ML. 1999. The development of live attenuated cold-adapted influenza virus vaccine for humans. Rev. Med. Virol. 9:237–244 [DOI] [PubMed] [Google Scholar]
- 22. Maassab HF, et al. 1969. Laboratory and clinical characteristics of attenuated strains of influenza virus. Bull. World Health Organ. 41:589–594 [PMC free article] [PubMed] [Google Scholar]
- 23. Mallory RM, Yi T, Ambrose C. 2011. Shedding of Ann Arbor strain live attenuated influenza vaccine virus in children 6-59 months of age. Vaccine 29:4322–4327 [DOI] [PubMed] [Google Scholar]
- 24. Matrosovich MN, et al. 1993. Probing of the receptor-binding sites of the H1 and H3 influenza virus hemagglutinins by synthetic and natural sialosides. Virology 196:111–123 [DOI] [PubMed] [Google Scholar]
- 25. Matrosovich MN, Matrosovich TY, Gray T, Roberts NA, Klenk H-D. 2004. Human and avian influenza viruses target different cell types in cultures of human airway epithelium. Proc. Natl. Acad. Sci. U. S. A. 101:4620–4624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mochalova L, et al. 2003. Receptor-binding properties of modern human influenza viruses primarily isolated in Vero and MDCK cells and chicken embryonated eggs. Virology 313:473–480 [DOI] [PubMed] [Google Scholar]
- 27. Monto AS, et al. 2009. Comparative efficacy of inactivated and live attenuated influenza vaccines. N. Engl. J. Med. 361:1260–1267 [DOI] [PubMed] [Google Scholar]
- 28. Murphy BR, et al. 1981. Evaluation of A/Alaska/6/77 (H3N2) cold-adapted recombinant viruses derived from A/Ann Arbor/6/60 cold-adapted donor virus in adult seronegative volunteers. Infect. Immun. 32:693–697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Murphy BR, et al. 1984. Dose response of cold-adapted, reassortant influenza A/California/10/78 virus (H1N1) in adult volunteers. J. Infect. Dis. 149:816. [DOI] [PubMed] [Google Scholar]
- 30. Murphy BR, Coelingh K. 2002. Principles underlying the development and use of live attenuated cold-adapted influenza A and B virus vaccines. Viral Immunol. 15:295–323 [DOI] [PubMed] [Google Scholar]
- 31. Murphy BR. 1993. Use of live attenuated cold-adapted influenza reassortant virus vaccines in infants, children, young adults, and elderly adults. Infect. Dis. Clin. Pract. 2:174–181 [Google Scholar]
- 32. Nair H, et al. 2011. Global burden of respiratory infections due to seasonal influenza in young children: a systematic review and meta-analysis. Lancet 378:1917–1930 [DOI] [PubMed] [Google Scholar]
- 33. Nichol KL, et al. 1999. Effectiveness of live, attenuated intranasal influenza virus vaccine in healthy, working adults: a randomized controlled trial. JAMA 282:137–144 [DOI] [PubMed] [Google Scholar]
- 34. Nicholson KG, et al. 1987. Infectivity and reactogenicity of reassortant cold-adapted influenza A/Korea/1/82 vaccines obtained from the USA and USSR. Bull. World Health Organ. 65:295–301 [PMC free article] [PubMed] [Google Scholar]
- 35. Nicholson KG, Wood JM, Zambon M. 2003. Influenza. Lancet 362:1733–1745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ohmit SE, et al. 2006. Prevention of antigenically drifted influenza by inactivated and live attenuated vaccines. N. Engl. J. Med. 355:2513–2522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Oshansky CM, et al. 2011. Avian influenza viruses infect primary human bronchial epithelial cells unconstrained by sialic acid α2,3 residues. PLoS One 6:e21183 doi:10.1371/journal.pone.0021183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Osterholm MT, Kelley NS, Sommer A, Belongia EA. 2012. Efficacy and effectiveness of influenza vaccines: a systematic review and meta-analysis. Lancet Infect. Dis. 12:36–44 [DOI] [PubMed] [Google Scholar]
- 39. Parrish CR, Kawaoka Y. 2005. The origins of new pandemic viruses: the acquisition of new host ranges by canine parvovirus and influenza A viruses. Annu. Rev. Microbiol. 59:553–586 [DOI] [PubMed] [Google Scholar]
- 40. Reed LJ, Muench H. 1938. A simple method for estimating fifty percent endpoints. Am. J. Hyg. (Lond.) 27:493–497 [Google Scholar]
- 41. Rota PA, et al. 1989. Laboratory characterization of a swine influenza virus isolated from a fatal case of human influenza. J. Clin. Microbiol. 27:1413–1416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sears SD, et al. 1988. Comparison of live, attenuated H1N1 and H3N2 cold-adapted and avian-human influenza A reassortant viruses and inactivated virus vaccine in adults. J. Infect. Dis. 158:1209–1219 [DOI] [PubMed] [Google Scholar]
- 43. Shinya K, et al. 2006. Avian flu: influenza virus receptors in the human airway. Nature 440:435–436 [DOI] [PubMed] [Google Scholar]
- 44. Stevens J, et al. 2006. Structure and receptor specificity of the hemagglutinin from an H5N1 influenza virus. Science 312:404–410 [DOI] [PubMed] [Google Scholar]
- 45. Subbarao K, Joseph T. 2007. Scientific barriers to developing vaccines against avian influenza viruses. Nat. Rev. Immunol. 7:267–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Subbarao K, Murphy B, Fauci AS. 2006. Development of effective vaccines against pandemic influenza. Immunity 24:5–9 [DOI] [PubMed] [Google Scholar]
- 47. Suguitan AL, et al. 2006. Live, attenuated influenza A H5N1 candidate vaccines provide broad cross-protection in mice and ferrets. PLoS Med. 3:1541–1554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Suzuki Y, et al. 2000. Sialic acid species as a determinant of the host range of influenza A viruses. J. Virol. 74:11825–11831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Talbot R, et al. 2005. Duration of virus shedding after trivalent intranasal live attenuated influenza vaccination in adults. Infect. Control Hosp. Epidemiol. 26:494–500 [DOI] [PubMed] [Google Scholar]
- 50. Thompson CI, Barclay WS, Zambon MC, Pickles RJ. 2006. Infection of human airway epithelium by human and avian strains of influenza A virus. J. Virol. 80:8060–8068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Wareing MD, Tannock GA. 2001. Live attenuated vaccines against influenza; an historical review. Vaccine 19:3320–3330 [DOI] [PubMed] [Google Scholar]
- 52. Wareing MD, Watson JM, Brooks MJ, Tannock GA. 2001. Immunogenic and isotype-specific responses to Russian and US cold-adapted influenza A vaccine donor strains A/Leningrad/134/17/57, A/Leningrad/134/47/57, and A/Ann Arbor/6/60 (H2N2) in mice. J. Med. Virol. 65:171–177 [PubMed] [Google Scholar]
- 53. Wright PF, et al. 2005. Growth of respiratory syncytial virus in primary epithelial cells from the human respiratory tract. J. Virol. 79:8651–8654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Wright PF, Neumann G, Kawaoka Y. 2006. Orthomyxoviruses, p 1641–1740 In Knipe DM, et al. (ed), Fields virology, 5th ed Lippincott Williams and Wilkins, Philadelphia, PA [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.