Abstract
Herpes simplex virus 1 (HSV-1) was shown to contain catalase, an enzyme able to detoxify hydrogen peroxide by converting it to water and oxygen. Studies with a catalase inhibitor indicated that virus-associated catalase can have a role in protecting the virus from oxidative inactivation. HSV-1 was found to be more sensitive to killing by hydrogen peroxide in the presence of a catalase inhibitor than in its absence. The results suggest a protective role for catalase during the time HSV-1 spends in the oxidizing environment outside a host cell.
TEXT
Viruses experience quite different environments depending on whether they are replicating inside a host cell or in transit from one host to another. Within a cell, the virus and virus components are exposed to a reducing environment, where the redox potential is determined primarily by glutathione (18). In contrast, outside a cell, the virus is exposed to oxygen and toxic products derived from oxygen, such as hydrogen peroxide, superoxide, and hydroxyl radical, reactive species that have the potential to inactivate the virus. To cope with such highly reactive compounds, plants and animals express enzymes able to convert them to nontoxic products. Examples of such enzymes are catalase, peroxidases, and superoxide dismutase (28). Here, we describe the results of studies that demonstrate the presence of catalase inside the purified herpes simplex virion. Tests were then carried out to determine whether internal catalase could protect the virus from inactivation by H2O2.
Studies of catalase were carried out with herpes simplex virus 1 (HSV-1) that was grown on Vero cells in culture and purified by sucrose density gradient centrifugation. When virus suspensions were adjusted to 1% H2O2, bubbles of oxygen began to form promptly, indicating the presence of catalase (Fig. 1a, left tube). Bubbles became apparent visually after a few seconds of incubation at room temperature and continued to form and enlarge for at least 20 min. No bubbles formed, however, if the catalase inhibitor sodium azide was added to the virus suspension prior to H2O2 treatment (Fig. 1a, right). Assays were also negative when (i) the virus was removed from the solution by centrifugation prior to the addition of H2O2 or (ii) HSV-1 capsids (B capsids) were substituted for intact virus. In similar assays, bubbles indicating the presence of catalase were not observed with purified vesicular stomatitis virus or human adenovirus 2 (data not shown). Western blot analysis confirmed the presence of catalase associated with HSV-1 virus but not with adenovirus, vesicular stomatitis virus (VSV), or HSV-1 A capsids (Fig. 1b).
Fig 1.
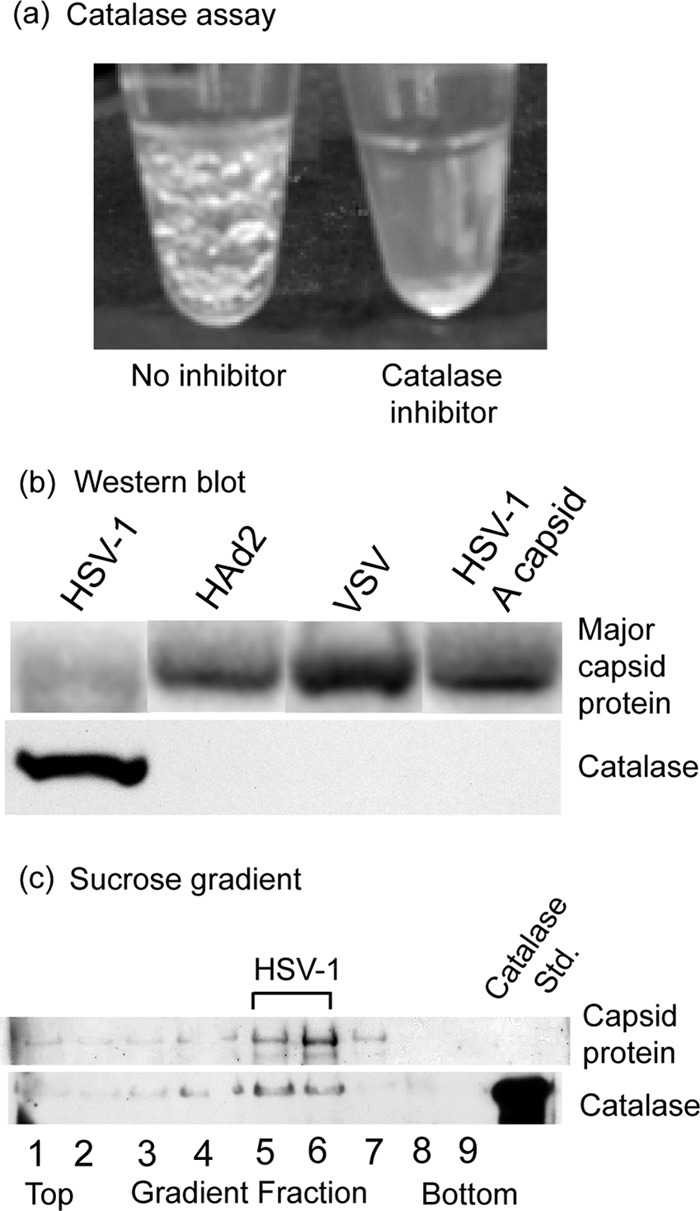
Identification of HSV-1-associated catalase by enzyme assay (a), Western blot analysis (b), and sucrose density gradient centrifugation (c). Panel a shows a tube containing purified HSV-1 20 min after the solution was adjusted to 1% H2O2 (left tube) and a similar incubation to which a catalase inhibitor (2 mM sodium azide) was added prior to H2O2 (right). Note the formation of bubbles in the left tube, indicating the presence of catalase in the virus. The virus concentration was 0.25 mg/ml in TNE (0.01 M Tris-HCl, 0.5 M NaCl, 1 mM EDTA, pH 7.5), and incubation was at room temperature. The virus was the KOS strain of HSV-1, which was grown on monolayer cultures of Vero cells and purified by sucrose density gradient centrifugation, and the titer was determined by endpoint dilution as previously described (14). Panel b shows the results of a Western blot test for the presence of catalase in HSV-1, human adenovirus 2 (HAd2), vesicular stomatitis virus (VSV), and HSV-1 A capsids. Human adenovirus 2 and vesicular stomatitis virus (Indiana; San Juan strain) were grown on monolayer cultures of HeLa and Vero cells, respectively, and purified by previously described procedures (12, 17). Published methods were also used for purification of HSV-1 A and B capsids (23). Major capsid proteins were detected by staining the blot with 1% Ponceau S (upper row), while catalase was detected by immunostaining with antibody specific for catalase (Calbiochem; rabbit polyclonal 219010; 1:5,000 dilution) (15). Note that catalase was detected in HSV-1 virus but not HAd2, VSV, or HSV-1 A capsids. Panel c shows the results of sucrose density gradient analysis. A total of 50 μg of purified HSV-1 in 50 μl TNE was centrifuged on a 600-μl gradient of 20% to 50% sucrose in TNE. Centrifugation was for 45 min at 22,000 rpm in a Beckman SW55 rotor at 4°C. The gradient was fractionated, and individual fractions were analyzed by SDS-PAGE followed by blotting onto polyvinylidene difluoride (PVDF) and staining with 1% Ponceau S. The blot was then destained and stained with antibody specific for catalase (which migrated coincidently with standard catalase; Worthington LS001872). Note that the position of the virus in the gradient (top row) coincides with that of catalase (bottom row), suggesting the two are associated.
Control experiments were carried out to confirm that catalase was associated with HSV-1 and not with impurities present in the virus preparation. Purified HSV-1 was centrifuged into a band on a sucrose density gradient, the gradient was fractionated, and Western blot analysis was used to test individual gradient fractions for the presence of catalase. The results showed that catalase was present in virus-containing fractions but not in flanking ones (Fig. 1c). The results are interpreted to indicate that catalase is associated with HSV-1 and not with contaminants, such as catalase-containing bacteria or host cell materials in the virus preparation. Since the HSV-1 genome does not encode catalase (22), the virus-associated enzyme must be derived from the host cell. Early studies of vaccinia virus demonstrated the presence of catalase inside the mature virion (8). Apart from this observation, we know of no other report of catalase as a component of a virus structure.
Further analysis of HSV-1-associated catalase was aimed at determining the location of the enzyme in the virion. Two types of experiments were done. First, purified virus was treated with the proteolytic enzyme pronase to degrade the virus glycoproteins and any proteins not contained within the virion membrane. Internal viral proteins were expected to be unaffected, as pronase does not cross the virus lipid bilayer (16). After pronase treatment, the virus was harvested by centrifugation, and the extent of catalase loss was determined by Western blot analysis. The results showed no evidence of catalase loss in two concentrations of virus tested (see the catalase band in Fig. 2a, lanes 1 and 2). Internal controls demonstrated that the virus surface glycoproteins gB and gH were cleaved as expected (Fig. 2a; compare lanes 2 and 3). Similarly, Western blot analysis demonstrated that HSV-1 glycoprotein D was digested by pronase or trypsin under conditions in which catalase was not affected (Fig. 2b). Purified catalase in solution was also digested under the same conditions. In contrast, no degradation of internal tegument (UL47, UL48, and UL49) or capsid proteins (UL19, UL38, UL18; Fig. 2a, lanes 1 and 2) was observed. The results are interpreted to indicate that catalase is located inside the HSV-1 envelope.
Fig 2.
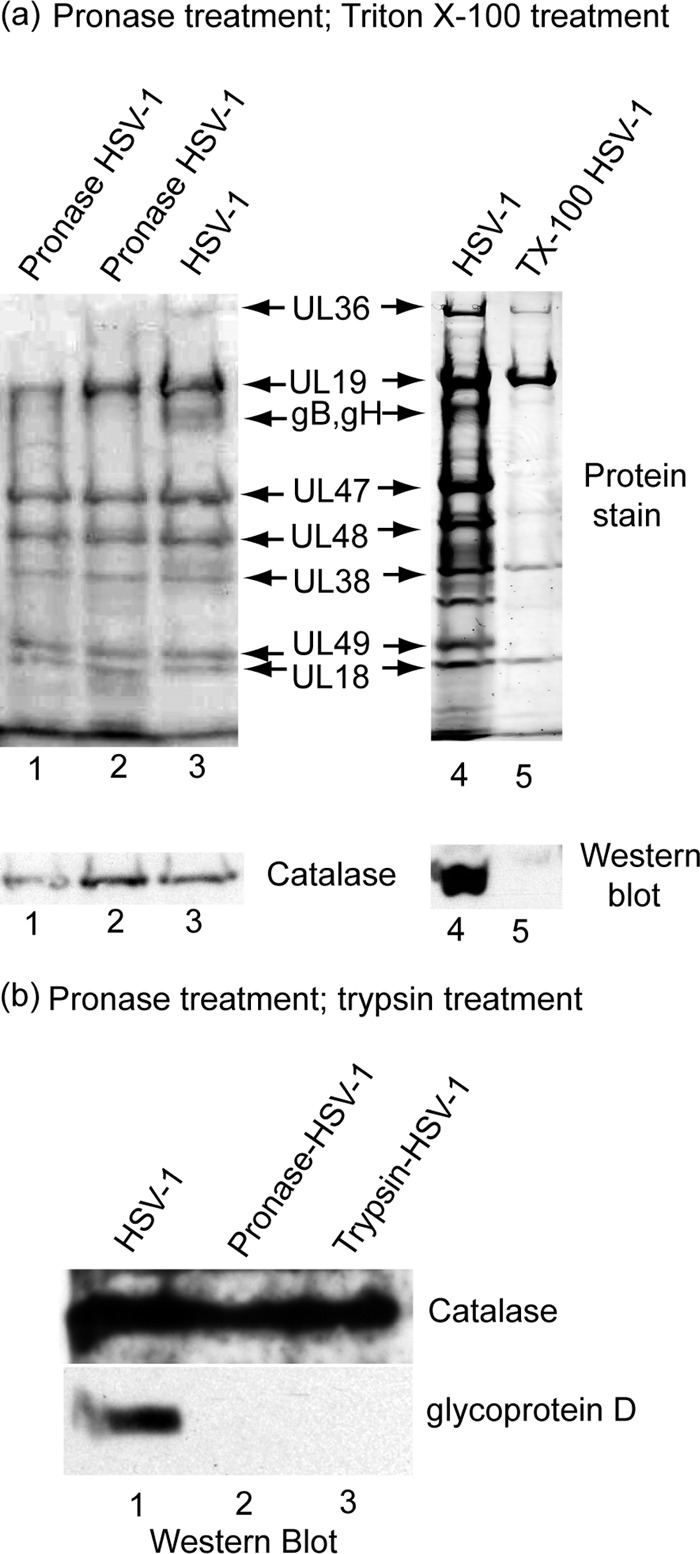
Localization of catalase in HSV-1 by treatment of purified virions with pronase (lanes 1 to 3) and Triton X-100 (a, lanes 4 and 5) and by pronase and trypsin treatment (b, lanes 1 to 3). Panel a shows SDS-PAGE and Western blot analysis of proteins present in intact HSV-1 (lanes 3 and 4) and in virions after treatment with pronase (lanes 1 and 2) and Triton X-100 (lane 5). Note that catalase was found to be present in intact virions (lanes 3 and 4) and in pronase-treated virions (lanes 1 and 2) but not in virions after treatment with 1% Triton X-100 (lane 5). Pronase treatment was carried out by digesting purified HSV-1 (0.25 mg/ml) with 1 mg/ml pronase (Streptomyces griseus; Calbiochem 537088) for 4 h at 37°C. After pronase treatment, the virus was reisolated by sucrose density gradient centrifugation prior to SDS-PAGE and Western blot analysis. Triton X-100 treatment was performed by adjusting purified virus to 1% Triton X-100 in TNE buffer containing 10 mM dithiothreitol (DTT) and incubating for 15 min on ice (4°C). The resulting capsids were then purified by sucrose gradient centrifugation before SDS-PAGE and Western blot analysis. (b) Western blot analysis of purified HSV-1 after treatment in vitro with pronase or trypsin. A total of 100 μl of purified HSV-1 (1 mg/ml) was adjusted to 2 mg/ml pronase or 2 mg/ml trypsin. A control incubation had no enzyme. Mixtures were incubated for 2 h at 37°C, and virions were purified away from enzymes by centrifugation on a 20% to 50% sucrose gradient as described in the legend to Fig. 1. Virus specimens were pelleted by centrifugation and examined by SDS-PAGE and Western blot analysis. Blots were stained for catalase (rabbit polyclonal; see above) and glycoprotein D (mouse monoclonal antibody DL11; a gift from Gary Cohen and Roselyn Eisenberg; 1:5,000). Note that protease treatment caused digestion of glycoprotein D, but not catalase, indicating that catalase is located inside the HSV-1 membrane.
A more precise definition for the location of catalase was obtained by treatment of purified virus with the nonionic detergent Triton X-100 (TX-100). When carried out with fresh virus, this treatment causes loss of the virus membrane, the membrane glycoproteins, and nearly all the 20 or more tegument proteins (all but UL36, UL37, and US3) (13, 21, 27). The capsid retains its integrity, however, and none of the major capsid proteins are lost. The virus DNA is retained inside the capsid. Experiments involved treatment of HSV-1 with 1% TX-100 and isolation of the resulting capsids by sucrose density gradient centrifugation. Western blot analysis was then used to test the capsids for the presence of catalase. The results demonstrated that the virus glycoproteins and tegument proteins were removed as expected and that catalase was removed as well (see Fig. 2a, lanes 4 and 5). This experiment is interpreted to indicate that catalase is present in the HSV-1 tegument.
Information such as that shown in Fig. 2 can be used to determine the number of catalase molecules per HSV-1 virion (15). This measurement was carried out beginning with two identical aliquots of purified virus. The two were used for determination of (i) the number of major capsid protein molecules (UL19) from a Coomassie blue-stained gel and (ii) the number of 60-kDa catalase molecules from a calibrated Western blot. In a representative determination, this analysis yielded a value of 1:207.5 for the molar ratio of catalase to UL19. Since there are 955 UL19 molecules per HSV-1 capsid, the number of catalase molecules per capsid was determined to be 955/207.5 or 4.6. A second similar determination yielded a value of 7.1 catalase molecules. As there are four 60-kDa subunits in an active catalase molecule (11, 20), the results indicate the presence of 1 to 2 catalase tetramers per virion.
The presence of catalase inside the HSV-1 virion suggests the possibility that it may be involved in protection of the virus from oxidative damage by H2O2. Alternatively, catalase may be passively incorporated into HSV-1 as the tegument is added in the host cell cytoplasm and not have any protective function. To distinguish between the two possibilities, we examined the sensitivity of purified HSV-1 to inactivation by H2O2 in vitro. HSV-1 was treated with H2O2 in the presence or absence of the catalase inhibitor sodium azide (NaN3), and the virus titer was determined thereafter (in the absence of catalase inhibitor). The results showed that while 50 mM H2O2 produced a modest decrease in titer (3- to 4-fold), a lethal effect of 106-fold or greater was observed if NaN3 was present (Table 1). Control experiments showed little killing of HSV-1 by NaN3 alone (Table 1). The results are interpreted to indicate that catalase provides a significant level of protection against inactivation of HSV-1 by H2O2.
Table 1.
Virus titer after treatment with H2O2 with or without a catalase inhibitor (NaN3)a
| Treatment | Titer |
|||||
|---|---|---|---|---|---|---|
| Time in 10 mM H2O2 |
Time in 50 mM H2O2 |
|||||
| None | 2 h | 20 h | None | 2 h | 20 h | |
| No NaN3 | 28 × 1010 | 60 × 1010 | 8 × 1010 | 28 × 1010 | 10 × 1010 | 8 × 1010 |
| 2 mM NaN3 | 30 × 1010 | 6 × 1010 | <104 | 30 × 1010 | 1 × 1010 | <104 |
Purified HSV-1 (200 μl; 0.25 mg/ml in TNE) was incubated at room temperature for the indicated times with H2O2 in the presence or absence of 2 mM NaN3, a catalase inhibitor. The virus titer was then determined on Vero cells (14) in the absence of H2O2 and NaN3.
It is not expected that HSV-1 would need to be protected from oxidative damage by H2O2 while it is associated with a host cell. As described above, a reducing environment is found within the cell, and that would prevent formation of H2O2. Outside the host cell, however, the environment is an oxidizing one, able to produce H2O2 both inside the virus and in the surrounding medium. HSV-1 would encounter this environment as it is transmitted from one host to another, and catalase may be involved in protection of HSV-1 infectivity during transit. HSV-1-associated catalase may also provide protection from H2O2 produced by commensal bacteria or other sources. H2O2 produced by lactobacilli, for instance, has been demonstrated to protect the female genital tract from disease due to microbial infection (4). Because of its high catalytic rate, virus-associated catalase could have a role in protection of HSV-1 despite its low copy number (1 to 2 copies per virion). Liver catalase, for instance, is able to detoxify tens of millions of H2O2 molecules per second, among the highest catalytic rates reported for any enzyme (2).
All viruses in the herpes family have a tegument, a layer of protein that lies between the virus capsid and the membrane (5, 6, 9, 13). The HSV-1 tegument is 40 to 50 nm in thickness and consists of approximately 20 distinct protein species, nearly all of which are encoded in the virus genome. Tegument proteins differ substantially in their abundance in the virion with 800 copies or more of the major species, such as UL47, UL48, and UL49 (see Fig. 2a, lane 3) (5, 16). Many tegument proteins are involved in early steps of HSV-1 replication, such as activation of early gene transcription and attenuation of host cell protein synthesis (13, 24, 25). The tegument is assembled into the nascent HSV-1 virion as a DNA-containing capsid buds into a vesicle of the trans-Golgi network (10). It is suggested that catalase is incorporated along with other tegument proteins during the budding process.
In uninfected cells, most catalase is found sequestered in peroxisomes (26). In order for it to be incorporated into progeny HSV-1 during tegumentation as described above, catalase would need to be released from peroxisomes. We suggest this may take place as a consequence of the large-scale rearrangement of cytoplasmic membranes that accompanies HSV-1 replication (1).
Palamara et al. (19) have demonstrated that a decrease in the cytoplasmic glutathione concentration occurs promptly after Vero cells are infected with HSV-1. A decrease in glutathione concentration is expected to cause a decrease in the cytosolic reducing potential, and this decrease appears to potentiate HSV-1 replication. Extra glutathione provided in the growth medium was found to antagonize HSV-1 growth (19). Like externally added glutathione, cytosolic catalase may increase the reducing potential of the cytoplasm by removing H2O2. It is suggested, therefore, that one consequence of catalase incorporation into progeny virions may be to potentiate virus growth by depriving infected cells of catalase.
The small number of catalase molecules per virion may explain why it was not detected in a mass spectrometric analysis of whole HSV-1 virions (7). There are several high-copy-number HSV-1 virion proteins that could obscure signal from catalase (for instance, there are thousands of copies of glycoprotein molecules and 955 copies of the major capsid protein [3]). The abundance of catalase does not meet the informal standard for detection by mass spectrometry (visibility on a Coomassie-stained SDS-PAGE gel). Peroxiredoxin, a peroxisomal protein with antioxidant activity, was detected in the mass spectrometric analysis of HSV-1 (7, 26).
In the future, it may be possible to exploit the sensitivity of HSV-1 to the cytotoxic effects of H2O2. It is expected that the virus would be especially vulnerable when it is in an oxidizing environment outside the host cell and when catalase is inhibited.
ACKNOWLEDGMENTS
We thank Tom Crowell, Dean Kedes, Lisa Jones, and Nick Sherman for help with interpretation of results, Djamila Harouaka for a gift of vesicular stomatitis virus, and Oneida Mason for help with experimental design.
This work was supported by NIH award AI041644.
Footnotes
Published ahead of print 22 August 2012
REFERENCES
- 1. Avitabile E, et al. 1995. Redistribution of microtubules and Golgi apparatus in herpes simplex virus-infected cells and their role in viral exocytosis. J. Virol. 69:7472–7482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Greenfield RE, Price VE. 1954. Liver catalase. I. A manometric determination of catalase activity. J. Biol. Chem. 209:355–361 [PubMed] [Google Scholar]
- 3. Handler CG, Eisenberg RJ, Cohen GH. 1996. Oligomeric structure of glycoproteins in herpes simplex virus type 1. J. Virol. 70:6067–6070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hawes SE, et al. 1996. Hydrogen peroxide-producing lactobacilli and acquisition of vaginal infections. J. Infect. Dis. 174:1058–1063 [DOI] [PubMed] [Google Scholar]
- 5. Heine JW, Honess RW, Cassai E, Roizman B. 1974. Proteins specified by herpes simplex virus XII. The virion polypeptides of type 1 strains. J. Virol. 14:640–651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kelly BJ, Fraefel C, Cunningham AL, Diefenbach RJ. 2009. Functional roles of the tegument proteins of herpes simplex virus type 1. Virus Res. 145:173–186 [DOI] [PubMed] [Google Scholar]
- 7. Loret S, Guay G, Lippe R. 2008. Comprehensive characterization of extracellular herpes simplex virus type 1 virions. J. Virol. 82:8605–8618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Macfarlane MG, Salaman MH. 1938. The enzymic activity of vaccinial elementary bodies. Brit. J. Exptl. Path. 19:184–191 [Google Scholar]
- 9. Maurer UE, Sodeik B, Grunewald K. 2008. Native 3D intermediates of membrane fusion in herpes simplex virus 1 entry. Proc. Natl. Acad. Sci. U. S. A. 105:10559–10564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Mettenleiter TC, Klupp BG, Granzow H. 2009. Herpesvirus assembly: an update. Virus Res. 143:222–234 [DOI] [PubMed] [Google Scholar]
- 11. Murthy MR, Reid TJ, Sicignano A, III, Tanaka N, Rossmann MG. 1981. Structure of beef liver catalase. J. Mol. Biol. 152:465–499 [DOI] [PubMed] [Google Scholar]
- 12. Newcomb WW, Boring JW, Brown JC. 1984. Ion etching of human adenovirus 2: structure of the core. J. Virol. 51:52–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Newcomb WW, Brown JC. 2009. Time-dependent transformation of the herpesvirus tegument. J. Virol. 83:8082–8089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Newcomb WW, Brown JC. 2010. Structure and capsid association of the herpesvirus large tegument protein UL36. J. Virol. 84:9408–9414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Newcomb WW, Homa FL, Brown JC. 2006. Herpes simplex virus capsid structure: DNA packaging protein UL25 is located on the external surface of the capsid near the vertices. J. Virol. 80:6286–6294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Newcomb WW, Jones LM, Dee A, Chaudhry F, Brown JC. 2012. Role of a reducing environment in disassembly of the herpesvirus tegument. Virology 431:71–79 [DOI] [PubMed] [Google Scholar]
- 17. Newcomb WW, Tobin GJ, McGowan JJ, Brown JC. 1982. In vitro reassembly of vesicular stomatitis virus skeletons. J. Virol. 41:1055–1062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ostergaard H, Tachibana C, Winther JR. 2004. Monitoring disulfide bond formation in the eukaryotic cytosol. J. Cell Biol. 166:337–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Palamara AT, et al. 1995. Evidence for antiviral activity of glutathione: in vitro inhibition of herpes simplex virus type 1 replication. Antiviral Res. 27:237–253 [DOI] [PubMed] [Google Scholar]
- 20. Putnam CD, Arvai AS, Bourne Y, Tainer JA. 2000. Active and inhibited human catalase structures: ligand and NADPH binding and catalytic mechanism1. J. Mol. Biol. 296:295–309 [DOI] [PubMed] [Google Scholar]
- 21. Radtke K, et al. 2010. Plus- and minus-end directed microtubule motors bind simultaneously to herpes simplex virus capsids using different inner tegument structures. PLoS Pathog. 6:e1000991 doi:10.1371/journal.ppat.1000991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Roizman B. 1996. The function of herpes simplex virus genes: a primer for genetic engineering of novel vectors. Proc. Natl. Acad. Sci. U. S. A. 93:11307–11312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sheaffer AK, et al. 2001. Herpes simplex virus DNA cleavage and packaging proteins associate with the procapsid prior to its maturation. J. Virol. 75:687–698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Stern S, Tanaka M, Herr W. 1989. The Oct-1 homeodomain directs formation of a multiprotein-DNA complex with the HSV transactivator VP16. Nature 341:624–630 [DOI] [PubMed] [Google Scholar]
- 25. Taddeo B, Roizman B. 2006. The virion host shutoff protein (UL41) of herpes simplex virus 1 is an endoribonuclease with a substrate specificity similar to that of RNase A. J. Virol. 80:9341–9345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wanders RJ, Waterham HR. 2006. Biochemistry of mammalian peroxisomes revisited. Annu. Rev. Biochem. 75:295–332 [DOI] [PubMed] [Google Scholar]
- 27. Wolfstein A, et al. 2006. The inner tegument promotes herpes simplex virus capsid motility along microtubules in vitro. Traffic 7:227–237 [DOI] [PubMed] [Google Scholar]
- 28. Zamocky M, Furtmuller PG, Obinger C. 2008. Evolution of catalases from bacteria to humans. Antioxid. Redox Signal. 10:1527–1548 [DOI] [PMC free article] [PubMed] [Google Scholar]


