Abstract
The first line of defense against viral infection is the interferon (IFN) response, which culminates in the expression of hundreds of proteins with presumed antiviral activity, and must be overcome by a virus for successful replication. The nonstructural NSs protein is the primary IFN antagonist encoded by Bunyamwera virus (BUNV), the prototype of the Orthobunyavirus genus and the family Bunyaviridae. The NSs protein interferes with RNA polymerase II-mediated transcription, thereby inhibiting cellular mRNA production, including IFN mRNAs. A recombinant virus, rBUNdelNSs, that is unable to express the NSs protein does not inhibit cellular transcription and is a strong IFN inducer. We report here that cells stimulated into the antiviral state by IFN-β treatment were protected against wild-type BUNV and rBUNdelNSs infection but addition of IFN-β after infection had little effect on the replication cycle of either virus. By screening a panel of cell lines that overexpressed individual IFN-stimulated genes, we found that protein kinase R (PKR), MTAP44, and particularly viperin appreciably restricted BUNV replication. The enzymatic activities of PKR and viperin were required for their inhibitory activities. Taken together, our data show that the restriction of BUNV replication mediated by IFN is an accumulated effect of at least three IFN-stimulated genes that probably act on different stages of the viral replication cycle.
INTRODUCTION
A major component of vertebrate innate immunity is the type I interferon (IFN) system. Type I IFNs (IFN-α and -β) are glycoproteins synthesized and secreted by cells in response to a virus infection. Pattern recognition receptors detect the presence of virus, and the resultant IFN-α/β secreted by the infected cell binds to the IFN-α/β receptor (IFNAR) in an autocrine and paracrine manner. This triggers the JAK/STAT signal transduction pathways resulting in the upregulation of hundreds of IFN-stimulated genes (ISGs) and the establishment of the so-called “antiviral state” that limits the spread of infection (3, 20). Well-characterized ISGs include the 2′-5′ oligoadenylate synthetase (OAS)/RNase L system, the Mx proteins, ISG15, the double-stranded RNA (dsRNA)-dependent protein kinase R (PKR), and viperin (36, 38). However, little is known about the antiviral activities of most ISG products, though they are probably able to inhibit viral replication at many stages in the virus life cycle. The IFN system is a powerful response to virus infection, and most, if not all, viruses have evolved mechanisms to overcome it to some extent in order to replicate (34). Thus, for any particular virus it is of interest not only to study the viral IFN antagonist but also to identify and elucidate the mechanism by which individual ISGs inhibit viral replication, as this may help in the development of new antiviral therapies.
The family Bunyaviridae contains more than 350 serologically distinct viruses that share morphological and molecular characteristics (32). Important pathogens in this family include Crimean-Congo hemorrhagic fever virus (CCHFV), Hantaan virus, Rift Valley fever virus (RVFV), Oropouche virus, and tomato spotted wilt virus, and together with other bunyaviruses, they cause significant socioeconomic costs annually through disease of animals, humans, and plants. Few vaccines are available to prevent the majority of bunyavirus diseases (33). Bunyamwera virus (BUNV) is the prototype of both the family Bunyaviridae and the Orthobunyavirus genus. BUNV has a single-stranded, negative-sense RNA genome comprising three differently sized segments designated large (L), medium (M), and small (S). The L segment encodes the viral RNA-dependent RNA polymerase (or L protein), the M segment encodes the two envelope glycoproteins Gn and Gc and a nonstructural protein called NSm, and the S segment codes for the nucleocapsid (N) protein and, in an overlapping reading frame, a second nonstructural protein termed NSs. Each genome segment is encapsidated by the N protein to form ribonucleoprotein (RNP) complexes that are the templates for RNA synthesis by the L protein (reviewed in reference 9).
The NSs protein has multiple functions in the virus life cycle. NSs was shown to regulate viral polymerase activity in a minireplicon system (48). A recombinant virus lacking NSs, rBUNdelNSs, is attenuated in IFN-competent cells, activates the IFN-β promoter, and therefore is a strong IFN-α/β inducer (5, 19). Although NSs predominantly localizes to the cytoplasm, a proportion enters the nucleus, where it inhibits phosphorylation of the C-terminal domain (CTD) of RNA polymerase II, resulting in cessation of all RNA polymerase II-driven transcription, including synthesis of IFN mRNAs. This is mediated by interaction of NSs with the cellular protein MED8, a component of the Mediator complex, which is involved in control of mRNA synthesis (23). Consequently, NSs is the primary IFN antagonist. In addition, NSs inhibits translation of cellular mRNAs (2, 14). However, NSs is not able to dismantle a preexisting antiviral state, as Streitenfeld et al. (43) demonstrated that IFN-treated cells had decreased susceptibility to BUNV infection.
To understand more about the IFN-induced inhibition of BUNV replication, we screened a panel of cell lines expressing 20 individual ISGs (18) for the ability to support BUNV multiplication. We found that a number of ISGs had modest inhibitory activity but that PKR, MTAP44, and particularly viperin caused a significant reduction in virus replication.
MATERIALS AND METHODS
Cells and viruses.
BHK-21 cells were grown in Glasgow modified Eagle's medium (GMEM) supplemented with 8% tryptose phosphate broth and 10% newborn calf serum (NCS; Invitrogen), and Vero E6 and A549 cells were maintained in Dulbecco's modified Eagle's medium (DMEM; Invitrogen) supplemented with 10% fetal bovine serum (FBS; Lonza). A panel of HEK293-derived cell lines based on the FLP-IN T Rex system (Invitrogen) and containing single integrations of individual ISG cDNAs, with expression under the control of a tetracycline-responsive (TET-on) promoter (18), were generously provided by Ju-Tao Guo (Drexel Institute for Biotechnology and Virology Research, PA). The cells were maintained in DMEM supplemented with 10% tetracycline-free FBS (Invitrogen), 250 μg/ml hygromycin B (Invitrogen), and 5 μg/ml blasticidin (Invivogen). ISG expression was induced by addition of 1 μg/ml tetracycline (Sigma) for 48 h. Wild-type Bunyamwera virus (wtBUNV) and a recombinant virus lacking the NSs gene, rBUNdelNSs (14), were amplified in BHK-21 cells at 33°C, and titers were determined by plaque assay on BHK-21 cells, as described previously (47).
Antibodies, interferon, and plasmids.
Rabbit anti-viperin and mouse anti-PKR antisera were from Abcam, rabbit anti-MxA antiserum was from Santa Cruz Biotechnology, and mouse anti-tubulin antibody was from Sigma. Rabbit antisera against purified BUNV and BUNV N protein were described previously (22, 48). Recombinant human IFN-β-1a was purchased from PBL Interferon Source. A plasmid containing the luciferase gene under the control of the human viperin promoter (42) was kindly provided by K. A. Fitzgerald (University of Massachusetts Medical School, MA).
Growth curves and virus yields.
To monitor the effects of IFN on virus growth, untreated or IFN-β-treated (1,000 IU/ml) Vero E6 cells were infected at a multiplicity of infection (MOI) of 1 PFU per cell, and the supernatants were harvested 24 h after infection. To measure virus yield in ISG-expressing HEK cells, uninduced or tetracycline-induced cells were infected at an MOI of 0.01 PFU per cell and the supernatants were harvested at various times after infection. Virus titers were determined by plaque formation on Vero E6 cells.
Metabolic radiolabeling and immunoprecipitation.
Cells grown in 35-mm-diameter dishes were incubated in starvation medium that lacks methionine for 1 h prior to radiolabeling with medium containing 50 μCi/ml [35S]methionine for 1 h at various times postinfection. Cells were then lysed on ice with 150 μl of RIPA buffer (1% Triton X-100, 50 mM Tris [pH 7.4], 300 mM NaCl, 5 mM EDTA) containing a cocktail of protease inhibitors (Complete; Roche). The cell lysates were centrifuged at 16,000 × g at 4°C for 10 min. For immunoprecipitation, 50 μl of the supernatant was incubated with anti-BUNV or anti-viperin antibodies conjugated to protein A-Sepharose beads (Sigma) for 24 h at 4°C. The beads were washed with RIPA buffer containing 0.1% Triton X-100 four times and once with ice-cold phosphate-buffered saline (PBS) and then analyzed by SDS-PAGE on a 12% gel.
Western blotting.
Unlabeled cells were lysed as described above, and proteins were separated by SDS-PAGE on a 12% gel. The proteins were transferred to a nitrocellulose membrane (Hybond C; Amersham), followed by incubation in blocking solution (PBS containing 0.05% Tween, 5% nonfat dry milk) for 1 h. The membrane was incubated for 1 h with the primary antibody, washed 3 times with PBS-0.05% Tween, and then incubated with horseradish peroxidase-conjugated secondary antibody for 1 h, followed by 3 further washes. Protein detection was performed using the Supersignal West Pico chemiluminescent substrate kit (Pierce) as per the manufacturer's instructions, and the membrane was exposed to X-ray film.
Northern blotting.
Total cellular RNA was extracted using TRIzol (Invitrogen) according to the manufacturer's instructions and then separated by agarose electrophoresis using TAE buffer (27). Following transfer to a positively charged nylon membrane (Sigma), viral RNAs were detected by hybridization with strand-specific digoxigenin-labeled RNA probes as previously described (26).
Reporter gene assay.
Subconfluent monolayers of HEK293 cells were transfected with 50 ng of the viperin promoter reporter plasmid DNA in 100 μl of Opti-MEM (Gibco-BRL) containing 3 μl of Lipofectamine 2000 (Invitrogen). Twenty-four hours posttransfection, the cells were infected at an MOI of 5 PFU/cell and either stimulated or not with 1,000 IU/ml IFN-β, and they were then further incubated at 37°C. After 24 h, the cells were harvested and lysed, and luciferase activity was measured using the dual luciferase reporter assay kit (Promega) as per the manufacturer's instructions. The data shown are from a representative experiment that was repeated three times in triplicate.
RESULTS
Effect of IFN on virus replication.
Vero cells do not produce IFN but respond to exogenously applied IFN (7, 29), and previously it was shown that BUNV replication was severely impaired in cells treated with 1,000 IU/ml for 24 h prior to infection (43). In this study, we examined the effect on virus replication when IFN was applied after infection (Fig. 1). Firstly, we confirmed the previous result that both wtBUNV and rBUNdelNSs growth was inhibited 1,000-fold in cells pretreated with IFN. However, addition of IFN to cells immediately after adsorption of virus (0 h) or 6 h or 12 h after infection had no effect on the yield of either virus 24 h after infection.
Fig 1.
Effect of IFN treatments on BUNV titer. Vero cells were treated with 1,000 IU/ml IFN-β for 24 h before infection (+IFN −24), treated immediately after infection (+IFN 0) or at 6 or 12 hpi (+IFN + 6 and +IFN + 12) or left untreated (− IFN), and mock infected or infected with either wtBUNV or rBUNdelNSs at an MOI of 0.01 PFU/cell as indicated. At 24 hpi, the titer of virus released into the growth medium was determined by plaque assay on Vero cells.
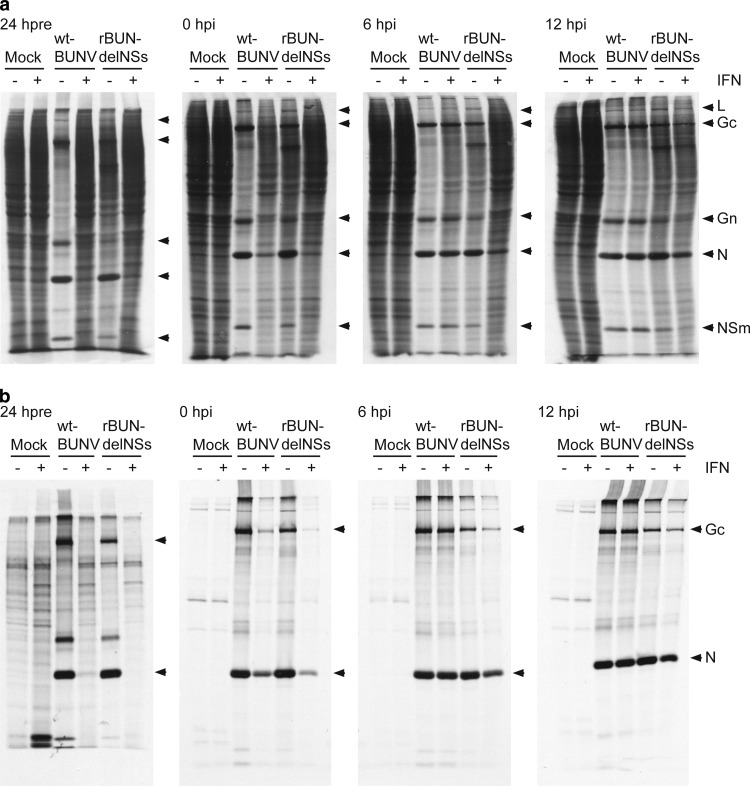
We next investigated the effect of IFN treatment on viral protein synthesis. Infected cells, subjected to IFN treatments, were pulse-labeled with [35S]methionine for 1 h at 24 h postinfection (hpi) (Fig. 2). In untreated cells, wtBUNV causes significant shutoff of host cell protein synthesis, whereas this effect is much reduced in untreated cells infected with rBUNdelNSs. As expected from the virus yield experiment, viral protein synthesis was markedly reduced in cells pretreated with IFN, and for wtBUNV, there was no detectable shutoff of host cell protein synthesis compared to that in untreated cells (Fig. 2a). There was some reduction in the incorporation of label into viral proteins when IFN was added immediately after infection, and shutoff of host cell protein synthesis was less obvious in wtBUNV-infected cells. IFN treatment at later times after infection had minimal effects on wtBUNV protein synthesis, though that of rBUNdelNSs seemed more sensitive when IFN was added at 6 hpi.
Fig 2.
Effect of IFN treatement on viral protein synthesis. Vero cells were treated with 1,000 IU/ml IFN-β for 24 h before infection (24 hpre), treated immediately after infection (0 hpi) or at 6 or 12 hpi, and mock infected or infected with either wtBUNV or rBUNdelNSs at an MOI of 1 PFU/cell as indicated. At 24 hpi, the cells were labeled for 1 h with [35S]methionine and cell lysates prepared. Radiolabeled proteins were directly analyzed by SDS-PAGE on a 12% gel (a), or the viral proteins were precipitated using anti-BUNV antibodies before analysis by SDS-PAGE on a 12% gel (b). Virus proteins are labeled on the right.
The corresponding immunoprecipitates obtained using an antibody prepared against purified virions (47) are shown in Fig. 2b. These confirm the lack of detectable viral protein synthesis in pretreated cells, reduced uptake of radiolabel into viral proteins when IFN was added immediately after infection, and slightly reduced incorporation of radiolabel into viral proteins in cells infected with rBUNdelNSs when IFN was added at 6 hpi.
The effect of adding IFN immediately after the adsorption period was studied in more detail for wtBUNV (Fig. 3). There was no difference in the synthesis of viral proteins at 4, 8, or 12 hpi; a difference was observed only at the 24-hpi labeling time point, where less viral protein was produced and host cell protein shutoff was less obvious. To determine whether the difference in protein synthesis at 24 hpi reflected a difference in RNA synthesis, total cell RNA was extracted and analyzed by Northern blotting, with single-stranded, digoxigenin-labeled segment- and polarity-specific RNA probes to detect negative-sense (genome) or positive-sense (antigenome and mRNA [Fig. 3c]) RNA species. As described previously, only S segment antigenome and mRNA are sufficiently different in size to be resolved by gel electrophoresis (26). There seemed little difference in the accumulation of genomic RNAs between IFN-treated and untreated cells (Fig. 3c, left). Similarly, there was no marked difference in the accumulation of positive-sense L and M segment RNAs (Fig. 3c, right). For the S segment, there appeared to be slightly less mRNA in the IFN-treated cells, but this could not be accurately quantified by Northern blotting. Taken together, these data reinforce the notions that (i) BUNV is unable to overcome the IFN-induced antiviral state, and (ii) IFN has little effect on either wtBUNV or virus from which NSs has been deleted if the virus is added to Vero cells once the viral replication cycle has begun.
Fig 3.
Analysis of BUNV proteins and RNAs in IFN-β-treated cells. Vero cells were mock infected or infected with BUNV at an MOI of 1 PFU/cell and immediately treated with 1,000 IU/ml IFN-β. Proteins were labeled at the time points indicated with [35S]methionine and radiolabeled proteins were analyzed directly by SDS-PAGE on a 12% gel (a), or the viral proteins were precipitated using anti-BUNV antibodies before analysis on a 12% gel (b). Total cell RNA was extracted from replicate cell cultures by using TRIzol reagent and analyzed by Northern blotting using strand-specific digoxigenin-labeled probes to detect viral genomic RNA (c) or antigenomic RNA and mRNA (d). Only for the S segment was antigenomic (AG) RNA separated from mRNA.
Identification of ISGs that restrict BUNV infection.
To begin to determine which of the many hundreds of ISGs are responsible for inhibiting BUNV replication, we screened a panel of HEK293-derived cell lines (18) that inducibly express 20 individual ISGs for the ability to support BUNV replication. A cell line that expressed the chloramphenicol acetyltransferase (CAT) gene was used as a control. The cells were incubated in medium containing tetracycline to induce ISG expression for 48 h prior to infection with wtBUNV or rBUNdelNSs. As the ISGs and CAT gene were tagged with the FLAG epitope, expression of the appropriate protein following induction was confirmed by Western blotting with an anti-FLAG antibody (data not shown). The supernatants from infected uninduced or induced cells were collected at intervals up to 72 hpi, and the amount of released virus was titrated by plaque assay (see Fig. S1 in the supplemental material). The results are summarized in Table 1. By titrating virus yields at different time points, we could determine consistent differences in yields rather than the possibility of an aberrant result by examining only a single time point. As expected, expression of the control protein CAT had no effect on the titer of wtBUNV or rBUNdelNSs. No difference in titer of wtBUNV or rBUNdelNSs was seen in 14 cell lines when comparing induced to uninduced cultures (Table 1), while 5 cell lines showed a small decrease (<10-fold) in titer of wtBUNV following tetracycline induction compared to the titer from the uninduced cells (cells expressing ISG9-27, ISG15, MAPK8, BST2, and OAS1). rBUNdelNSs titers were also slightly reduced in the same 5 cell lines and additionally in cells expressing ISG56. More marked inhibition of wtBUNV replication was observed in the cells expressing PKR (Fig. 4a), MTAP44 (approximately 10-fold reduction), and viperin (approximately 100-fold reduction [Fig. 5a]). rBUNdelNSs also showed an approximately 100-fold decrease in titer in viperin-expressing cells (Fig. 5c), though the virus was less inhibited in cells expressing MTAP44 and PKR. These data suggest that a number of ISGs may have small inhibitory effects on BUNV replication, while significant inhibition is mediated by viperin.
Table 1.
Summary of the replication of wtBUNV and rBUNdelNSs in 20 cell lines expressing individual ISGs
| Cell line | Inhibition ofa: |
GenBank accession no.b | |
|---|---|---|---|
| wtBUNV | rBUNdelNSs | ||
| CAT | − | − | |
| ISG 9–27 | + | − | 003641 |
| ISG15 | + | + | 005101 |
| ISG56 | − | + | 001548 |
| 1-8D | − | − | 006435 |
| ADAR1 | − | − | 001111 |
| MTAP44 | ++ | + | D28915 |
| GBP1 | − | − | 002053 |
| MAPK8 | + | + | 002750 |
| Viperin | +++ | +++ | AF442151 |
| VPM1 | − | − | |
| PKR | ++ | + | AH008429 |
| PKRM | − | − | |
| STAF50 | − | − | X82200 |
| BST2 | + | + | 004335 |
| FLJ38348 | − | − | AK095667 |
| FLJ20035 | − | − | AK000042 |
| PLSCR1 | − | − | 021105 |
| PLSCR2 | − | − | 020359 |
| UBE2L6 | − | − | 004223 |
| USP18 | − | − | 017414 |
| OAS1V1 | + | + | 016816 |
| OAS1V2 | − | + | 002534 |
Inhibition of yield was based on a consistent difference in virus titer between induced and uninduced cells over the time points presented in Fig. S1 in the supplemental material. −, no difference; +, <10-fold difference; ++, ≥10-fold difference; +++, ≥100-fold difference.
From reference 18.
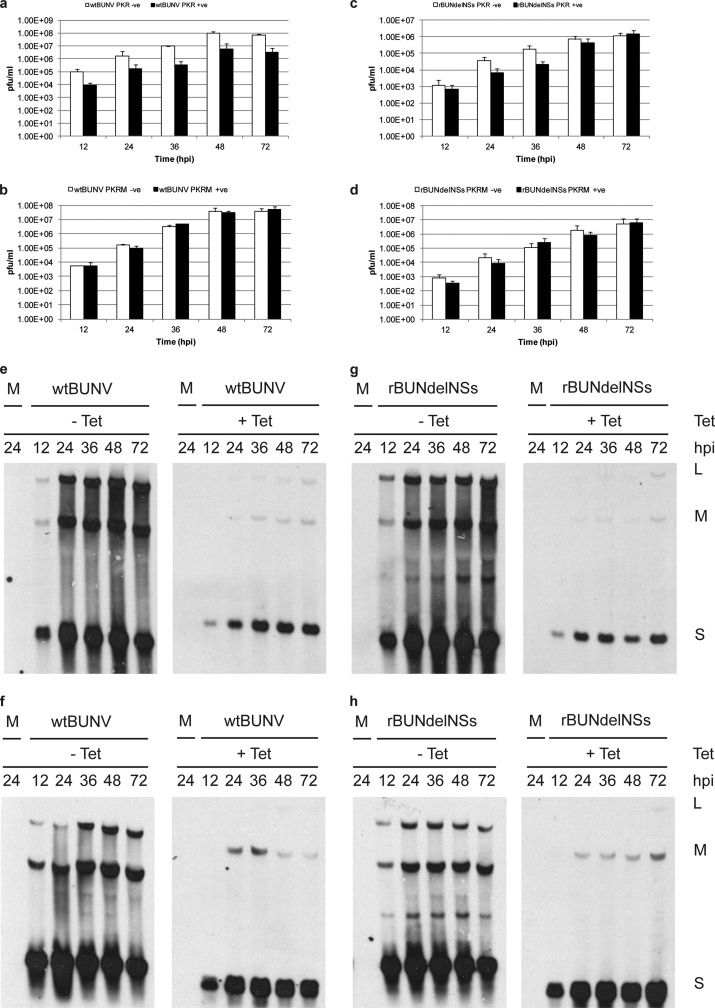
Fig 4.
PKR restriction of BUNV is dependent on its enzymatic activity. HEK PKR cells and HEK PKRM cells were infected with either wtBUNV or rBUNdelNSs at an MOI of 0.01 PFU/cell in the absence or presence of tetracycline. At the time points indicated, the titer of released virus was determined by plaque assay on Vero cells (a to d). Total cell RNA was extracted from duplicate cells by using TRIzol reagent and analyzed by Northern blotting using strand-specific digoxigenin-labeled probes to detect each virus segment genomic RNA (e and g) or antigenomic RNA (f and h). Note that the S segment antigenome and mRNA were not resolved in this experiment.
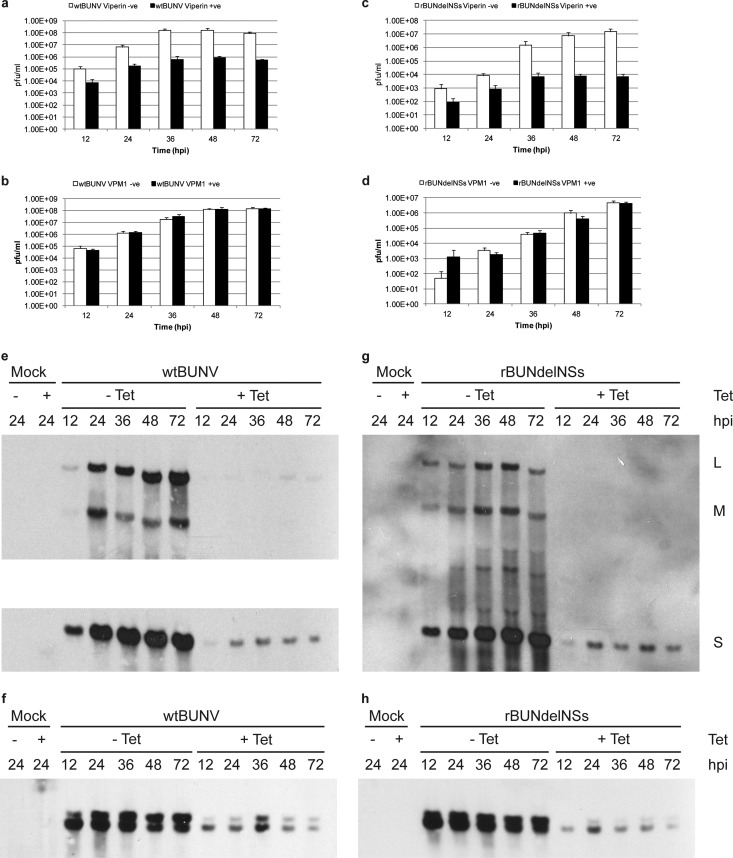
Fig 5.
Viperin restriction of BUNV is dependent on its CX3CX2C motif. HEK viperin cells and HEK VPM1 cells were infected with either wtBUNV or rBUNdelNSs at an MOI of 0.01 PFU/cell in the absence or presence of tetracycline. At the time points indicated, the titer of released virus was determined by plaque assay on Vero cells (a to d). Total cell RNA was extracted from duplicate cells using TRIzol reagent and analyzed by Northern blotting using strand-specific digoxigenin-labeled probes to detect each virus segment genomic RNA (e and g) or S segment antigenomic RNA and mRNA (f and h).
The inhibition exhibited by PKR and viperin is dependent on their enzymatic activities.
PKR is activated by dsRNA, which results in its autophosphorylation, and phosphorylated PKR then phosphorylates eIF2α, which, in turn, blocks cellular protein synthesis (11). HEK PKRM cells express a mutant form of PKR in which the conserved lysine residue in the ATP-binding pocket has been replaced with an arginine residue, generating a dominant negative PKR that is deficient in its protein kinase activity (18). HEK VPM1 cells, when induced, express a viperin mutant (VPM1) in which the three cysteine residues in conserved motif I (CX3CX2C) that are necessary for the reductive cleavage of S-adenosylmethionine have been replaced by alanine residues, thereby abrogating viperin's enzymatic capability (8, 18). We compared the abilities of wtBUNV and rBUNdelNSs to replicate in the mutant and parental cell lines following induction with tetracycline. Virus titers in supernatant medium were determined at various time points over a 72-h period.
There was essentially no difference in titer of wtBUNV released from induced and uninduced PKRM cells (Fig. 4b), in contrast to the 10- to 25-fold difference seen in cells expressing wild-type PKR (Fig. 4a). rBUNdelNSs similarly was not restricted in PKRM cells, though interestingly, in parental PKR cells, restriction of rBUNdelNSs was only significant at earlier time points, whereas by 48 hpi, there was no difference between induced and uninduced cells (Fig. 4c and d).
We further analyzed viral RNA synthesis in cells expressing PKR. Equal amounts of total cell RNA were fractionated on an agarose gel, blotted to a nylon membrane, and then probed with strand-specific digoxigenin-labeled RNAs. Blots from the corresponding induced and uninduced cells were hybridized at the same time with the same amount of probe. There was a clear reduction in the synthesis of genomic RNA species and antigenomic RNA or mRNA species in cells expressing PKR compared to uninduced cells when infected with wtBUNV (Fig. 4e and f) or rBUNdelNSs (Fig. 4g and h).
Neither wtBUNV nor rBUNdelNSs was restricted in cells expressing the mutant form of viperin (Fig. 5b and d), in contrast to the marked reduction in virus yield from cells expressing the parental form (Fig. 5a and c). Similarly, the amounts of both viral genomic and antigenomic RNAs or mRNAs were severely reduced in cells expressing viperin compared to uninduced cells for both viruses (Fig. 5e to h).
Taken together, these results demonstrate that expression of PKR and viperin restricts BUNV replication, that the enzymatic activities of both these ISGs are required for their inhibitory activity, and that in cells expressing either of these ISGs, viral RNA synthesis is drastically reduced.
rBUNdelNSs, but not wtBUNV, induces viperin expression in human cells.
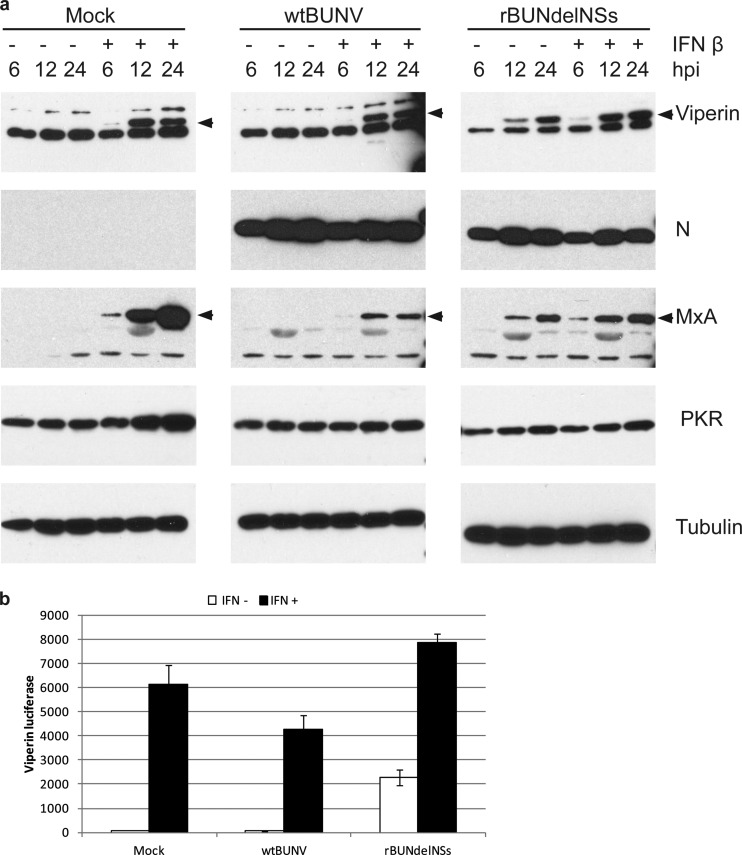
Previously it was shown that wtBUNV did not activate PKR in mouse cells, while the observed activation of PKR by rBUNdelNSs was due to IFN induced by infection with this virus (43). In this study, we investigated whether viperin behaved similarly. Human lung carcinoma (A549) cells were used, as they have a fully intact IFN system. A549 cells were either mock infected or infected with wtBUNV or rBUNdelNSs at an MOI of 5 PFU/cell, and then IFN-β was added to half of the cultures. At 6, 12, and 24 hpi, the cells were lysed and examined by Western blotting. Viperin was detected only in mock-infected cells following IFN treatment (Fig. 6a). No viperin was detected in cells infected with wtBUNV, presumably because of the inhibitory activity of NSs. However, IFN treatment did result in viperin induction in wtBUNV-infected cells. In contrast, viperin was detected in cells infected with rBUNdelNSs whether or not they were treated with IFN. Expression of the viral N protein was used to confirm that the cells were infected, while induction of MxA served to show that the IFN system was functional.
Fig 6.
Viperin expression is directly induced by rBUNdelNSs. (a) A549 cells were mock infected or infected with BUNV or rBUNdelNSs at an MOI of 5 and either left untreated or treated with IFN-β. At the time points indicated, cell lysates were prepared and analyzed by SDS-PAGE, followed by Western blotting with antibodies for viperin, N protein, MxA, PKR, and tubulin as indicated. (b) HEK293 cells were transfected with a viperin-promoter reporter plasmid and 24 h later mock infected or infected with BUNV or rBUNdelNSs, followed by either no treatment or treatment with IFN-β. After incubation for a further 24 h, the luciferase activity in cell lysates was measured. The error bars represent the standard deviations of a representative experiment done in triplicate.
To corroborate these results, a plasmid transfection assay was used. HEK293 cells were transfected with a reporter plasmid that contains the firefly luciferase gene under the control of the viperin promoter (42). Twenty-four hours later, the cells were either mock infected or infected with BUNV or rBUNdelNSs, followed by addition, or not, of IFN-β to the culture medium; luciferase activity was measured 24 h later. Mock-infected cells stimulated with IFN-β showed a marked increase in luciferase activity, indicating that the viperin promoter had been activated (Fig. 6b). No activation occurred in wtBUNV-infected cells unless IFN was applied, whereas rBUNdelNSs infection activated the viperin promoter independently of IFN. Addition of IFN to rBUNdelNSs-infected cells further enhanced activation of the viperin promoter to a level higher than that seen in IFN-treated mock-infected cells.
DISCUSSION
The IFN response is a powerful cellular defense against viral infection, resulting in the upregulation of many hundreds of genes whose products are presumed to have antiviral activity. However, the mechanism of inhibitory function is known for relatively few ISGs, and indeed, cataloguing what particular ISGs are active against particular viruses is a rather recent pursuit (25, 38). In this paper, we describe the IFN activity against the prototypic bunyavirus, BUNV. We showed that priming cells with IFN-β before infection appeared to fully restrict BUNV replication, whereas treating the cells with IFN-β once infection was established apparently had little effect on virus replication. The NSs protein is the BUNV IFN antagonist (9), but NSs is unable to dismantle a preexisting antiviral state. However, once virus replication is under way, IFN-β treatment of Vero cells (which do not synthesize their own IFN but respond to exogenously applied IFN) did not block the replication of either wtBUNV or rBUNdelNSs. This suggests that the relatively fast replication of BUNV outcompetes the rate of IFN signaling, even when IFN is applied at the end of the virus adsorption period. The slight reduction in viral protein synthesis and shutoff of host cell proteins observed at 24 hpi in such IFN-treated cells (Fig. 2 and 3a) would hence reflect the effects of IFN signaling at this time, though these are largely controlled by the robust virus replication.
The BUNV NSs protein acts by globally inhibiting all RNA polymerase II-mediated transcription (44), including IFN mRNA synthesis, and to date, NSs has not been shown to affect specifically steps in the IFN induction or signaling pathways or to directly affect ISG action, as seen for some other viral IFN antagonists (34). A number of ISGs individually had small effects on virus replication (Table 1; see also Fig. S1 in the supplemental material), though three ISGs, MTAP44, PKR, and especially viperin, showed rather more inhibitory effects. Presumably, in concert their combined effects would result in the overall inhibition mediated by IFN treatment (39). Little is known about MTAP44 (also known as IFN-induced protein 44). MTAP44 has been shown to aggregate microtubules (13), and therefore, it could affect early intracellular viral transport. This requires further investigation.
PKR is a critical molecule in antiviral immunity, since it acts both as a pattern recognition receptor that is able to detect viral dsRNA and as an ISG that inhibits host cell protein synthesis by phosphorylating eIF2α (11). This mechanism can be an extremely potent way of blocking viral protein synthesis, and several viruses target PKR in order to permit replication, such as direct interaction with and/or degradation of PKR, sequestration of dsRNA, and the dephosphorylation of eIF2α (11, 21). Therefore, overexpression of PKR should make cells highly sensitive to dsRNA and enhance the effects of subsequent PKR activation on host cell protein synthesis. Streitenfeld et al. (43) demonstrated that in murine embryonic fibroblasts, both BUNV and rBUNdelNSs activated PKR, but this activation did not confer any resistance to either virus in cell culture. However, using PKR knockout mice, they found that PKR did afford some weak protection to BUNV infection in vivo. Thus, it is conceivable that expression of physiological levels of PKR could be beneficial to BUNV replication by enhancing the effect of host cell protein synthesis shutoff by blocking further cellular protein translation. This PKR-induced translational inhibition, along with virus-mediated cap snatching, may allow for increased viral protein translation, as NSs blocks de novo cellular mRNA synthesis and cap snatching reduces cellular levels of mRNA. At the same time, translation of viral mRNAs increases because the cellular translational machinery has been liberated from its burden of translating cellular mRNAs. However, in the presence of supraphysiological amounts of PKR, as in the overexpressing HEK cells used in this study, the viruses are unable to outcompete the inhibitory effects of PKR, and viral titers and RNA synthesis are reduced (Fig. 4) but not abrogated. Further, even in the absence of NSs, there is some viral replication, suggesting that there is more to BUNV inhibition of the IFN response than just NSs.
The phlebovirus RVFV also encodes an NSs protein on the S segment but uses an ambisense coding strategy. RVFV NSs is about three times larger than that of BUNV and does not show amino acid similarity, but it acts similarly to block IFN induction at a transcriptional level, though via a different mechanism (4). RVFV mutants deficient in NSs expression are IFN inducers and are thus attenuated. Habjan et al. (12) and Ikegami et al. (17) also demonstrated that not only does RVFV inhibit PKR expression but also the virus targets PKR for proteasomal degradation. This is specific to RVFV NSs, as recombinant viruses expressing sandfly fever Sicilian phlebovirus or La Crosse orthobunyavirus NSs proteins in place of the homologous NSs did not degrade PKR (12), as also observed in this study for BUNV.
Research investigating the mechanisms behind the action of PKR, both as a dsRNA sensor and as an antiviral protein, is continually updating the complex pathways and cellular proteins involved in the PKR-dependent antiviral response (31). The ISG ADAR1 has been shown to actually inhibit PKR activation, thereby suppressing the phosphorylation of eIF2α and enhancing the replication of vesicular stomatitis virus (VSV) (24). Also, PKR-mediated inhibition of human immunodeficiency virus (HIV) replication can be returned to normal by the expression of ADAR1, and inhibition of ADAR1 expression results in a decrease of HIV production (6). Thus, some viruses are able to inhibit the PKR response but also are helped by cellular factors that are most likely there to regulate the activity of PKR. As yet, it is unknown whether ADAR1 or other ISGs enhance BUNV replication by inhibiting PKR.
Viperin (virus inhibitory protein, endoplasmic reticulum-associated interferon-inducible) has attracted much interest as an ISG with antiviral properties and has been the subject of several recent reviews (10, 28, 40). When expressed prior to infection, viperin has been shown to inhibit the replication of a range of positive- and negative-strand RNA viruses, HIV, and human cytomegalovirus (HCMV). The antiviral mechanism of viperin has yet to be completely elucidated, although insights into the action of viperin-mediated restriction on some specific viruses have been obtained. Wang et al. (46) demonstrated that viperin interacts with farnesyl diphosphate synthase (FPPS) and inhibits the enzymatic function of FPPS in lipid metabolism, which results in the disruption of lipid raft microdomain formation. Lipid rafts play a vital role in the replication cycle of influenza A virus (IAV) as the site of virus budding from the plasma membrane (37). Thus, viperin restricts IAV by blocking viral release. Viperin also associates with lipid droplets (16), vital components in the hepatitis C virus (HCV) life cycle. Viperin appears to disrupt the formation of the HCV replication complex on lipid droplets by its interaction with hVAP-33 (vesicle-associated membrane protein-associated protein of 33 kDa), which interferes with binding of the viral NS5A protein to hVAP-33 (15, 45). Besides being induced by IFN, viperin expression is also induced following virus infection, and viperin actually aids HCMV replication in fibroblasts (41). In HCMV-infected cells, viperin is induced independently of IFN and relocalizes from the endoplasmic reticulum to mitochondria via an association with the viral protein vMIA. This results in a reduction of ATP production, leading to disruption of the actin cytoskeleton that benefits virus infection.
Bunyaviruses assemble at the Golgi, where the viral glycoproteins accumulate, and budding of new virus particles is triggered by the arrival of RNPs at the cytoplasmic side of vesicular membranes. Progeny virions are presumably transported to the cell surface within vesicles analogous to those in the secretory pathway, and release of virus from infected cells presumably occurs when the virus-containing vesicles fuse with the plasma membrane as in normal exocytosis (30). The role of lipids or lipid raft domains in bunyavirus assembly and maturation is unknown and is under investigation. For some bunyaviruses, it has been reported that the N protein binds actin (1, 35), and treating infected cells with cytochalasin D to disrupt the actin network reduced assembly of infectious CCHFV (1). Thus, considering the known activities of viperin, a number of steps in the bunyavirus life cycle could be inhibited. In addition, viral RNA synthesis was inhibited in cells overexpressing viperin or PKR. Whether this is because of a direct effect on the viral polymerase or indirect effects in the cellular environment is under study.
In summary, we have identified three ISGs that have marked inhibitory activity against the prototype bunyavirus. It will be important to determine whether these ISGs are also active against other members of the family, not only other orthobunyaviruses but also viruses in other genera. An understanding of the mechanisms by which these ISGs exert their inhibitory activities will help in the design of new antiviral therapies against this significant group of viruses.
Supplementary Material
ACKNOWLEDGMENTS
We thank Ju-Tao Guo for generously providing the cell lines, Katherine Fitzgerald for providing the viperin reporter plasmid, and Angela McLees for expert technical assistance.
C.C.-S. received a postgraduate studentship from the University of St. Andrews, and work in R.M.E.'s laboratory is supported by grants from the BBSRC, MRC, and Wellcome Trust.
Footnotes
Published ahead of print 15 August 2012
Supplementary material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1. Andersson I, et al. 2004. Role of actin filaments in targeting of Crimean Congo hemorrhagic fever virus nucleocapsid protein to perinuclear regions of mammalian cells. J. Med. Virol. 72:83–93 [DOI] [PubMed] [Google Scholar]
- 2. Blakqori G, van Knippenberg I, Elliott RM. 2009. Bunyamwera orthobunyavirus S-segment untranslated regions mediate poly(A) tail-independent translation. J. Virol. 83:3637–3646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bonjardim CA, Ferreira PC, Kroon EG. 2009. Interferons: signaling, antiviral and viral evasion. Immunol. Lett. 122:1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bouloy M. 2011. Molecular biology of phleboviruses, p 95–128 In Plyusnin A, Elliott RM. (ed), Bunyaviridae: molecular and cellular biology. Caister Academic Press, Norfolk, United Kingdom [Google Scholar]
- 5. Bridgen A, Weber F, Fazakerley JK, Elliott RM. 2001. Bunyamwera bunyavirus nonstructural protein NSs is a nonessential gene product that contributes to viral pathogenesis. Proc. Natl. Acad. Sci. U. S. A. 98:664–669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Clerzius G, et al. 2009. ADAR1 interacts with PKR during human immunodeficiency virus infection of lymphocytes and contributes to viral replication. J. Virol. 83:10119–10128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Desmyter J, Melnick JL, Rawls WE. 1968. Defectiveness of interferon production and of rubella virus interference in a line of African green monkey kidney cells (Vero). J. Virol. 2:955–961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Duschene KS, Broderick JB. 2010. The antiviral protein viperin is a radical SAM enzyme. FEBS Lett. 584:1263–1267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Elliott RM, Blakqori G. 2011. Molecular biology of orthobunyaviruses, p 1–39 In Plyusnin A, Elliott RM. (ed), Bunyaviridae: molecular and cellular biology. Caister Academic Press, Norfolk, United Kingdom [Google Scholar]
- 10. Fitzgerald KA. 2011. The interferon inducible gene: viperin. J. Interferon Cytokine Res. 31:131–135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. García MA, et al. 2006. Impact of protein kinase PKR in cell biology: from antiviral to antiproliferative action. Microbiol. Mol. Biol. Rev. 70:1032–1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Habjan M, et al. 2009. NSs protein of Rift Valley fever virus induces the specific degradation of the double-stranded RNA-dependent protein kinase. J. Virol. 83:4365–4375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hallen LC, et al. 2007. Antiproliferative activity of the human IFN-alpha-inducible protein IFI44. J. Interferon Cytokine Res. 27:675–680 [DOI] [PubMed] [Google Scholar]
- 14. Hart TJ, Kohl A, Elliott RM. 2009. Role of the NSs protein in the zoonotic capacity of orthobunyaviruses. Zoonoses Public Health 56:285–296 [DOI] [PubMed] [Google Scholar]
- 15. Helbig KJ, et al. 2011. The antiviral protein viperin inhibits hepatitis C virus replication via interaction with nonstructural protein 5A. Hepatology 54:1506–1517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hinson ER, Cresswell P. 2009. The N-terminal amphipathic alpha-helix of viperin mediates localization to the cytosolic face of the endoplasmic reticulum and inhibits protein secretion. J. Biol. Chem. 284:4705–4712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ikegami T, et al. 2009. Rift Valley fever virus NSs protein promotes post-transcriptional downregulation of protein kinase PKR and inhibits eIF2alpha phosphorylation. PLoS Pathog. 5:e1000287 doi:10.1371/journal.ppat.1000287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Jiang D, et al. 2008. Identification of three interferon-inducible cellular enzymes that inhibit the replication of hepatitis C virus. J. Virol. 82:1665–1678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kohl A, et al. 2003. Bunyamwera virus nonstructural protein NSs counteracts interferon regulatory factor 3-mediated induction of early cell death. J. Virol. 77:7999–8008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Koyama S, Ishii KJ, Coban C, Akira S. 2008. Innate immune response to viral infection. Cytokine 43:336–341 [DOI] [PubMed] [Google Scholar]
- 21. Langland JO, Cameron JM, Heck MC, Jancovich JK, Jacobs BL. 2006. Inhibition of PKR by RNA and DNA viruses. Virus Res. 119:100–110 [DOI] [PubMed] [Google Scholar]
- 22. Lappin DF, Nakitare GW, Palfreyman JW, Elliott RM. 1994. Localisation of Bunyamwera bunyavirus G1 glycoprotein to the Golgi requires association with G2 but not NSm. J. Gen. Virol. 75:3441–3451 [DOI] [PubMed] [Google Scholar]
- 23. Léonard VH, Kohl A, Hart TJ, Elliott RM. 2006. Interaction of Bunyamwera orthobunyavirus NSs protein with mediator protein MED8: a mechanism for inhibiting the interferon response. J. Virol. 80:9667–9675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Li Z, Wolff KC, Samuel CE. 2010. RNA adenosine deaminase ADAR1 deficiency leads to increased activation of protein kinase PKR and reduced vesicular stomatitis virus growth following interferon treatment. Virology 396:316–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Liu SY, Sanchez DJ, Aliyari R, Lu S, Cheng G. 2012. Systematic identification of type I and type II interferon-induced antiviral factors. Proc. Natl. Acad. Sci. U. S. A. 109:4239–4244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lowen AC, Boyd A, Fazakerley JK, Elliott RM. 2005. Attenuation of bunyavirus replication by rearrangement of viral coding and noncoding sequences. J. Virol. 79:6940–6946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Masek T, Vopalensky V, Suchomelova P, Pospisek M. 2005. Denaturing RNA electrophoresis in TAE agarose gels. Anal. Biochem. 336:46–50 [DOI] [PubMed] [Google Scholar]
- 28. Mattijssen S, Pruijn GJ. 2012. Viperin, a key player in the antiviral response. Microbes Infect. 14:419–426 [DOI] [PubMed] [Google Scholar]
- 29. Mosca JD, Pitha PM. 1986. Transcriptional and posttranscriptional regulation of exogenous human beta interferon gene in simian cells defective in interferon synthesis. Mol. Cell. Biol. 6:2279–2283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Pettersson RF, Melin L. 1996. Synthesis, assembly and intracellular transport of Bunyaviridae membrane proteins, p 159–188 In Elliott RM. (ed), The Bunyaviridae. Plenum Press, New York, NY [Google Scholar]
- 31. Pfaller CK, Li Z, George CX, Samuel CE. 2011. Protein kinase PKR and RNA adenosine deaminase ADAR1: new roles for old players as modulators of the interferon response. Curr. Opin. Immunol. 23:573–582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Plyusnin A, et al. 2012. Bunyaviridae, p 725–741 In King AMQ, Adams MJ, Carstens EB, LEJ (ed), Virus taxonomy: ninth report of the International Committee on Taxonomy of Viruses. Elsevier Academic Press, London, United Kingdom [Google Scholar]
- 33. Plyusnin A, Elliott RM. (ed). 2011. Bunyaviridae: molecular and cellular biology. Caister Academic Press, Norfolk, United Kingdom [Google Scholar]
- 34. Randall R, Goodbourn S. 2008. Interferons and viruses: an interplay between induction, signalling, antiviral responses and virus countermeasures. J. Gen. Virol. 89:1–27 [DOI] [PubMed] [Google Scholar]
- 35. Ravkov EV, Nichol ST, Peters CJ, Compans RW. 1998. Role of actin microfilaments in Black Creek Canal virus morphogenesis. J. Virol. 72:2865–2870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sadler AJ, Williams BR. 2008. Interferon-inducible antiviral effectors. Nat. Rev. Immunol. 8:559–568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Scheiffele P, Rietveld A, Wilk T, Simons K. 1999. Influenza viruses select ordered lipid domains during budding from the plasma membrane. J. Biol. Chem. 274:2038–2044 [DOI] [PubMed] [Google Scholar]
- 38. Schoggins JW, Rice CM. 2011. Interferon-stimulated genes and their antiviral effector functions. Curr. Opin. Virol. 1:519–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Schoggins JW, et al. 2011. A diverse range of gene products are effectors of the type I interferon antiviral response. Nature 472:481–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Seo JY, Yaneva R, Cresswell P. 2011. Viperin: a multifunctional, interferon-inducible protein that regulates virus replication. Cell Host Microbe 10:534–539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Seo JY, Yaneva R, Hinson ER, Cresswell P. 2011. Human cytomegalovirus directly induces the antiviral protein viperin to enhance infectivity. Science 332:1093–1097 [DOI] [PubMed] [Google Scholar]
- 42. Severa M, Coccia EM, Fitzgerald KA. 2006. Toll-like receptor-dependent and -independent viperin gene expression and counter-regulation by PRDI-binding factor-1/BLIMP1. J. Biol. Chem. 281:26188–26195 [DOI] [PubMed] [Google Scholar]
- 43. Streitenfeld H, et al. 2003. Activation of PKR by Bunyamwera virus is independent of the viral interferon antagonist NSs. J. Virol. 77:5507–5511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Thomas D, et al. 2004. Inhibition of RNA polymerase II phosphorylation by a viral interferon antagonist. J. Biol. Chem. 279:31471–31477 [DOI] [PubMed] [Google Scholar]
- 45. Wang S, et al. 2012. Viperin inhibits hepatitis C virus replication by interfering with binding of NS5A to host protein hVAP-33. J. Gen. Virol. 93:83–92 [DOI] [PubMed] [Google Scholar]
- 46. Wang X, Hinson ER, Cresswell P. 2007. The interferon-inducible protein viperin inhibits influenza virus release by perturbing lipid rafts. Cell Host Microbe 2:96–105 [DOI] [PubMed] [Google Scholar]
- 47. Watret GE, Pringle CR, Elliott RM. 1985. Synthesis of bunyavirus-specific proteins in a continuous cell line (XTC-2) derived from Xenopus laevis. J. Gen. Virol. 66:473–482 [DOI] [PubMed] [Google Scholar]
- 48. Weber F, Dunn EF, Bridgen A, Elliott RM. 2001. The Bunyamwera virus nonstructural protein NSs inhibits viral RNA synthesis in a minireplicon system. Virology 281:67–74 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.