Abstract
Gammaherpesviruses are important pathogens in human and animal populations. During early events of infection, these viruses manipulate preexisting host cell signaling pathways to allow successful infection. The different proteins that compose viral particles are therefore likely to have critical functions not only in viral structures and in entry into target cell but also in evasion of the host's antiviral response. In this study, we analyzed the protein composition of bovine herpesvirus 4 (BoHV-4), a close relative of the human Kaposi's sarcoma-associated herpesvirus. Using mass spectrometry-based approaches, we identified 37 viral proteins associated with extracellular virions, among which 24 were resistant to proteinase K treatment of intact virions. Analysis of proteins associated with purified capsid-tegument preparations allowed us to define protein localization. In parallel, in order to identify some previously undefined open reading frames, we mapped peptides detected in whole virion lysates onto the six frames of the BoHV-4 genome to generate a proteogenomic map of BoHV-4 virions. Furthermore, we detected important glycosylation of three envelope proteins: gB, gH, and gp180. Finally, we identified 38 host proteins associated with BoHV-4 virions; 15 of these proteins were resistant to proteinase K treatment of intact virions. Many of these have important functions in different cellular pathways involved in virus infection. This study extends our knowledge of gammaherpesvirus virions composition and provides new insights for understanding the life cycle of these viruses.
INTRODUCTION
Kaposi's sarcoma-associated herpesvirus (KSHV) and Epstein-Barr virus (EBV) are important human pathogens associated with numerous cancers (52, 56). Although much effort has been invested in these viruses, their limited lytic growth in vitro makes studies of these viruses particularly challenging. Related animal gammaherpesviruses are therefore an important source of information. bovine herpesvirus 4 (BoHV-4) belongs to the Gammaherpesvirinae subfamily and to the Rhadinovirus genus (11). Similar to its human counterparts, BoHV-4 is widespread in natural populations and BoHV-4 infection persists in the vast majority of individuals as a lifelong, asymptomatic infection (50).
The BoHV-4 genome is estimated to encode at least 79 open reading frames (ORFs) (42, 59). Like all herpesviruses, BoHV-4 virions are composed of a large (diameter > 100 nm) icosahedral nucleocapsid, which is assembled in the nucleus from at least eight different conserved proteins (1). The capsid is surrounded by a proteinaceous layer called tegument that is acquired in both the nucleus and the cytoplasm. More than 15 viral proteins have been reported in the tegument of herpesviruses; however, the organization and function of tegument proteins is still largely unknown (18). The cytoplasmic capsids with tegument are finally enclosed by a lipid envelope bearing proteins to form mature infectious virus particles (diameter, ∼200 nm). Herpesviruses encode multiple envelope glycoproteins, among which glycoproteins gB, gH, gL, gM, and gN are shared by all members of the Herpesviridae family (23).
In addition to virally encoded structural proteins, a variety of host proteins has been found in herpesvirus virions. Although some of these proteins have only been reported in a given virus, others are shared by several members of the Herpesviridae family. However, whether or not they are specifically incorporated and what function they may have during infection is still largely unknown. Complete identification of viral and cellular proteins that compose virions will help elucidate processes such as virus production, virion entry, or immune evasion. It could also allow defining new antiviral drug targets.
In recent years, mass spectrometry (MS)-based analysis has been used to identify the composition of different herpesviruses (2, 3, 7, 13, 25, 27, 28, 31, 41, 53, 55, 58). Within rhadinoviruses, KSHV (3, 58), rhesus monkey rhadinovirus (RRV) (41), and murid herpesvirus 4 (MuHV-4) (7) extracellular virions have been analyzed using this approach. However, while analysis of RRV virions identified 33 structural proteins, only 25 and 14 were revealed in KSHV and MuHV-4 virions, respectively. Moreover, only 10 proteins were common to the different analyses. In contrast, analyses of herpes simplex virus 1 (HSV-1) and pseudorabies virus (PRV) virions identified, respectively, 44 and 47 structural proteins, among which 39 were shared by both viruses.
In this study, we used complementary proteomic approaches to characterize the protein content of purified BoHV-4 virions. We identified 37 viral proteins associated with virion-enriched preparations. Of these proteins, 24 were resistant to proteinase K treatment of BoHV-4 intact virions, and 16 were found in at least two other rhadinoviruses. Moreover, we identified the glycoproteins gB, gH, and gp180 as the most glycosylated proteins of the virion. Finally, analysis of the host protein content revealed the potential incorporation of at least 15 host cellular proteins in BoHV-4 extracellular virions.
MATERIALS AND METHODS
Cells and virus.
Madin-Darby bovine kidney cells (MDBK [ATCC CCL-22]) were cultured in minimum essential medium (Invitrogen) containing 10% fetal calf serum, 2% penicillin-streptomycin (Invitrogen) and 1% nonessential amino acids (Invitrogen). The BoHV-4 v.test strain was used throughout the present study (16, 51).
Production and purification of BoHV-4 virions.
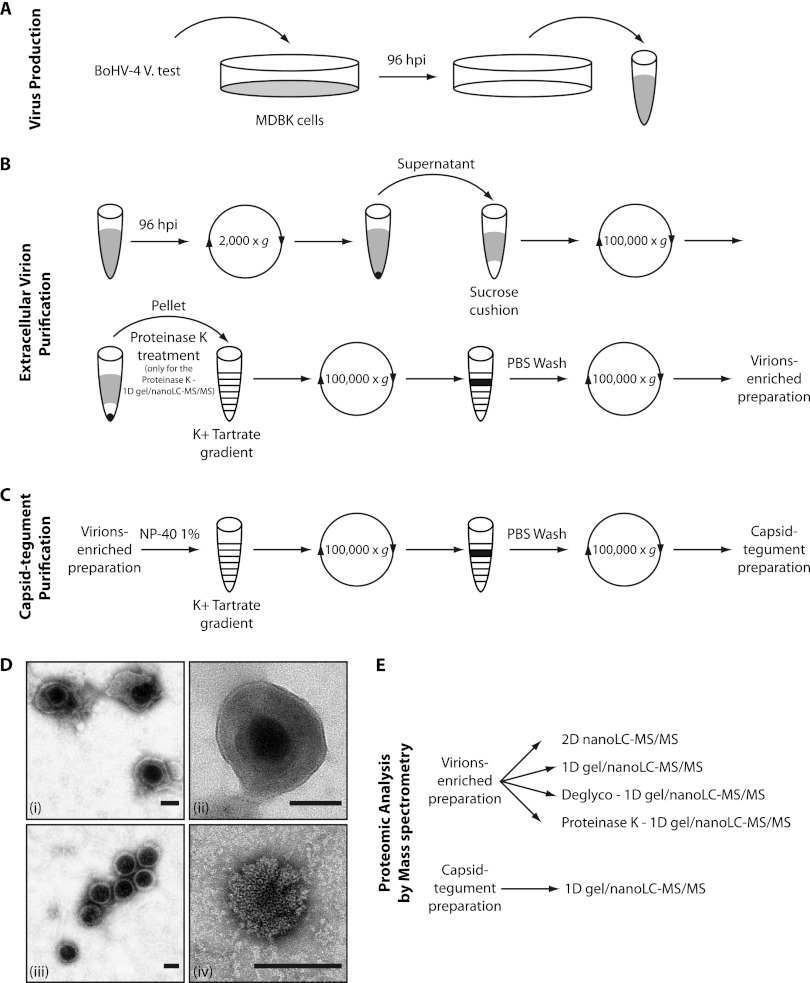
MDBK cells were infected with the BoHV-4 v.test strain at a multiplicity of infection (MOI) of 0.05 PFU/cell. To reduce cellular contaminants, the supernatant was harvested after 96 h postinfection (hpi) before complete cell lysis. Extracellular virions were purified from the cell supernatant as described previously (29, 38), with minor modifications (Fig. 1). Briefly, after removal of the cell debris by low-speed centrifugation (1,000 × g, 10 min, 4°C), virions present in the infected cell supernatant (ca. 1 × 106 to 5 × 106 PFU/ml) were harvested by ultracentrifugation (100,000 × g, 2 h, 4°C) through a 30% (wt/vol) sucrose cushion. Virions were then banded by isopycnic gradient ultracentrifugation in a continuous 20 to 50% (wt/vol) potassium tartrate gradient in phosphate-buffered saline (PBS; 100,000 × g, 2 h, 4°C). The band containing virions was collected (∼3 ml), diluted 10-fold in PBS, and pelleted by ultracentrifugation (100,000 × g, 2 h, 4°C). The pellet was finally resuspended in PBS, and virus-enriched preparations (ca. 1 × 108 to 5 × 108 PFU/ml) were stored at −80°C.
Fig 1.
Strategy for BoHV-4 purification and proteomic analysis by mass spectrometry (MS). (A) MDBK cells were infected with the v.test strain of BoHV-4 at an MOI of 0.05. At 96 hpi, the extracellular medium was harvested. (B and C) BoHV-4 extracellular virions (B) and BoHV-4 capsids (C) were enriched as described in Materials and Methods. (D) The purity of the virions (i and ii) and capsids (iii and iv) preparations was assessed by EM. Bar, 50 nm. Original magnifications, ×30,000 for panels i and iii and ×49,000 for panels ii and iv. (E) Virion-enriched and capsid-tegument preparations were subjected to proteomic analysis by MS using different complementary approaches.
Protease treatment.
Virions were treated with proteinase K as described previously (28). Briefly, after ultracentrifugation through the sucrose cushion described above (Fig. 1), the viral pellet was resuspended in 1 ml of MNT buffer (30 mM morpholineethanesulfonic acid [MES], 10 mM NaCl, and 20 mM Tris-HCl [pH 7.4]) containing 10 μg of proteinase K (Roche, Mannheim, Germany)/ml for 45 min at room temperature and subsequently treated with 2 mM phenylmethylsulfonyl fluoride (Roche) prior to density gradient centrifugation on a 20 to 50% (wt/vol) potassium tartrate gradient in PBS (100,000 × g, 2 h, 4°C). The band containing virions was collected (∼3 ml), diluted 10-fold in PBS, and pelleted by ultracentrifugation (100,000 × g, 2 h, 4°C). Proteinase K-treated virions were finally resuspended in PBS and stored at −80°C.
Fractionation of BoHV-4 virions.
Lipid envelopes were removed from capsids and teguments by incubation with a nonionic detergent as described previously (10). Briefly, virions enriched preparations were lysed in PBS containing 1% (vol/vol) Triton X-100 (Fig. 1C). Capsids associated with tegument were banded by isopycnic gradient ultracentrifugation in a continuous 20 to 50% (wt/vol) potassium tartrate gradient in PBS (100,000 × g, 2 h, 4°C). The resulting band was collected, washed in PBS, and finally pelleted by ultracentrifugation (100,000 × g, 2 h, 4°C). The supernatant was discarded, and the capsid-tegument pellet was resuspended in PBS and stored at −80°C until further use.
Negative staining and electron microscopy (EM).
Copper grids covered by a thin film of Formvar (400 mesh; Agar Scientific) were incubated for 10 min with 1% Alcian blue 8G solution (Gurr Microscopy Materials; BHD) to add positive charges. After washing, virion-enriched or capsid-tegument preparations were adsorbed to the grids for 10 min. Viral particles were then stained for contrast by incubation in 2% uranyl acetate solution for 10 s (Agar Scientific). Samples were observed using a transmission electron microscope (FEI Tecnai Spirit).
1D gel/nano-LC-MS/MS.
Proteins from virion-enriched (treated or not with proteinase K) and from capsid-tegument preparations were extracted in Laemmli sample buffer and separated by SDS-PAGE on 4 to 20% acrylamide 7-cm gels (Invitrogen). Separated proteins in the gel were excised in 31 serial slices along the lane. Gel slices were submitted to in-gel digestion with sequencing grade modified trypsin as described previously (53). Briefly, gels were washed successively with 50 mM ammonium bicarbonate (ABC) buffer and ABC buffer–50% (vol/vol) acetonitrile (ACN). Proteins were reduced and alkylated using dithiothreitol (DTT) and iodoacetamide followed by washing with ABC and ABC-ACN. The resulting peptides were analyzed by one-dimensional gel/nano-liquid chromatography-tandem mass spectrometry (1D gel/nano-LC-MS/MS) using a 40-min ACN gradient as described previously (34).
2D gel/nano-LC-MS/MS.
Proteins of virion-enriched preparations were extracted from complete virions using guanidine chloride (GC) as described previously (38). Briefly, virions were suspended in 6 M GC, sonicated for 5 min, and shaken at 900 rpm for 30 min at room temperature. After centrifugation, the proteins were reduced with 10 mM dithiothreitol at 50°C for 20 min and alkylated with 25 mM iodoacetamide at 25°C for 20 min in the dark. Proteins were recovered by acetone precipitation and dissolved in ABC buffer. The proteins were digested overnight at 37°C with trypsin (enzyme/substrate ratio, 1:50). Tryptic peptides were cleaned using spin tips (Thermo Fisher Scientific, Waltham, MA) according to the manufacturer's instructions. The resulting peptides were analyzed by 2D (strong cation exchange [SCX], reversed phase) chromatography and online MS/MS as described by Mastroleo et al. (34) except that only three salt plugs of 25, 100, and 800 mM NH4Cl were analyzed in addition to the SCX flowthrough.
Oligosaccharide digestion.
The deglycosylated extract was obtained by treating virion-enriched preparations with an enzymatic protein deglycosylation kit (Sigma) according to the instructions of the manufacturer. Viral proteins were successively denatured for 5 min at 100°C in a denaturation solution and then treated with Triton X-100, peptide–N-glycosidase F, O-glycosidase, α(2,3,6,8,9)-neuraminidase, β(1,4)-galactosidase, and β-N-acetylglucosaminidase for 3 h at 37°C. This extract was then subjected to the 1D gel/nano-LC-MS/MS approach.
MS/MS analyses.
Peptides were analyzed using the “peptide scan” option of the HCT Ultra ion trap (Bruker), consisting of a full-scan MS and MS/MS scan spectrum acquisitions in ultrascan mode (26,000 m/z s−1). Peptide fragment mass spectra were acquired in data-dependent AutoMS (2) mode with a scan range of 100 to 2,800 m/z, three means, and five precursor ions selected from the MS scan 300 to 1,500 m/z. Precursors were actively excluded within a 0.5 min window, and all singly charged ions were excluded. Peptide peaks were detected and deconvoluted automatically using Mascot distiller 2.3.3 and submitted to database search using an in-house Mascot search engine (version 2.2). The default search parameters used were as follows: enzyme = trypsin; maximum missed cleavages = 2; fixed modifications = carbamidomethyl (C); variable modifications = oxidation (M); peptide tolerance ± 1.5 Da (Da); MS/MS tolerance ± 0.5 Da; peptide charge = 2+ and 3+; and instrument = ESI-TRAP. All data were also searched against the NCBI Bos taurus database in order to detect host proteins host proteins or against a BoHV-4 v.test database (42) to detect viral proteins. Only sequences identified with a Mascot score greater than 30 were considered, and single peptide identification was systematically evaluated manually. For each approach, the exponentially modified protein abundance index (emPAI) (24) was calculated to estimate protein relative abundance for the complete virion extracts. The protein abundance index (PAI) is defined as the number of observed peptides divided by the number of observable peptides per protein. The exponentially modified PAI (10PAI − 1) is proportional to protein content in a protein mixture in LC-MS/MS experiments.
Proteogenomic mapping.
The complete nucleotide sequence of BoHV-4 (42) was translated in silico in all six frames resulting in six peptidic sequences. These sequences were compiled in a FASTA-style database that had frame position embedded into each header tag for a given sequence. The mass lists obtained in the different complete virions analyses were searched against this database using the criteria stated above. Detected peptides were graphically mapped onto the BoHV-4 v.test genome which also displayed the predicted ORFs as defined previously (42). The results were generated and visualized using R (49) and the SeqinR package (9).
RESULTS
Purification of extracellular BoHV-4 virions.
The major hurdle to characterization of virion components by MS is purification of extracellular mature virions with both quality and quantity. In the present study, we harvested and purified extracellular virions from the medium of MDBK cells infected with the BoHV-4 v.test strain as described in Materials and Methods (Fig. 1A and B). Virion-enriched preparations were checked for quality by negative-stain EM. Most of the observed particles were complete virions with intact or disrupted envelopes (Fig. 1D). We also observed small amounts of isolated capsids (1 to 5% of the particles) probably resulting from the loss of the viral envelope during purification or sample preparation for EM. In contrast, we did not observe any contamination by cell debris. This therefore indicates that our virion purification strategy could be considered as successful, at least as evaluated by EM.
Viral protein composition of BoHV-4 virions.
Three complementary MS approaches were used to analyze BoHV-4 virion composition (Fig. 1E). First, BoHV-4 virion proteins were separated by 1D SDS-PAGE, digested in gel, and analyzed by MS (1D gel/nano-LC-MS/MS). This approach allows association of the identified protein with the apparent molecular mass (MM) assessed by 1D SDS-PAGE. In parallel, virion proteins were analyzed by a gel-free approach. After protein extraction and trypsin digestion, sample is submitted to sequential two-dimensional liquid chromatography coupled on-line to MS/MS analysis (2D gel/nano-LC-MS/MS). This protocol therefore avoids some problems associated with proteins poorly entering gel, proteins present in very low abundance, or proteins extracted from gel slices inefficiently after in-gel digestion. Finally, as protein glycans could interfere with MS/MS analysis, proteins of virion-enriched preparations were treated with different glycosidases before analysis by 1D gel/nano-LC-MS/MS (Deglyco-1D gel/nano-LC-MS/MS).
Altogether, these three complementary approaches enabled us to identify 34 virally encoded proteins in BoHV-4 viral particle. A total of 21 of these proteins were detected by the three protocols. This number is consistent with the numbers previously reported for other members of the Herpesviridae family. These proteins are listed in Table 1 according to their position in the viral genome.
Table 1.
Viral content of BoHV-4 extracellular virionsa
| ORF | Protein description | Predicted MM (kDa) | pKb | 2D gel/nano-LC-MS/MS |
1D gel/nano-LC-MS/MS |
Deglyco-1D gel/nano-LC-MS/MS |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No. of peptides | Prot matches | Coverage (%) | emPAI | emPAI (%) | No. of peptides | Prot matches | Coverage (%) | emPAI | emPAI (%) | No. of peptides | Prot matches | Coverage (%) | emPAI | emPAI (%) | ||||
| ORF3 | BORFA1; v-FGAM-synthase | 144.18 | + | 16 | 21 | 0.18 | 0.54 | 2.21 | 17 | 76 | 0.20 | 0.62 | 0.71 | 8 | 11 | 0.10 | 0.22 | 0.69 |
| ORF6 | Single-stranded DNA-binding protein MDBP | 129.68 | – | 2 | 2 | 0.03 | 0.06 | 0.25 | 1 | 1 | 0.01 | 0.03 | 0.03 | |||||
| ORF8 | Glycoprotein B | 100.08 | + | 21 | 33 | 0.31 | 1.48 | 6.05 | 25 | 110 | 0.37 | 3.11 | 3.55 | 21 | 84 | 0.30 | 1.97 | 6.22 |
| Bo5 | Homolog of KSHV K5-encoded E3 ubiquitin ligase | 35.48 | – | 4 | 6 | 0.20 | 0.65 | 2.66 | ||||||||||
| ORF16 | v-Bcl-2 protein | 26.06 | – | 1 | 1 | 0.10 | 0.15 | 0.61 | 1 | 1 | 0.04 | 0.15 | 0.17 | |||||
| ORF17 | Minor scaffold protein (protease) | 57.74 | + | 9 | 23 | 0.22 | 1.25 | 5.11 | 9 | 47 | 0.22 | 1.25 | 1.43 | 5 | 9 | 0.13 | 0.37 | 1.17 |
| ORF19 | Capsid vertex-specific complex protein (CVSC) | 63.39 | + | 5 | 6 | 0.15 | 0.33 | 1.35 | 7 | 14 | 0.16 | 0.49 | 0.56 | 1 | 1 | 0.02 | 0.06 | 0.19 |
| ORF22 | Glycoprotein H | 82.23 | + | 6 | 8 | 0.11 | 0.3 | 1.23 | 13 | 54 | 0.25 | 0.94 | 1.07 | 13 | 38 | 0.26 | 0.94 | 2.97 |
| ORF25 | Major capsid protein | 155.12 | + | 56 | 296 | 0.58 | 8.52 | 34.80 | 63 | 1208 | 0.60 | 13.88 | 15.86 | 51 | 480 | 0.54 | 7.27 | 22.94 |
| ORF26 | Triplex component | 34.3 | + | 8 | 26 | 0.39 | 1.84 | 7.52 | 12 | 155 | 0.56 | 5.53 | 6.32 | 11 | 57 | 0.54 | 5.53 | 17.45 |
| ORF32 | Viral DNA cleavage/packaging protein (CVSC) | 59.9 | + | 3 | 3 | 0.10 | 0.23 | 0.94 | 6 | 28 | 0.19 | 0.72 | 0.82 | 3 | 6 | 0.10 | 0.23 | 0.73 |
| ORF33 | Tegument protein | 39.36 | + | 7 | 16 | 0.36 | 1.07 | 4.37 | 13 | 107 | 0.47 | 3.7 | 4.23 | 7 | 48 | 0.33 | 1.48 | 4.67 |
| ORF36 | Kinase | 45.16 | – | 1 | 1 | 0.04 | 0.08 | 0.09 | ||||||||||
| ORF38 | Myristylated tegument protein | 7.7 | + | 1 | 1 | 0.50 | 0.54 | 0.62 | 1 | 1 | 0.50 | 0.54 | 1.70 | |||||
| ORF39 | Glycoprotein M | 43.36 | + | 3 | 4 | 0.15 | 0.28 | 1.14 | 1 | 20 | 0.04 | 0.09 | 0.10 | 1 | 8 | 0.04 | 0.09 | 0.28 |
| ORF42 | Potential tegument protein (UL7 homolog) | 33.65 | + | 1 | 1 | 0.06 | 0.11 | 0.35 | ||||||||||
| ORF43 | Portal protein | 71.11 | + | 3 | 5 | 0.09 | 0.16 | 0.65 | 2 | 6 | 0.05 | 0.11 | 0.13 | |||||
| ORF45 | Tegument protein, IRF-7 blocking protein | 27.18 | + | 8 | 21 | 0.37 | 3.21 | 13.11 | 9 | 36 | 0.48 | 3.21 | 3.67 | 4 | 11 | 0.25 | 0.69 | 2.18 |
| ORF46 | Uracil DNA glycosylase | 29.07 | – | 1 | 1 | 0.04 | 0.13 | 0.15 | ||||||||||
| ORF47 | Glycoprotein L | 16.28 | – | 2 | 2 | 0.20 | 0.53 | 2.17 | 1 | 4 | 0.06 | 0.24 | 0.27 | 1 | 1 | 0.06 | 0.24 | 0.76 |
| ORF48 | 59.17 | + | 11 | 18 | 0.32 | 1.21 | 4.94 | 13 | 79 | 0.36 | 1.82 | 2.08 | 14 | 59 | 0.34 | 1.65 | 5.21 | |
| Bo10 | Glycoprotein gp180 | 28.51 | + | 1 | 1 | 0.05 | 0.13 | 0.53 | 2 | 12 | 0.08 | 0.45 | 0.51 | 3 | 6 | 0.12 | 0.45 | 1.42 |
| ORF52 | Tegument protein | 13.85 | + | 7 | 103 | 0.55 | 24.48 | 100.0 | 8 | 140 | 0.55 | 87.53 | 100.0 | 6 | 61 | 0.55 | 31.69 | 100.0 |
| ORF54 | dUTPase | 31.14 | – | 1 | 1 | 0.06 | 0.12 | 0.14 | ||||||||||
| ORF55 | Tegument protein (UL51 homolog) | 22.23 | + | 2 | 4 | 0.14 | 0.37 | 1.17 | ||||||||||
| ORF57 | Posttranscriptional regulatory protein | 48.17 | – | 1 | 1 | 0.04 | 0.08 | 0.25 | ||||||||||
| ORF59 | DNA replication protein | 44.06 | – | 6 | 8 | 0.22 | 0.77 | 3.15 | 7 | 12 | 0.24 | 0.77 | 0.88 | 3 | 7 | 0.12 | 0.39 | 1.23 |
| ORF60 | Ribonucleotide reductase small subunit | 35.65 | – | 1 | 1 | 0.04 | 0.11 | 0.13 | ||||||||||
| ORF62 | Capsid triplex component | 38.71 | + | 6 | 17 | 0.22 | 1.1 | 4.49 | 11 | 74 | 0.38 | 3.39 | 3.87 | 8 | 27 | 0.25 | 1.77 | 5.59 |
| ORF63 | Tegument protein | 109 | + | 5 | 6 | 0.08 | 0.18 | 0.74 | 18 | 30 | 0.26 | 0.95 | 1.09 | 6 | 11 | 0.10 | 0.22 | 0.69 |
| ORF64 | Tegument protein | 288.8 | + | 11 | 15 | 0.08 | 0.18 | 0.74 | 37 | 77 | 0.19 | 0.64 | 0.73 | 1 | 1 | 0.01 | 0.01 | 0.03 |
| ORF65 | Small capsomer interacting protein | 14.61 | + | 1 | 1 | 0.11 | 0.27 | 1.10 | 2 | 15 | 0.35 | 1.58 | 1.81 | 1 | 4 | 0.25 | 0.61 | 1.92 |
| ORF67 | Tegument protein | 30.33 | – | 2 | 2 | 0.13 | 0.27 | 0.31 | ||||||||||
| ORF75 | v-FGAM-synthase | 125.3 | + | 18 | 25 | 0.22 | 0.64 | 2.61 | 23 | 122 | 0.29 | 1.19 | 1.36 | 22 | 95 | 0.27 | 1.25 | 3.94 |
2D gel/nano-LC-MS/MS and 1D gel/nano-LC-MS/MS were accomplished in technical replicates, and the results were pooled. The “number of peptides” indicates the number of unique peptides identified per protein. “Prot matches” indicates the number of peptides detected per protein. “Coverage” indicates the percentages of coverage of proteins by peptides. emPAI values were calculated as described by Ishihama et al. (24). Relative emPAI values [emPAI (%)] were calculated as percentages of the pORF52 maximum abundance.
pK, proteinase K treatment. +, Proteins detected in the proteinase K–1D gel/nano-LC-MS/MS.
Despite our multistep purification protocol, detection of some proteins could result from nonspecific sticking to the virion rather than true integration into the particle. This is especially true for cellular proteins (see below). Moreover, some of these proteins could be highly abundant and could therefore impede the detection of proteins that are truly virion associated but present in only a few copies per virion. To address this issue, we therefore treated virions with proteinase K, in the absence of detergent, prior to density centrifugation (Fig. 1B).
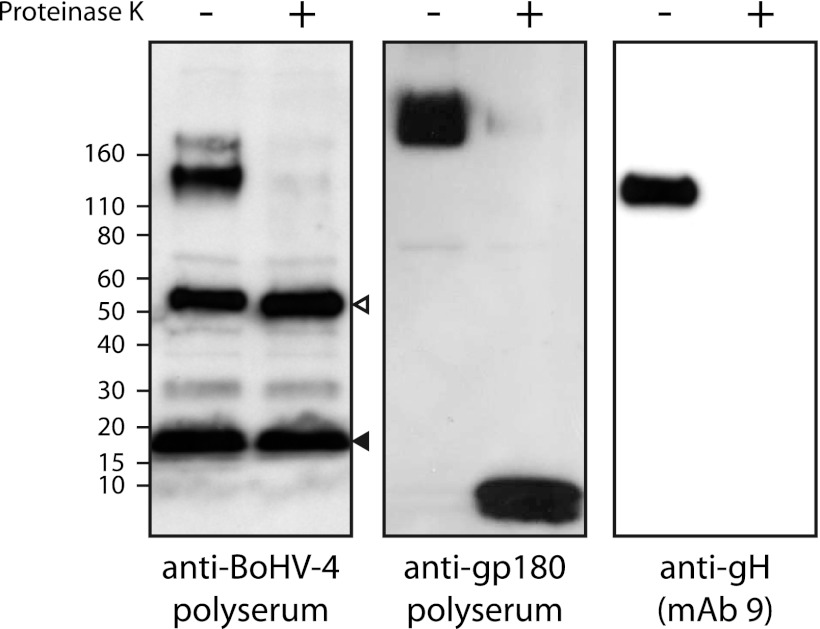
We validated this treatment by Western blotting (Fig. 2). As BoHV-4 virions are enveloped within a phospholipid bilayer, only viral and host proteins contained within the tegument and capsid are protected from protease digestion (28, 41). Immunoblotting with anti-BoHV-4 immune serum confirmed that some proteins disappeared after proteinase K treatment, while some others (likely capsid proteins) were not affected (Fig. 2). In addition, the full-length form of BoHV-4 type I membrane proteins gp180 and gH were identified only in untreated virions, while they were either truncated (for gp180, since the anti-gp180 polyserum is raised against the “tegumental tail” of the protein [32]) or undetectable (gH) in proteinase K-treated virions (Fig. 2). Finally, as expected, this treatment increased the proportion of detection of viral peptides (data not shown).
Fig 2.
Sensitivity of viral proteins to proteinase K treatment of virions. Western blot analysis of viral proteins in purified virions from mock (−)- and proteinase K (+)-treated samples. gp180 and gH are type I transmembrane proteins that have an N-terminal domain predicted to be sensitive to proteinase K digestion. Open and black triangles indicate bands corresponding to VP7 and VP24 proteins, respectively. These bands correspond likely to capsid proteins.
Among the 34 proteins described in Table 1, 11 were not detected after proteinase K treatment. These are the proteins encoded by ORF6, Bo5, ORF16, ORF36, ORF46, ORF47, ORF54, ORF57, ORF59, ORF60, and ORF67. Unexpectedly, although BoHV-4 envelope glycoproteins were sensitive to proteinase K digestion (Fig. 2), most of them remained sufficiently intact to allow continued detection. This was the case for gB, gH, gM, and gp180. No additional protein was specifically detected by this protocol.
Estimation of protein abundance in BoHV-4 virion.
To estimate absolute protein contents in complex mixtures, Rappsilber et al. previously defined a protein abundance index (PAI) as the number of observed peptides in LC-MS/MS experiments divided by the number of theoretically observable peptides per protein (46). Then, Ishihama et al. (24) showed that PAI values show a linear relationship with the logarithm of protein concentration. For absolute quantitation, these researchers thus converted PAI to exponentially modified PAI (emPAI = 10PAI − 1), which is proportional to protein content in a protein mixture. In order to relatively quantify viral proteins in virions, emPAI values were expressed as percentages of the maximal emPAI value obtained by one approach (Table 1). Surprisingly, the tegument protein encoded by ORF52 was the most abundant protein detected in BoHV-4 virions. It was around four times more abundant than the major capsid protein (pORF25). This apparent abundance could not be biased through the enhanced detection of a single peptide since seven different peptides were detected in similar proportions by our analyses (data not shown).
Identification of viral capsid and capsid associated tegument components.
To identify capsid proteins and tegument proteins specifically associated with capsids, we purified viral capsid-tegument preparations as described in Materials and Methods (Fig. 1C). The preparation of purified capsid-tegument was checked for quality by transmission EM (Fig. 1D). As expected, the sample contained only isolated capsids and no trace of intact virions or envelope debris.
MS analysis revealed the presence of 24 viral proteins in this sample (Table 2). As expected, no predicted viral envelope glycoprotein (i.e., gB, gH, gM, gL, or gp180) was detected in this sample confirming the quality of our purification procedure. Moreover, proteins encoded by ORF16, ORF36, ORF38, ORF42, ORF46, ORF57, and ORF60 were also not detected in this sample, although they had been detected in complete virions before proteinase K treatment. Among these proteins, only the proteins encoded by ORF38 and ORF42 were detected after proteinase K treatment of intact virions. This experiment suggests therefore that they could be considered as outer tegument proteins. In contrast, proteins encoded by ORF16, ORF36, ORF46, ORF57, and ORF60 are likely not incorporated in BoHV-4 virions. Finally, peptides corresponding to pORF10 and pORF35 were detected in the capsid-tegument preparations, whereas they were not detected in extracellular virions.
Table 2.
Proteins of BoHV-4 virions identified by 1D gel/nano-LC-MS/MS as associated with purified capsid-tegument samples
| ORF | Protein description | Predicted MM (kDa) | pKa | No. of peptidesb | Prot matchesc | Coverage (%)d | emPAIe | emPAI (%)f | Expected relative abundance in capsid (%)g |
|---|---|---|---|---|---|---|---|---|---|
| ORF3 | BORFA1; v-FGAM-synthase | 144.18 | + | 7 | 11 | 11.18 | 0.13 | 3.04 | |
| ORF6 | Single-stranded DNA-binding protein MDBP | 129.68 | – | 1 | 1 | 1.32 | 0.08 | 1.87 | |
| Bo5 | Homolog of KSHV K5-encoded E3 ubiquitin ligase | 35.48 | – | 2 | 5 | 17.61 | 0.15 | 3.50 | |
| ORF10 | Derived from herpesvirus dUTPase | 48.23 | – | 1 | 1 | 3.99 | 0.05 | 1.17 | |
| ORF17h | Minor scaffold protein (protease) | 57.74 | + | 7 | 20 | 18.99 | 0.55 | 12.85 | 15.7 |
| ORF19 | Capsid vertex-specific complex protein | 63.39 | + | 4 | 4 | 10.57 | 0.13 | 3.04 | Up to 6.28 |
| ORF25 | Major capsid protein | 155.12 | + | 54 | 636 | 55.13 | 4.28 | 100 | 100 |
| ORF26 | Capsid triplex component | 34.3 | + | 9 | 35 | 37.83 | 1.39 | 32.48 | 67 |
| ORF32 | Viral DNA cleavage/packaging protein (capsid vertex-specific complex) | 59.9 | + | 1 | 2 | 3.07 | 0.05 | 1.17 | Up to 6.28 |
| ORF33 | Tegument protein | 39.36 | + | 4 | 12 | 14.46 | 0.29 | 6.78 | |
| ORF35 | Tegument protein (homolog to herpesvirus core gene UL14) | 17.87 | – | 1 | 2 | 22.29 | 0.15 | 3.50 | |
| ORF43 | Portal protein | 71.11 | + | 5 | 6 | 15.1 | 0.19 | 4.44 | 1.25 |
| ORF45 | Tegument protein, IRF-7 blocking protein | 27.18 | + | 1 | 3 | 6.22 | 0.09 | 2.10 | |
| ORF48 | 59.17 | + | 15 | 61 | 40.47 | 1.35 | 31.54 | ||
| ORF52 | Tegument protein | 13.85 | + | 6 | 22 | 62.5 | 2.95 | 68.93 | |
| ORF54 | dUTPase | 31.14 | – | 1 | 2 | 5.67 | 0.08 | 1.87 | |
| ORF55 | Tegument protein (homolog to herpesvirus core gene UL51) | 22.23 | + | 4 | 10 | 26 | 0.55 | 12.85 | |
| ORF59 | DNA replication protein | 44.06 | – | 8 | 56 | 27.62 | 0.87 | 20.33 | |
| ORF62 | Capsid triplex component | 38.71 | + | 12 | 23 | 39.23 | 1.47 | 34.35 | 33.5 |
| ORF63 | Tegument protein | 109 | + | 14 | 20 | 20.13 | 0.42 | 9.81 | |
| ORF64 | Tegument protein | 288.8 | + | 13 | 16 | 8.99 | 0.12 | 2.80 | |
| ORF65 | Small capsomer interacting protein | 14.61 | + | 1 | 23 | 24.62 | 0.63 | 14.72 | 94.2i |
| ORF67 | Tegument protein | 30.33 | – | 1 | 1 | 8.2 | 0.09 | 2.10 | |
| ORF75 | Tegument protein/v-FGAM synthetase | 125.3 | + | 4 | 5 | 6.58 | 0.09 | 2.10 |
pK, proteinase K treatment. +, Proteins detected in the proteinase K–1D gel/nano-LC-MS/MS analysis of virions.
Number of unique peptides identified per protein.
“Prot matches” indicates the number of peptides detected per protein.
The values shown are the percentages of coverage of proteins by peptides.
emPAI values were calculated as described by Ishihama et al. (24).
Relative emPAI values [emPAI (%)] were calculated as percentages of the maximum pORF25 abundance.
As described for HSV-1B capsids by Baines (1).
Predicted capsid proteins are highlighted in gray.
On the basis of full occupancy, i.e., one copy decorating each of six hexon tips.
Proteogenomic mapping.
Experimental identification of protein composition by MS is based on the quality of the ORFs annotations in the genomes. After genome sequencing, ORFs are primarily identified using computational gene prediction algorithms. However, these methods often produce false-positive and false-negative predictions. Moreover, as virus genomes have evolved with size constraints, a unique sequence can encode several ORFs distributed in different reading frames. Therefore, experimental identification of expressed proteins by proteomic techniques constitutes an interesting and reliable approach to identify genomic location and structure of protein-coding genes.
In order to identify possible BoHV-4 virions proteins that had not been annotated, we generated a database containing the entire genome of the BoHV-4 v.test strain translated in the six frames. The mass lists obtained in the different complete virions analyses were searched against this database as described in Materials and Methods. Finally, the detected peptides were graphically mapped onto the BoHV-4 V. test genome (Fig. 3). The results obtained showed that all identified peptides mapped into previously annotated ORFs. Moreover, this approach allowed us to analyze the localization of detected peptides within annotated protein sequences. This improved our analysis for proteins encoded by overlapping sequences. This was particularly the case for the ORF17-17.5 proteins (see Fig. S1 in the supplemental material). The majority of the inner shelf of all herpesvirus capsids is made of copies of a scaffold protein. In rhadinoviruses, this protein is encoded by ORF17.5. Similar to observations in other herpesviruses, the coding sequence of this gene is entirely contained within and in frame with a larger ORF, called ORF17 in rhadinoviruses. This larger ORF encodes a protease involved in capsid maturation. Distribution of identified peptides in ORF17-17.5 showed that most of these peptides were located into the 3′-coterminal region. However, few peptides were also identified into the ORF17 specific N-terminal region. These results suggest therefore that, as expected, peptides belonging to pORF17.5 are the most largely expressed. However, the pORF17 protease was also found in our virion preparation.
Fig 3.
Proteogenomic map of BoHV-4. The six possible frames of the BoHV-4 v.test strain genome translation are shown with rectangle indicating the predicted ORFs. Red and blue ORFs represent forward and reverse frames, respectively. Detected peptides are indicated by bars whose height is proportional to the number of detections. Genomic positions are indicated in base pairs.
Glycosylation of virion components.
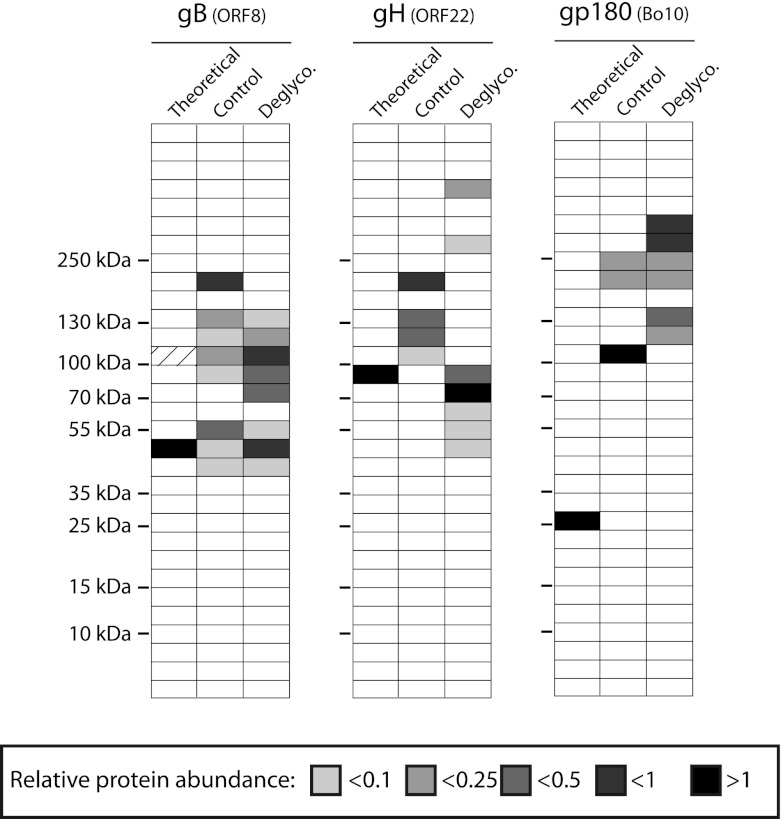
Although our study was primarily designed to identify the protein content of BoHV-4 virions, we were also able to predict glycosylation of several virion proteins by comparing results obtained by the 1D gel/nano-LC-MS/MS approach performed on untreated and deglycosylated samples run in parallel. Briefly, both samples were submitted to 1D gel electrophoresis as described in Materials and Methods. After protein migration, the gels were cut in 31 slices, and the protein composition of each of the 31 slices was determined as described in Materials and Methods. Distribution across the gel was then determined for each protein after deglycosylation or not. Surprisingly, only three proteins showed gel distribution change after glycosidase treatment (Fig. 4). Interestingly, these proteins were gB, gH, and gp180. BoHV-4 gB has a theoretical MM of 100 kDa. However, like its homologues in other herpesviruses, BoHV-4 gB possesses a putative protease cleavage site (30). Two polypeptides with calculated MMs of 51 and 46 kDa corresponding to the N- and C-terminal parts of gB, respectively, should arise from this cleavage. Experimentally, it has been shown that two gB polypeptides are incorporated in the viral particle, one of 128 kDa, corresponding to the N-terminal part of gB and the other of 56 kDa, corresponding to the C-terminal part of gB. It has been assumed that the difference between the predicted MM of the unglycosylated products and the apparent MM of both parts is mainly due to the addition of N- and O-linked sugars (30). Our results confirmed this hypothesis since the content of the 50-kDa slice was increased after deglycosylation (Fig. 4). Surprisingly, a 200-kDa gel slice contained most of the gB polypeptides in the control lane. This is in accordance with the observed MM of uncleaved gB (30). In the deglycosylated sample, no gB peptides were observed at that position in the gel. In contrast, the gB content of slices around 100 kDa increased. These results suggest therefore that cleaved and uncleaved gB were incorporated in our virion preparation and that these different fragments are glycosylated. Similarly, our results confirmed that gH is glycosylated in BoHV-4 virions. We recently showed that these are mainly N-linked glycans (29). Finally, although we have previously shown that the Bo10 encoded glycoprotein, gp180, is massively glycosylated (33), we observed that removing glycans increased the apparent molecular mass of the protein. This is likely due to aggregates formation as we previously observed for this protein after deglycosylation (unpublished results).
Fig 4.
Analysis of BoHV-4 structural protein glycosylation. Control or deglycosylated proteins of purified BoHV-4 virions were separated by SDS-PAGE. After migration, each sample was divided in 31 serial slices along the lane, and proteins of each slice were identified as described in Materials and Methods. For the two treatments, slices containing gB, gH, or gp180 proteins are shown with color intensity indicating the mean relative abundance (emPAI). For each protein, the predicted molecular mass (MM) is shown (theoretical lane). For the gB protein, the dashed slice indicated the predicted position of gB proteins before potential cleavage by cellular furin. The position of an MM standard is shown on the left.
Host proteins associated with BoHV-4 extracellular virions.
All of the different proteomic analyses of herpesvirus virions detected host proteins as virion constituents. Similarly, we detected host proteins in our samples. A total of 15 of these proteins were systematically detected by all of the different approaches, even after proteinase K treatment of intact virions (Table 3). Among these proteins, actin, cofilin-1, and annexin 2 were particularly abundant. Interestingly, several of these cellular proteins had previously been associated with other herpesvirus virions. This is mainly the case for actin and annexin 2. The presence of annexin 2 within BoHV-4 virions was further confirmed by immunoblotting (see Fig. S2 in the supplemental material). In addition, 23 proteins were systematically detected by our different approaches but were not detected after proteinase K treatment of intact virions (these proteins are shown in Table S1 in the supplemental material).
Table 3.
Cellular proteins detected in BoHV-4 virions and comparison to other herpesvirusesa
| Protein description | Accession no. | Relative emPAI (%)b |
Gammaherpesvirinae |
Alphaherpesvirinae |
Beta- |
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| CTRL | pK | Rhadinoviruses |
Maca- |
Lymph- |
Simpl- |
Varicel- |
Cyto- |
|||
| KSHV | MuHV-4 | AlHV-1 | EBV | HSV-1 | PRV | HCMV | ||||
| Signaling | ||||||||||
| 14-3-3 protein theta | gi¦5803227 | 0.25 | 0.07 | + | + | |||||
| 14-3-3 protein gamma | gi¦9507245 | 0.25 | 0.07 | + | + | |||||
| Cytoskeleton | ||||||||||
| Profilin-1 | gi¦62751593 | 1.11 | 0.14 | + | ||||||
| Ezrin | gi¦27806351 | 0.70 | 0.03 | + | ||||||
| Actin | gi¦4501885 | 4.30 | 0.61 | + | + | + | + | + | + | |
| KRT18 protein | gi¦148878161 | 0.47 | 0.08 | + | ||||||
| Cofilin-1 | gi¦51592135 | 2.10 | 1.11 | + | + | + | ||||
| mRNA binding and processing | ||||||||||
| Elongation factor 1-alpha 1 | gi¦4503471 | 0.45 | 0.13 | + | + | |||||
| Membrane organization and trafficking | ||||||||||
| Annexin 1 | gi¦74 | 0.83 | 0.31 | + | + | + | + | |||
| Annexin 2 | gi¦27807289 | 6.40 | 2.04 | + | + | + | + | + | ||
| Metabolism | ||||||||||
| Triosephosphate isomerase | gi¦61888856 | 0.57 | 0.26 | + | + | |||||
| Bov mitochondrial F1-ATPase (A) | gi¦1827809 | 1.10 | 0.08 | |||||||
| Chain B, Bov cytochrome Bc1 | gi¦3891849 | 0.15 | 0.04 | |||||||
| Ion channel | ||||||||||
| Chloride intracellular channel 1 | gi¦62751970 | 0.57 | 0.16 | |||||||
| Phosphate carrier protein | gi¦27807185 | 0.17 | 0.05 | |||||||
Abbreviations: Beta-, Betaherpesvirinae; Maca-, macaviruses; Lymph-, lymphocryptoviruses; Simpl-, simplexviruses; Varicel-, varicelloviruses; Cyto-, cytomegaloviruses; Bov, bovine.
Relative emPAI values were calculated as the emPAI values calculated for each analysis relative to the abundance of pORF52 taken as 100%. Values > 0.5 are highlighted in boldface. CTRL, values obtained with virion-enriched preparations analyzed by the 1D gel/nano-LC-MS/MS procedure. pK, values obtained with virion-enriched preparations (same as in CTRL) analyzed by the 1D gel/nano-LC-MS/MS procedure after proteinase K treatment. Data indicated for Gammaherpesvirinae, Alphaherpesvirinae, and Betaherpesvirinae are based on previously published studies (2, 3, 5, 8, 17, 20, 23, 30, 42, 45).
Finally, manual categorization of the identified proteins according to their previously known molecular function was performed as recently described for cellular proteins associated with PRV virions (28). As for PRV, many proteins were involved in cellular signaling, cytoskeleton organization, and membrane organization and trafficking (Table 3 and see Table S1 in the supplemental material).
DISCUSSION
MS studies have previously characterized viral and host protein content of several herpesvirus virions. All infectious herpesvirus virions are composed of an icosahedral capsid surrounded by a proteinaceous matrix called tegument, which in turn is wrapped in a lipid envelope. The large size of these particles (∼200 nm in diameter) gives the potential for packaging numerous viral and host proteins. In the present study, we performed a comprehensive analysis of the protein content of BoHV-4 virion, including both viral and host proteins.
Capsid proteins.
Although the names of the proteins differ between herpesvirus families, the structure and arrangement of the capsid proteins is remarkably conserved across Herpesviridae (1, 8). Herpesviruses have a T16 icosahedral capsid composed of 11 pentons containing 5 copies of the major capsid protein (MCP), encoded by ORF25 in rhadinoviruses, and 150 hexons, each comprising 6 copies of MCP. While each penton is one of the 12 vertices of the icosahedron, one is unique, consisting of 12 copies of a portal protein (ORF43 in rhadinoviruses). The pentons and hexons are linked together by 320 triplexes which are composed, in rhadinoviruses, of two copies of pORF26 and one copy of pORF62. A small protein, pORF65, decorates the capsid shell by virtue of its interaction with MCP. The internal space is occupied by the scaffold protein (pORF17.5) and to a lesser extent by the maturational protease (pORF17). During DNA packaging, these two proteins are degraded and replaced by the viral genome. Finally, two minor capsid proteins, pORF19 and pORF32, associate with capsid triplexes and form the capsid vertex-specific complex (CVSC). These proteins are necessary for viral DNA cleavage and packaging.
Interestingly, our MS analysis identified all nine of these proteins (Tables 2 and 4). In contrast, none of the viral terminase components—pORF7, pORF29, and pORF67.5—were detected in our analysis. Although terminase subunits have been detected in other viruses (Table 5) (7, 55, 58) and although we cannot rule out that the terminase is below the detection threshold of our MS approach, these results are in accordance with observations in other viruses such as RRV (41), HSV-1 (31), and PRV (28). This therefore implies that the terminase complex dissociates from mature capsid once monomeric viral genome has been packaged.
Table 4.
Comparison of BoHV-4 proteins identified in virions to other herpesvirusesa

Proteins identified in other virions are highlighted in black. Proteins highlighted in gray were detected in very low abundance.
bBeta-, Betaherpesvirinae; Maca-, macaviruses; Lymph-, lymphocryptoviruses; Simplex-, simplexviruses; Varicel-, varicelloviruses; Cyto-, cytomegaloviruses.
cProteinase K treatment. +, Proteins detected in the proteinase K–1D gel/nano-LC-MS/MS analysis.
Table 5.
Comparison of BoHV-4 proteins not identified in virions to observations in other herpesvirusesa
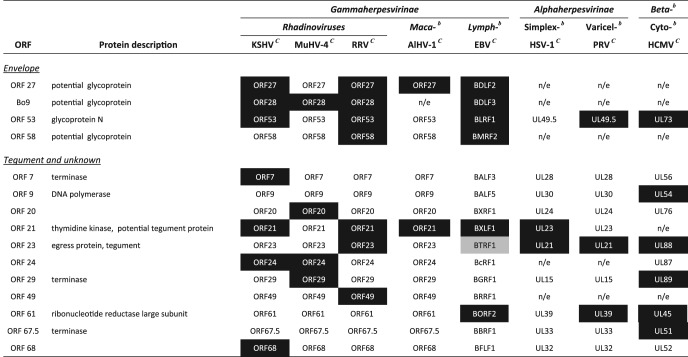
Proteins identified in other virions are highlighted in black. Proteins highlighted in gray were detected in very low abundance.
bBeta-, Betaherpesvirinae; Maca-, macaviruses; Lymph-, lymphocryptoviruses; Simplex-, simplexviruses; Varicel-, varicelloviruses; Cyto-, cytomegaloviruses.
It is commonly acknowledged that ORF17 encodes a N-terminal protease domain. The ORF17 C-terminal oligomerization and capsid protein-binding domains are identical to those of ORF17.5. The pORF17 protease cleaves itself, releasing minor scaffold protein homologous to HSV-1 VP24 and VP21, and the more abundant pORF17.5 protein, releasing the major scaffold protein homologous to HSV-1 VP22a (see Fig. S1 in the supplemental material). In HSV-1 it has been shown that VP21 and VP22a are removed from capsids upon DNA packaging while VP24 (containing the protease domain) is quantitatively retained (48). Surprisingly, we detected both peptides corresponding to VP24 and VP21-22a BoHV-4 homologous proteins.
Interestingly, the VP22a homologous protein has also been detected in KSHV, RRV, AlHV-1, EBV, and HCMV virions (Table 4). This observation may reflect a consequence of the biology of these viruses, such that the scaffold leaves inefficiently during DNA packaging. However, this could seem unlikely given the tight packing of DNA which requires the entire internal content of the capsid (5). B capsids could also be released from dying cells or particles that lack DNA could occasionally bud, as previously observed for KSHV and RRV (39, 40). This would explain the detection of the BoHV-4 pORF17.5 in our samples (see Fig. S1 in the supplemental material).
Finally, since the relative abundances of these different capsids proteins are known (1), we were able to evaluate the quality of our abundance assessment by determining the emPAI. The abundances of the capsid proteins were calculated relative to the abundance of the MCP (Table 2). Interestingly, relative emPAI values were in accordance with predicted values for all capsid proteins excepted for pORF26 and pORF65. However, when calculated on whole virion samples, the relative abundance of pORF26 increased and was ∼2-fold higher than that observed for pORF62, the other triplex component, as expected. In contrast, the relative abundance of pORF65 was even lower in whole virion samples, confirming our observation on the capsid-tegument preparation. ORF65 encodes the small capsid protein of rhadinoviruses which is found on the hexon configuration of the major capsid protein. Although its homologues are nonessential in Alphaherpesvirinae and Betaherpesvirinae, it has recently been shown to be required for KSHV capsid formation (43). On the basis of full occupancy, it should be the second most abundant capsid protein. However, our results suggest that it is much less.
Tegument proteins.
As an indispensable part of mature virions, tegument plays important roles in virion assembly, egress, and entry (18). Based on comparisons with other Gammaherpesvirinae, BoHV-4 tegument is predicted to contain ∼15 proteins. Proteomic analysis of BoHV-4 virions identified 13 of these proteins (the products of ORFs 3, 33, 35, 38, 42, 45, 48, 52, 55, 63, 64, 67, and 75 [Tables 1 and 2]). In contrast, the BoHV-4 thymidine kinase (TK), encoded by ORF21, was not detected in virions (Table 1). This differs from other herpesviruses, where TK is expressed as a tegument protein (3, 13, 25, 31, 41, 58). However, TK was also not detected in the alloherpesvirus CyHV-3 virions (38). Similarly, we did not detect BoHV-4 pORF23 in our samples. The pORF23 homologue in HSV-1 has been shown to be involved in capsid transport to the trans-Golgi network during virion egress (21). However, that study also suggested that the virus could need to release it to enable proper transport during subsequent virus entry events (i.e., toward the nucleus). According to that hypothesis, pORF23 was not identified in KSHV, MuHV-4, and AlHV-1, and only very low levels were observed in EBV virions (25).
Surprisingly, the most abundant BoHV-4 virion protein is encoded by ORF52 (Table 1). Based on their abundance in mature virions, some tegument proteins can be classified as major, structurally significant components, whereas some others are minor (22). Subfamily-specific major tegument proteins have been described previously (18). While ORF52 has no homologue in the Alphaherpesvirinae or Betaherpesvirinae, it is conserved in the Gammaherpesvirinae. In these viruses, ORF52 encodes a small protein of ∼20 kDa. Most of the knowledge about gammaherpesvirus ORF52 has been obtained with MuHV-4 (6, 7, 19, 57). It appears that, MuHV-4 ORF52 is essential for tegumentation and secondary envelopment (6, 57). Interestingly, transcomplementation assays showed that the homologous proteins in KSHV and EBV share similar functions (57). According to structural analysis (4), pORF52 functions as a dimer and the N-terminal α-helix is likely involved in interactions with other components. Immunoprecipitation assays revealed that pORF52 interacts with pORF33, pORF75, gM, and gN in KSHV (47) and with pORF42 in MuHV-4 (57). The results obtained in the present study suggest that it is also a very abundant and important component of BoHV-4 virion. Further studies will be needed to determine whether it shares a similar function.
Finally, all of these potential tegument proteins except two were copurified with capsids (Table 2). The two proteins that did not show any association with capsids are pORF38 and pORF42. pORF42 has recently been shown to associate with pORF52 (57), which associates with capsid. Further studies are therefore required to determine whether it is associated with capsid or not. In contrast, pUL11, the HSV-1 homologue of pORF38, is dependent on some envelope glycoproteins to be packaged in virion (20). Our results suggest therefore that pORF38 could be considered an “outer tegument” viral protein (37).
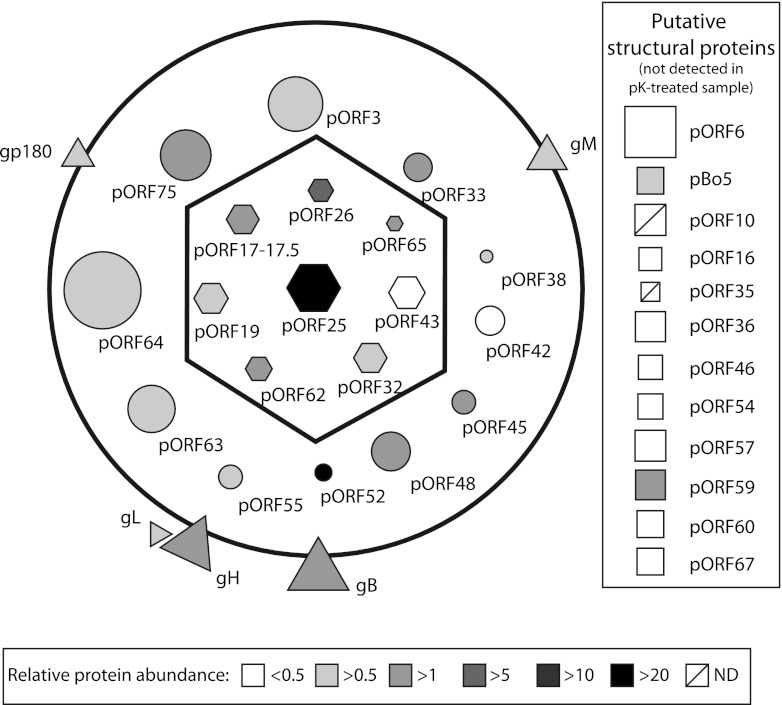
In addition to these proteins, our analyses also detected peptides encoded by ORF6, Bo5, ORF10, ORF16, ORF36, ORF46, ORF54, ORF57, ORF59, and ORF60. Although nearly all of them had previously been associated with some herpesvirus virions (Table 4), most of these proteins were only detected by one or two peptides (Tables 1 and 2). Moreover, none of them was detected after proteinase K treatment of virions (Table 1 and Fig. 5). Therefore, they are potential contaminants of virion preparations. However, specific experiments will need to be undertaken to provide a definitive answer. pORF35 and pORF67 were also not detected after proteinase K treatment. However, they are described as tegument proteins in herpesviruses. Therefore, they are likely rare BoHV-4 virion components (Fig. 5). We interpreted their difficult detection as a marker of their very low abundance. However, specific experiments will be required to confirm their presence in BoHV-4 virions.
Fig 5.
Schematic representation of the protein composition of mature extracellular BoHV-4 virions. Belonging of the proteins to the typical herpesvirus is indicated by shapes. Capsid proteins are represented as hexagons, tegument proteins as circles, envelope proteins as triangles. Proteins detected in some samples but not in the proteinase K-treated sample are represented as putative structural proteins (squares). The predicted protein masses are directly proportional to their surfaces. The mean relative abundance (emPAI) determined by the different analyses of complete virions is indicated in color intensity (see scale). ND, not determined.
Envelope proteins.
Our MS analysis identified five envelope proteins associated with BoHV-4 virions (gB, gH, gL, gp180, and gM). This is rather low in comparison to other herpesviruses. However, in contrast with other rhadinoviruses, BoHV-4 does not encode any ORF4 or ORF74 homologues, and only four other potential envelope glycoproteins were predicted in the BoHV-4 genome: ORF27, ORF53, ORF58, and Bo9. The proteins encoded by ORF27 and ORF58 are involved in intercellular viral spread in MuHV-4 (35, 36). One hypothesis could therefore be that different virions are produced following the major way of dissemination. Bo9 is the positional homologue of KSHV, MuHV-4, and RRV ORF28. However, Bo9 is much smaller than these genes (42), which might be an indication of the loss of selective pressure. Interestingly, the gene is absent from AlHV-1 genome. The absence of detection of pBo9 is therefore not surprising. Finally, we did not detect any peptide corresponding to gN, which is encoded by ORF53 (Table 5). The detection of gM but the absence of gN in mature BoHV-4 virions is relatively surprising since both proteins form a complex in herpesviruses (26) and since gN is needed for the proper processing of gM. Although we cannot rule out that gN was present but undetected by our MS approach, gN could also dissociate from gM in mature virions, as suggested for HCMV (55). The absence of gN detection in both HSV-1 (31) and AlHV-1 (13) virions is in agreement with this hypothesis.
Unexpectedly, gB, gH, gM, and gp180 remained detected after proteinase K treatment, although at least gp180 and gH were digested (Fig. 2). However, similar observations had already been made before for other herpesviruses (28, 41). In contrast, gL was not detected after proteinase K treatment. However, gL is a well-known component of herpesvirus virions (Table 4), and we recently showed that the absence of BoHV-4 gL is associated with an entry deficit (29). We therefore considered it a BoHV-4 structural protein. The disappearance of gL after protease treatment could be linked to its accessibility at the viral surface and to the fact that it has no transmembrane domain. Indeed, gL is incorporated in herpesvirus virions as a heterodimer with gH. However, this interaction is loose as we have shown with MuHV-4 that gL dissociates during entry, allowing gH to change its conformation and mediate hemifusion (15). Similarly, PRV gL is not detected after protease treatment (28).
In addition to identifying viral components, we showed that three BoHV-4 envelope glycoproteins are massively glycosylated (Fig. 4). This is in accordance with our recent results showing that gp180 bears numerous O-glycans (33), whereas BoHV-4 gH is N-glycosylated (29). Moreover, previous results suggested that BoHV-4 gB is O- and N-glycosylated (30). Interestingly, no other viral product appeared to be glycosylated. As shown for other viruses, massive glycosylation of envelope protein could be involved in the protection of BoHV-4 virion from neutralizing antibody. Thus, gp180 O-glycans provides part of a glycan shield for otherwise vulnerable viral epitopes, some of which are gL dependent (33). Similarly, the O-glycans of the N terminus (NT) of MuHV-4 gB provide a protective cover for a vulnerable part of gH/gL (17). Our results show that BoHV-4 gB-NT is also massively glycosylated (Fig. 4). Moreover, sequence analysis reveals that BoHV-4 gB-NT contains 40 potential O-glycosylation sites, whereas only 9 are already sufficient to protect MuHV-4 from neutralization (17). This is particularly interesting as BoHV-4 Bo17 gene encodes a mucin-type β-1,6-N-acetylglucosaminyltransferase (54) that is central to the O-glycosylation process. Since BoHV-4 seems particularly resistant to antibody neutralization (14), we hypothesize that it could use gB-NT, gp180, and its mucin-type β-1,6-N-acetylglucosaminyltransferase to form a glycan shield at the virion surface that could be particularly important in the context of virus transmission between hosts.
For each analysis of complete virions, the emPAI was calculated to estimate protein relative abundance (Table 1). The mean relative emPAI was then calculated as described above, and a schematic representation of BoHV-4 virion was made (Fig. 5). Viral structural proteins were represented according to their predicted protein mass, location, and relative abundance. Viral proteins detected in some samples but not detected after proteinase K treatment were represented as putative structural proteins (except gL, which has been represented in virions).
Host proteins.
The present study revealed a heteroclite collection of cellular proteins associated with BoHV-4 virions (Table 3 and see Table S1 in the supplemental material). The presence of these proteins in virion preparation may be attributed to different reasons, including copurification of cellular components in virion preparation. However, proteinase K treatment of virions showed that at least 15 of these proteins seem to be incorporated in BoHV-4 virions (Table 3). Even if most of the detected proteins have low relative emPAI values, some are much more abundant. This is particularly the case for annexin 2 (Table 3), which is nearly as abundant in BoHV-4 virions as some capsid proteins (Table 3). Annexin 2 has been associated with numerous virus species. In HCMV, it has been shown to be associated with gB (45), although its role during entry is still controversial (12, 44). In the future, it will be important to validate the presence of these proteins and ultimately address their putative function for the virion.
In summary, we described here the first comprehensive analysis of BoHV-4 virion composition. We identified 37 viral proteins associated with virion-enriched preparations. Among these proteins, 24 were resistant to proteinase K treatment of BoHV-4 intact virions, and 16 were found in at least two other rhadinoviruses. Moreover, we identified at least 15 cellular proteins as structural components of BoHV-4 virions. In the future, these results could enhance our understanding of gammaherpesvirus life cycle, including the processes of viral assembly, egress, entry, and immune evasion.
Supplementary Material
ACKNOWLEDGMENTS
C.L., B.M., and L.G. are a research fellow, postdoctoral researcher, and research associate for the Fonds de la Recherche Scientifique-Fonds National Belge de la Recherche Scientifique (FRS-FNRS), respectively. L.P. is supported by a postdoctoral fellowship from the University of Liège. This study was supported by the following grants: ARC GLYVIR, a starting grant of the University of Liège (D-09/11), a scientific impulse grant of the FRS-FNRS (F.4510.10), and an FRFC grant from the FRS-FNRS (2.4622.10).
Footnotes
Published ahead of print 15 August 2012
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1. Baines JD. 2011. Herpes simplex virus capsid assembly and DNA packaging: a present and future antiviral drug target. Trends Microbiol. 19:606–613 [DOI] [PubMed] [Google Scholar]
- 2. Baldick CJ, Jr, Shenk T. 1996. Proteins associated with purified human cytomegalovirus particles. J. Virol. 70:6097–6105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bechtel JT, Winant RC, Ganem D. 2005. Host and viral proteins in the virion of Kaposi's sarcoma-associated herpesvirus. J. Virol. 79:4952–4964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Benach J, et al. 2007. Structural and functional studies of the abundant tegument protein ORF52 from murine gammaherpesvirus 68. J. Biol. Chem. 282:31534–31541 [DOI] [PubMed] [Google Scholar]
- 5. Booy FP, et al. 1991. Liquid-crystalline, phage-like packing of encapsidated DNA in herpes simplex virus. Cell 64:1007–1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bortz E, et al. 2007. Murine gammaherpesvirus 68 ORF52 encodes a tegument protein required for virion morphogenesis in the cytoplasm. J. Virol. 81:10137–10150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bortz E, et al. 2003. Identification of proteins associated with murine gammaherpesvirus 68 virions. J. Virol. 77:13425–13432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Brown JC, Newcomb WW. 2011. Herpesvirus capsid assembly: insights from structural analysis. Curr. Opin. Virol. 1:142–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Charif D, Lobry J. 2007. SeqinR 1.0-2: a contributed package to the R project for statistical computing devoted to biological sequences retrieval and analysis. Springer Verlag, New York, NY [Google Scholar]
- 10. Davison AJ, Davison MD. 1995. Identification of structural proteins of channel catfish virus by mass spectrometry. Virology 206:1035–1043 [DOI] [PubMed] [Google Scholar]
- 11. Davison AJ, et al. 2009. The order Herpesvirales. Arch. Virol. 154:171–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Derry MC, Sutherland MR, Restall CM, Waisman DM, Pryzdial EL. 2007. Annexin 2-mediated enhancement of cytomegalovirus infection opposes inhibition by annexin 1 or annexin 5. J. Gen. Virol. 88:19–27 [DOI] [PubMed] [Google Scholar]
- 13. Dry I, et al. 2008. Proteomic analysis of pathogenic and attenuated alcelaphine herpesvirus 1. J. Virol. 82:5390–5397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Dubuisson J, et al. 1990. Neutralization of bovine herpesvirus type 4 by pairs of monoclonal antibodies raised against two glycoproteins and identification of antigenic determinants involved in neutralization. J. Gen. Virol. 71(Pt 3):647–653 [DOI] [PubMed] [Google Scholar]
- 15. Gillet L, Colaco S, Stevenson PG. 2008. The murid herpesvirus-4 gL regulates an entry-associated conformation change in gH. PLoS One 3:e2811 doi:10.1371/journal.pone.0002811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gillet L, et al. 2005. Development of bovine herpesvirus 4 as an expression vector using bacterial artificial chromosome cloning. J. Gen. Virol. 86:907–917 [DOI] [PubMed] [Google Scholar]
- 17. Gillet L, Stevenson PG. 2007. Antibody evasion by the N terminus of murid herpesvirus-4 glycoprotein B. EMBO J. 26:5131–5142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Guo H, Shen S, Wang L, Deng H. 2010. Role of tegument proteins in herpesvirus assembly and egress. Protein Cell 1:987–998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Guo H, Wang L, Peng L, Zhou ZH, Deng H. 2009. Open reading frame 33 of a gammaherpesvirus encodes a tegument protein essential for virion morphogenesis and egress. J. Virol. 83:10582–10595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Han J, Chadha P, Meckes DG, Jr, Baird NL, Wills JW. 2011. Interaction and interdependent packaging of tegument protein UL11 and glycoprotein E of herpes simplex virus. J. Virol. 85:9437–9446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Harper AL, et al. 2010. Interaction domains of the UL16 and UL21 tegument proteins of herpes simplex virus. J. Virol. 84:2963–2971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Heine JW, Honess RW, Cassai E, Roizman B. 1974. Proteins specified by herpes simplex virus. XII. The virion polypeptides of type 1 strains. J. Virol. 14:640–651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Heldwein EE, Krummenacher C. 2008. Entry of herpesviruses into mammalian cells. Cell. Mol. Life Sci. 65:1653–1668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ishihama Y, et al. 2005. Exponentially modified protein abundance index (emPAI) for estimation of absolute protein amount in proteomics by the number of sequenced peptides per protein. Mol. Cell. Proteomics 4:1265–1272 [DOI] [PubMed] [Google Scholar]
- 25. Johannsen E, et al. 2004. Proteins of purified Epstein-Barr virus. Proc. Natl. Acad. Sci. U. S. A. 101:16286–16291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Jons A, Dijkstra JM, Mettenleiter TC. 1998. Glycoproteins M and N of pseudorabies virus form a disulfide-linked complex. J. Virol. 72:550–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kattenhorn LM, et al. 2004. Identification of proteins associated with murine cytomegalovirus virions. J. Virol. 78:11187–11197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kramer T, Greco TM, Enquist LW, Cristea IM. 2011. Proteomic characterization of pseudorabies virus extracellular virions. J. Virol. 85:6427–6441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lete C, Machiels B, Stevenson PG, Vanderplasschen A, Gillet L. 2012. Bovine herpesvirus type 4 glycoprotein L is nonessential for infectivity but triggers virion endocytosis during entry. J. Virol. 86:2653–2664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lomonte P, et al. 1997. Glycoprotein B of bovine herpesvirus 4 is a major component of the virion, unlike that of two other gammaherpesviruses, Epstein-Barr virus and murine gammaherpesvirus 68. J. Virol. 71:3332–3335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Loret S, Guay G, Lippe R. 2008. Comprehensive characterization of extracellular herpes simplex virus type 1 virions. J. Virol. 82:8605–8618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Machiels B, et al. 2011. The bovine herpesvirus 4 Bo10 gene encodes a nonessential viral envelope protein that regulates viral tropism through both positive and negative effects. J. Virol. 85:1011–1024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Machiels B, et al. 2011. Antibody evasion by a gammaherpesvirus O-glycan shield. PLoS Pathog. 7:e1002387 doi:10.1371/journal.ppat.1002387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mastroleo F, et al. 2009. Shotgun proteome analysis of Rhodospirillum rubrum S1H: integrating data from gel-free and gel-based peptides fractionation methods. J. Proteome Res. 8:2530–2541 [DOI] [PubMed] [Google Scholar]
- 35. May JS, de Lima BD, Colaco S, Stevenson PG. 2005. Intercellular gammaherpesvirus dissemination involves coordinated intracellular membrane protein transport. Traffic 6:780–793 [DOI] [PubMed] [Google Scholar]
- 36. May JS, Walker J, Colaco S, Stevenson PG. 2005. The murine gammaherpesvirus 68 ORF27 gene product contributes to intercellular viral spread. J. Virol. 79:5059–5068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Mettenleiter TC, Klupp BG, Granzow H. 2009. Herpesvirus assembly: an update. Virus Res. 143:222–234 [DOI] [PubMed] [Google Scholar]
- 38. Michel B, et al. 2010. The genome of cyprinid herpesvirus 3 encodes 40 proteins incorporated in mature virions. J. Gen. Virol. 91:452–462 [DOI] [PubMed] [Google Scholar]
- 39. Nealon K, et al. 2001. Lytic replication of Kaposi's sarcoma-associated herpesvirus results in the formation of multiple capsid species: isolation and molecular characterization of A, B, and C capsids from a gammaherpesvirus. J. Virol. 75:2866–2878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. O'Connor CM, Damania B, Kedes DH. 2003. De novo infection with rhesus monkey rhadinovirus leads to the accumulation of multiple intranuclear capsid species during lytic replication but favors the release of genome-containing virions. J. Virol. 77:13439–13447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. O'Connor CM, Kedes DH. 2006. Mass spectrometric analyses of purified rhesus monkey rhadinovirus reveal 33 virion-associated proteins. J. Virol. 80:1574–1583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Palmeira L, Machiels B, Lete C, Vanderplasschen A, Gillet L. 2011. Sequencing of bovine herpesvirus 4 v.test strain reveals important genome features. Virol. J. 8:406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Perkins EM, et al. 2008. Small capsid protein pORF65 is essential for assembly of Kaposi's sarcoma-associated herpesvirus capsids. J. Virol. 82:7201–7211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Pietropaolo R, Compton T. 1999. Interference with annexin II has no effect on entry of human cytomegalovirus into fibroblast cells. J. Gen. Virol. 80(Pt 7):1807–1816 [DOI] [PubMed] [Google Scholar]
- 45. Pietropaolo RL, Compton T. 1997. Direct interaction between human cytomegalovirus glycoprotein B and cellular annexin II. J. Virol. 71:9803–9807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Rappsilber J, Ryder U, Lamond AI, Mann M. 2002. Large-scale proteomic analysis of the human spliceosome. Genome Res. 12:1231–1245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Rozen R, Sathish N, Li Y, Yuan Y. 2008. Virion-wide protein interactions of Kaposi's sarcoma-associated herpesvirus. J. Virol. 82:4742–4750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Sheaffer AK, et al. 2000. Evidence for controlled incorporation of herpes simplex virus type 1 UL26 protease into capsids. J. Virol. 74:6838–6848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Team RDC. 2010. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria [Google Scholar]
- 50. Thiry E, et al. 1992. Molecular biology of bovine herpesvirus type 4. Vet. Microbiol. 33:79–92 [DOI] [PubMed] [Google Scholar]
- 51. Thiry E, Pastoret PP, Dessy-Doizé C, Hanzen C, Calberg-Bacq CM. 1981. Herpesvirus in infertile bull's testicle. Vet. Rec. 108:426. [DOI] [PubMed] [Google Scholar]
- 52. Thorley-Lawson DA, Gross A. 2004. Persistence of the Epstein-Barr virus and the origins of associated lymphomas. N. Engl. J. Med. 350:1328–1337 [DOI] [PubMed] [Google Scholar]
- 53. van Beurden SJ, et al. 2011. Identification and localization of the structural proteins of anguillid herpesvirus 1. Vet. Res. 42:105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Vanderplasschen A, et al. 2000. A multipotential β-1,6-N-acetylglucosaminyl-transferase is encoded by bovine herpesvirus type 4. Proc. Natl. Acad. Sci. U. S. A. 97:5756–5761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Varnum SM, et al. 2004. Identification of proteins in human cytomegalovirus (HCMV) particles: the HCMV proteome. J. Virol. 78:10960–10966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Verma SC, Robertson ES. 2003. Molecular biology and pathogenesis of Kaposi sarcoma-associated herpesvirus. FEMS Microbiol. Lett. 222:155–163 [DOI] [PubMed] [Google Scholar]
- 57. Wang L, et al. 2012. Distinct domains in ORF52 tegument protein mediate essential functions in murine gammaherpesvirus 68 virion tegumentation and secondary envelopment. J. Virol. 86:1348–1357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Zhu FX, Chong JM, Wu L, Yuan Y. 2005. Virion proteins of Kaposi's sarcoma-associated herpesvirus. J. Virol. 79:800–811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Zimmermann W, et al. 2001. Genome sequence of bovine herpesvirus 4, a bovine rhadinovirus, and identification of an origin of DNA replication. J. Virol. 75:1186–1194 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.