Abstract
Koi herpesvirus (KHV) (species Cyprinid herpesvirus 3) ORF134 was shown to transcribe a spliced transcript encoding a 179-amino-acid (aa) interleukin-10 (IL-10) homolog (khvIL-10) in koi fin (KF-1) cells. Pairwise sequence alignment indicated that the expressed product shares 25% identity with carp IL-10, 22 to 24% identity with mammalian (including primate) IL-10s, and 19.1% identity with European eel herpesvirus IL-10 (ahvIL-10). In phylogenetic analyses, khvIL-10 fell in a divergent position from all host IL-10 sequences, indicating extensive structural divergence following capture from the host. In KHV-infected fish, khvIL-10 transcripts were observed to be highly expressed during the acute and reactivation phases but to be expressed at very low levels during low-temperature-induced persistence. Similarly, KHV early (helicase [Hel] and DNA polymerase [DNAP]) and late (intercapsomeric triplex protein [ITP] and major capsid protein [MCP]) genes were also expressed at high levels during the acute and reactivation phases, but only low-level expression of the ITP gene was detected during the persistent phase. Injection of khvIL-10 mRNA into zebrafish (Danio rerio) embryos increased the number of lysozyme-positive cells to a similar degree as zebrafish IL-10. Downregulation of the IL-10 receptor long chain (IL-10R1) using a specific morpholino abrogated the response to both khvIL-10 and zebrafish IL-10 transcripts, indicating that, despite the structural divergence, khvIL-10 functions via this receptor. This is the first report describing the characteristics of a functional viral IL-10 gene in the Alloherpesviridae.
INTRODUCTION
Koi herpesvirus (KHV), classified taxonomically as the type species Cyprinid herpesvirus 3 of the genus Cyprinivirus, family Alloherpesviridae, has a 295-kb genome, the largest of the Herpesvirales (3, 9). KHV is an emerging pathogen of koi and common carp (Cyprinus carpio L.), threatening aquaculture production and the worldwide trade in ornamental fish (16, 18, 54). It has been suggested that international trade in subclinically infected fish is responsible for the rapid spread of koi herpesvirus disease (KHVD) (12, 46, 52), which has been declared as notifiable by the World Organization for Animal Health (40).
The mechanism by which KHV persists subclinically in fish has not yet been elucidated (11, 22). However, the ability to establish a lifelong latent infection is the hallmark of herpesviruses, which employ many different strategies to evade host immunity, including the expression of major histocompatibility complex (MHC) and cytokine genes that have been captured from the host (4, 14, 19, 29, 38). Cellular interleukin-10 (IL-10) is a pleiotropic immunomodulatory cytokine that occurs in a wide range of vertebrate species, including fish (49). IL-10 suppresses expression of a number of other cytokines and chemokines, including tumor necrosis factor alpha (TNF-α), gamma interferon (IFN-γ), IL-1β, IL-2, IL-3, IL-6, and MHC class II (36, 37, 43). Many viruses utilize the immunosuppressive effects of IL-10 to evade immune recognition, either by upregulation of host IL-10 or by expression of a virus-encoded IL-10 homolog (44). Among mammalian herpesviruses, IL-10 homologs have been reported in members of the Betaherpesvirinae (e.g., human cytomegalovirus [HCMV] and rhesus cytomegalovirus) and Gammaherpesvirinae (e.g., Epstein-Barr virus [EBV] and equine herpesvirus 2) but not in the Alphaherpesvirinae (49). Although they are broadly immunomodulatory, herpesvirus IL-10s vary in their structural homology to the host IL-10s, functional activities, and expression patterns. For example, while the EBV-encoded IL-10 (ebvIL-10) shares high sequence identity with human cellular IL-10 and retains many of its immunosuppressive and immunostimulatory properties, it has much lower binding affinity for the cellular IL-10 receptor (IL-10R) and is unable to stimulate thymocyte or mast cell proliferation or to upregulate MHC class II surface expression on B cells (33, 49). In contrast, HCMV-encoded IL-10 (cmvIL-10) shares quite low sequence identity with human IL-10 but also functions via the IL-10R receptor, to which it binds strongly, and, like human IL-10, triggers the Jak1/STAT3 pathway (27, 29, 50). Also, unlike other viral IL-10s, HCMV IL-10 transcripts occur as two major splice variants, cmvIL-10 and LAcmvIL-10, the latter of which is expressed during latent and productive phases of infection and does not appear to share the full range of immunomodulatory functions of cmvIL-10 (25, 26, 49).
Among the Alloherpesviridae, genes with homology to IL-10 have been identified in both KHV and the eel herpesvirus Anguillid herpesvirus 1 (AngHV1), although none has yet been reported in the amphibian alloherpesviruses (3, 56, 57). In this study, we analyzed expression of the KHV IL-10 ortholog (khvIL-10) in koi fin (KF-1) cells and showed that it is highly expressed in infected carp tissue during the acute and reactivation phases of infection and at significantly lower levels during virus persistence at low temperature. We also demonstrated that khvIL-10 is functionally similar to piscine IL-10 in zebrafish embryos and acts through the cognate IL-10 receptor.
MATERIALS AND METHODS
Origins and cultivation of viruses.
The Indonesian KHV isolate (C07) used in this study was isolated from common carp during a disease outbreak in West Java, Indonesia, in 2007 (53). U.S. isolate F9850 was provided by Ronald Hedrick (University of California, Davis, CA), and United Kingdom isolate G406 was supplied by Keith Way (Centre for Environment, Fisheries and Aquaculture Science, United Kingdom). KHV was cultured at 25°C in the koi fin cell line (KF-1) maintained in Leibovitz L-15 medium supplemented with 10% fetal bovine serum, 2 mM l-glutamine, 100 IU/ml penicillin, and 100 μg/ml streptomycin (18).
Experimental infection of carp.
An experimental temperature shift model for acute, latent, and reactivation phases of KHV infection in common carp has been described elsewhere (52). Juvenile common carp (mean length ± standard deviation, 12.1 ± 1.0 cm) were supplied by Fisheries Victoria. Upon arrival at the laboratory, the carp were acclimatized for 8 days, during which they were treated for 2 h in a bath containing 11 ppm formalin and 3 ppm Virkon Aquatic (DuPont) to remove adventitious ectoparasites, bacteria, or viruses, with the formalin bath repeated over five successive days using the procedure described previously (30). The fish were held in 100-liter tanks on a 12-h/12-h daylight cycle and were fed with commercial feed at a rate of 1% body weight per day throughout the experiment. Carp were infected with KHV by immersion at a dose of 100 50% tissue culture infective doses (TCID50)/ml for 2 h at 22°C. After exposure, fish were briefly rinsed twice in freshwater and then separated into two groups: tank 1, in which fish were held at a water temperature of 22°C until there was evidence of clinical signs of KHVD with mortalities reaching 90% by 6 to 9 days postinfection (acute phase), and tank 2, in which fish were held initially at 22°C for 24 h and then the water temperature was decreased to 11°C over a period of 4 days, following which there was no evidence of disease (persistent phase). At 28 days postinfection (dpi), the water temperature in tank 2 was increased from 11 to 22°C in increments of 2 to 3°C per day, resulting in 45% mortality by day 10 (reactivation phase). A tank of untreated carp from the same batch served as a control. Gill, kidney, and spleen tissue samples were collected from four fish in the control group and from four fish at 7 days (acute), 28 days (persistent), and 37 days (reactivation) postinfection. All experiments with carp were approved by the CSIRO Australian Animal Health Laboratory Animal Ethics Committee.
Nucleic acid extraction and PCR amplification.
Total DNA and RNA were extracted from carp tissues and infected cell cultures using the AllPrep DNA/RNA extraction kit (Qiagen). Contaminating DNA in total RNA extracts was removed by on-column digestion with RNase-free DNase I (Qiagen). ORF134 was amplified from KHV DNA using either forward primer F (5′TCTCGACGGATTGGAAGACG) or Fb (5′CAGAAAGTCTCCACAGTTAAC) and reverse primer R (5′CTAACCGCGACCATCTTCTTCG). PCR was conducted using the HotStarTaq master mix (Qiagen) according to the manufacturer's protocols and using the following cycling conditions: one cycle at 95°C for 15 min; 30 cycles at 95°C for 30 s, 52°C for 30 s, and 72°C for 30 s; and one cycle at 72°C for 10 min. Viral transcripts were detected using the same primers and protocols, but with an initial reverse transcription step at 50°C for 30 min, using a OneStep reverse transcription-PCR (RT-PCR) kit (Qiagen). Amplified products were analyzed by electrophoresis using 2% agarose gels in 1× Tris-acetate-EDTA (TAE) buffer and visualized by staining with SYBR green (Invitrogen).
Sequencing and phylogenetic analysis.
Purified amplicons were sequenced using BigDye terminator sequencing kits (Applied Biosystems). Phylogenetic analyses were conducted using MEGA 5 software from amino acid sequences obtained by using the BLASTp algorithm (2). Multiple-sequence alignments were performed in MUSCLE using a neighbor-joining clustering method for each iteration. Phylogenetic trees were constructed using the maximum-likelihood (ML) statistical method and the close-neighbor interchange (CNI) heuristic searching strategy, with the initial ML tree constructed automatically. The Jones-Taylor-Thornton substitution model with gamma-distributed rates among sites was selected as the best substitution model (lowest Bayesian information criterion scores). The reliability of inferred trees was tested by the bootstrap method using 1,000 resamplings.
Quantification of gene expression.
Gene expression was quantified using a TaqMan quantitative RT-PCR (qRT-PCR) assay with the forward primers, reverse primers, and probes shown in Table S1 in the supplemental material. Expression levels were normalized against expression of carp 18S rRNA and quantified based on comparative threshold cycle (ΔΔCT) using the 7500 Fast SDS software version 2.0.3 and relative expression software tools in REST 2009 V2.0.13 (41).
Zebrafish maintenance and manipulation.
Adult zebrafish (Danio rerio) were maintained at 28°C on a 13-h/11-h light/dark cycle and fed twice daily. Embryos from wild-type fish were manually spawned and maintained on a heat block in a petri dish containing egg water and 0.00005% (wt/vol) methylene blue until 16 h postfertilization (hpf), when it was replaced with egg water containing 0.003% (wt/vol) 1-phenyl-2-thiourea to inhibit pigmentation. Embryos were anesthetized at 56 hpf with 0.4 mg/ml benozocaine before fixation with 4% (wt/vol) paraformaldehyde (PFA) in phosphate-buffered saline. All experiments were performed under appropriate Deakin University Animal Welfare Committee guidelines.
Morpholino and mRNA injection.
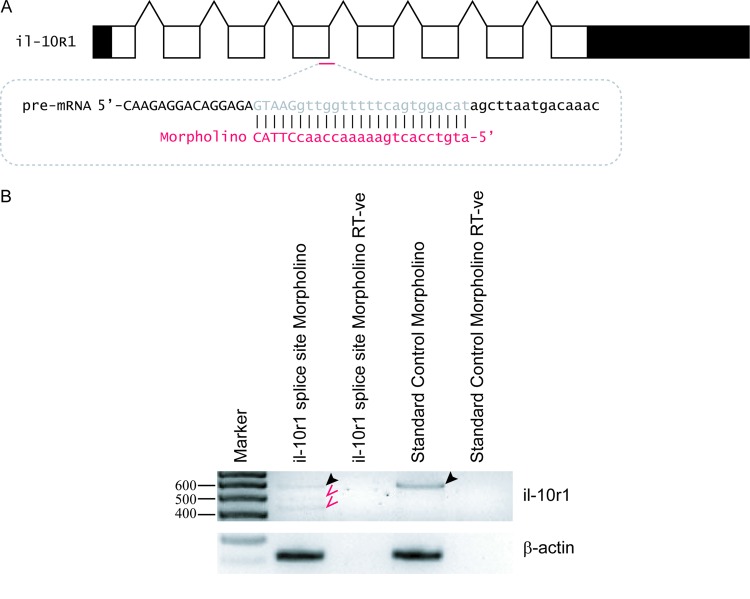
Two morpholinos (Gene Tools) were obtained: a standard control morpholino (StdCon, 5′CCTCTTACCTCAGTTACAATTTATA), and a previously published morpholino complementary to the exon 4/intron 4 splice junction of the zebrafish il-10r1 gene (il-10r1SSMo, 5′ATGTCCACTGAAAAACCAACCTTAC) (31) (Fig. 1A). These were used at a concentration of 1 mM in Danieau solution [58 mM NaCl, 0.7 mM. KCl, 0.4 mM MgSO4, 0.6 mM Ca(NO3)2, 5.0 mM HEPES pH 7.6]. The khvIL-10 open reading frame (ORF) was cloned from RNA extracted from infected gills using primers 5′ATGTTCCTTGCAGTGCTACTAAC and 5′TCAATGTTTGCGCTTGGTTTTC. Danio rerio IL-10 (drIL-10) was amplified from 50-hpf embryos by RT-PCR using primers 5′CTCGAGAATTAACCCTCACTAAAGGGAAATATGATTTTCTCTGGAGTCATCC and 5′CTCGAGGTTTTCAGAATGTTCAGACAGAGTG, with the product subsequently ligated into p-GEM-T Easy. mRNAs encoding khvIL-10 and drIL-10 were generated from linearized plasmids using an mMESSAGEmMACHINE kit (Ambion) and diluted to 100 ng/μl in Danieau solution. Aliquots of morpholino or morpholino and mRNA with 1% (wt/vol) phenol red were injected into embryos between the 1-cell and 4-cell developmental stages. Confirmation that the il-10r1SSMo had suppressed the normal splicing of the endogenous il-10r1 gene was achieved by RT-PCR using the primers 5′GCACTAGACACTGGTCCTCATGC and 5′AACAGCGGGCATTTTACCAG.
Fig 1.
Targeted knockdown of the zebrafish il-10r1 gene using a splice site-blocking morpholino. (A) Sequence targeted by the il-10r1 morpholino. The splice site structure of the zebrafish il-10r1 gene, showing exons (rectangles) and intervening introns (chevrons) with the noncoding regions shaded black, is shown. The il-10r1 splice site-blocking morpholino targets the exon 4/intron 4 splice-site (red line), with the target sequence on the pre-mRNA (gray) and morpholino sequence (red) shown. The exon sequence is in uppercase and the intron sequence in lowercase. (B) Effectiveness of the il-10r1 morpholino in vivo, showing RT-PCR of RNA extracted from 56-hpf zebrafish embryos injected with il-10r1 splice site-blocking or control morpholinos, as indicated, including samples not treated with reverse transcriptase (RT-ve). The black arrowhead indicates the wild-type transcript in the standard control embryos that is robustly ablated in il-10r1 morpholino-injected embryos, with alternate splicing products indicated with red arrowheads.
Whole-mount visualization.
Whole-mount in situ hybridization (WISH) was performed using digoxigenin-labeled antisense RNA probes transcribed from appropriately linearized plasmids as described previously (32). Statistical analyses of cell counts after WISH were performed using GraphPad Prism (version 4) software. The unpaired independent t test was employed to determine the statistical significance of various treatments, typically with sample populations of approximately 20 to 30 embryos obtained from independent manipulation.
RESULTS AND DISCUSSION
Characterization of khvIL-10 transcripts.
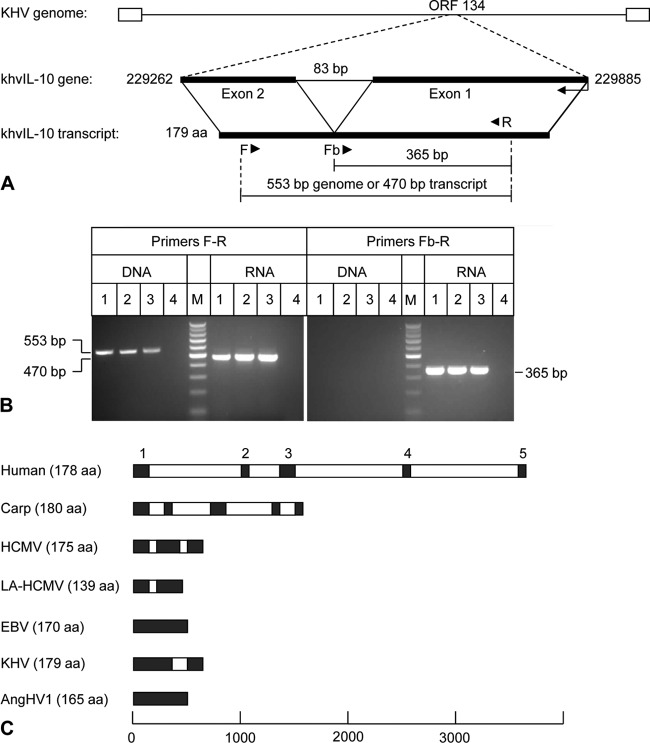
ORF134 (624 bp) is located in an antisense orientation at nucleotides 229262 to 229885 in the U.S. strain of the KHV genome (GenBank accession number DQ657948, RefSeq NC_009127). It has been predicted to contain an 83-bp intron flanked by 2 exons encoding a 179-amino-acid (aa) product with sequence homology to cellular IL-10 (Fig. 2A) (3). PCR primers F (located within putative exon 2) and R (located within putative exon 1) amplified the predicted 553-bp product from DNA extracted from KHV-infected KF-1 cells harvested at 5 dpi and the predicted 470-bp product from reverse-transcribed total RNA (Fig. 2B, left panel). PCR primer Fb (spanning the putative splice site) and primer R amplified the predicted 365-bp product from total RNA, but no product was amplified from genomic DNA (Fig. 2B, right panel). Sequence analysis of these PCR products confirmed the presence of a single 83-bp intron with typical splice motifs at the 5′ (GT) and 3′ (AG) termini (data not shown). Comparison of splicing patterns indicated that although herpesvirus IL-10 genes are more compact than those in their mammalian and fish hosts (Fig. 2C), the encoded polypeptides are similar in size (e.g., human, 178 aa; carp, 180 aa; HCMV, 175 aa; EBV, 170 aa; KHV, 179 aa; AngHV1, 165 aa) (3, 29, 47, 56, 58). It was concluded that spliced KHV transcripts encoding a viral IL-10 homolog are expressed during productive infection of KF-1 cells.
Fig 2.
PCR and RT-PCR analysis of the khvIL-10 gene and comparison of splicing patterns of host and viral IL-10 genes. (A) Predicted structure of KHV ORF134 and locations of primer binding sites. (B) Agarose gel electrophoresis of products amplified from DNA and total RNA extracted from KHV-infected cells using PCR primer sets F-R and Fb-R. Lanes 1, KHV U.S. strain F9850; lanes 2, United Kingdom strain G406; lanes 3, Indonesian strain C07; lanes 4, mock infected; lanes M, 100-bp DNA ladder. (C) Structures of host IL-10 and viral IL-10 genes. Introns (white boxes) and exons (black boxes) are indicated, and exon numbers for human IL-10 are provided.
Sequence analysis of khvIL-10.
Based on the conserved domain structure, khvIL-10 is a member of the IL-10 superfamily (35). BLASTp analysis indicated that it shares low but significant amino acid sequence identity with other viral, mammalian, piscine, amphibian, and avian IL-10s. In pairwise sequence alignments, khvIL-10 is most closely related to IL-10 of its natural host, the common carp, and those of other piscine species; it shares 25 to 26% identity with carp IL-10 and those of rainbow trout (Oncorhynchus mykiss) and zebrafish, 22 to 24% identity with mammalian (including primate) IL-10 sequences, and 19.1% identity with Anguillid herpesvirus 1 IL-10 (ahvIL-10). In contrast, ahvIL-10 shares 26 to 35% identity with available piscine IL-10 sequences, all of which are very distant phylogenetically from its natural host, the European eel. The level of identity between khvIL-10 and carp IL-10 is comparable to that between cmvIL-10 and human IL-10 sequences (27% identity) and is in keeping with the equally low levels among other fish cytokines (48) but far lower than that between ebvIL-10 and human IL-10 (80% identity) (29, 38).
The neural network and hidden Markov model (5, 39; H. Nielsen and A. Krogh, presented at the Sixth International Conference on Intelligent Systems for Molecular Biology, Menlo Park, CA, 1998) predicts that the khvIL-10 signal peptide cleavage site is located between G17 and A18. Four cysteine residues that are universally conserved in the IL-10 superfamily are preserved in khvIL-10. There are two potential N-glycosylation sites (N95 and N113), only one of which occurs in ebvIL-10 and cmvIL-10 (23, 61, 62). Although four conserved cysteine residues are present, ahvIL-10 contains no predicted N-glycosylation sites (57). Like cellular IL-10s and other vIL-10s, khvIL-10 and ahvIL-10 are each predicted to contain six α-helices (hA to hF) and appear to adopt a similar folded structure (57). Interestingly, despite the modest amino acid sequence identity and differences in interdomain angles, host IL-10s and most herpesvirus-encoded IL-10s have been reported to bind to and induce transduction signals through the same host IL-10 receptor complex (27, 29, 33, 49). Binding of viral IL-10s to the cellular IL-10 receptor has been attributed to helix hA, the AB loop, and helix hF structures (28, 42). The similarities in the three-dimensional (3D) structures (57) of khvIL-10 and carp IL-10 suggest they may bind to and function through the same receptor. Identification of the carp IL-10 receptor has been hindered by the lack of genome sequence data (23, 47), but the IL-10 receptor (IL-10R1) has been recently identified and characterized for the other cyprinid species, zebrafish (Danio rerio), and goldfish (Carassius auratus) (13).
Phylogenetic analysis of khvIL-10.
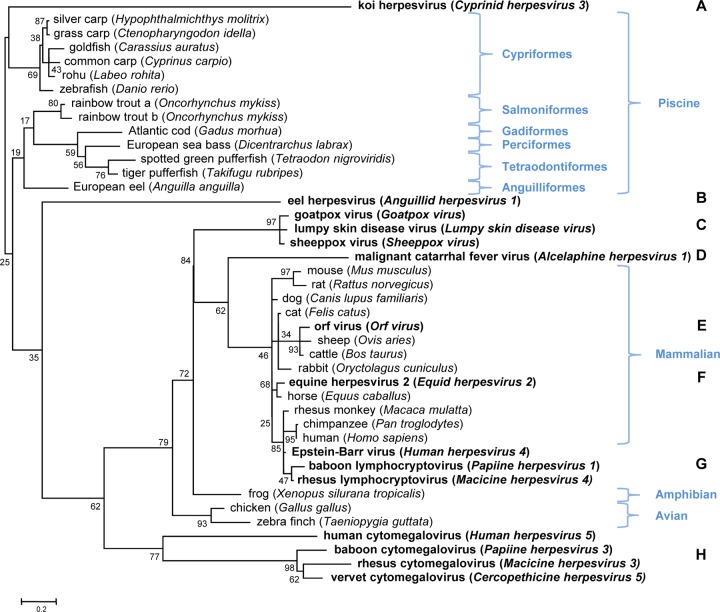
Phylogenetic analysis was conducted using mammalian, avian, amphibian, and piscine IL-10 sequences and IL-10 homologs from poxviruses, herpesviruses, and alloherpesviruses, including KHV and AngHV1 (Fig. 3). The analysis indicated that khvIL-10 is highly divergent from all host IL-10 sequences, such that its evolutionary origins remain unclear. As reported previously (57), ahvIL-10 is also very distantly related to all piscine IL-10s and to khvIL-10. This, and their very different locations in the viral genomes (3, 56), suggests that alloherpesvirus IL-10s may have arisen from independent gene capture events and that each has undergone rapid divergent evolution since capture from the host genome. Similar patterns of divergence from host IL-10 sequences have been observed among certain mammalian herpesvirus and poxvirus IL-10 orthologs (49), and this appears to have been driven by functional adaptations that modify their interaction with the host IL-10 receptor, resulting in altered signaling profiles (15, 60). There is abundant evidence for a very dynamic genetic interaction between large DNA viruses and their hosts in evolutionary time scales through processes that can result in both the capture and loss of genes (20, 24, 49). The phylogeny illustrated in Fig. 3 and the comparative genome locations of viral IL-10 genes suggest that at least eight independent capture events may have occurred during the evolutionary history of herpesviruses and poxviruses.
Fig 3.
Phylogenetic tree inferred by using host- and virus-encoded IL-10 proteins. Numbers on the branches are bootstrap values for 1,000 replicates expressed as percentage. Virus-encoded IL-10s are shown in bold. Clusters of host (piscine, amphibian, avian, and mammalian) IL-10 genes and eight (A to H) putatively independent host gene capture events are indicated. The systematic assignment of fish species at the level of taxonomic order is also indicated.
Expression of khvIL-10 during acute, persistent, and reactivation phases of infection.
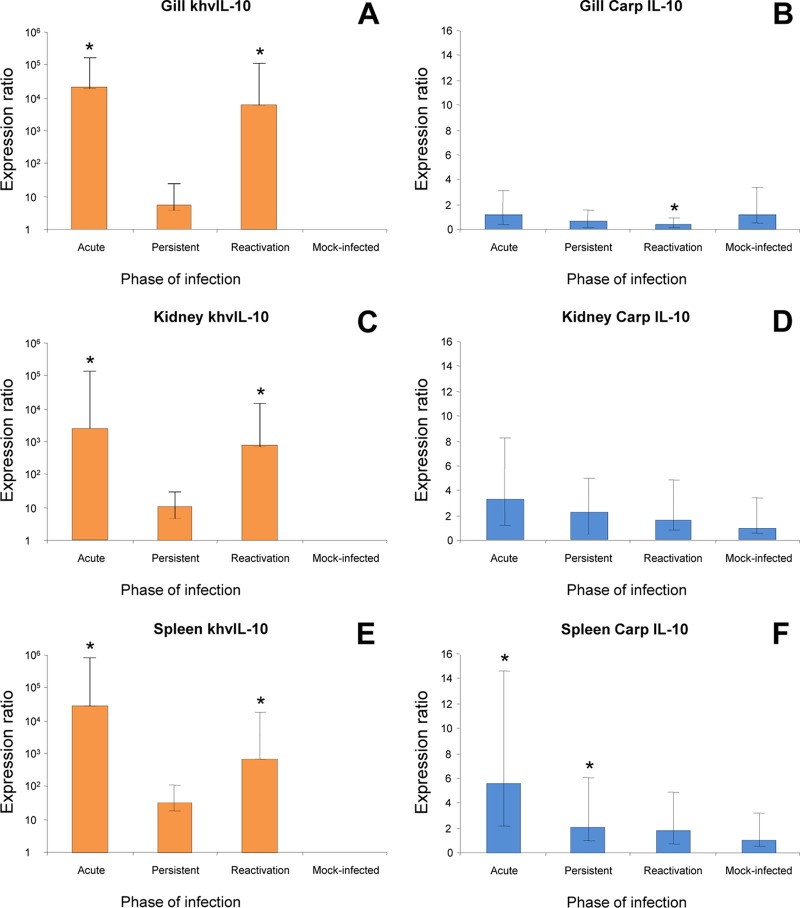
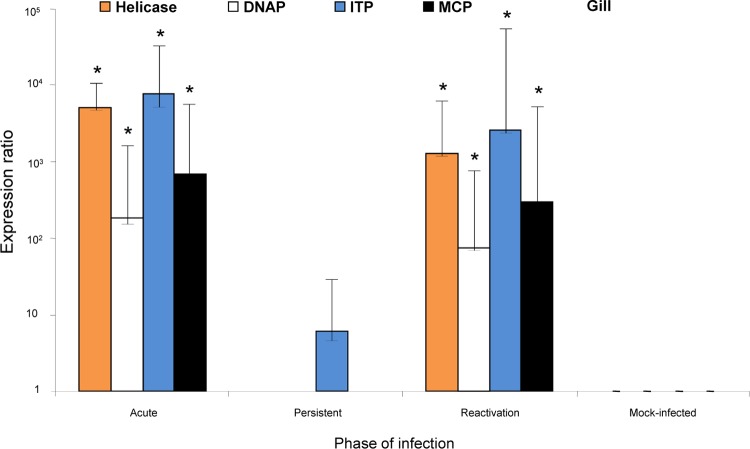
Expression of KHV and host IL-10 genes was assessed by qRT-PCR using RNA extracted from carp tissues during the acute, persistent, and reactivation phases of infection in a temperature shift experimental model (52). khvIL-10 was expressed at high levels in gill, kidney, and spleen tissues during the acute and reactivation phases of infection and at significantly lower levels (P < 0.05) during the persistent phase (Fig. 4A, C, and E). KHV early gene (helicase [Hel] [ORF71] and DNA polymerase [DNAP] [ORF79]) and late gene (intercapsomeric triplex protein [ITP] [ORF72] and major capsid protein [MCP]) expression was also quantified by qRT-PCR assay of gill tissue collected during each phase of infection (Fig. 5). Like khvIL-10, each of the viral early and late genes was highly significantly expressed (P < 0.05) in the acute and reactivation phases. However, there was no evidence of Hel, DNAP, or MCP gene expression during the persistent phase, and although low-level ITP gene expression was detected, transcripts were present at significantly lower levels (P < 0.05) than in the acute or reactivation phase.
Fig 4.
Expression of the khvIL-10 gene (A, C, and E) and carp IL-10 gene (B, D, and F) in carp tissues during the acute, persistent, and reactivation phases of KHV infection. Transcripts were quantified using a TaqMan qRT-PCR assay based on comparative threshold cycle (ΔΔCT) and normalized against expression of the 18S rRNA gene. Data are presented as the means ± standard errors for four individual fish per group. Asterisks indicate significant differences (P < 0.05) in expression between acute/reactivation and persistent phases of KHV infection. Note the difference in scale for KHV versus carp genes.
Fig 5.
Expression of KHV early (helicase and DNA polymerase [DNAP]) and late (intercapsomeric triplex protein [ITP] and major capsid protein [MCP]) genes in carp tissues during the acute, persistent, and reactivation phases of KHV infection. Transcripts were quantified using a TaqMan qRT-PCR assay based on comparative threshold cycle (ΔΔCT) and normalized against expression of the 18S rRNA gene. Data are presented as the means ± standard errors for four individual fish per group. Asterisks indicate significant differences (P < 0.05) in expression between acute/reactivation and persistent phases of KHV infection.
The results for khvIL-10 expression are similar to those reported for expression of cmvIL-10 with respect to the relative levels of expression during different phases of infection (25) but differ from those for ebvIL-10, which is expressed only during productive infection (19). The khvIL-10 expression patterns suggest that its function may be important for KHV survival at each stage of infection (14, 34). High-level expression during the acute and reactivation phases may mediate suppression of the carp immune response to establish a productive infection. As reported for cmvIL-10 (6, 8), low-level khvIL-10 expression during the persistent phase may serve to counter host immune surveillance and help maintain the persistent (or latent) state. In contrast, carp IL-10 was expressed at similar, very low levels during all phases of KHV infection and in uninfected fish (Fig. 4B, D, and F), suggesting that its expression is not induced in response to KHV infection. However, as acute-phase samples were collected from moribund fish, we cannot exclude the possibility that KHV induces a transient increase in carp IL-10 early after infection.
In vivo functionality of khvIL-10 in Danio rerio.
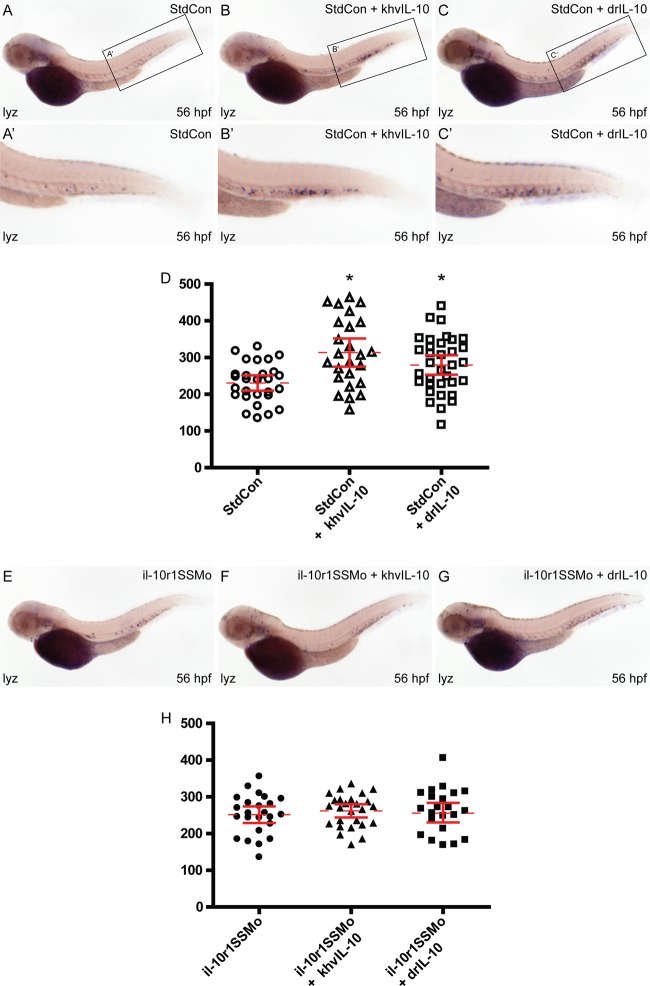
IL-10 is known to stimulate transient neutrophilia and monocytosis in addition to suppression of T cells (7). Therefore, the in vivo functionality of the khvIL-10 was assessed by injection of zebrafish embryos with mRNA encoding khvIL-10 and analysis by whole-mount in situ hybridization using the pan-leukocyte marker lysozyme (lyz) (17) at 56 hpf, before T cell development commences (55). A slight but statistically significant increase in the number of lyz+ cells was observed in khvIL-10 injected embryos compared to control embryos (Fig. 6A, B, and D). Injection of Danio rerio IL-10 (drIL-10) mRNA resulted in a similar increase in the lyz+ cells compared to controls (Fig. 6A, C, and D), suggesting functional equivalence.
Fig 6.
Functional conservation between khvIL-10 and Danio rerio IL-10. (A to D) Both khvIL-10 and drIL-10 increase embryonic leukocyte numbers. Embryos were injected with standard control morpholino (StdCon) alone (A) or coinjected with mRNA encoding either khvIL-10 (B) or Danio rerio IL-10 (drIL-10) (C) and subjected to WISH using the pan-leukocytic marker (lyz). The number of individual blue-stained lyz+ cells was quantified by manual counting (D), showing the mean (dashed red line), 95% confidence interval (red lines), and level of statistical significance (P < 0.01, *). (E to H) Both khvIL-10 and drIL-10 require IL-10R1. Embryos were injected with il-10r1SSMo alone (E) or coinjected with either khvIL-10 (F) or dril-10 (G), analyzed by WISH using lyz, and quantified (H), as described above.
IL-10 signals via a heterodimeric class II cytokine receptor consisting of a ligand-specific “long” chain, IL-10R1, and a shared “short” chain, IL-10R2 (45). To confirm that khvIL-10 was acting via the heterologous IL-10 receptor, the Danio rerio il-10r1 gene was targeted for knockdown using a morpholino directed at the exon 4/intron 4 splice site (il-10r1SSMo) (Fig. 1A), which was confirmed using RT-PCR (Fig. 1B). Consistent with a previous study (31), embryos injected using this morpholino displayed no overt developmental phenotype (Fig. 6A and E). Importantly, however, coinjection with either khvIL-10 or drIL-10 mRNA blocked their ability to increase the number of lyz+ cells (Fig. 6E to H), indicating that both ligands require IL-10R1 to mediate their effects on leukocyte numbers.
Danio rerio IL-10R1 (also known as crfb7) has previously been shown to have no overt role in development or in the signal transduction for Danio rerio phi and gamma interferons (1, 31). Our data confirm that IL-10R1 is functional during embryonic development in response to its cognate ligand, causing an increase in leukocytes. Similarly, drIL-10 overexpression also causes an increase in leukocytes, which is consistent with its role in other systems (7, 21). Importantly, we have shown that khvIL-10 could also increase leukocyte numbers in this heterologous setting, acting via the IL-10 receptor subunit, IL-10R1. This provides definitive evidence of the in vivo functionality of the virus-derived protein.
Virus survival requires evasion of the host immune response (14), and many DNA viruses have evolved effective immunosuppressive mechanisms by capturing and exploiting host IL-10 genes (44, 49, 51, 59). Molecular studies suggest there has been adaptive evolution of viral IL-10s following capture through positive selection to retain properties most beneficial for virus survival (10, 20). In mammalian herpesviruses, the evolved functions of viral IL-10s have been shown to be associated with both the productive and latent phases of infection (49). The nature of persistence in piscine herpesviruses and its relationship to true latency, as it is defined for mammalian herpesviruses, are poorly understood. Functional analysis of khvIL-10, with emphasis on immunomodulatory strategies, is likely to assist in understanding KHV infection and pathogenesis in carp and the evolutionary origins of temperature-induced persistence in poikilothermic organisms.
Supplementary Material
ACKNOWLEDGMENTS
We thank Tony Pye for DNA sequencing and Adam Foord for assistance with real-time PCR. We also thank Edward C. Holmes for advice on phylogenetic analysis methods.
Agus Sunarto was the recipient of a John Allwright Fellowship from the Australian Centre for International Agricultural Research (ACIAR).
Footnotes
Published ahead of print 15 August 2012
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1. Aggad D, et al. 2010. In vivo analysis of IFN-γ1 and IFN-γ2 signaling in zebrafish. J. Immunol. 185:6774–6782 [DOI] [PubMed] [Google Scholar]
- 2. Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410 [DOI] [PubMed] [Google Scholar]
- 3. Aoki T, et al. 2007. Genome sequences of three koi herpesvirus isolates representing the expanding distribution of an emerging disease threatening koi and common carp worldwide. J. Virol. 81:5058–5065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Beck S, Barrell BG. 1988. Human cytomegalovirus encodes a glycoprotein homologous to MHC class-I antigens. Nature 331:269–272 [DOI] [PubMed] [Google Scholar]
- 5. Bendtsen JD, Nielsen H, von Heijne G, Brunak S. 2004. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340:783–795 [DOI] [PubMed] [Google Scholar]
- 6. Chang WLW, Barry PA, Szubin R, Wang D, Baumgarth N. 2009. Human cytomegalovirus suppresses type I interferon secretion by plasmacytoid dendritic cells through its interleukin 10 homolog. Virology 390:330–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chernoff AE, et al. 1995. A randomized, controlled trial of IL-10 in humans. Inhibition of inflammatory cytokine production and immune responses. J. Immunol. 154:5492–5499 [PubMed] [Google Scholar]
- 8. Cheung AK, et al. 2009. The role of the human cytomegalovirus UL111A gene in down-regulating CD4+ T-cell recognition of latently infected cells: implications for virus elimination during latency. Blood 114:4128–4137 [DOI] [PubMed] [Google Scholar]
- 9. Davison AJ. 2010. Herpesvirus systematics. Vet. Microbiol. 142:52–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ding Y, Qin L, Kotenko SV, Pestka S, Bromberg JS. 2000. A single amino acid determines the immunostimulatory activity of interleukin 10. J. Exp. Med. 191:213–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dishon A, Davidovich M, Ilouze M, Kotler M. 2007. Persistence of cyprinid herpesvirus 3 in infected cultured carp cells. J. Virol. 81:4828–4836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gilad O, et al. 2003. Molecular comparison of isolates of an emerging fish pathogen, koi herpesvirus, and the effect of water temperature on mortality of experimentally infected koi. J. Gen. Virol. 84:2661–2667 [DOI] [PubMed] [Google Scholar]
- 13. Grayfer L, Belosevic M. 2012. Identification and molecular characterization of the interleukin-10 receptor 1 of the zebrafish (Danio rerio) and the goldfish (Carassius auratus L.). Dev. Comp. Immunol. 36:408–417 [DOI] [PubMed] [Google Scholar]
- 14. Griffin BD, Verweij MC, Wiertz EJHJ. 2010. Herpesviruses and immunity: the art of evasion. Vet. Microbiol. 143:89–100 [DOI] [PubMed] [Google Scholar]
- 15. Gruber SG, Gloria Luciani M, Grundtner P, Zdanov A, Gasche C. 2008. Differential signaling of cmvIL-10 through common variants of the IL-10 receptor 1. Eur. J. Immunol. 38:3365–3375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Haenen OLM, Way K, Bergmann SM, Ariel E. 2004. The emergence of koi herpesvirus and its significance to European aquaculture. Bull. Eur. Assoc. Fish Pathol. 24:293–307 [Google Scholar]
- 17. Hall C, Flores MV, Storm T, Crosier K, Crosier P. 2007. The zebrafish lysozyme C promoter drives myeloid-specific expression in transgenic fish. BMC Dev. Biol. 7:e42 doi:10.1186/1471-213X-7-42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hedrick RP, et al. 2000. A herpesvirus associated with mass mortality of juvenile and adult koi, a strain of common carp. J. Aquat. Anim. Health 12:44–55 [DOI] [PubMed] [Google Scholar]
- 19. Hsu DH, et al. 1990. Expression of interleukin-10 activity by Epstein-Barr virus protein BCRF1. Science 250:830–832 [DOI] [PubMed] [Google Scholar]
- 20. Hughes AL. 2002. Origin and evolution of viral interleukin-10 and other DNA virus genes with vertebrate homologues. J. Mol. Evol. 54:90–101 [DOI] [PubMed] [Google Scholar]
- 21. Huhn RD, et al. 1997. Pharmacodynamics of subcutaneous recombinant human interleukin-10 in healthy volunteers. Clin. Pharmacol. Ther. 62:171–180 [DOI] [PubMed] [Google Scholar]
- 22. Ilouze M, Dishon A, Kotler M. 2006. Characterization of a novel virus causing a lethal disease in carp and koi. Microbiol. Mol. Biol. Rev. 70:147–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Inoue Y, et al. 2005. Molecular cloning and expression analysis of rainbow trout (Oncorhynchus mykiss) interleukin-10 cDNAs. Fish Shellfish Immunol. 18:335–344 [DOI] [PubMed] [Google Scholar]
- 24. Jayawardane G, et al. 2008. A captured viral interleukin 10 gene with cellular exon structure. J. Gen. Virol. 89:2447–2455 [DOI] [PubMed] [Google Scholar]
- 25. Jenkins C, Abendroth A, Slobedman B. 2004. A novel viral transcript with homology to human interleukin-10 is expressed during latent human cytomegalovirus infection. J. Virol. 78:1440–1447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Jenkins C, et al. 2008. Immunomodulatory properties of a viral homolog of human interleukin-10 expressed by human cytomegalovirus during the latent phase of infection. J. Virol. 82:3736–3750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Jones BC, et al. 2002. Crystal structure of human cytomegalovirus IL-10 bound to soluble human IL-10R1. Proc. Natl. Acad. Sci. U. S. A. 99:9404–9409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Josephson K, Logsdon NJ, Walter MR. 2001. Crystal structure of the IL-10/IL-10R1 complex reveals a shared receptor binding site. Immunity 15:35–46 [DOI] [PubMed] [Google Scholar]
- 29. Kotenko SV, Saccani S, Izotova LS, Mirochnitchenko OV, Pestka S. 2000. Human cytomegalovirus harbors its own unique IL-10 homolog (cmvIL-10). Proc. Natl. Acad. Sci. U. S. A. 97:1695–1700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lahnsteiner F, Weismann T. 2007. Treatment of Ichthyophthiriasis in rainbow trout and common carp with common and alternative therapeutics. J. Aquat. Anim. Health 19:186–194 [DOI] [PubMed] [Google Scholar]
- 31. Levraud JP, et al. 2007. Identification of the zebrafish IFN receptor: implications for the origin of the vertebrate IFN system. J. Immunol. 178:4385–4394 [DOI] [PubMed] [Google Scholar]
- 32. Liongue C, Hall CJ, O'Connell BA, Crosier P, Ward AC. 2009. Zebrafish granulocyte colony-stimulating factor receptor signaling promotes myelopoiesis and myeloid cell migration. Blood 113:2535–2546 [DOI] [PubMed] [Google Scholar]
- 33. Liu Y, et al. 1997. The EBV IL-10 homologue is a selective agonist with impaired binding to the IL-10 receptor. J. Immunol. 158:604–613 [PubMed] [Google Scholar]
- 34. Lockridge KM, et al. 2000. Primate cytomegaloviruses encode and express an IL-10-like protein. Virology 268:272–280 [DOI] [PubMed] [Google Scholar]
- 35. Marchler-Bauer A, et al. 2009. CDD: specific functional annotation with the conserved domain database. Nucleic Acids Res. 37:D205–D210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Moore KW, de Waal Malefyt R, Coffman RL, O'Garra A. 2001. Interleukin-10 and the interleukin-10 receptor. Annu. Rev. Immunol. 19:683–765 [DOI] [PubMed] [Google Scholar]
- 37. Moore KW, O'Garra A, de Waal Malefyt R, Vieira P, Mosmann TR. 1993. Interleukin-10. Annu. Rev. Immunol. 11:165–190 [DOI] [PubMed] [Google Scholar]
- 38. Moore KW, et al. 1990. Homology of cytokine synthesis inhibitory factor (IL-10) to the Epstein-Barr virus gene BCRFI. Science 248:1230–1234 [DOI] [PubMed] [Google Scholar]
- 39. Nielsen H, Engelbrecht J, Brunak S, von Heijne G. 1997. Improved identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 10:1–6 [DOI] [PubMed] [Google Scholar]
- 40. OIE 2009. Manual of diagnostic tests for aquatic animals, p 236–250 World Animal Health Organisation, Paris, France [Google Scholar]
- 41. Pfaffl MW, Horgan GW, Dempfle L. 2002. Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 30:36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Pletnev S, Magracheva E, Wlodawer A, Zdanov A. 2005. A model of the ternary complex of interleukin-10 with its soluble receptors. BMC Struct. Biol. 5:e10 doi:10.1186/1472-6807-5-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Redpath S, Angulo A, Gascoigne NR, Ghazal P. 1999. Murine cytomegalovirus infection down-regulates MHC class II expression on macrophages by induction of IL-10. J. Immunol. 162:6701–6707 [PubMed] [Google Scholar]
- 44. Redpath S, Ghazal P, Gascoigne NR. 2001. Hijacking and exploitation of IL-10 by intracellular pathogens. Trends Microbiol. 9:86–92 [DOI] [PubMed] [Google Scholar]
- 45. Renauld JC. 2003. Class II cytokine receptors and their ligands: key antiviral and inflammatory modulators. Nat. Rev. Immunol. 3:667–676 [DOI] [PubMed] [Google Scholar]
- 46. Sadler J, Marecaux E, Goodwin AE. 2008. Detection of koi herpesvirus (CyHV-3) in goldfish, Carrasius auratus (L.), exposed to infected koi. J. Fish Dis. 31:71–72 [DOI] [PubMed] [Google Scholar]
- 47. Savan R, Igawa D, Sakai M. 2003. Cloning, characterization and expression analysis of interleukin-10 from the common carp, Cyprinus caprio L. Eur. J. Biochem. 270:4647–4654 [DOI] [PubMed] [Google Scholar]
- 48. Secombes CJ, Wang T, Bird S. 2011. The interleukins of fish. Dev. Comp. Immunol. 35:1336–1345 [DOI] [PubMed] [Google Scholar]
- 49. Slobedman B, Barry PA, Spencer JV, Avdic S, Abendroth A. 2009. Virus-encoded homologs of cellular interleukin-10 and their control of host immune function. J. Virol. 83:9618–9629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Spencer JV. 2007. The cytomegalovirus homolog of interleukin-10 requires phosphatidylinositol 3-kinase activity for inhibition of cytokine synthesis in monocytes. J. Virol. 81:2083–2086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Spencer JV, et al. 2002. Potent immunosuppressive activities of cytomegalovirus-encoded interleukin-10. J. Virol. 76:1285–1292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. St. Hilaire S, et al. 2005. Reactivation of koi herpesvirus infections in common carp Cyprinus carpio. Dis. Aquat. Organ. 67:15–23 [DOI] [PubMed] [Google Scholar]
- 53. Sunarto A, et al. 2011. Isolation and characterization of koi herpesvirus (KHV) in Indonesia: identification of a new genetic lineage. J. Fish Dis. 34:87–101 [DOI] [PubMed] [Google Scholar]
- 54. Sunarto A, Taukhid, et al. 2005. Field investigations on a serious disease outbreak among koi and common carp (Cyprinus carpio) in Indonesia, p 125–136 In Walker PJ, Lester RG, Bondad-Reantaso MG. (ed), Diseases in Asian aquaculture V. Asian Fisheries Society, Fish Health Section, Manila, Philippines [Google Scholar]
- 55. Trede NS, Langenau DM, Traver D, Look AT, Zon LI. 2004. The use of zebrafish to understand immunity. Immunity 20:367–379 [DOI] [PubMed] [Google Scholar]
- 56. van Beurden SJ, et al. 2010. Complete genome sequence and taxonomic position of anguillid herpesvirus 1. J. Gen. Virol. 91:880–887 [DOI] [PubMed] [Google Scholar]
- 57. van Beurden SJ, et al. 2011. The alloherpesviral counterparts of interleukin 10 in European eel and common carp. Fish Shellfish Immunol. 31:1211–1217 [DOI] [PubMed] [Google Scholar]
- 58. Vieira P, et al. 1991. Isolation and expression of human cytokine synthesis inhibitory factor cDNA clones: homology to Epstein-Barr virus open reading frame BCRFI. Proc. Natl. Acad. Sci. U. S. A. 88:1172–1776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Xu ZG, et al. 2001. The latency pattern of Epstein-Barr virus infection and viral IL-10 expression in cutaneous natural killer/T-cell lymphomas. Br. J. Cancer 84:920–925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Yoon S, et al. 2012. Epstein-Barr virus IL-10 engages IL-10R1 by a two-step mechanism leading to altered signaling properties. J. Biol. Chem. doi:10.1074/jbc.M112.376707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Zdanov A, et al. 1995. Crystal structure of interleukin-10 reveals the functional dimer with an unexpected topological similarity to interferon γ. Structure 3:591–601 [DOI] [PubMed] [Google Scholar]
- 62. Zdanov A, Schalk-Hihi C, Menon AS, Moore KE, Wlodawer A. 1997. Crystal structure of Epstein-Barr virus protein BCRF1, a homolog of cellular interleukin-10. J. Mol. Biol. 268:460–467 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.