Abstract
Bacteriophage ϕ6 is a double-stranded RNA (dsRNA) virus whose genome is packaged sequentially as three single-stranded RNA (ssRNA) segments into an icosahedral procapsid which serves as a compartment for genome replication and transcription. The procapsid shell consists of 60 copies each of P1A and P1B, two nonequivalent conformers of the P1 protein. Hexamers of the packaging ATPase P4 are mounted over the 5-fold vertices, and monomers of the RNA-dependent RNA polymerase (P2) attach to the inner surface, near the 3-fold axes. A fourth protein, P7, is needed for packaging and also promotes assembly. We used cryo-electron microscopy to localize P7 by difference mapping of procapsids with different protein compositions. We found that P7 resides on the interior surface of the P1 shell and appears to be monomeric. Its binding sites are arranged around the 3-fold axes, straddling the interface between two P1A subunits. Thus, P7 may promote assembly by stabilizing an initiation complex. Only about 20% of the 60 P7 binding sites were occupied in our preparations. P7 density overlaps P2 density similarly mapped, implying mutual occlusion. The known structure of the ϕ12 homolog fits snugly into the P7 density. Both termini—which have been implicated in RNA binding—are oriented toward the adjacent 5-fold vertex, the entry pathway of ssRNA segments. Thus, P7 may promote packaging either by interacting directly with incoming RNA or by modulating the structure of the translocation pore.
INTRODUCTION
It is widely accepted that the packaging of segmented viral genomes tends to be rather precise, resulting in the presence of one copy of each segment per virion. However, the mechanisms responsible for this feat are unknown for the Reoviridae, which house 10 to 12 double-stranded RNA (dsRNA) segments (16), or for the influenza viruses, with their 8 segments of single-stranded RNA (ssRNA) (23). It is only for the Cystoviridae that a model has been advanced for the specific packaging of the three genomic segments of RNA (17). Derivation of this model drew on the finding that faithful in vitro genome packaging can be accomplished with isolated procapsids. Procapsids can be obtained from cells expressing their proteins from plasmids (17) or can be assembled in vitro from purified proteins (31). In vitro-packaged particles could be used to produce viable phage, confirming that the procapsids were functional (25).
Bacteriophage ϕ6 is the prototype member of the Cystoviridae (18, 31). Its tripartite genome is packaged as single-stranded segments into a procapsid. Packaging induces major conformational changes in the procapsid, in a process called “maturation” (3, 18, 20). The mature capsid goes on to acquire an additional layer of protein (P8) and a lipid bilayer containing the P3 and P6 proteins as it develops into an infectious virion (17, 28). Viruses that function as biosynthetic compartments as well as delivery vehicles face additional architectural requirements in that they must incorporate the proteins needed for replication and transcription as well as those needed for packaging of the genome. The goal of this study was to address how these requirements are met in the ϕ6 system.
The procapsid has an unusual structure, with deeply recessed vertices that give it a dodecahedral morphology (3). In addition to the major capsid protein, P1 (85 kDa), which forms the surface shell, the procapsid has three other proteins. P4, on the outer surface (5), translocates the ssRNA segments into the procapsid, and P2, on the inside (34), replicates and transcribes the genome. High-resolution structures have been determined for both proteins (15, 33). The third protein, P7 (17 kDa), is less well characterized. Although not absolutely required for assembly, P7 enhances assembly efficiency: if it is present, the assembly rate is doubled (30). P7 is also required for packaging (14), as truncating its C terminus severely suppresses the output of infectious particles (29). Although P7 is present in substantial amounts—an estimated 30 to 60 copies per virion (13)—its location in the procapsid has not been established, although this information is essential to understanding how the protein may function.
Nevertheless, some clues have been forthcoming. P7 is reported to form dimers in solution (13), and it has been proposed, on the basis of small-angle neutron scattering data, to be positioned at a radius of about 160 Å from the procapsid center (11). The structure of P7 from the related phage ϕ12 has been solved by X-ray crystallography and nuclear magnetic resonance (NMR) spectroscopy, also as a dimer (6), and it has been proposed, based on interpretation of a cryo-electron microscopy (cryo-EM) reconstruction of mature ϕ12 virions, that P7 is situated next to the P4 hexamer, outside the P1 shell (36).
In the present study, we set out to localize P7 and to confirm the location of P2 in the procapsid by cryo-electron microscopy. By comparing procapsids with different protein compositions, we sought to identify potential interactions between these proteins and other procapsid components. This approach was intended to exploit the property that both proteins may be omitted while still maintaining a good yield of correctly formed procapsids. The term P1247 refers to procapsids composed of P1, P2, P4, and P7. In addition, we prepared and imaged P124, P147, and P14 procapsids. Three-dimensional reconstructions were calculated and used to define the positions of the targeted proteins by difference mapping. In parallel, we assessed the protein compositions of the respective preparations of procapsids by quantitative SDS-PAGE.
MATERIALS AND METHODS
Preparation and purification of ϕ6 procapsids.
P1247 procapsids were produced in Escherichia coli strain JM109 with the plasmid pLM687 to coexpress the wild-type P1, P2, P4, and P7 subunits (P1247). The plasmids pLM574 and pLM1906 had deletions in genes 2 and 7, respectively, and produced procapsids with only P1, P4, and P7 subunits (P147) or procapsids with only P1, P2, and P4 subunits (P124). Procapsids without P2 and P7 subunits (P14) were produced using the plasmid pLM358, from which both genes 2 and 7 had been deleted. The same procedure was used to purify all types of procapsids. Cells were grown overnight in LB medium with 0.5 mM IPTG (isopropyl-β-d-thiogalactopyranoside), harvested, lysed with a French press, and centrifuged at 26,000 rpm for 90 min in a 10 to 30% sucrose gradient. The procapsid band was isolated, concentrated, treated with RNase A, and then centrifuged overnight in a 30 to 60% sucrose gradient at 23,000 rpm. The procapsid band was extracted, concentrated by centrifugation, and resuspended in buffer P (10 mM potassium phosphate, 1 mM MgCl2, pH 8.0) with 50% sucrose. Prior to electron microscopy, the samples were buffer exchanged into buffer P by use of Zeba-Midi buffer exchange columns with a 7-kDa cutoff (Thermo Scientific, Rockford, IL).
Cryo-electron microscopy.
Samples of procapsids were diluted to a protein concentration of ∼10 mg/ml, applied to glow-discharged C-flat grids (Protochips, Inc., Raleigh, NC), blotted, and plunge-frozen in liquid ethane by use of a Vitrobot (FEI, Hillsboro, OR) that had been equilibrated at 25°C and 90% humidity. The vitrified specimens were imaged at a magnification of ×38,880 (calibrated) with a CM200 FEG microscope (FEI, Eindhoven, The Netherlands) fitted with a Gatan 626 cryoholder and operating at an accelerating voltage of 120 kV. Micrographs were recorded on Kodak SO163 film at ∼15 e−/Å2 per exposure, with a 1.0- to 3.5-μm underfocus.
Image processing.
Micrographs were digitized using a Nikon Super Coolscan 9000ED camera at 4,000 dots per inch (1.633 Å/pixel). Image processing was done with the Bsoft package (8). The contrast transfer function (CTF) parameters were estimated and particles picked in bshow. These images were CTF corrected by phase flipping in the initial stages and by phase and baseline compensation in later stages to enhance high-frequency shells. Origins and orientations were determined by projection matching using borient in a global search with 1° steps and then refined to a final step size of 0.1°, using brefine. The magnifications of individual particles were fine-tuned in the final iterations as well. Two density maps were reconstructed from the halves of each data set by use of breconstruct and then masked with a tight soft mask of the procapsid shell. These maps were used to estimate the resolution by Fourier shell correlation at a cutoff of 0.3, using bresolve (Table 1). The two maps were then averaged.
Table 1.
Statistics for reconstructions
| Procapsid | No. of micrographs | Defocus range (μm) | No. of particles | FSC0.3a (Å) |
|---|---|---|---|---|
| P1247 | 84 | 0.65–3.50 | 6,839 | 8.1 |
| P124 | 62 | 1.23–3.28 | 4,023 | 9.7 |
| P147 | 69 | 1.22–2.80 | 9,625 | 9.6 |
| P14 | 84 | 1.25–2.58 | 4,047 | 12.4 |
Fourier shell correlation with a cutoff of 0.3.
To calculate difference maps, the maps were adjusted to the same reciprocal-space amplitudes by use of bampweigh and then low-pass filtered to 8-Å resolution and rescaled to the same real-space intensity average (0) and standard deviation (1) by using bfilter. Finally, the adjusted maps were subtracted with bop to generate the difference map.
Segmentation of the P1 shell.
The reconstruction of the P1247 procapsid was band-pass filtered to 7-Å resolution and segmented in Chimera (26). To simplify interpretation of the icosahedral map, segmentation was performed on a subset of asymmetric units that constituted one 5-fold vertex. The density was thresholded at 4-sigma to visualize the characteristic rod-like densities of α-helices. The vertex was segmented with SEGGER into many small volumes (27). These segments were merged manually based on their connectivity and the 5-fold symmetry of the vertex. During this procedure, similar features in the two non-symmetry-related subunits of the shell became obvious and guided further merging of individual segments. Finally, the two P1 subunits were compared to deduce the boundaries. The P1 subunit at the 5-fold axis is denoted P1A and that at the 3-fold axis is denoted P1B, according to the nomenclature of Huiskonen et al. (9).
Segmentation of the P7 and P2 densities.
Boundaries of the P7 subunits were derived from the difference map for the P147 and P14 procapsids. The difference map was filtered to 25-Å resolution and converted into a mask with the program bmask, using an intensity threshold that clearly separated P7 density from the background noise. The mask was then dilated to enclose all features of the P7 density at 7-Å resolution and corrected to exclude any protruding density from the P1 shell. The mask for the P2 subunit was derived in a similar way from the difference map for the P124 and P14 procapsids. The P2 and P7 masks were then used to extract densities of the P2 and P7 subunits, respectively, from the high-resolution map of the P1247 procapsid.
Fitting of P7 structure into the electron density.
The X-ray structure of the partial P7 structure from bacteriophage ϕ12 (Protein Data Bank [PDB] ID 2Q82) was converted into a density map with 1.633-Å/voxel sampling and filtered to 7-Å resolution by using the program bsf. This density was fitted into the density assigned to P7 (extracted from the P1247-P124 difference map). The orientation was determined through a global search over all Euler angles with 3° steps and followed by local refinement within 1° accuracy, using the program bfind.
Fitting of P2 structure into the electron density.
A monomer was taken from the crystal structure of P2 (33) (PDB ID 1UVJ) molecules and converted into a density map with 1.633-Å/voxel sampling by using the program bgex. To find the best alignment of the P2 structure to the P2 density extracted from the P1247-P147 and P124-P14 difference maps, both crystal structure and EM densities were oriented and centered along the 3-fold axis and band-pass filtered to 15-Å resolution. Optimal displacement of the P2 density from the 3-fold axis was searched within ±10 pixels in all x, y, and z coordinates. The density was 3-fold symmetrized and its best alignment to the EM density found in a grid search over the three Euler angles by use of bsymcomp. The solution with the best cross correlation was selected, transformed back to the orientation in the procapsid, and depicted in one of the three equivalent positions at the 3-fold axis.
Stoichiometry.
The amount of protein in each band from a Coomassie-stained SDS-PAGE gel was estimated by fitting a Gaussian curve, integrating under this curve, and correcting for the background. Loading different amounts of sample on SDS-PAGE gels and quantitating P1 indicated that its response curve is nonlinear and may give high relative estimates for the other components when they are present in small amounts. To obtain accurate estimates, all densitometry results for the four proteins were plotted and the amounts calculated from the initial, linear parts of the curves. These values were further corrected for the disparities in arginine-plus-lysine content between the proteins, as indicated previously (1). Errors were estimated from the deviations in the linear fits.
The amounts of P2 and P7 in the procapsids were estimated from maps that were low-pass filtered to 15-Å resolution. Densities for P2, P4, or P7 subunits were extracted from each map, using tight masks for each subunit, averaged, and compared with the pertinent background density and that of the P1 shell, representing zero and full occupancy, respectively. To compensate for residual CTF effects around the P1 shell that diminished the apparent P2 and P7 densities, the background level was taken from a region under the 5-fold vertices. For the P1 shell and P4 densities, the background was determined from an external spherical shell.
RESULTS
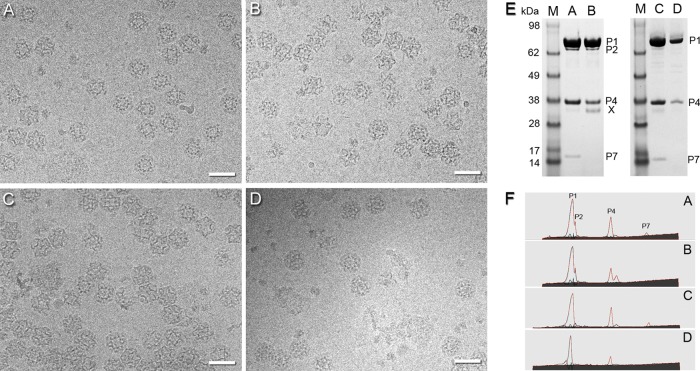
For this study, we refined our previous purification protocol (34) to produce larger quantities of purer and better-preserved procapsids (see Materials and Methods). Cryo-EM grids were prepared from freshly purified material immediately after removing the sucrose used in the gradient step. Representative micrographs of all four procapsids—P1247, P124, P147, and P14—are shown in Fig. 1A to D. From these data, three-dimensional reconstructions were calculated (Table 1). Their protein compositions as conveyed by SDS-PAGE are given in Fig. 1E and F.
Fig 1.
(A to D) Cryo-electron micrographs of P1247 (A), P124 (B), P147 (C), and P14 (D) procapsids taken with an ∼2.1-μm defocus. Bars, 500 Å. The procapsids are randomly oriented. As a result, the images show markedly varying projections of the icosahedral shell with its deeply inverted vertices. (E) SDS-PAGE gel containing P1247 (lane A), P124 (lane B), P147 (lane C), and P14 (lane D) procapsids. The P124 sample also contained a minor contaminant of ∼35 kDa (X). (F) Densitometry profiles (cyan lines) obtained from the SDS-PAGE gels shown in panel E were fitted with asymmetric Gaussian curves (red lines). Peaks of the P1, P2, P4, and P7 subunits are indicated in the sample of P1247 procapsids (profile A).
Assignment of proteins P1A and P1B in the procapsid surface lattice.
A surface rendering of the P1247 procapsid is shown in Fig. 2A. The map reveals numerous rod-like densities that we take to represent α-helices (Fig. 2B and C). This interpretation is consistent with the observation by Raman spectroscopy that the capsid protein is mostly α-helical (2). In earlier reconstructions of the procapsid at lower resolution, it was not possible to delineate the molecular boundaries between the two P1 conformers (3, 5, 34). However, the substructural detail visualized in the present reconstructions allowed an unambiguous solution. Matching features were located in the P1A and P1B subunits and used for delineation of individual P1A subunits around the 5-fold axis. The interface between the P1A and P1B subunits was further refined according to correspondence of the respective rod-like densities in the P1A and P1B subunits (see Materials and Methods). Our solution is consistent with that of Huiskonen et al., which was based on a cryo-EM reconstruction of the mature RNA-containing capsid at about the same resolution (9).
Fig 2.
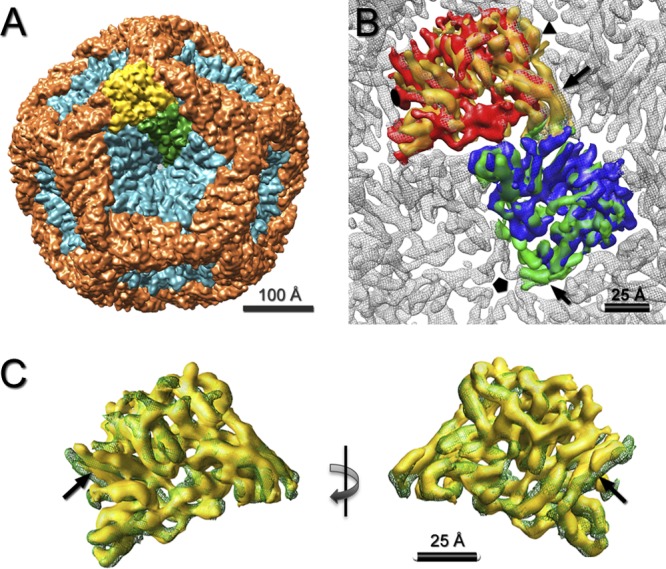
(A) Segmentation of P1 subunits in the procapsid shell. The P1A subunits (teal) form inverted pentamers located at the 5-fold axes. The P1B subunits (orange) connect neighboring 3-fold vertices via 2-fold axes. One P1A subunit and one P1B subunit are depicted in green and yellow, respectively. (B) Fit of the segmented P1A (green) and P1B (yellow) subunits from the procapsid into the mature shell of the RNA-packaged capsid (gray) (9) (EMDataBank [EMDB] ID 1206). The unassigned densities in the previously described segmentation (12), near the 5-fold and 3-fold axes, are included in the P1A and P1B subunits, respectively (black arrows). (C) Overlay of the segmented P1A (green mesh) and P1B (yellow surface) subunits as viewed from the procapsid exterior (left) and interior (right). Most of the rod-like densities (putative α-helices) are superimposable, with only a few at the edges having relative shifts (black arrows).
The P1A subunits (colored teal in Fig. 2A) are organized in pentameric rings centered on the 5-fold vertices, whereas the P1B subunits (colored orange in Fig. 2A) meet at the 3-fold vertices, forming the dodecahedral framework. The structures of the two subunits are very similar (Fig. 2B and C), indicating that their folds are affected only slightly by their different, nonequivalent packing environments. Their close resemblance to P1 in the mature capsid (9) indicates that the subunits essentially pivot as rigid bodies in the maturation transformation and undergo little, if any, refolding.
Mapping of other procapsid proteins.
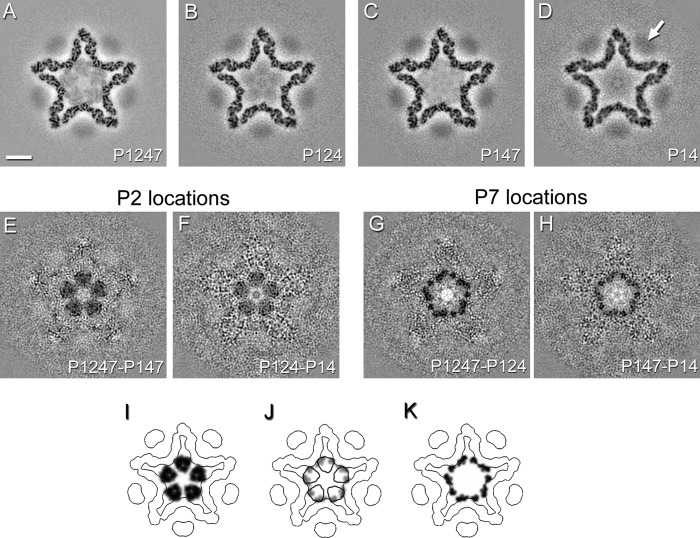
In cryo-EM reconstructions of the procapsid, P4 hexamers are perceived as blurred disk-like densities overlying the 5-fold vertices (3, 5, 34), as shown in Fig. 3D (arrow). The blurring arises from the symmetry mismatch between the 6-fold hexamer and the 5-fold vertex structure (10) and because the respective symmetry axes are offset about 20 Å, on average (21). P4-related density is typically lower than that in the P1 shell because procapsids tend to shed P4 during purification. All four procapsids in this study contained P4 at about 50% occupancy, i.e., 6 hexamers per capsid, on average, as assessed by quantitative SDS-PAGE (Table 2).
Fig 3.
(A to D) Slices through icosahedral reconstructions of ϕ6 procapsids filtered to 8-Å resolution and viewed along the 5-fold icosahedral axis ∼73 Å from the procapsid center. Bar, 100 Å. The P4 density is indicated in the map of the P14 procapsid (white arrow). (E to H) Difference maps calculated between P1247 and P147 procapsids or P124 and P14 procapsids show locations of P2, while the difference maps calculated between P1247 and P124 procapsids or P147 and P14 procapsids show locations of P7. (I to K) Corresponding slices through models of P2 and P7 in the procapsid, determined by fitting the P2 structure (PDB ID 1UVJ) and the P7 homology model into the difference maps (see the text). The densities are shown for P2 (I) and P7 (K) subunits. An overlay of the P2 (black contour) and P7 (gray density) locations is shown in panel J.
Table 2.
Copy numbers of P2, P4, and P7 subunits per procapsid, based on quantitation by SDS-PAGE and integration of densities in maps
| Procapsid | No. of copies |
|||||
|---|---|---|---|---|---|---|
| P2 |
P4 |
P7 |
||||
| SDS-PAGEa | Mapb | SDS-PAGEa | Mapb | SDS-PAGEa | Mapb | |
| P1247 | 4 ± 1 | 7 ± 1c | 29 ± 1 | 25 ± 1 | 12 ± 4 | 10 ± 2c |
| P124 | 4 ± 1 | 6 ± 1 | 40 ± 2 | 31 ± 1 | ||
| P147 | 35 ± 1 | 33 ± 1 | 12 ± 6 | 3 ± 1 | ||
Data are means with standard deviations, which were calculated from linear fits to the densitometry data.
Data are means with standard deviations, which were calculated from maps reconstructed from half data sets.
P2 occupancy was determined from the density that does not overlap with P7. This contribution of P2 density was then subtracted in the calculation of P7 occupancy.
P2 contributes internal densities in cryo-EM density maps of the procapsid (34). These densities are discernible in cross sections of the present reconstructions of P2-containing procapsids, i.e., P1247 and P124 (Fig. 3A and B), but they are faint, suggesting low occupancy. This inference was confirmed by quantitative SDS-PAGE, which indicated only about 20% occupancy, on average, of the 20 P2 binding sites (Table 2 and data not shown). These data also indicated that P7 was present in much smaller amounts than would be expected for wild-type virions, which are reported to have 30 to 60 copies of P7 (13, 35a). These shortfalls presumably reflect the fact that our procapsids were produced by expressing the proteins from plasmids in a nonnatural host.
Because of the low copy numbers, we concluded that the best prospects for identifying densities associated with P2 and P7 would be seen with difference density maps. In both cases, two independent difference maps could be calculated from our data. P2 could be detected by subtracting the P147 map from the P1247 map and by subtracting the P14 map from the P124 map, and the method was used similarly for P7. The results are shown in gray-scale sections in a plane perpendicular to a 5-fold axis in Fig. 3E to H. In both cases, the positive difference densities (shown as dark areas) in the two independent determinations are fully consistent. The dimensions of the five P2-related densities are consistent with monomers of a 75-kDa protein. The densities deriving from P7 (17-kDa monomer) in this section are smaller and reflect contributions from more than five subunits (see below). Interestingly, the respective densities—those for P2 and P7—overlap, indicating that a bound P2 structure will occlude adjacent binding by P7 and vice versa (Fig. 3I to K and 4). Because of the low occupancies (see above), this does not invoke a contradiction. These data show that P7, like P2, binds to the inner surface of the P1 shell, not far from the 3-fold axes.
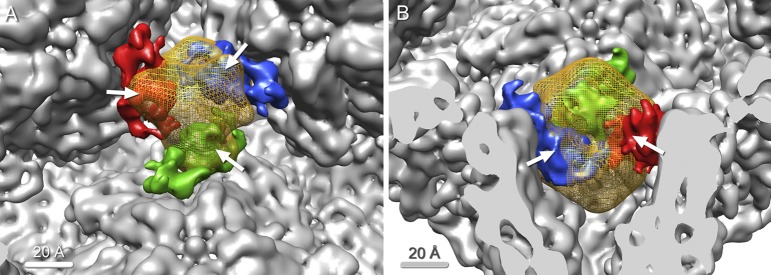
Fig 4.
Overlap of P2 and P7 as shown by difference density maps. (A) Inside view of the procapsid along the 3-fold axis, showing one P2 subunit (yellow mesh) and three neighboring P7 subunits (red, blue, and green). P2 significantly overlaps all three P7 subunits (white arrows). (B) Large overlap between P2 and P7 (white arrows), as seen in a side view.
We defined the P7 molecules in three dimensions by a mask generated from the P147-P14 difference map. These densities are consistent in size with a 17-kDa monomer. They are arranged in threes around the 3-fold axis (Fig. 5A and B).
Fig 5.
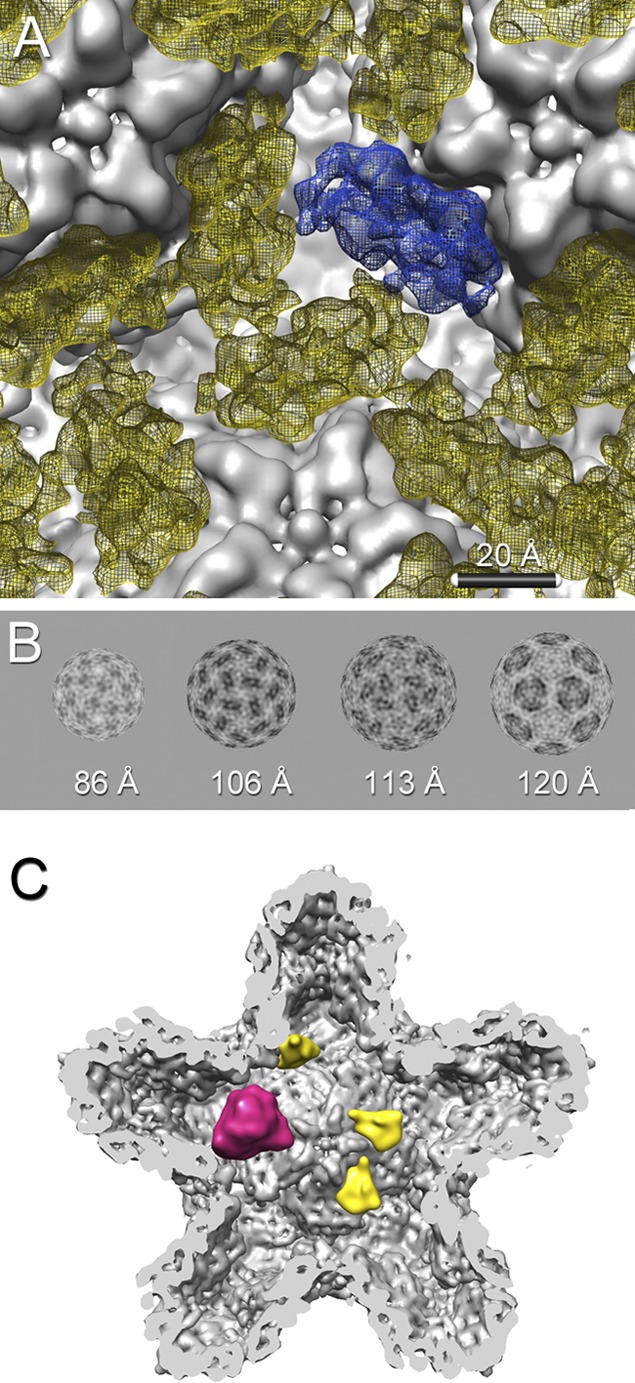
(A) In the icosahedrally averaged map, the P7 densities (yellow mesh) appear as triplets surrounding the 3-fold axis between the 5-fold vertices of the procapsid shell (gray). One of the three P7 densities is shown in blue. (B) Radial shells of the P1247-P124 difference map at radii of 86, 106, 113, and 120 Å. The P7 density (dark) is organized around the 3-fold axes. (C) Due to the small amounts of P7 in the procapsid and to partial overlap of the P7 and P2 binding sites, most locations at the 3-fold axes are occupied by only one P7 (yellow) or one P2 (magenta) molecule.
As with P7, the P2 densities were defined by a mask derived from a difference map, in this case from the P124-P14 difference map. The P2 density exhibits significant overlap with the P7 density and occludes all three P7 binding sites (Fig. 4). Conversely, occupancy of one P7 binding site is sufficient to occlude the P2 site. The procapsid thus cannot bind 20 P2s and 60 P7s simultaneously. Figure 5C illustrates the binding of small amounts of P2 and P7, consistent with our estimation of their copy numbers. Because of the (artifactual) 3-fold averaging of P2 density, we could not make a more precise specification of the location and orientation of P2 in the procapsid.
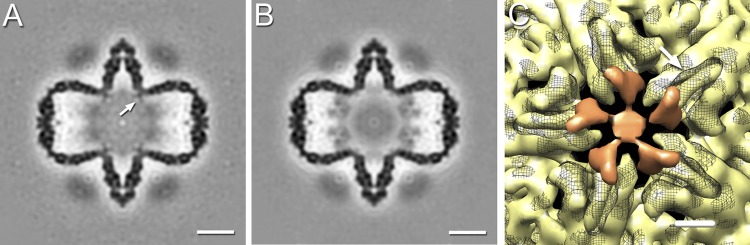
Close inspection of the density maps revealed an additional density under the tip of the 5-fold vertex in the P124 map (Fig. 6A) compared with the P1247 map (Fig. 6B). We infer that this density is part of the P1 lattice, whose conformation is affected by the presence of P7 in its proximity (Fig. 6C).
Fig 6.
Central slices through P124 (A) and P1247 (B) procapsids filtered to 15-Å resolution and oriented along the 2-fold icosahedral axis. The P124 procapsid exhibits additional density under the 5-fold vertex (arrow). Bars, 100 Å. (C) Inside view of the 5-fold vertex of the P1247 (gray mesh) and P124 (yellow) procapsids filtered to 8-Å resolution and depicted at a 3-sigma threshold. The P124 procapsid shows decreased density in the short helices at the tip of P1A subunits at the 5-fold vertex (arrow). The additional density in the P124 procapsid (orange) has a similar size and shape to those of the short helix and may represent its alternative conformation in the absence of P7. Bar, 10 Å.
Orientation of the P7 subunit.
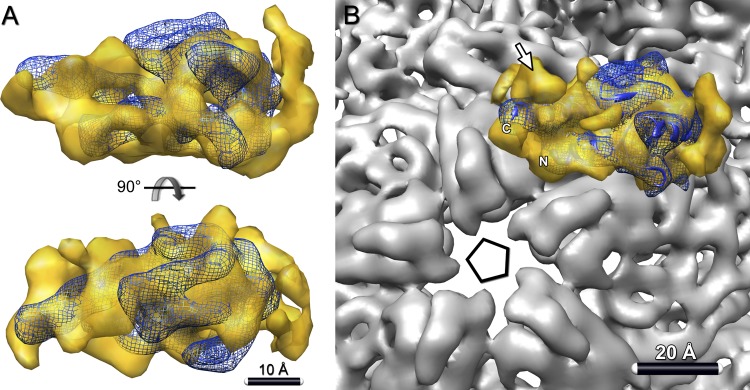
A high-resolution structure has been determined for the N-terminal part (129 of the 169 residues) of P7 from ϕ12 (6). Although its sequence identity with P7 of ϕ6 is only ∼10% (29), the two proteins have similar sizes. The other procapsid subunits (P1, P2, and P4) also have low sequence identity (10 to 15%) among cystoviruses; nonetheless, the subunits are thought to have similar folds (10, 12). Moreover, there are many examples of homologs in different bacteriophages retaining similar structures in the absence of sequence similarity (e.g., capsid proteins [7] or portal subunits [24]). Accordingly, we attempted a fit of ϕ12 P7 into the P7-related density from the difference density map of ϕ6 procapsids (Fig. 7A). The two molecular representations are quite compatible, showing similar sizes and an elongated shape. Notably, the N- and C-terminal helices of ϕ12 P7 (and hence, we posit, of ϕ6 P7 as well), as fitted, point toward the adjacent 5-fold axis in the procapsid (Fig. 7B).
Fig 7.
Orientation of P7 in the procapsid. (A) Crystal structure of ϕ12 P7 (blue ribbon) (PDB ID 2Q82) (6) fitted into the P7 density map (yellow) from the P1247-P124 difference map. The fitting, which involved a global search over all orientations, is described in Materials and Methods. (B) In this setting, the C- and N-terminal helices of P7 both lie close to the 5-fold vertex of the procapsid. Both P7 termini were implicated in RNA binding and could thus interact with packaged RNA in this orientation. Additionally, the EM map rendered at a lower threshold shows an unassigned density near the vertex (arrow) that may correspond to a part of the C-terminal region of P7 that is missing from the crystal structure.
P7 stoichiometry.
The cryo-EM reconstructions afforded an opportunity to obtain estimates of P7 and P2 occupancy independent of those obtained by SDS-PAGE (Table 2). To do so, the densities above background associated with these two proteins were calibrated against the density in the P1 shell, taken to represent 100% occupancy. For P2, the resulting numbers are consistent with each other and with the SDS-PAGE data. For P7, the cryo-EM data yielded a result consistent with the SDS-PAGE result for P1247 procapsids but a smaller number for P147 procapsids (Table 2). To account for this discrepancy, we hypothesize that some of the P7 in the latter particles became detached from the P1 shell and was therefore unseen in the reconstruction but remained inside and consequently was detected by SDS-PAGE.
DISCUSSION
P7 is inside the procapsid.
P7 is required for effective packaging of the ϕ6 genome (14) and also contributes to procapsid assembly (30). Insight into these modes of action has been hampered by a lack of information on the location and interactions of P7. Its inferred status as a dimer (13, 29, 30) and estimated copy number of 60 led to the proposal that it binds on the 2-fold axes, of which there are 30. Wei et al. assigned P7 to the external surface of the ϕ12 virion (36), and Ikonen et al. estimated that P7 is about 160 Å from the center (11). Our analysis by difference imaging differs in consistently visualizing P7 as monomer-sized densities bound around the 3-fold axes on the inner surface of the P1 shell, at a radius of ∼110 Å from the procapsid center (the particle has an outer radius of 240 Å). Our preparations have a lower content of P7 than that in native ϕ6 particles (32), but there is no reason to suppose that the binding sites differ.
Oligomeric status: monomers, dimers, and trimers.
The three P7-related densities around each 3-fold axis appear to make contact or to be close to doing so (Fig. 5A and B). This does not necessarily mean that the protein is trimeric, as the intersubunit contact area may not be large enough to support a stable interaction. Moreover, the triplets of subunits seen in the difference maps may be generated from monomers randomly distributed over all possible sites at 20% average occupancy. On the other hand, these observations are hard to reconcile with a dimer. Accordingly, we suggest that incorporation of P7 into the procapsid involves dissociation of the dimer and attachment to P1 as monomers.
Both P7 termini are vertex oriented.
The crystal structure of the N-terminal 129 residues of P7 from ϕ12 features a β-sheet core surrounded by helices (6). Its elongated shape matches quite well with the molecular envelope assigned to P7 in our difference maps (Fig. 7) and supports the idea that the ϕ6 and ϕ12 proteins have generally similar structures, despite low sequence identity.
The C-terminal tail is important for both assembly and packaging and is deemed flexible until bound to P1 (29). In our fit, the last residue visualized in the ϕ12 P7 structure (residue 129) is located next to a density not accounted for by the structure (Fig. 7B). We speculate that this density may be part of the missing C-terminal region (residues 130 to 169). It is located close to the 5-fold vertex, and the pore through the vertex is the likely entry conduit for RNA strands. As the P4 motor threads the RNA strand through the pore, the RNA could interact with P7 inside the shell. Both the N-terminal core and the C-terminal tail have been implicated in RNA binding (6). One possibility is that P7 may provide a catch to prevent retraction of incoming RNA strands during packaging.
P7 may facilitate RNA packaging by stabilizing a vertex-proximal P1 motif.
Our reconstructions consistently show a small density extending from the 5-fold vertex in procapsids that lack P7 (Fig. 6). We interpret this density as part of the P1 tip that is less ordered than when P7 is present. P7 interacts directly with a short helix at the vertex tip that is less defined in the P7-lacking procapsids and has a similar size to that of the extended density. The density is better defined in the P124 procapsid, and P2 molecules appear to make contact with it (Fig. 6C). However, the structure surrounding the packaging pore at the vertex may have functional importance for packaging.
P7 may promote assembly by stabilizing P1A interactions.
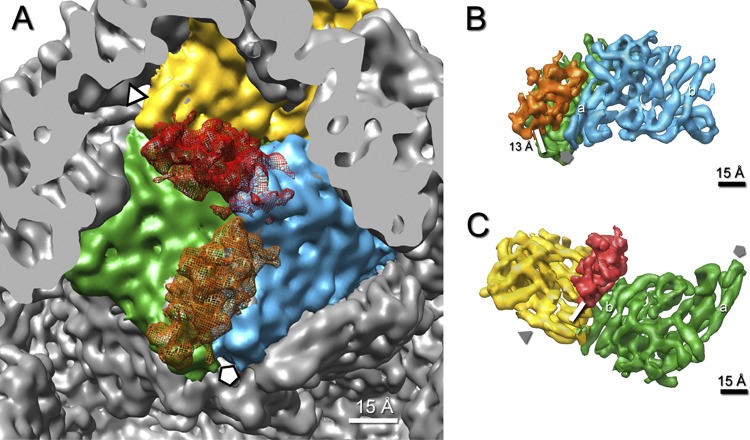
P7 subunits bind only to the P1A subunits, not to P1B (Fig. 8A). P7 interacts with two P1A subunits, bridging the interface between them. Assuming that the tubular densities in the reconstructions represent α-helices, P7 binds to short helices at the tip of one P1A subunit and to a long helix of the adjacent P1A subunit (Fig. 8B). At the current resolution, this region already shows significant differences between P1A and P1B, which provides an explanation for its nonbinding to the corresponding P1A-P1B interface (Fig. 8C).
Fig 8.
Interaction of P7 with P1 subunits in the procapsid. (A) Interior view of the 5-fold vertex (marked by a pentagon) in the procapsid. P7 (orange mesh) is attached to an interface that comprises two P1A subunits (green and blue). A hypothetical attachment of P7 (red mesh) at the corresponding site between P1B (yellow) and P1A (green) subunits produces a clash between P7 and the adjacent P1A subunit (blue). (B) Densities of bound P7 (orange) and two P1A (green and blue) subunits are shown at a high-density threshold. The binding interface is defined by the distance between centers of neighboring α-helices, which falls within 10 to 15 Å. The interface comprises a long α-helix (a) of the blue P1A subunit that points toward the 5-fold vertex. (C) The hypothetical binding interface between P7 (red) and the P1B (yellow) and P1A (green) subunits does not comprise the same α-helix (a) of P1A as that in panel B, but another α-helix (b) on the opposite site of P1A.
It transpires that all three minor procapsid proteins—P2, P4, and P7—bind only to P1A subunits. Accordingly, P1A appears to play a role in nucleating procapsid assembly, while P1B may be less important in this context. According to Poranen et al., the nucleation complex in the absence of P7 is P14P46, while that with P7 is P14P412P72 (30). One arrangement for the latter complex is a P1 tetramer with a P4 hexamer and one P7 molecule bound to each pair of P1 subunits. In fact, it has been proposed that a dimer of dimers serves as a common starting point for assembly of several dsRNA viruses (28). However, in the procapsid, P1A molecules bound to different P4 hexamers do not interact. An alternative possibility is for a P1A5P46 complex, equivalent to a vertex of the procapsid, with up to five P7 molecules bound at the interfaces between P1A subunits. This would be consistent with an involvement of the P7 C-terminal tail in promoting assembly (29) and with the possibility, raised here, that P7 stabilizes the 5-fold vertex. A large excess of P4 would result in complexes with P1 only in the A conformation, hindering proper assembly (35a). Excess P7 would not be detrimental, because it binds only to the P1A subunit interface, presumably already in the P1A5P46 complex. Subsequent steps in assembly would then involve the association of the P1B subunits and neighboring P1A5P46P7n complexes.
Mutual occlusion of P2 and P7 and its implications.
The two difference density maps—one disclosing P2 and the other disclosing P7 (Fig. 3E to H)—demonstrate that the respective binding sites on the inner surface of the P1 shell are mutually occluding (Fig. 4), i.e., if a P2 binding site is occupied, P7 subunits are blocked off from its adjacent sites, and vice versa. While P7 is directly represented in our difference density maps, the P2 monomer has been subjected to 3-fold symmetrization, precluding a very precise placement. To address this uncertainty, we tried modeling with the P2 monomer in different positions but did not find any that would allow even one P7 subunit access to a binding site while retaining consistency with the symmetrized P2 density. We conclude that all three P7 sites around a given 3-fold axis are occluded by a bound P2 molecule and, conversely, that binding of a single P7 subunit occludes the P2 site. The upshot for stoichiometry is that the procapsid cannot simultaneously incorporate 60 copies of P7 and 20 copies of P2 but can accommodate either a trio of P7 or a monomer of P2 at each of the 20 3-fold sites. From a functional point of view, less-than-full stoichiometry for these proteins may not be detrimental. The key requirement is that both P2 and P7 are incorporated in sufficient numbers into the assembling procapsid.
The inference from this structural study, i.e., that P2 and P7 are mutually occluding, is consistent with biochemical data from in vitro procapsid assemblies with progressively increasing amounts of one or the other protein, which eventually showed mutual interference (35a).
Functional implications of mutual occlusion by competing proteins in capsid assembly.
Steric interference between adjacent binding sites has been documented as affecting the binding of antibodies to the outer surfaces of capsids, e.g., hepatitis B virus (HBV) capsids (35). Also, in principle, there is competition between the portal protein and the major capsid protein for occupancy of vertex sites in the procapsids of tailed bacteriophages and herpesvirus (19, 22). This appears to be managed, at least in part, by synthesizing much larger amounts of the latter molecule, and perhaps by kinetics (4). However, we are unaware of a precedent for two viral proteins competing to bind to the inner surface of a procapsid (or a capsid). Concerning its incidence in ϕ6, we note that the area available for binding proteins on the inner surface of the procapsid, with its convoluted, deeply indented structure, is quite limited and subdivides into 60 copies of a much smaller subarea. Consequently, given the need to package two distinct proteins—P2 and P7—mutual occlusion between their binding sites may have emerged as a geometrical inevitability for this system.
ACKNOWLEDGMENTS
We thank M. Poranen and D. Bamford for discussion and for sharing their data prior to publication and D. Winkler and N. Cheng for expert support with EM resources.
This work was supported by the Intramural Research Program of NIAMS and by grant GM34352 to L.M. from the National Institutes of Health.
Footnotes
Published ahead of print 15 August 2012
REFERENCES
- 1. Atherton BA, Cunningham EL, Splittgerber AG. 1996. A mathematical model for the description of the Coomassie brilliant blue protein assay. Anal. Biochem. 233:160–168 [DOI] [PubMed] [Google Scholar]
- 2. Bamford JK, Bamford DH, Li T, Thomas GJ., Jr 1993. Structural studies of the enveloped dsRNA bacteriophage phi 6 of Pseudomonas syringae by Raman spectroscopy. II. Nucleocapsid structure and thermostability of the virion, nucleocapsid and polymerase complex. J. Mol. Biol. 230:473–482 [DOI] [PubMed] [Google Scholar]
- 3. Butcher SJ, Dokland T, Ojala PM, Bamford DH, Fuller SD. 1997. Intermediates in the assembly pathway of the double-stranded RNA virus phi6. EMBO J. 16:4477–4487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cardone G, Heymann JB, Cheng N, Trus BL, Steven AC. 2012. Procapsid assembly, maturation, nuclear exit: dynamic steps in the production of infectious herpesvirions. Adv. Exp. Med. Biol. 726:423–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. de Haas F, Paatero AO, Mindich L, Bamford DH, Fuller SD. 1999. A symmetry mismatch at the site of RNA packaging in the polymerase complex of dsRNA bacteriophage phi6. J. Mol. Biol. 294:357–372 [DOI] [PubMed] [Google Scholar]
- 6. Eryilmaz E, et al. 2008. Structure and dynamics of the P7 protein from the bacteriophage phi 12. J. Mol. Biol. 382:402–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Fokine A, et al. 2005. Structural and functional similarities between the capsid proteins of bacteriophages T4 and HK97 point to a common ancestry. Proc. Natl. Acad. Sci. U. S. A. 102:7163–7168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Heymann JB, Belnap DM. 2007. Bsoft: image processing and molecular modeling for electron microscopy. J. Struct. Biol. 157:3–18 [DOI] [PubMed] [Google Scholar]
- 9. Huiskonen JT, et al. 2006. Structure of the bacteriophage phi6 nucleocapsid suggests a mechanism for sequential RNA packaging. Structure 14:1039–1048 [DOI] [PubMed] [Google Scholar]
- 10. Huiskonen JT, Jaalinoja HT, Briggs JA, Fuller SD, Butcher SJ. 2007. Structure of a hexameric RNA packaging motor in a viral polymerase complex. J. Struct. Biol. 158:156–164 [DOI] [PubMed] [Google Scholar]
- 11. Ikonen T, Kainov D, Timmins P, Serimaa R, Tuma R. 2003. Locating the minor components of double-stranded RNA bacteriophage phi 6 by neutron scattering. J. Appl. Crystallogr. 36:525–529 [Google Scholar]
- 12. Jaalinoja HT, Huiskonen JT, Butcher SJ. 2007. Electron cryomicroscopy comparison of the architectures of the enveloped bacteriophages phi6 and phi8. Structure 15:157–167 [DOI] [PubMed] [Google Scholar]
- 13. Juuti JT, Bamford DH. 1997. Protein P7 of phage phi6 RNA polymerase complex, acquiring of RNA packaging activity by in vitro assembly of the purified protein onto deficient particles. J. Mol. Biol. 266:891–900 [DOI] [PubMed] [Google Scholar]
- 14. Juuti JT, Bamford DH. 1995. RNA binding, packaging and polymerase activities of the different incomplete polymerase complex particles of dsRNA bacteriophage phi 6. J. Mol. Biol. 249:545–554 [DOI] [PubMed] [Google Scholar]
- 15. Mancini EJ, et al. 2004. Atomic snapshots of an RNA packaging motor reveal conformational changes linking ATP hydrolysis to RNA translocation. Cell 118:743–755 [DOI] [PubMed] [Google Scholar]
- 16. McDonald SM, Patton JT. 2011. Assortment and packaging of the segmented rotavirus genome. Trends Microbiol. 19:136–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Mindich L. 2012. Packaging in dsRNA viruses. Adv. Exp. Med. Biol. 726:601–608 [DOI] [PubMed] [Google Scholar]
- 18. Mindich L. 2004. Packaging, replication and recombination of the segmented genome of bacteriophage Phi6 and its relatives. Virus Res. 101:83–92 [DOI] [PubMed] [Google Scholar]
- 19. Moore SD, Prevelige PE., Jr 2002. Bacteriophage p22 portal vertex formation in vivo. J. Mol. Biol. 315:975–994 [DOI] [PubMed] [Google Scholar]
- 20. Nemecek D, et al. 2011. Stepwise expansion of the bacteriophage varphi6 procapsid: possible packaging intermediates. J. Mol. Biol. 414:260–271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Nemecek D, Heymann JB, Qiao J, Mindich L, Steven AC. 2010. Cryo-electron tomography of bacteriophage phi6 procapsids shows random occupancy of the binding sites for RNA polymerase and packaging NTPase. J. Struct. Biol. 171:389–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Newcomb WW, Thomsen DR, Homa FL, Brown JC. 2003. Assembly of the herpes simplex virus capsid: identification of soluble scaffold-portal complexes and their role in formation of portal-containing capsids. J. Virol. 77:9862–9871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Noda T, et al. 2012. Three-dimensional analysis of ribonucleoprotein complexes in influenza A virus. Nat. Commun. 3:639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Olia AS, Prevelige PE, Jr, Johnson JE, Cingolani G. 2011. Three-dimensional structure of a viral genome-delivery portal vertex. Nat. Struct. Mol. Biol. 18:597–603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Olkkonen VM, et al. 1990. In vitro assembly of infectious nucleocapsids of bacteriophage phi 6: formation of a recombinant double-stranded RNA virus. Proc. Natl. Acad. Sci. U. S. A. 87:9173–9177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Pettersen EF, et al. 2004. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25:1605–1612 [DOI] [PubMed] [Google Scholar]
- 27. Pintilie GD, Zhang J, Goddard TD, Chiu W, Gossard DC. 2010. Quantitative analysis of cryo-EM density map segmentation by watershed and scale-space filtering, and fitting of structures by alignment to regions. J. Struct. Biol. 170:427–438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Poranen MM, Bamford DH. 2012. Assembly of large icosahedral double-stranded RNA viruses. Adv. Exp. Med. Biol. 726:379–402 [DOI] [PubMed] [Google Scholar]
- 29. Poranen MM, Butcher SJ, Simonov VM, Laurinmaki P, Bamford DH. 2008. Roles of the minor capsid protein P7 in the assembly and replication of double-stranded RNA bacteriophage phi6. J. Mol. Biol. 383:529–538 [DOI] [PubMed] [Google Scholar]
- 30. Poranen MM, Paatero AO, Tuma R, Bamford DH. 2001. Self-assembly of a viral molecular machine from purified protein and RNA constituents. Mol. Cell 7:845–854 [DOI] [PubMed] [Google Scholar]
- 31. Poranen MM, Tuma R. 2004. Self-assembly of double-stranded RNA bacteriophages. Virus Res. 101:93–100 [DOI] [PubMed] [Google Scholar]
- 32. Poranen MM, Tuma R, Bamford DH. 2005. Assembly of double-stranded RNA bacteriophages. Adv. Virus Res. 64:15–43 [DOI] [PubMed] [Google Scholar]
- 33. Salgado PS, et al. 2004. The structural basis for RNA specificity and Ca2+ inhibition of an RNA-dependent RNA polymerase. Structure 12:307–316 [DOI] [PubMed] [Google Scholar]
- 34. Sen A, et al. 2008. Initial location of the RNA-dependent RNA polymerase in the bacteriophage Phi6 procapsid determined by cryo-electron microscopy. J. Biol. Chem. 283:12227–12231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Steven AC, et al. 2005. Structure, assembly, and antigenicity of hepatitis B virus capsid proteins. Adv. Virus Res. 64:127–165 [DOI] [PubMed] [Google Scholar]
- 35a. Sun X, Bamford DH, Poranen MM. Probing, by self-assembly, the number of potential binding sites for minor protein subunits in the procapsid of double-stranded RNA bacteriophage phi6. J. Virol., in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wei H, et al. 2009. Three-dimensional structure of the enveloped bacteriophage phi12: an incomplete T=13 lattice is superposed on an enclosed T=1 shell. PLoS One 4:e6850 doi:10.1371/journal.pone.0006850 [DOI] [PMC free article] [PubMed] [Google Scholar]