Abstract
The immune system targets virus-infected cells by different means. One of the essential antiviral mechanisms is apoptosis induced by ligation of tumor necrosis factor receptor 1 (TNFR1). This receptor can be activated by tumor necrosis factor alpha (TNF-α), which upon binding to TNFR1 induces the assembly of first an inflammatory and later a proapoptotic signaling complex. Here, we report that infection by human herpesvirus 6B (HHV-6B) inhibited poly(ADP-ribose) polymerase (PARP) cleavage, caspase 3 and 8 activation, and IκBα Ser-32 phosphorylation downstream of TNFR1, indicating inhibition of both the inflammatory and apoptotic signaling pathways. We identified a hitherto uncharacterized viral protein, U20, as sufficient for mediating this inhibition. U20 was shown to locate to the cell membrane, and overexpression inhibited PARP cleavage, caspase 3 and 8 activation, IκBα Ser-32 phosphorylation, and NF-κB transcriptional activity. Moreover, small interfering RNA (siRNA) knockdown of U20 demonstrated that the protein is necessary for HHV-6B-mediated inhibition of TNFR signaling during infection. These results suggest an important novel function of U20 as a viral immune evasion protein during HHV-6B infection.
INTRODUCTION
Human herpesvirus 6B (HHV-6B) is a ubiquitous human betaherpesvirus with a seroprevalence of >95% in the adult population in the Western world (28, 45). HHV-6B shares approximately 90% nucleotide similarity with HHV-6A but differs from it in biological and clinical characteristics (9, 15, 16, 35, 36). Primary infection with HHV-6B causes a mild febrile illness called exanthem subitum, which is characterized by a few days with high fever followed by the appearance of a rash (47). Primary infection is most often seen within the first 2 years of life and is normally an uncomplicated infection (28, 45). Following primary infection, HHV-6B, like other herpesviruses, establishes lifelong latency in the host (19, 25). The mechanism of latency is controversial, since it has been suggested to occur by integration in the telomeric repeat region rather than by establishing a permanent episome, as is known for other herpesviruses (2). In any case, latent HHV-6B may be reactivated later in life and can cause severe disease, particularly in immunocompromised individuals. In addition to the common lateral infection, HHV-6B may also be transmitted vertically by congenital infection. The majority of these congenital infections are the result of chromosomally integrated HHV-6B. The frequency of this chromosomal integration may be as high as 1% of all newborns (12).
HHV-6B has been shown to infect a large range of cell types both in vivo and in vitro, although not all infected cell types sustain a productive infection. Productive HHV-6B infection occurs primarily in CD4+ T cells but may also occur in CD8+ T cells and B cells, although only to a mild extent in the latter case (26). Little is known about the effects of HHV-6B in nonproductively infected cells, but cytoplasmic transport from cell to cell is possible, circumventing tissue barriers (3).
Both HHV-6A and -6B are neurotropic, and both viruses have been linked to diseases of the central nervous system, such as encephalitis (48). In addition, evidence exists for an association between HHV-6B and mesial temporal lobe epilepsy (10, 11). A number of studies have associated HHV-6A or -6B with different T- and B-cell lymphomas (20, 21), although evidence for a direct role of the viruses is complicated by their ubiquitous nature and ability to integrate chromosomally (14). Possibly, HHV-6A or -6B might act as a cofactor in certain oncogenic diseases by promoting replication of other viruses, thereby helping the double-infected cells to evade the immune system (8).
Upon viral infection, the host cell initiates a number of antiviral responses, the most definitive being programmed cell death. This program occurs through intracellular or extracellular induced pathways. Whereas intracellular pathways often originate from the mitochondria, the extracellular pathway is induced by cytokines. One of these is tumor necrosis factor alpha (TNF-α), which signals through TNF receptor 1 (TNFR1). The consequences of the interaction between TNF-α and TNFR1 are complex and may result in cell survival or cell death signaling, depending on the binding of different cytoplasmic adaptor proteins to TNFR1. Ligation of soluble trimerized TNF-α to the TNFR1 trimer recruits the TNFR-associated death domain protein (TRADD) to the death domain of TNFR1 (4, 27). TRADD functions as a scaffold to assemble a signaling complex called complex I (23, 43). This complex causes phosphorylation of IκBs, leading to NF-κB translocation to the nucleus and thereby transcription of a wide array of proinflammatory genes, as well as genes involved in both cell survival and cell death (24, 44). Approximately 60 min after TNFR1 ligation, a new signaling complex, known as complex II, is formed in the cytoplasm (27, 37). Complex II includes FADD, which recruits procaspase 8, which is then cleaved to form active caspase 8. This caspase activates the effector caspase 3, which leads to the onset of apoptosis (37).
Death receptors are likely to be targeted by viruses in order to prevent cell death and proinflammatory responses. As HHV-6B has evolved mechanisms to persist in host cells, we speculated whether HHV-6B infection was able to prevent signaling through death receptors. Here, we demonstrate that HHV-6B is able to prevent apoptosis by interfering with TNFR1 signaling. We identify and characterize an HHV-6B gene product, U20, which is sufficient to exert this effect. We show that U20 interferes with downstream signaling of TNFR1 after ligand stimulation. Finally, we demonstrate that U20 is necessary for HHV-6B-induced inhibition of TNFR1 signaling, suggesting an immune evasive function of the hitherto uncharacterized U20 during HHV-6B infection.
MATERIALS AND METHODS
Cell cultures and virus.
The human epithelial colon cancer cell line HCT116 was grown in McCoy's 5A medium, and the human acute T-lymphoblastic leukemia cell line MOLT3 was grown in Iscove's modified Dulbecco's medium (IMDM). All media were supplemented with 10% fetal calf serum (FCS), glutamine (0.2 g/liter), streptomycin (0.2 g/liter), penicillin (0.2 g/liter), and HEPES (10 mM). Cell-free HHV-6B, strain PL-1, was used for all experiments. The virus was propagated in MOLT3 cells, and the cells were freeze-thawed and centrifuged for 1 h at 300 × g. The supernatants were centrifuged for 4 h at 20,000 × g. The pellet was resuspended in medium, and the viral titer was determined by the 50% tissue culture infective dose (TCID50), as previously described (30, 42). For HHV-6B infection studies, a titer of 120 TCID50 was used.
Antibodies and reagents.
Apoptosis analysis by Western blotting (WB) was performed using the following antibodies: rabbit polyclonal IgG anti-poly(ADP-ribose) polymerase (PARP) (Cell Signaling Technology, Beverly MA), rabbit polyclonal IgG anti-caspase 3 active form (Abcam, Cambridge, United Kingdom), rabbit polyclonal IgG1 anti-caspase 8 (Cell Signaling Technology), and mouse monoclonal IgG2a anti-phosphor-IκBα (Santa Cruz Biotechnology, CA). Flow cytometry analysis of TNFR1 was performed using a mouse monoclonal IgG2a anti-TNFR1 antibody (Santa Cruz Biotechnonolgy). Scrambled and U20 small interfering RNAs (siRNAs) were On-Targedplus four-pool siRNAs (Dharmacon, Thermo Scientific, Denmark). The siRNAs were introduced into the cells using an Amaxa Nucleofector Device I with reagent V and program D-32 according to the manufacturers' instructions. Camptothecin (CPT), cycloheximide (CHX), and recombinant TNF-α were purchased from Sigma-Aldrich (Copenhagen, Denmark).
Plasmids and clones.
All PCRs for cloning were performed on cDNA generated from mRNA of HHV-6B-infected MOLT3 cells, as previously described (18). The U20 open reading frame (ORF) was amplified with the forward primer 5′-ATGATAACTGTTTTTGTCGC-3′ and the reverse primer 5′-TTACAAAGGCAACATTTCTA-3′. Generation of the pcDNA3.1/U20 plasmid was performed with Topo cloning (Life Technologies Europe BV, Naerum, Denmark), and generation of the pcDNA3/U20-FLAG plasmid was performed with HindIII and EcoRI cloning (Roche). The stable U20-expressing HCT116 cell line was generated by transfection with pcDNA3.1/U20, followed by Geneticin G418 selection for 2 weeks and subsequent screening of U20 mRNA by reverse transcription (RT)-PCR. Transfections used for confocal microscopy were performed using FuGene-6 according to the manufacturer's instructions. All other transfections were performed in suspension using the Amaxa Nucleofector Device I with nucleofector solution V (Lonza Group Ltd., Basel, Switzerland) and program D-32. The transfections were performed according to the manufacturer's instructions.
Western blotting.
Cells used for WB were lysed in a 1% Triton lysis buffer (LB) (Roche) supplemented with 1% phenylmethylsulfonyl fluoride (PMSF), 1% NaF, and the Complete-Mini protease inhibitor cocktail (Roche). Cell lysate fractionations were generated using the ProteoExtract Subcellular Proteome Extraction Kit according to the manufacturer's instructions (Merck KGaA, Damstadt, Germany). Quantification was performed using Image J software (1). Regions of interest (ROIs) of a fixed size were placed around each band in the gel and measured. Background was measured outside the stained area and subtracted from the measurements. The percentage of the band representing cleaved PARP was determined relative to the total amount in both bands.
Luciferase measurements.
HCT116 cells (wt) and U20-S cells were transfected with the WWP-Luciferase plasmid using the Amaxa transfection system. The cells were treated 48 h posttransfection (p.t.) with TNF-α for 3 h, and luciferase intensity was measured with the Luciferase 1000 assay system (Promega, Madison, WI) on an Ascent Luminoskan.
Confocal microscopy.
Cells used for confocal microscopy were grown on poly-l-lysine-coated 12-mm glass slides (0.17-mm thickness) for the indicated time periods. The cells were washed twice in phosphate-buffered saline (PBS) and fixed in 4% formalin. After fixation, the cells were washed twice in PBS, permeabilized in 0.2% Triton X-100, and blocked in 5% bovine serum albumin (BSA) in PBS. Antibodies were dissolved in PBS and incubated for 1 h at room temperature. All images were taken with a 63× oil immersion objective on a Zeiss LSM-710 confocal microscope using the 405-nm line of a purple diode laser and the 543-nm line of a green helium neon laser. Each experiment was performed with fixed laser power, gain, and offset to ensure comparability.
Flow cytometry.
Cells analyzed for surface expression of TNFR1 were washed in 2% FCS in PBS, resuspended in AccuMax (Millipore, Billerica, MA) for 5 min, washed in 2% FCS in PBS, stained with IgG2a isotype-specific (UPC-10; Sigma) or TNFR1-specific (550514; BD Pharmingen, Albertslund, Denmark) antibody for 30 min at 4°C, and washed in 2% FCS in PBS. The cells were stained with anti-mouse biotin-conjugated secondary antibody and washed in 2% FCS in PBS (B2763; Invitrogen), followed by staining with phycoerythrin (PE)-conjugated streptavidin for 20 min at room temperature. After staining, the cells were resuspended in 1% paraformaldehyde and analyzed immediately on a Beckman Coulter FC500 flow cytometer. Cells used for apoptosis analysis were washed in 2% FCS in PBS, resuspended in AccuMax for 5 min, washed in 2% FCS in PBS, resuspended in 20 μl 7-aminoactinomycin D (7-AAD), washed in 2% FCS in PBS, and analyzed on a Beckman Coulter FC500 flow cytometer.
RESULTS
HHV-6B infection inhibits TNFR1 signaling.
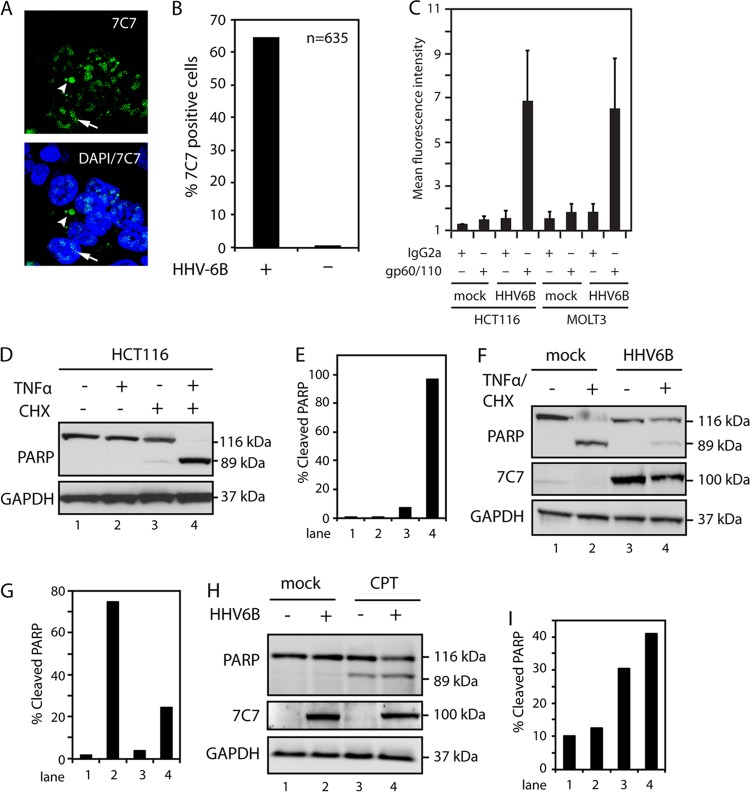
We have previously reported that the cell line HCT116 can be nonproductively infected by HHV-6B (31). HHV-6B-infected HCT116 cells (at 120 TCID50) were examined by confocal microscopy at 48 h postinfection (p.i.) using the 7C7 antibody, which recognizes an uncharacterized protein from HHV-6A and -6B. This gave rise to nuclear and cytoplasmic staining (Fig. 1A). Approximately two-thirds of the cells were infected, as measured by identification of 7C7-positive cells (n = 635), whereas no staining was visible in uninfected cells (Fig. 1B). To further address the level of infection in this cell system, HHV-6B-infected HCT116 cells were compared with infected MOLT3 cells by flow cytometry with an antibody against the viral glycoprotein gp60/110. HCT116 cells showed gp60/110 expression similar to that of MOLT3 cells, indicating that the number of infected HCT116 cells is similar to this well-established infection model (Fig. 1C).
Fig 1.
HHV-6B infection inhibits TNF-α-induced apoptosis. (A) Confocal microscopy of HCT116 cells infected with HHV-6B for 48 h and stained with 7C7, an HHV-6A/B-specific antibody. The arrows point to nuclear viral proteins, and the arrowheads point to cytoplasmic proteins. DAPI, 4′,6-diamidino-2-phenylindole. (B) Quantification of 7C7-positive cells from confocal microscopy of HCT116 cells mock treated or infected with HHV-6B for 48 h and stained with 7C7 antibody. (C) HCT116 and MOLT3 cells were infected with HHV-6B for 48 h and stained with isotype- or gp60/110-specific antibodies, followed by flow cytometry analyses. The graph represents an average of four independent experiments. The error bars indicate standard deviations (SD). (D) Western blot analyses with antibodies against PARP or GAPDH (glyceraldehyde-3-phosphate dehydrogenase) (loading control). HCT116 cells were treated with TNF-α (25 ng/ml), CHX (10 μM), or a combination of both for 4 h, followed by Western blot analyses with antibodies against PARP or GAPDH. (E) Quantifications of cleaved PARP from panel D. (F) Western blot analysis of lysates from mock-treated or HHV-6B-infected (48 h) HCT116 cells, followed by treatment with TNF-α and CHX for 4 h. The Western blots were probed with antibodies against PARP or GAPDH. (G) Quantifications of cleaved PARP from panel F. (H) Western blot analyses of lysates from mock-treated or HHV-6B-infected (48 h) HCT116 cells, followed by treatment with CPT for 18 h. The Western blots were probed with antibodies against PARP or GAPDH. (I) Quantifications of cleaved PARP from panel H. Representatives of at least two experiments are shown.
We have previously observed that HHV-6B-infected cells appeared to escape apoptosis (29). To determine whether HHV-6B is able to rescue cells from extrinsically induced apoptosis, the ability of HCT116 cells to undergo extrinsic apoptosis through TNFR1 was first examined. HCT116 cells were treated with TNF-α in combination with CHX. The addition of CHX blocks NF-κB transcriptional activity, allowing TNF-α/CHX to stimulate complex II formation, which leads to rapid apoptosis. Cell lysates were subsequently examined by WB analyses of PARP cleavage. This demonstrated that apoptosis can be induced rapidly in HCT116 cells by TNF-α/CHX treatment (Fig. 1D and E). Importantly, when the cells were infected with HHV-6B, a significant reduction in TNF-α/CHX-induced apoptosis 48 h p.i. was observed (Fig. 1F and G). To determine if the infection also inhibited intrinsically induced apoptosis, cells were treated with CPT, a topoisomerase I inhibitor known to induce intrinsic apoptosis. Treatment with CPT caused apoptosis in both wt cells and HHV-6B-infected cells (Fig. 1H and I). Together, these data suggested the possibility that HHV-6B inhibited TNF-α-induced apoptosis by interfering with the signal transduction from TNFR1.
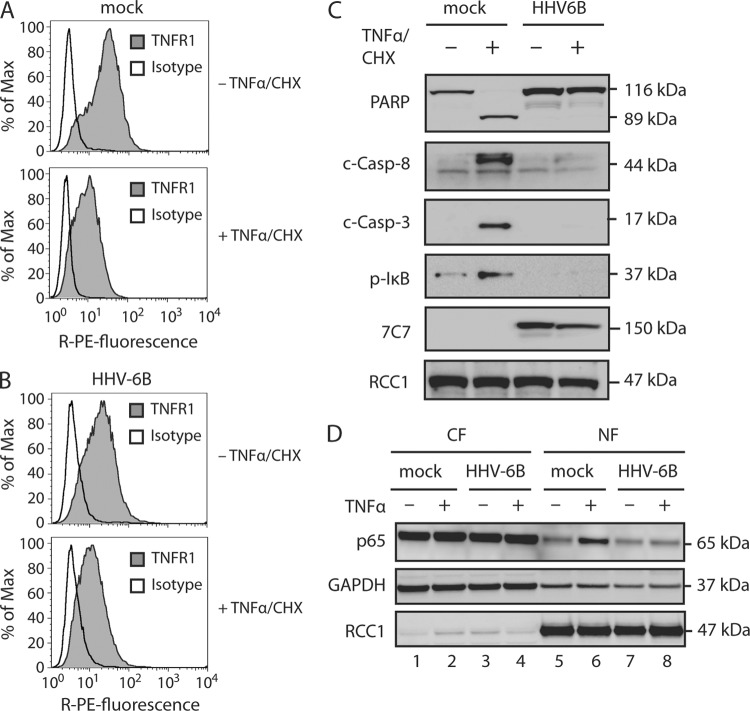
To rule out the possibility that the observed effects were due to downregulation or rapid internalization of TNFR1, its surface expression was examined in mock-treated or HHV-6B-infected cells 48 h p.i. Flow cytometry analyses demonstrated similar levels of surface-expressed TNFR1 when comparing mock-treated and HHV-6B-infected cells (Fig. 2A). This indicated that the lack of TNF-α response in HHV-6B-infected cells was not caused by HHV-6B-induced TNFR1 downregulation. When the cells were pretreated with TNF-α and CHX for 30 min prior to staining for TNFR1 expression, the amount of receptor was reduced in both wt and HHV-6B-infected (48 h p.i.) HCT116 cells (Fig. 2A and B). This indicated that TNF-α did bind TNFR1 in both uninfected and HHV-6B-infected cells. The decrease in TNFR1 staining could be explained either by internalization of the receptor or by TNF-α blockage, since the antibody and TNF-α compete for binding to the receptor.
Fig 2.
HHV-6B infection inhibits proinflammatory and apoptotic TNFR1 signaling pathways. (A to C) HCT116 cells were mock treated or HHV-6B infected (48 h) and then incubated in the presence or absence of TNF-α (25 ng/ml) and CHX (10 μM) for 4 h, followed by flow cytometry analyses with TNFR1- and isotype-specific antibodies (A and B) or Western blot analyses with antibodies against PARP, cleaved caspase 8 (c-Casp-8), cleaved caspase 3 (c-Casp-3), p-IκB, 7C7 (HHV-6B infection control), and RCC1 (loading control) (C). (D) HCT116 cells were mock treated or infected with HHV-6B (48 h) and then incubated in the presence or absence of TNF-α (25 ng/ml) for 30 min. Shown are Western blot analyses of cytoplasmic (CF) and nuclear (NF) fractions with antibodies against p65 or the loading controls GAPDH and RCC1. Representatives of at least two experiments are shown.
To further understand how HHV-6B infection inhibited the signaling cascade, we examined key proteins downstream of TNFR1. Signals mediated through complex II cause cleavage of procaspase 3 into active caspase 3, which is essential for cleavage of a large number of proteins destined for degradation during apoptosis. As expected, caspase 3 was cleaved in wt cells upon TNF-α/CHX treatment. Importantly, after this treatment, HHV-6B-infected cells had no detectable caspase 3 cleavage 24 h p.i. (Fig. 2C). The first caspase to be activated in this pathway is caspase 8, which upon activation cleaves caspase 3. When cells were treated with TNF-α/CHX, HHV-6B infection inhibited the cleavage of procaspase 8 to active caspase 8, but no inhibition was observed in mock-treated cells (Fig. 2C).
Signaling through the other TNFR1-induced pathway (complex I) leads to IκBα phosphorylation by IKKα, which results in NF-κB translocation to the nucleus and subsequent transcription of antiapoptotic genes. To examine whether signaling through complex I was also inhibited by HHV-6B infection, IκBα phosphorylation was analyzed following TNF-α/CHX treatment. In HHV-6B-infected cells, IκBα phosphorylation was no longer detectable, suggesting that HHV-6B infection inhibited signaling through complex I (Fig. 2C). Together, these results indicate that HHV-6B infection inhibits the TNFR1 signaling pathway.
To further analyze the activation of NF-κB following TNF-α treatment, wt and HHV-6B-infected HCT116 cells were treated with TNF-α for 30 min and fractionated into cytoplasmic and nuclear lysates. WB analysis for the presence of the NF-κB subunit p65 demonstrated that wt cells accumulated p65 in the nuclear fraction after TNF-α treatment (Fig. 2D, lane 6), whereas accumulation was significantly reduced in the HHV-6B-infected cells (Fig. 2D, lane 8).
Viral glycoprotein U20 is sufficient for inhibiting TNF-α-induced apoptosis.
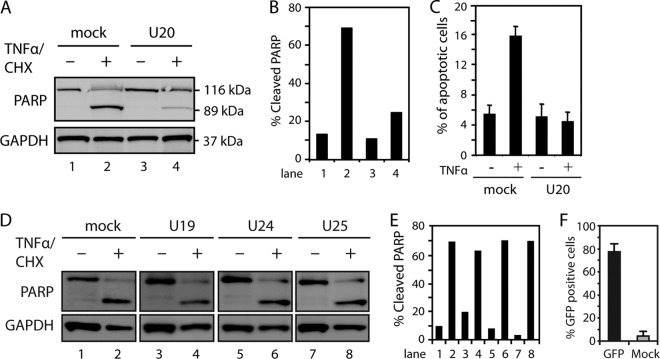
To address which HHV-6B proteins might be responsible for inhibiting TNFR1 signaling, we cloned four hypothetical antiapoptotic genes (U19, U20, U24, and U25) from a region in HHV-6B that is homologous to a region in human cytomegalovirus (HCMV) known to encode several antiapoptotic proteins (5, 40, 46). To analyze if either of these proteins could rescue cells from TNF-α-induced apoptosis, HCT116 cells were transfected with plasmids encoding U19, U20, U24, U25, or mock plasmid and treated with TNF-α/CHX for 4 h. Importantly, HCT116 cells transiently transfected with a U20-expressing plasmid showed reduced resistance to TNF-α-induced apoptosis, as measured by PARP cleavage (Fig. 3A and B). To corroborate these observations, transiently transfected U20-expressing HCT116 cells were examined by flow cytometry for the apoptosis marker 7-AAD. These data indicated a marked resistance to TNF-α-induced apoptosis in U20-expressing cells (Fig. 3C). In contrast, expression of U19, U24, and U25 did not inhibit TNF-α/CHX-induced PARP cleavage (Fig. 3D and E), although they all expressed mRNA upon transfection, as measured by PCR amplification (data not shown). To estimate the efficiency of transfection, four independent transfections with a green fluorescent protein (GFP) plasmid were performed. This indicated that the transfection efficiency was consistently around 75% (Fig. 3F).
Fig 3.
HHV-6B-encoded protein U20 inhibits TNF-α-induced apoptosis. (A) HCT116 cells were transfected with plasmid alone (mock) or a plasmid carrying U20 and left untreated or treated with TNF-α (25 ng/ml) and CHX (10 μM) for 4 h, followed by Western blot analysis with antibodies against PARP or GAPDH (loading control). (B) Quantification of cleaved PARP from panel A. (C) HCT116 cells were transfected with plasmid alone or a plasmid carrying U20 and treated with TNF-α and CHX for 4 h, followed by 7-AAD staining and flow cytometry analysis. The average percentages of apoptotic cells from three experiments are shown. The error bars indicate SD. (D) HCT116 cells were transfected with plasmid alone or a plasmid carrying U19, U24, or U25 and left untreated or treated with TNF-α and CHX for 4 h, followed by Western blot analysis with antibodies against PARP or GAPDH (loading control). (E) Quantification of cleaved PARP from panel D. Representatives of at least two experiments are shown. (F) HCT116 cells were transfected with plasmid alone or a plasmid encoding GFP for 24 h, followed by flow cytometry analyses. An average of four independent experiments is shown. The error bars indicate SD.
U20 is a 75-kDa membrane-spanning early protein.
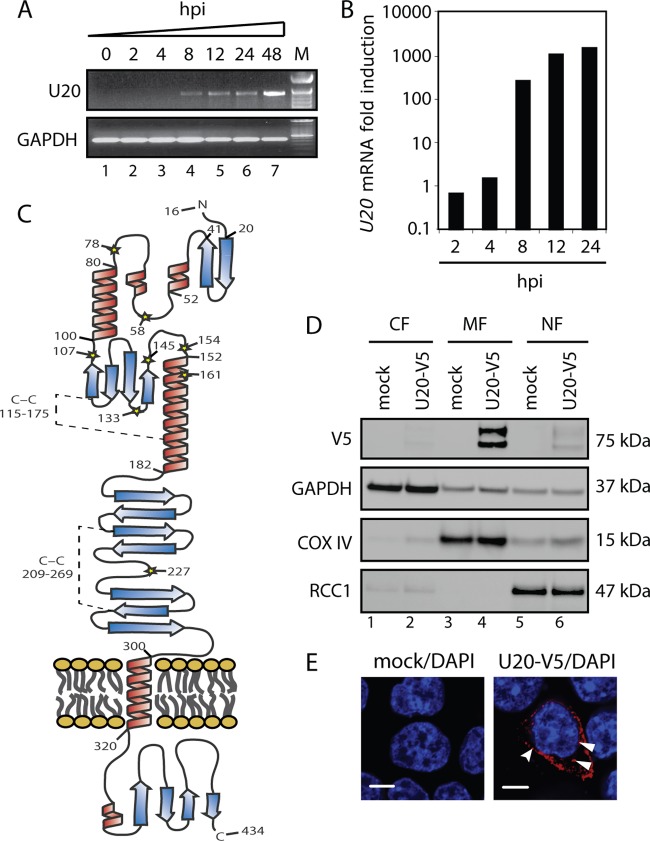
To determine the kinetics of U20 ORF transcription during productive HHV-6B infection, PCR using U20-specific primers amplifying the entire U20 ORF was performed on cDNA generated from HHV-6B-infected MOLT3 cells at various time points. Agarose gel electrophoresis of the PCR product yielded a single band of the expected size (Fig. 4A) that was verified to be U20 by sequencing. To determine whether U20 is also transcribed during a nonproductive infection, HCT116 cells were HHV-6B infected and examined at different time points postinfection. These analyses demonstrated that U20 was expressed in HCT116 cells with early expression kinetics (Fig. 4B). Thus, U20 was transcribed during both productive and nonproductive infections.
Fig 4.
U20 is a 75-kDa membrane-associated protein. (A) PCR with primers spanning the U20 ORF on cDNA from MOLT3 cells infected with HHV-6B for 0, 2, 4, 8, 12, 24, or 48 h. GAPDH served as a control. M, base pair marker. (B) Real-time PCR with U20 ORF-specific primers on cDNA from HCT116 cells infected with HHV-6B for 0, 2, 4, 8, 12, or 24 h. The amount of U20 was quantified relative to β-actin and is presented as fold induction relative the 0-h measurement. (C) Hypothetical model of U20. Shown are signal peptides (aa 1 to 15), the immunoglobulin-like domain (aa 182 to 288), and the transmembrane α-helix (aa 300 to 320). Predicted α-helices are shown in red, and predicted β-sheets are shown in blue. Predicted C-C bridges between Cys-115 and Cys-175 and between Cys-209 and Cys 269 are indicated by broken lines. (D) HCT116 cells were transfected with plasmid alone or a plasmid carrying N-terminally V5-tagged U20. Shown are Western blot analyses of cytoplasmic (CF), membrane (MF), and nuclear (NF) fractions with antibodies against V5 epitope or the loading controls GAPDH, COX IV, and RCC1. (E) HCT116 cells were transfected with plasmid alone or a plasmid carrying N-terminally V5-tagged U20 and subjected to confocal microscopy with an antibody against the V5 epitope (red) and DAPI (blue). Scale bars, 10 μm. The curved arrowhead points to U20 associated with the surface membrane, and the full arrowheads point to U20 associated with intracellular membranes.
The U20 ORF exclusively appears in HHV-6A, -6B, and -7 and has no known homologue in any of the other herpesviruses, including the betaherpesvirus HCMV. Studies on the protein product of the U20 ORF have not been reported for any of the three viruses. Bioinformatic analysis (CLC-Bio, Aarhus, Denmark) (6, 7, 32–34) predicted U20 to be a membrane protein with a single membrane-spanning α-helix (amino acids [aa] 319 to 340) and an N-terminal signal peptide (aa 1 to 15) for endoplasmic reticulum (ER) sorting to the plasma membrane (Fig. 4C). The prediction of several highly probable N-glycosylation sites on the N-terminal side of the membrane and none on the C-terminal side further suggested that U20 is transported to the membrane with an N-terminal extracellular part and a C-terminal intracellular part. Bioinformatic analysis also identified a very likely immunoglobulin chain C-like fold spanning aa 182 to 288 on the extracellular side and, furthermore, indicated a highly organized N-terminal part and a highly disordered C-terminal part.
To examine the cellular localization of U20, HCT116 cells were transfected with either mock plasmid or a plasmid expressing U20 with a C-terminal V5 epitope tag. Fractionation of the cell lysate identified U20-V5 as a double band in the membrane/organelle fraction, supporting the predicted localization of U20 as a membrane protein (Fig. 4D). Confocal microscopy using an anti-V5 antibody demonstrated staining of ER/Golgi apparatus-like structures 24 h p.t., as well as distinct staining of small clusters in close proximity to the outer cell membrane (Fig. 4E). Taken together, these data indicate that U20 is a glycoprotein expressed on both intracellular and extracellular membranes and oriented with a large N-terminal extracellular part and a small C-terminal intracellular part.
U20 is sufficient for inhibiting TNFR1 signaling.
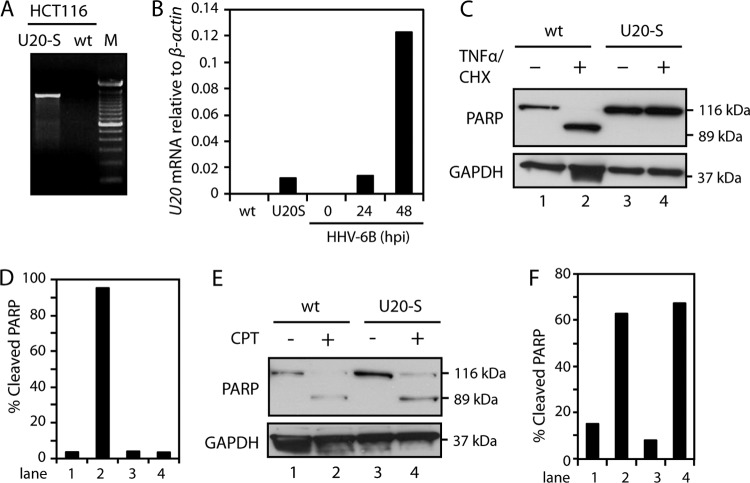
To extend the data obtained with transient expression, a stably U20-expressing HCT116 cell line was generated (termed U20-S cells). The stable clone expressed U20 mRNA (Fig. 5A) at a level comparable with HHV-6B infection at 24 h p.i. (Fig. 5B). When treated with TNF-α/CHX for 4 h followed by WB analysis of PARP cleavage, the U20-S clone was resistant to TNF-α-induced apoptosis (Fig. 5C and D). To determine if the U20-S cells were able to block intrinsic apoptosis, cells were treated with CPT for 24 h. Similar to HHV-6B-infected cells, the U20-expressing clone was sensitive to CPT-induced apoptosis (Fig. 5E and F).
Fig 5.
Stable expression of U20 inhibits TNF-α-induced apoptosis. (A) PCR with the U20 ORF spanning primers on cDNA from HCT116 wt or HCT116-U20S cells. M, base pair markers. (B) Real-time PCR with primers against U20 or β-actin on mRNA extracted from HCT116 wt, U20-S, and HHV-6B-infected HCT116 wt (0, 24, and 48 h p.i.) cells. (C) HCT116 wt and U20-S cells were left untreated or treated with TNF-α (25 ng/ml) and CHX (10 μM) for 4 h, followed by Western blotting with antibodies against PARP or GAPDH (loading control). (D) Quantification of cleaved PARP from panel C. (E) HCT116 wt and U20-S cells were left untreated or treated with CPT for 18 h, followed by Western blot analyses with antibodies against PARP or GAPDH (loading control). (F) Quantification of cleaved PARP from panel E.
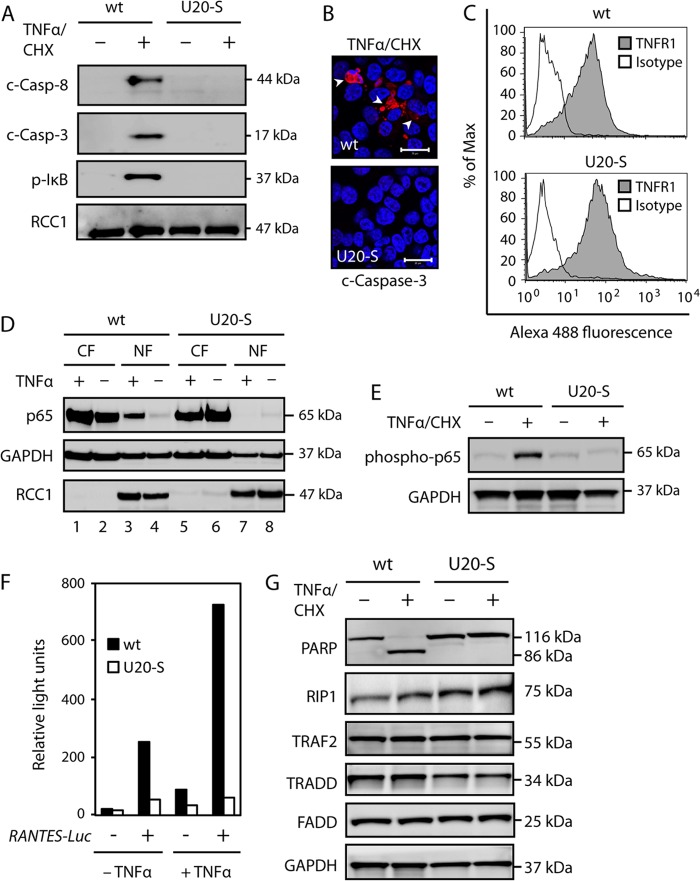
To address the level at which the TNFR1 signaling pathway was inhibited by U20, the presence of activated caspase 3 and 8, as well as phosphorylated IκB (p-IκB), was examined following TNF-α/CHX treatment. These analyses gave results similar to those obtained with HHV-6B-infected cells, that is, neither caspase 3 or 8 cleavage nor IκBα Ser-32 phosphorylation was observed in cells expressing U20 (Fig. 6A). Potential activation of caspase 3 was also examined by confocal microscopy using an antibody specific for cleaved caspase 3 (Fig. 6B). This further corroborated the finding that U20 prevented caspase 3 cleavage and activation. To ensure that the abrogation of TNF-α-induced signaling was not due to loss of TNFR1, wt and U20-S cells were analyzed by flow cytometry with antibodies against TNFR1. Wt and U20-S cells expressed similar levels of TNFR1 (Fig. 6C).
Fig 6.
U20 inhibits proinflammatory and apoptotic TNFR1 signaling pathways. (A) HCT116 wt and U20-S cells were incubated in the presence or absence of TNF-α (25 ng/ml) and CHX (10 μM) for 4 h, followed by Western blot analyses with antibodies against PARP, cleaved caspase 8 (c-Casp-8), cleaved caspase 3 (c-Casp-3), p-IκB, or RCC1 (loading control). (B) Confocal microscopy with antibodies against cleaved caspase 3 (red) and DAPI (blue). Scale bars, 20 μm. (C) HCT116 wt and U20-S cells were analyzed by flow cytometry with TNFR1- or isotype-specific antibodies. (D) HCT116 wt and U20-S cells were treated with TNF-α (25 ng/μl) for 30 min, followed by Western blot analyses of cytoplasmic (CF) and nuclear (NF) fractions with antibodies against p65 or the loading controls GAPDH and RCC1. (E) HCT116 wt and U20-S cells were incubated in the presence or absence of TNF-α (25 ng/ml) and CHX (10 μM) for 1 h, followed by Western blot analyses with antibodies against phosphorylated p65 and GAPDH (loading control). (F) HCT116 wt and U20-S cells were transfected with a RANTES-Luc reporter plasmid and treated with or without TNF-α (25 ng/ml) for 4 h, followed by luciferase measurement. (G) HCT116 wt and U20-S cells were incubated in the presence or absence of TNF-α and CHX for 4 h, followed by Western blot analyses with antibodies against PARP, RIP1, TRAF2, TRADD, FADD, or GAPDH (loading control). All data are representative of at least two experiments.
To determine if U20 also inhibited NF-κB activation, similar to HHV-6B infection, we treated HCT116 wt and U20-S cells with TNF-α for 30 min and fractionated the cells into cytoplasmic and nuclear lysates. As expected, the p65 subunit of NF-κB accumulated in the nuclear fraction after TNF-α treatment (Fig. 6D, lane 3). Similar to our observations with HHV-6B, U20-expressing cells did not accumulate p65 after treatment (Fig. 6D, lane 7). Furthermore, p65 was not phosphorylated in U20-S cells, as observed in wt cells after TNF-α/CHX treatment (Fig. 6E). To examine whether U20 downregulated NF-κB activity in the nucleus, a luciferase expression plasmid driven by an NF-κB-dependent promoter construct from RANTES was transfected into U20-S and wt cells. In this system, TNF-α induced Luciferase transcription in wt cells, indicating NF-κB signaling (Fig. 6F). This response was, however, virtually abrogated in U20-S cells in the presence or absence of TNF-α treatment (Fig. 6F). Thus, U20-S was unable to assemble a signaling platform to activate NF-κB. Taken together, these results demonstrated that U20 expression inhibited TNFR1 signaling, similar to the data observed in HHV-6B-infected cells.
A number of adaptor proteins are required for complex I formation and thus for TNFR1 signaling. To determine if the adaptors TRADD, RIP1, TRAF2, and FADD were downregulated in the U20-S cells, HCT116 wt and U20-S cells were examined by WB in the presence or absence of TNF-α/CHX treatment. This demonstrated that the expression levels of these adaptor proteins were similar in HCT116 wt and U20-S cells (Fig. 6G).
U20 is sufficient for inhibiting TNF-α-induced apoptosis during HHV-6B infection.
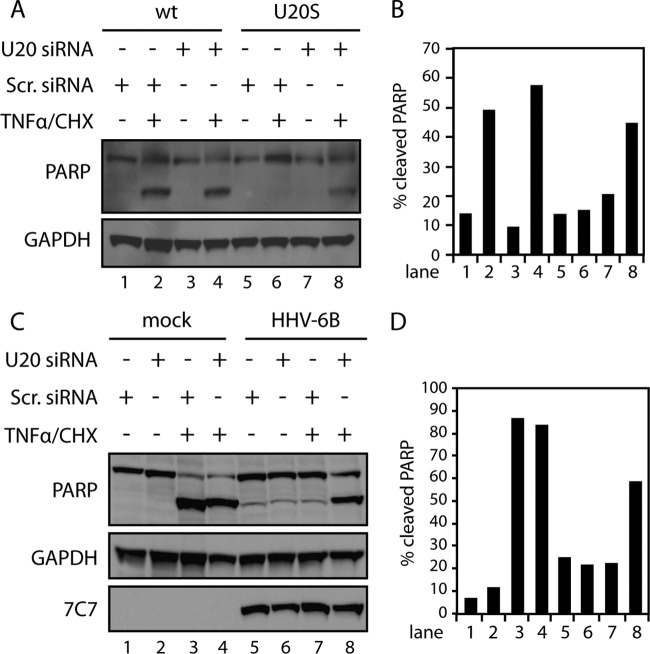
To determine if U20 was predominantly responsible for TNF-α inhibition during HHV-6B infection, we used siRNA to knock down U20. To ensure that the siRNA was able to inhibit U20, we first treated wt and U20-S cells with U20 siRNA or scrambled siRNA as a control for 48 h, followed by TNF-α/CHX treatment. The presence of siRNA against U20 reversed the resistance of U20-S cells and made them sensitive to TNF-α/CHX-induced PARP cleavage (Fig. 7A and B), indicating that the siRNA was able to knock down U20 to a level where its functions were abrogated. Next HCT116 cells were transfected with U20 siRNA or scrambled siRNA. After 24 h, the cells were infected with HHV-6B and incubated for an additional 48 h, followed by TNF-α/CHX treatment. Expression of U20 siRNA during infection clearly reduced the amount of cleaved PARP, indicating that U20 was the primary protein responsible for the inhibition of TNFR1 signaling (Fig. 7C and D).
Fig 7.
U20 reverses HHV-6B-induced resistance to apoptosis. (A) HCT116 wt and U20-S cells were transfected with U20 siRNA (200 nM) or scrambled (Scr.) siRNA (200 nM) for 48 h and incubated in the presence or absence of TNF-α (25 ng/ml) and CHX (10 μM) for 4 h, followed by Western blot analyses with antibodies against PARP or GAPDH (loading control). (B) Quantification of cleaved PARP from panel A. (C) HCT116 cells were transfected with U20 siRNA (200 nM) or scrambled siRNA (200 nM) for 24 h and mock treated or HHV-6B infected for 48 h and then incubated in the presence or absence of TNF-α and CHX for 4 h, followed by Western blot analyses with antibodies against PARP, GAPDH (loading control), or 7C7 (infection control). (D) Quantification of cleaved PARP from panel C. Representatives of at least two experiments are shown.
DISCUSSION
Signaling through death receptors is one of the major mechanisms used by the immune system to target and eliminate infected cells. Disruption of death receptor signaling is therefore important for viral immune evasion and continued progression of infection. Although the mechanisms for signaling through TNFR1 have been well described, it is still unclear exactly how persistent pathogens, like herpesviruses, can circumvent this signaling.
We showed that TNFR1-mediated apoptosis measured by PARP cleavage or 7-AAD exclusion was significantly inhibited by infection with HHV-6B. The infection blocked signaling through the proinflammatory pathway, as well as through the proapoptotic pathway. We identified a protein encoded by the U20 ORF that was necessary and sufficient for inhibiting TNFR1 downstream signaling. U20 mRNA appeared during HHV-6B infections with an early-like expression. This is in agreement with a recently published microarray and PCR study of the expression profiles of HHV-6B genes (41), which also identified U20 as an early gene. U20 has not previously been investigated, but bioinformatic predictions suggested it was a membrane-spanning protein. We were able to demonstrate this by confocal microscopy, as well as fractionation of cell lysate. Interestingly, we observed a U20 double band in the membrane fraction on WB. We do not know the reason for this but speculate that it may represent U20 in the outer cell membrane in a glycosylated form and U20 in the internal membrane in a less glycosylated or unglycosylated form. Whether U20 may have a natural ligand is an interesting possibility. It possesses an immunoglobulin-like domain, which may suggest the possibility of interaction with other proteins.
A possible mechanism for U20-mediated inhibition of TNFR1 signaling is interference due to physical interaction between the proteins. Alternatively, U20 may bind one or more adaptor proteins needed to form the TNFR1 signaling complexes. In favor of the latter hypothesis, the Epstein-Barr virus (EBV)-encoded membrane protein LMP-1 binds TRADD and assembles an alternative signaling complex (17). It is thus possible that HHV-6B has evolved a similar functional mechanism for TNFR1 inactivation and alternative signaling. In an attempt to address this possibility, we expressed U20 with a V5-tagged C terminus to be able to monitor its presence. However, we have not been able to coimmunoprecipitate either TNFR1 or any adaptor proteins together with U20-V5 (data not shown). This could be explained by interference of the V5 tag with the binding capacity of the U20 C terminus. Speaking against the possibility that U20 assembles an alternative signaling complex, we did not observe p65 translocation to the nucleus in U20-expressing cells, nor did we observe any increased transcription from the RANTES reporter construct.
TNF-α-induced signaling through TNFR1 plays a major role in inflammation and is therefore a significant factor in autoimmune disease (39). Infection with HHV-6B may impair proinflammatory responses through molecules like U20. Similar to other herpesviruses, HHV-6B persists in the host in a latent form throughout life. It is unknown to what extent viral proteins are expressed during latent HHV-6B infection, but a low level of U20 expression could lead to a reduced impact of cytokines, like TNF-α, in infected tissue. While this may reduce inflammation, it may also increase the risk of immune evasion of infected and transformed cells. The role of immunoevasins, like U20, might therefore be deleterious or advantageous, depending on the precise conditions. Besides its role during natural infection, the anti-TNFR1 function of U20 may be exploited as a possible therapeutic use.
HHV-6B has been suggested to participate as a cofactor together with EBV in the development of Hodgkin's and Burkitt's lymphomas, although a clear relationship remains controversial. Our data suggest a mechanism by which infection with HHV-6B might increase cell survival. Infection of B cells with EBV can lead to immortalization of the infected cell through LMP-1 and EBNA proteins 1 to 4 (13, 17, 22, 38, 49). LMP-1 is known to prevent death receptor signaling through binding of TRADD and assembly of complex I on LMP-1 in intracellular vesicles, thereby stimulating cell survival. In vivo coinfection of B cells with HHV-6B and EBV might therefore increase the survival of immortalized clones and increase the risk of developing lymphomas due to a cooperative effect of LMP-1 and U20.
In summary, we have identified a novel viral immune evasion strategy by HHV-6B. We have shown the HHV-6B protein U20 to be responsible for the ability of the viral infection to interfere with TNFR1 signaling. Our results demonstrate a mechanism by which a virus prevents programmed death induced through the extrinsic pathway. We suggest that HHV-6B infection through the activities of U20 may interfere with the proinflammatory response and the cellular defense against the virus.
ACKNOWLEDGMENTS
This study was supported by a Doctor Sofus Carl Emil Friis and Wife Olga Doris Friis Award; the Danish Medical Research Council; the Technology Transfer Office, Aarhus University, Aarhus, Denmark; and the AU Ideas Program, Aarhus University, Aarhus, Denmark.
We thank Paolo Lusso for the PL-1 strain of HHV-6B and Bert Vogelstein for the WWP-Luciferase plasmid.
Footnotes
Published ahead of print 15 August 2012
REFERENCES
- 1. Abramoff MD, Magalhaes PJ, Ram SJ. 2004. Image processing with ImageJ. Biophoton Int. 11:36–42 [Google Scholar]
- 2. Arbuckle JH, et al. 2010. The latent human herpesvirus-6A genome specifically integrates in telomeres of human chromosomes in vivo and in vitro. Proc. Natl. Acad. Sci. U. S. A. 107:5563–5568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cassiani-Ingoni R, et al. 2005. CD46 on glial cells can function as a receptor for viral glycoprotein-mediated cell-cell fusion. Glia 52:252–258 [DOI] [PubMed] [Google Scholar]
- 4. Chan FK, et al. 2000. A domain in TNF receptors that mediates ligand-independent receptor assembly and signaling. Science 288:2351–2354 [DOI] [PubMed] [Google Scholar]
- 5. Chen Z, Knutson E, Wang S, Martinez LA, Albrecht T. 2007. Stabilization of p53 in human cytomegalovirus-initiated cells is associated with sequestration of HDM2 and decreased p53 ubiquitination. J. Biol. Chem. 282:29284–29295 [DOI] [PubMed] [Google Scholar]
- 6. Cole C, Barber JD, Barton GJ. 2008. The JPRED 3 secondary structure prediction server. Nucleic Acids Res. 36:W197–W201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Combet C, Blanchet C, Geourjon C, Deléage G. 2000. NPS@: network protein sequence analysis. Trends Biochem. Sci. 25:147–150 [DOI] [PubMed] [Google Scholar]
- 8. Cuomo L, et al. 1998. Upregulation of Epstein-Barr virus-encoded latent membrane protein by human herpesvirus 6 superinfection of EBV-carrying Burkitt lymphoma cells. J. Med. Virol. 55:219–226 [PubMed] [Google Scholar]
- 9. Dominguez G, et al. 1999. Human herpesvirus 6B genome sequence: coding content and comparison with human herpesvirus 6A. J. Virol. 73:8040–8052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Fotheringham J, et al. 2007. Association of human herpesvirus-6B with mesial temporal lobe epilepsy. PLoS Med. 4:e180 doi:10.1371/journal.pmed.0040180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Fotheringham J, Williams EL, Akhyani N, Jacobson S. 2008. Human herpesvirus 6 (HHV-6) induces dysregulation of glutamate uptake and transporter expression in astrocytes. J. Neuroimmune Pharmacol. 3:105–116 [DOI] [PubMed] [Google Scholar]
- 12. Hall CB, et al. 2008. Chromosomal integration of human herpesvirus 6 is the major mode of congenital human herpesvirus 6 infection. Pediatrics 122:513–520 [DOI] [PubMed] [Google Scholar]
- 13. Hong M, et al. 2006. Suppression of Epstein-Barr nuclear antigen 1 (EBNA1) by RNA interference inhibits proliferation of EBV-positive Burkitt's lymphoma cells. J. Cancer Res. Clin. Oncol. 132:1–8 [DOI] [PubMed] [Google Scholar]
- 14. Hubacek P, et al. 2009. Prevalence of HHV-6 integrated chromosomally among children treated for acute lymphoblastic or myeloid leukemia in the Czech Republic. J. Med. Virol. 81:258–263 [DOI] [PubMed] [Google Scholar]
- 15. Isegawa Y, et al. 1999. Comparison of the complete DNA sequences of human herpesvirus 6 variants A and B. J. Virol. 73:8053–8063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Josephs SF, et al. 1986. Genomic analysis of the human B-lymphotropic virus (HBLV). Science 234:601–603 [DOI] [PubMed] [Google Scholar]
- 17. Kieser A. 2008. Pursuing different ‘TRADDes’: TRADD signaling induced by TNF-receptor 1 and the Epstein-Barr virus oncoprotein LMP1. Biol. Chem. 389:1261–1271 [DOI] [PubMed] [Google Scholar]
- 18. Kofod-Olsen E, Ross-Hansen K, Mikkelsen JG, Hollsberg P. 2008. Human herpesvirus 6B U19 protein is a PML-regulated transcriptional activator that localizes to nuclear foci in a PML-independent manner. J. Gen. Virol. 89:106–116 [DOI] [PubMed] [Google Scholar]
- 19. Kondo K, Kondo T, Okuno T, Takahashi M, Yamanishi K. 1991. Latent human herpesvirus 6 infection of human monocytes/macrophages. J. Gen. Virol. 72:1401–1408 [DOI] [PubMed] [Google Scholar]
- 20. Krueger GR, Huetter ML, Rojo J, Romero M, Cruz-Ortiz H. 2001. Human herpesviruses HHV-4 (EBV) and HHV-6 in Hodgkin's and Kikuchi's diseases and their relation to proliferation and apoptosis. Anticancer Res. 21:2155–2161 [PubMed] [Google Scholar]
- 21. Lacroix A, et al. 2007. HHV-6 and EBV DNA quantitation in lymph nodes of 86 patients with Hodgkin's lymphoma. J. Med. Virol. 79:1349–1356 [DOI] [PubMed] [Google Scholar]
- 22. Lee JM, et al. 2004. EBNA2 is required for protection of latently Epstein-Barr virus-infected B cells against specific apoptotic stimuli. J. Virol. 78:12694–12697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Legler DF, Micheau O, Doucey MA, Tschopp J, Bron C. 2003. Recruitment of TNF receptor 1 to lipid rafts is essential for TNFalpha-mediated NF-kappaB activation. Immunity 18:655–664 [DOI] [PubMed] [Google Scholar]
- 24. Li H, Kobayashi M, Blonska M, You Y, Lin X. 2006. Ubiquitination of RIP is required for tumor necrosis factor alpha-induced NF-kappaB activation. J. Biol. Chem. 281:13636–13643 [DOI] [PubMed] [Google Scholar]
- 25. Luppi M, et al. 1999. Human herpesvirus 6 latently infects early bone marrow progenitors in vivo. J. Virol. 73:754–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lusso P, et al. 1988. In vitro cellular tropism of human B-lymphotropic virus (human herpesvirus-6). J. Exp. Med. 167:1659–1670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Micheau O, Tschopp J. 2003. Induction of TNF receptor I-mediated apoptosis via two sequential signaling complexes. Cell 114:181–190 [DOI] [PubMed] [Google Scholar]
- 28. Okuno T, et al. 1989. Seroepidemiology of human herpesvirus 6 infection in normal children and adults. J. Clin. Microbiol. 27:651–653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Oster B, Bundgaard B, Hollsberg P. 2005. Human herpesvirus 6B induces cell cycle arrest concomitant with p53 phosphorylation and accumulation in T cells. J. Virol. 79:1961–1965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Oster B, Hollsberg P. 2002. Viral gene expression patterns in human herpesvirus 6B-infected T cells. J. Virol. 76:7578–7586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Oster B, Kofod-Olsen E, Bundgaard B, Hollsberg P. 2008. Restriction of human herpesvirus 6B replication by p53. J. Gen. Virol. 89:1106–1113 [DOI] [PubMed] [Google Scholar]
- 32. Petersen B, Petersen TN, Andersen P, Nielsen M, Lundegaard C. 2009. A generic method for assignment of reliability scores applied to solvent accessibility predictions. BMC Struct. Biol. 9:51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Petersen TN, Brunak S, Heijne GV, Nielsen H. 2011. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods 8:785–786 [DOI] [PubMed] [Google Scholar]
- 34. Pollastri G, McLysaght A. 2005. Porter: a new, accurate server for protein secondary structure prediction. Bioinformatics 21:1719–1720 [DOI] [PubMed] [Google Scholar]
- 35. Salahuddin SZ, et al. 1986. Isolation of a new virus, HBLV, in patients with lymphoproliferative disorders. Science 234:596–601 [DOI] [PubMed] [Google Scholar]
- 36. Schirmer EC, Wyatt LS, Yamanishi K, Rodriguez WJ, Frenkel N. 1991. Differentiation between two distinct classes of viruses now classified as human herpesvirus 6. Proc. Natl. Acad. Sci. U. S. A. 88:5922–5926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Schneider-Brachert W, et al. 2004. Compartmentalization of TNF receptor 1 signaling: internalized TNF receptosomes as death signaling vesicles. Immunity 21:415–428 [DOI] [PubMed] [Google Scholar]
- 38. Silins SL, Sculley TB. 1995. Burkitt's lymphoma cells are resistant to programmed cell death in the presence of the Epstein-Barr virus latent antigen EBNA-4. Int. J. Cancer 60:65–72 [DOI] [PubMed] [Google Scholar]
- 39. Silva LC, Ortigosa LC, Benard G. 2010. Anti-TNF-alpha agents in the treatment of immune-mediated inflammatory diseases: mechanisms of action and pitfalls. Immunotherapy 2:817–833 [DOI] [PubMed] [Google Scholar]
- 40. Skaletskaya A, et al. 2001. A cytomegalovirus-encoded inhibitor of apoptosis that suppresses caspase-8 activation. Proc. Natl. Acad. Sci. U. S. A. 98:7829–7834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Tsao EH, et al. 2009. Microarray-based determination of the lytic cascade of human herpesvirus 6B. J. Gen. Virol. 90:2581–2591 [DOI] [PubMed] [Google Scholar]
- 42. Turcanova VL, Bundgaard B, Hollsberg P. 2009. Human herpesvirus-6B induces expression of the human endogenous retrovirus K18-encoded superantigen. J. Clin. Virol. 46:15–19 [DOI] [PubMed] [Google Scholar]
- 43. Wajant H, Scheurich P. 2001. Tumor necrosis factor receptor-associated factor (TRAF) 2 and its role in TNF signaling. Int. J. Biochem. Cell Biol. 33:19–32 [DOI] [PubMed] [Google Scholar]
- 44. Wang CY, Mayo MW, Korneluk RG, Goeddel DV, Baldwin AS., Jr 1998. NF-kappaB antiapoptosis: induction of TRAF1 and TRAF2 and c-IAP1 and c-IAP2 to suppress caspase-8 activation. Science 281:1680–1683 [DOI] [PubMed] [Google Scholar]
- 45. Ward KN, Gray JJ, Fotheringham MW, Sheldon MJ. 1993. IgG antibodies to human herpesvirus-6 in young children: changes in avidity of antibody correlate with time after infection. J. Med. Virol. 39:131–138 [DOI] [PubMed] [Google Scholar]
- 46. Xuan B, Qian Z, Torigoi E, Yu D. 2009. Human cytomegalovirus protein pUL38 induces ATF4 expression, inhibits persistent JNK phosphorylation, and suppresses endoplasmic reticulum stress-induced cell death. J. Virol. 83:3463–3474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Yamanishi K, et al. 1988. Identification of human herpesvirus-6 as a causal agent for exanthem subitum. Lancet 331:1065–1067 [DOI] [PubMed] [Google Scholar]
- 48. Yao K, et al. 2009. Detection of human herpesvirus-6 in cerebrospinal fluid of patients with encephalitis. Ann. Neurol. 65:257–267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Yi F, et al. 2009. Epstein-Barr virus nuclear antigen 3C targets p53 and modulates its transcriptional and apoptotic activities. Virology 388:236–247 [DOI] [PMC free article] [PubMed] [Google Scholar]