Abstract
MgtC is a virulence factor of unknown function important for survival inside macrophages in several intracellular bacterial pathogens, including Mycobacterium tuberculosis. It is also involved in adaptation to Mg2+ deprivation, but previous work suggested that MgtC is not a Mg2+ transporter. In this study, we demonstrated that the amount of the M. tuberculosis MgtC protein is not significantly increased by Mg2+ deprivation. Members of the MgtC protein family share a conserved membrane N-terminal domain and a more divergent cytoplasmic C-terminal domain. To get insights into MgtC functional and structural organization, we have determined the nuclear magnetic resonance (NMR) structure of the C-terminal domain of M. tuberculosis MgtC. This structure is not affected by the Mg2+ concentration, indicating that it does not bind Mg2+. The structure of the C-terminal domain forms a βαββαβ fold found in small molecule binding domains called ACT domains. However, the M. tuberculosis MgtC ACT domain differs from canonical ACT domains because it appears to lack the ability to dimerize and to bind small molecules. We have shown, using a bacterial two-hybrid system, that the M. tuberculosis MgtC protein can dimerize and that the C-terminal domain somehow facilitates this dimerization. Taken together, these results indicate that M. tuberculosis MgtC does not have an intrinsic function related to Mg2+ uptake or binding but could act as a regulatory factor based on protein-protein interaction that could be facilitated by its ACT domain.
INTRODUCTION
MgtC is a virulence factor common to several intracellular pathogens (2). MgtC was first described for Salmonella enterica serovar Typhimurium, where it is required for intramacrophage multiplication and long-term systemic infection in mice (3, 23, 36). Later, it was described as a critical factor for the intramacrophage growth of Mycobacterium tuberculosis, Brucella suis, Yersinia pestis, Burkholderia cenocepacia, and Salmonella enterica serovar Typhi (5, 12, 22, 24, 31).
Despite its importance in virulence of various bacterial pathogens, the biochemical function of MgtC, which is a membrane-associated protein, remains unknown, and members of the MgtC family show only low homology to proteins of known function. MgtC has been involved in adaptation to low-Mg2+ environments in liquid culture media in the various pathogens mentioned above: the growth of mgtC mutants is impaired when the Mg2+ concentration is low (2). The role of MgtC in adaptation to Mg2+ limitation has suggested that MgtC plays a role in Mg2+ uptake. Moreover, a role for MgtC in Mg2+ homeostasis is supported by the fact that expression of the mgtC gene is highly induced by deprivation of the external Mg2+ concentration in S. Typhimurium and Y. pestis (10, 41). Studies carried out with the Salmonella protein demonstrated that MgtC is not a Mg2+ transporter (15, 26) and led to alternative hypothesis. It was proposed that MgtC might act by sequestering Mg2+ and/or by facilitating transport by specific transporters. An indirect role in Mg2+ transport is strengthened by the fact that the mgtC gene is adjacent to mgtB, a gene encoding a P-type ATPase Mg2+ transporter, in several bacterial species, such as S. Typhimurium, Y. pestis, and B. suis (4, 35). In addition, heterologous expression of Salmonella MgtC in xenopus oocytes can activate the host Na+K+-P-ATPase (15). To reconcile all these data, we have proposed that MgtC might promote the activity of P-type ATPases, such as Mg2+ transporters or other cation transporters (2).
Alignment of the superfamily of MgtC proteins, as well as the hydrophobicity pattern, clearly defines two domains: a hydrophobic N-terminal part of the protein (of approximately 130 amino acids), and a soluble C-terminal part of the protein (of approximately 100 amino acids) (4). Surprisingly, whereas the N-terminal domain is well conserved in sequence, the C-terminal domain is highly variable within the MgtC superfamily (4). However, the sequence conservation of the C-terminal domain is stronger in a subfamily that includes MgtC proteins from intracellular pathogens (4). This particular evolution among MgtC proteins and the rapid divergence of the C-terminal domain may account for either a functional adaptation (to various environmental conditions) or a structural instability allowing unrestrained sequence evolution. To determine whether the C-terminal domain is folded and functional, we engaged in the biophysical characterization of the C-terminal region of the MgtC protein from M. tuberculosis. Phylogenetic analysis of MgtC proteins shows that the M. tuberculosis MgtC protein actually clustered in the subgroup of proteins that includes the S. Typhimurium protein as well as other proteins from pathogens that share the ability to survive in macrophages (4), being a good representative of this subfamily. First, we have examined the expression of the M. tuberculosis MgtC protein in the presence or absence of Mg2+ to gain insights into MgtC regulation in M. tuberculosis and provide a comparison with the regulation of other intracellular pathogens. We have solved the solution structure of the M. tuberculosis MgtC C-terminal domain using multidimensional nuclear magnetic resonance (NMR) spectroscopy. Our structural analyses show that the C-terminal domain of M. tuberculosis MgtC consists of a monomeric ACT domain that does not bind Mg2+. These results suggest a more subtle regulatory role of the C-terminal domain in the functioning of the whole MgtC protein.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Strains used for cloning and expression of recombinant protein were Escherichia coli DH5α and M15/pRep4 (Qiagen). E. coli strains were grown in LB medium supplemented with suitable antibiotics (ampicillin, 100 μg/ml; kanamycin, 25 μg/ml) at 37°C. The M. tuberculosis mc27000 strain (32) was routinely grown in Sauton's medium containing 0.025% tyloxapol (Sigma), supplemented with 24 μg/ml pantothenate. To monitor the effect of Mg2+ on MgtC expression, two flasks of 10 ml of M. tuberculosis culture were grown until the optical density at 600 nm (OD600) equaled 0.2. Bacteria were centrifuged for 5 min at 4,000 rpm and resuspended in 10 ml modified Sauton medium supplemented with 150 μM Mg2+ or without added Mg2+. After 48 h, 5 ml of bacteria were treated for RNA extraction and 5 ml were treated for bacterial lysate.
RT-PCR and q-RT-PCR.
RNA was prepared from 5 ml of mid-logarithmic-phase bacterial cultures. Bacteria were harvested, resuspended in 1 ml of RNA protect reagent (Qiagen), and incubated for 1 h at room temperature. Bacteria were centrifuged and resuspended in 1 ml of RLT buffer from the RNeasy kit (Qiagen), transferred in a lysing matrix B tube (MP Bio), and disrupted with a bead beater apparatus (3 times, 45 s, maximal speed). RNA was purified with the RNeasy kit according to the manufacturer's instructions. DNA was further removed using DNase I (Invitrogen). RNA integrity was analyzed on a 2100 bioanalyzer; Agilent). cDNA was produced using Superscript III reverse transcriptase (Invitrogen). Controls without reverse transcriptase were done for each RNA sample to rule out DNA contamination. Reverse transcription-PCR (RT-PCR) was carried out with the GoldStar mix (Eurogentec), and denaturation was followed by 24 (sigA), 25 (rv1811), or 26 (rv1806) cycles of 95°C for 30 s, 50°C for 30 s, and 72°C for 30 s. Quantitative real-time PCR (q-RT-PCR) was performed using an in-house SYBR green mix and a 480 LightCycler instrument (Roche). PCR conditions were as follows: 3 min of denaturation at 98°C, and 45 cycles of 98°C for 5 s, 70°C for 10 s, and 72°C for 10 s. The sigA gene was used as an internal control.
Preparation of bacterial lysates and Western blot analysis.
Five milliliters of bacterial culture was harvested, resuspended in 500 μl of phosphate-buffered saline (PBS), and disrupted using a bead beater (Retsch). The protein concentration was determined on total lysates using the bicinchoninic acid (BCA) protein assay reagent kit (Pierce). Expression of MgtC was investigated by Western blotting following migration of the crude extracts. Bacterial lysates equivalent to 40 μg of total proteins were separated by electrophoresis on a 15% SDS-PAGE gel, transferred to a membrane, probed with rabbit anti-MgtC antibodies (1:500 dilution), and subsequently incubated with antirabbit antibodies conjugated to alkaline phosphatase (1:7,000 dilution; Promega). An unrelated membrane protein, OmpATb, was used as a control and probed with rabbit anti-OmpATb antibodies (1:6,000 dilution) (37).
Purification of MgtC and production of polyclonal antiserum.
The region encoding the C-terminal domain of M. tuberculosis MgtC (amino acids 141 to 234; MgtC_Cter) was cloned into the expression vector pQE30 (Qiagen) to produce a recombinant protein with an N-terminal His tag. A PCR fragment was amplified using the Cter-Mtb-Bam-Fn and Cter-Mtb-Hind-Rn primers on H37Rv chromosomal DNA and cloned at the BamHI and HindIII sites of pQE30. The resulting plasmid, pQE30-MgtC-Cter, was transformed into the M15/pRep4 strain for protein production.
E coli M15/pREp4/pQE30-MgtC-Cter was grown to an OD600 of 0.6. Isopropyl-β-d-thiogalactopyranoside (IPTG) was added to a final concentration of 1 mM, and growth was continued at 37°C for 4 h. Cells were resuspended in phosphate buffer (20 mM NaPO4 [pH 7.4], 200 mM NaCl, and 2 mM dithiothreitol [DTT]) with 100 mM imidazole. After sonication, the supernatant was loaded onto Ni2+-nitrilotriacetic acid (NTA) agarose beads. After washing, the His-tagged MgtC_Cter protein was eluted with phosphate buffer containing 300 mM imidazole and then dialyzed in phosphate buffer. All steps were carried out at 4°C.
To produce a polyclonal antiserum against M. tuberculosis MgtC, two rabbits were immunized subcutaneously with 130 μl of recombinant MgtC (625 μg/ml in phosphate-buffered saline) in Freund's incomplete adjuvant (1:1 [vol/vol]). Similar booster injections were given at 14 and 28 days. Test bleeds were performed 7 days after each immunization, and the sera were checked for reactivity with M. tuberculosis MgtC. Animals were bled out, and sera were prepared 21 days after the last booster immunization.
NMR analysis.
All NMR experiments were carried out at 27°C on a Bruker Avance III 700 spectrometer equipped with a 5-mm z-gradient TCI CryoProbe instrument, using the standard pulse sequences (33). NMR samples consisted of approximately 0.5 mM 15N- or 15N,13C-labeled protein dissolved in 10 mM acetate buffer, 50 mM NaCl, pH 4.6, with 5% D2O for the lock. 1H chemical shifts were directly referenced to the methyl resonance of DSS (4,4-dimethyl-4-silapentane-1-sulfonic acid), while 13C and 15N chemical shifts were referenced indirectly to the absolute 15N/1H or 13C/1H frequency ratios. All NMR spectra were processed and analyzed with the software program Gifa (29).
Nuclear Overhauser effect (NOE) cross peaks identified by three-dimensional (3D) [1H,15N] and [1H,13C] NOE spectroscopy (NOESY)-heteronuclear single quantum coherence (HSQC) were converted into interproton distances: 2.4 Å, 2.8 Å, 3.6 Å,4.4 Å and 5.0 Å, corresponding to very strong, strong, medium, weak and very weak intensities, respectively. Backbone (Φ and Ψ) torsion angle constraints were obtained from a database search procedure on the basis of backbone (15N, HN, 13C′, 13Cα, Hα, and 13Cβ) chemical shifts using the software program TALOS (9). Hydrogen bond restraints were derived using standard criteria on the basis of the amide 1H/2H exchange experiments and NOE data. When identified, the hydrogen bond was enforced using the following restraints: ranges of 1.8 to 2.3 Å for d(N-H,O), and 2.7 to 3.3 Å for d(N,O). The final list of restraints (from which values redundant with the covalent geometry have been eliminated) for 200 structures was calculated using the software program CYANA-2.1 (14). The 30 best structures (based on the final target penalty function values) were minimized using CNS 1.2 software according to the RECOORD procedure (27). The final 15 structures of lowest energies were analyzed using the software program PROCHECK (21). Root mean square (RMS) deviations were calculated using the software program MOLMOL (19). All statistics are given in Table 1.
Table 1.
NMR structure parameters
| Parameter | Value |
|---|---|
| NMR distance and dihedral constraints | |
| Distance constraints | |
| Total NOE | 1,188 |
| Intraresidue | 240 |
| Interresidue | |
| Sequential (¦i − j¦ = 1) | 358 |
| Medium-range (¦1 < i − j¦ < 5) | 210 |
| Long-range (¦i − j¦ ≥ 5) | 380 |
| Hydrogen bonds | 0 |
| Total dihedral angle restraints | |
| ϕ | 69 |
| ψ | 66 |
| X1 | 17 |
| Structure statistics | |
| Violations (mean and SD) | |
| Max.a distance constraint violation (Å) | 0.3 |
| Max. dihedral angle violation (°) | 5.0 |
| Deviations from idealized geometry | |
| Bond lengths (Å) | 0.0117 ± 0.0001 |
| Bond angles (°) | 1.2167 ± 0.0072 |
| Impropers (°) | 1.3773 ± 0.0262 |
| Ramachandran plot (%) | |
| Most-favored region | 87.1 |
| Additionally allowed region | 12.1 |
| Generously allowed region | 0.8 |
| Disallowed region | 0.0 |
| Avg pairwise RMS deviationb (Å) (residues 143–229) | |
| Backbone | 0.84 ± 0.11 |
| Heavy | 1.56 ± 0.13 |
Max., maximal.
Pairwise RMS deviation was calculated among 15 refined structures.
Bacterial two-hybrid analysis.
The bacterial adenylate cyclase two-hybrid (BACTH) system (17) was used to evaluate M. tuberculosis MgtC dimerization. The mgtC (rv1811) gene was PCR amplified using H37Rv chromosomal DNA as the template and the primers MgtC-Mtb-Hind-F and MgtC-Mtb-Eco-R. The PCR fragment was cloned at the HindIII and EcoRI sites of the pUT18 vector to produce a fusion protein, MgtC-T18, and at those of the pKNT25 vector to produce the fusion protein MgtC-T25. Deletion of the N-terminal (amino acids 1 to 140) or C-terminal (amino acids 141 to 234) part was carried out by PCR amplification with primers flanking the region to be deleted (Table 2). Amplified fragments were phosphorylated and self-ligated. The two membrane proteins MalF and MalG were used as controls.
Table 2.
Primers used in this study
| Primer name | 5′-to-3′ sequence |
|---|---|
| Cter Mtb-Bam-Fn | CGGGATCCGTCGAAGACGAAGGGCTG |
| Cter Mtb-Hind-Rn | CCCAAGCTTTCATTCGGCCTGCGCGTGC |
| Rv1806-RT-F | GTCCAGATGCGCTGGCC |
| Rv1806-RT-R | CGCTGGCGCCCAACATC |
| Rv1811-RT2-Fn | CGTCCGCGGTCTGAACAC |
| Rv1811-RT3-R | GCACGTACATAGGTCTCTGC |
| SigA-Ln | CCTACGCTACGTGGTGGATTC |
| SigA-Rn | CTGGATTTCCAGCACCTTCTC |
| MgtC-Mtb-Hind-F | CCCAAGCTTGATGCAAACGCTGACTGTCGC |
| MgtC-Mtb-Eco-R | GGAATTCGATTCGGCCTGCGCGTGCTCAC |
| DelMgtCTb-Nter-R | CAAGCTTGGCGTAAT |
| DelMgtCTb-Nter-F | GTCGAAGACGAAGGG |
| DelMgtCTb-Cter-R | GGCGTTGTCGCGGTC |
| DelMgtCTb-Cter-F | GCCGAATCGAATTCA |
| MalG 18-Hind-F | CCCAAGCTTGATGGCTATGGTCCAACC |
| MalG 18-Bam-R | CGGGATCCGAACCTTTCACACCCCCTG |
| MalF 25N-Hind-F | CCCAAGCTTGATGGATGTCATTAAAAAG |
| MalF 25N-Bam-R | CGGGATCCGAATCAAACTTCATACGCGT |
Recombinant plasmids derived from pUT18 and pKNT25 genes were cotransformed into BTH101 bacteria. Transformants were plated on LB Amp Kan 5-bromo-4-chloro-3-indolyl-β-D-galactopyranoside (X-Gal) (50 μg/ml) medium at 30°C for 30 h. To quantify the interaction between hybrid proteins, bacteria were grown overnight at 30°C in NCE Amp Kan liquid medium supplemented with 0.1% Casamino Acids, 38 mM glycerol, 0.5 mM IPTG, and 10 μM or 10 mM Mg2+ (30). The assays were carried out as described previously (25), with activities expressed in arbitrary Miller units. Values are averages from at least four independent cultures. A level of β-galactosidase activity at least 5-fold higher than that measured for vectors indicates an interaction (18).
RESULTS
The levels of M. tuberculosis MgtC are similar under low- and high-Mg2+ growth conditions.
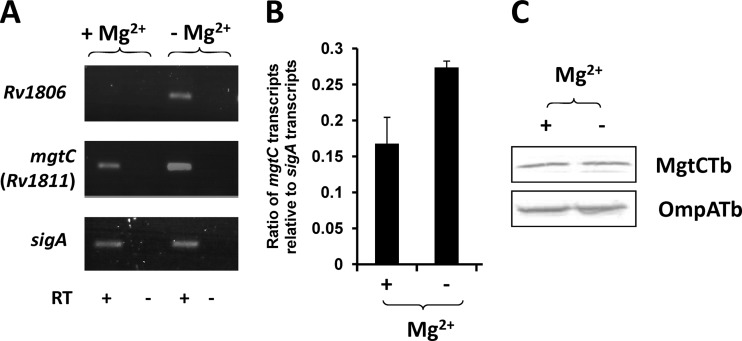
In M. tuberculosis, MgtC has been involved in the adaptation to the low-Mg2+ environment, because the growth of an mgtC mutant is impaired in liquid medium deprived of Mg2+ (5). However, the mgtC gene (rv1811) has not been considered to be significantly induced by Mg2+ deprivation in a microarray analysis, whereas genes immediately upstream of M. tuberculosis mgtC (rv1806 through rv1810) were found to be clearly upregulated following Mg2+ deprivation (38). To reinvestigate the effect of Mg2+ on M. tuberculosis mgtC expression and extend the study to expression of the M. tuberculosis MgtC protein, M. tuberculosis mc27000 was grown in Sauton medium with or without Mg2+. RNA extraction and bacterial lysates were assayed to monitor both mgtC gene and MgtC protein expression. RT-PCR indicated that, as previously reported, rv1806 was highly induced by Mg2+ deprivation (Fig. 1A) while the M. tuberculosis mgtC gene appears to be transcribed both with and without Mg2+, with a higher rate in low Mg2+. The control gene, sigA, was similarly transcribed under both conditions. Quantitative RT-PCR (Fig. 1B) confirmed that M. tuberculosis mgtC is slightly induced under low-Mg2+ conditions (1.6-fold), which is consistent with previous microarray data (38). Despite this moderate difference in the mRNA level, we failed to detect any difference in the amount of M. tuberculosis MgtC from cultures grown with or without Mg2+ as shown by Western blot analysis of crude bacterial extract using specific anti-MgtC antibodies (Fig. 1C). Taken together, the results indicate that Mg2+ does not significantly affect MgtC expression in M. tuberculosis.
Fig 1.
Expression of M. tuberculosis mgtC transcripts and the M. tuberculosis MgtC protein under high-Mg2+ and low-Mg2+ conditions. (A) RT-PCR experiment with RNA isolated from M. tuberculosis grown in high or low Mg2+. Reverse transcriptase was omitted in two lanes (as indicated by “-”) to verify the absence of genomic DNA contamination in the RNA sample. (B) Quantification of M. tuberculosis mgtC RNA by a q-RT-PCR experiment using RNA isolated from M. tuberculosis grown in high or low Mg2+. The sigma factor sigA was used as an internal standard. Results are expressed as means of the mgtC/sigA ratio ± standard deviations from three independent experiments. (C) Bacterial lysates equivalent to 40 μg of total proteins were separated by 15% SDS-PAGE. For the detection of M. tuberculosis MgtC, the membrane was incubated with rabbit anti-M. tuberculosis MgtC antibodies at a 1:500 dilution, washed, and probed with anti-rabbit antibodies conjugated to alkaline phosphatase used at a 1:7,000 dilution. The loading control was performed by probing the same amount of fractions with rabbit anti-OmpATb73–326 antibodies at a 1:6,000 dilution.
The C-terminal domain of M. tuberculosis MgtC is an ACT domain.
On the one hand, the hydrophobic N-terminal part of MgtC is well conserved in MgtC proteins from intracellular pathogens (about 56% identity and 73% similarity) and is also well conserved in proteins distantly related to MgtC (here called MgtC-like), which are not true orthologs based on complementation studies (such as the E. coli YhiD protein or Y. pestis YPO2332; about 40% identity and 62% similarity) (4, 30). On the other hand, the C-terminal domain sequence is much more variable within the MgtC proteins from intracellular pathogens (down to 20% identity and 43% similarity) and is not conserved in distantly related MgtC-like proteins. To investigate whether this loosely conserved domain is properly folded and stable, we engaged in the characterization of the C-terminal domain of mycobacterial MgtC by means of NMR.
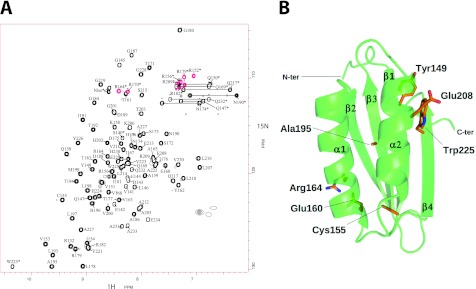
The soluble C-terminal region of the M. tuberculosis MgtC protein (93 amino acids) has been overexpressed in E. coli and then purified as a monomeric protein according to analytical gel filtration analysis. The assigned [1H,15N] HSQC spectrum of MgtC_Cter is shown in Fig. 2A. By combining the information from the double- and triple-resonance heteronuclear experiments, we were able to assign 100% of the amide group resonances for the nonproline residues (5 prolines), 98% of the other backbone resonances (Cα, C′, and Hα), 94% of the Cβ resonances, and 99% of the side chain protons. The chemical shift table was deposited in the BMRB databank (BMRB18316). No intermolecular NOE were detected suggesting that the protein is monomeric even at a relative high concentration (0.5 mM).
Fig 2.
NMR structure of M. tuberculosis MgtC_Cter. (A) [1H-15N] HSQC spectrum of MgtC_Cter from M. tuberculosis recorded at 27°C on an 15N-uniformly labeled sample. Cross-peak assignments are indicated using the one-letter amino acid and number. Red cross peaks corresponds to the “refolded” arginine side chains from the region at 80 to 90 ppm. Stars indicate cross peaks from side-chain atoms. (B) The overall three-dimensional structure of M. tuberculosis MgtC_Cter is shown as a green ribbon. Secondary structures are numbered following the nomenclature of ACT domains. The side chains of the seven residues strictly conserved among orthologous MgtC from intracellular pathogens (see alignment in Fig. 3A) are shown in wire frame representation (with carbon, nitrogen, oxygen, and sulfur atoms colored in orange, blue, red, and yellow, respectively). Residues are numbered following the mycobacterial sequence.
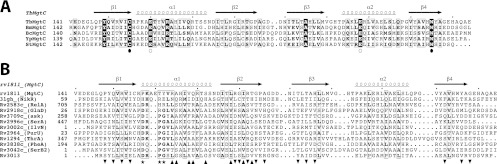
CYANA calculations performed for the C-terminal domain of MgtC revealed a βαββαβ motif with two α-helices on one side of a four-strand antiparallel β-sheet (Fig. 2B). The surface is mostly occupied by polar residues, and no particular patch (either charged or hydrophobic) is detected. The alignment of five known functional MgtC orthologs revealed only seven strictly conserved residues scattered along the whole sequence of the soluble C-terminal domain (Fig. 3A). Four conserved residues (corresponding to positions C155, E160, E208, and W225 in M. tuberculosis MgtC) were mutated during a previous analysis carried out with the S. Typhimurium MgtC protein, and two of them led to a loss of function when converted to alanine (Fig. 3A) (30). Despite the low level of amino acid conservation, sequence-structure comparison suggests a very good structural conservation among all five MgtC orthologs. A large hydrophobic core is conserved, including two strictly conserved residues (C155 and A195 in M. tuberculosis MgtC) that appear fully buried in the structure (Fig. 2B). In addition, two other conserved residues of opposite charge, E160 and R164, form a salt bridge at the surface of the first α-helix. With the buried but nearby cysteine (C155), these two polar residues form a conserved patch on one side of the protein (Fig. 2B). On the opposite side of the protein, a small cluster of strictly conserved residues is formed by Y149, E208, and W225. These residues point toward the solvent at the surface of the helix α2 and the strand β4. They might form a small binding site for an unknown ligand or partner. However, the lack of a sufficiently large and well-conserved patch paved with well-conserved residues argues against the tight recognition of a common and small partner. Furthermore, previous site-directed mutagenesis carried out with the S. Typhimurium MgtC protein indicated that a mutation of the conserved tryptophan (corresponding to W225 in M. tuberculosis MgtC) to alanine abolished the function whereas a conservative mutation to phenylalanine had no effect on the function (30). This result supports a structural role of the conserved tryptophan rather than a role in ligand interaction.
Fig 3.
Sequence alignment of MgtC proteins and ACT domains. (A) Sequence alignment of C-terminal domains of MgtC from pathogens which survive inside macrophages (M. tuberculosis, B. melitensis, B. cenocepacia, Y. pestis, and S. Typhimurium). Sequences were aligned manually, and the figure was drawn using the software program ESPript (11). Secondary structures were computed using the NMR structure of M. tuberculosis MgtC-Cter (this study). Four conserved residues that have been mutated to alanine during a previous analysis carried out with the S. Typhimurium MgtC protein (30) are indicated by circles. Black dots indicate residues that led to a loss of function when converted to alanine. (B) Sequence alignment of M. tuberculosis MgtC and the ACT domains identified in M. tuberculosis, as well as NikR from Helicobacter pylori. Mycobacterial proteins are identified by their gene names according to Tuberculist (http://genolist.pasteur.fr/TubercuList/). Sequences were aligned using the software program ViTO (6). We took advantage of the NMR structure of Rv1811 and the crystal structures available for Rv2996c (PDB 3DDN). For the other ACT domains, we modeled the 3D structure using close homologs detected with the server @TOME-2 (28) to help the refinement of our sequence-structure alignment. The figure was drawn using the program ESPript (11). The stars indicate the best-conserved residues that belong to the amino acid binding site of ACT domains. The positions buried by the dimerization of the ACT domain of NikR or of SerA (shown by up and down triangles, respectively) are highlighted in the alignment.
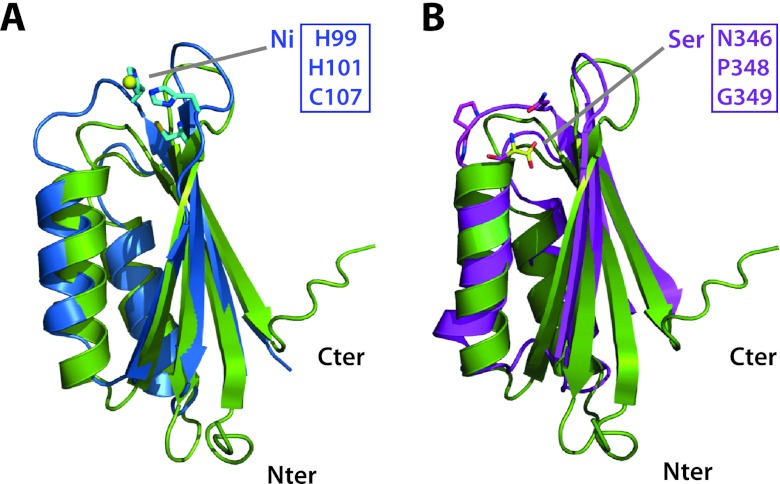
To gain new insight into the possible structure-function relationship, the new structure was compared to already-known structures using the DALI (16) and FATCAT (40) servers. The M. tuberculosis MgtC_Cter structure showed significant similarities to ACT (aspartokinase, chorismate mutase, and TyrA) domains from various enzymes (e.g., SerA; PDB 1PSD) (Fig. 4B), as well as that from the nickel-binding regulator NikR (PDB 3LGH) (Fig. 4A). ACT domains are small regulatory domains commonly involved in specifically binding amino acids or other small ligands (13). M. tuberculosis MgtC_Cter is also distantly related to other small modules adopting the same topology (βαββαβ), such as RRM (RNA recognition motif) and HMA (heavy metal-associated), which are involved in nucleic acid and metal binding, respectively (8). However, MgtC_Cter is more closely related in sequence and structure to the ACT domains than the RRM or HMA domains, since RRM and HMA are not found in the first hits by sequence-structure or structure-structure comparisons. Furthermore, MgtC lacks the motifs involved in RNA or metal binding that are found in RRM and HMA domains. Within the ACT family, the structure-structure comparison suggested a slightly better similarity to the ACT domain associated with ppGpp metabolism, since the closest structural homologue (PDB 2KO1; root mean square deviation [RMSD] = 1.9 Å) is the ACT domain from GTP pyrophosphokinase of Chlorobium tepidum (sequence identity, 18% for 77 residues). This protein is also detected as the most closely related using fold recognition (HHsearch E value and P value of 0.29 and 1.2E−05, respectively) and possesses a mycobacterial ortholog (Rv2583c). Other ACT domains are also detected with marginally weaker similarities, such as the small regulatory domains of the acetolactate synthase and the homoserine dehydrogenase (mycobacterial orthologs: Rv3002c and Rv1294, respectively) (Fig. 3B). Taken together, these results clearly classify the C terminus of M. tuberculosis MgtC as an ACT domain.
Fig 4.
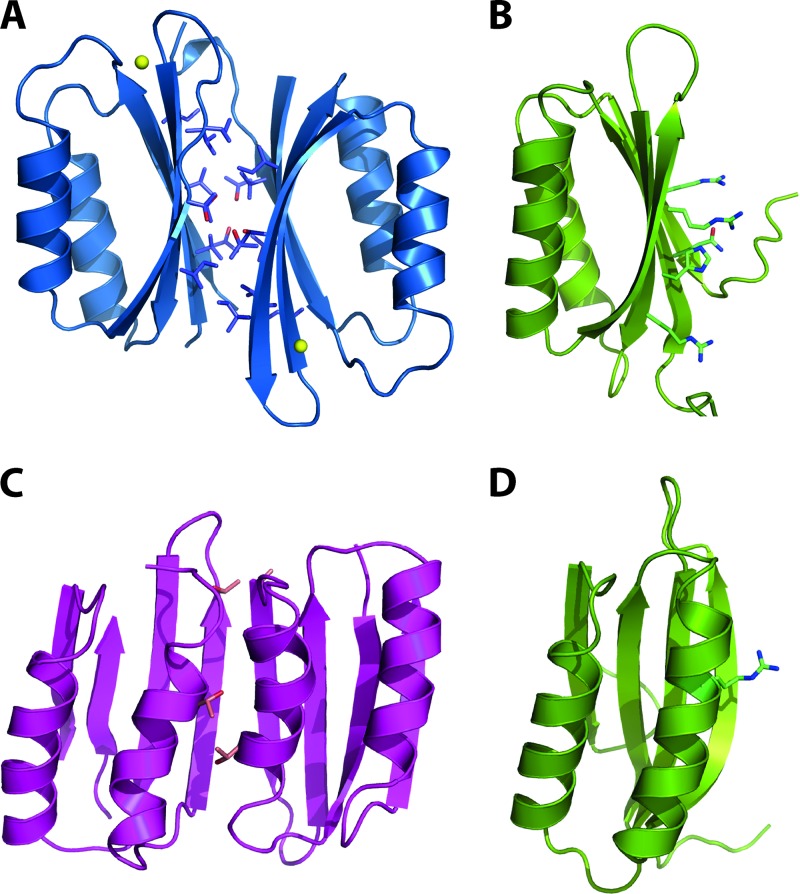
Superposition M. tuberculosis MgtC-Cter on other ACT domains. (A) Superposition of the ACT domains of M. tuberculosis MgtC (PDB 2LQJ; in green ribbon) and of nickel-bound NikR from Helicobacter pylori (PDB 3LGH; blue ribbon). The side chains involved in metal binding are shown as sticks. (B) Superposition of the ACT domains of M. tuberculosis MgtC (in green ribbon) and of serine-bound SerA from Escherichia coli (PDB 1PSD; pink ribbon). Three residues building up the amino acid binding site and the bound serine are shown as sticks. These residues are conserved in the amino acid binding site of ACT domains but are absent in members of the MgtC family and NikR (see Fig. 3). Two additional residues (H344 and N364; not as well conserved in M. tuberculosis MgtC) contacting the serine ligand through hydrogen-bound are not shown for clarity. The figure was prepared using the program Pymol (http://www.pymol.org).
The ACT domain of the MgtC protein diverges from the classical small molecule binding ACT domains.
ACT domains have been mainly involved in the binding of amino acids and in the regulation of amino acid metabolism (13). The knowledge of the M. tuberculosis MgtC_Cter high-resolution structure allows perfect sequence-structure alignment with representatives of the ACT domains solved in complex with an amino acid (as illustrated by SerA in Fig. 4B) (34). The ACT domain of M. tuberculosis MgtC appeared highly divergent from other ACT modules in the region involved in amino acid binding at the turn between the first β-strand and the first α-helix (residues 155 to 160 in M. tuberculosis MgtC) (13). This can be illustrated by comparing MgtC_Cter with canonical ACT domains present in other mycobacterial proteins (Fig. 3B). First, a two-residue insertion is present in the conserved motif in the loop connecting the first β-strand and the first α-helix, and second, and a tyrosine (Y162) is found instead of a conserved glycine residue known to be critical for amino acid binding (13). In addition, the C-terminus segment involved in the recognition of the amino acid of canonical ACT domains is apparently not conserved in Rv1811. Hence, the amino acid binding pocket in the M. tuberculosis MgtC ACT domain is likely abolished or totally reshaped by several substitutions and a two-residue insertion (Fig. 3B).
A few ACT domains have been proposed to bind other small ligands, such as thiamine (YkoF ACT domain) and cations (NikR ACT domain) (13). Again, the histidine and cysteine residues involved in nickel chelation in NikR are absent in MgtC_Cter (Fig. 3B and 4A) (39). Due to the phenotypic connections of MgtC proteins with Mg2+ uptake, magnesium was a suitable candidate as a small binding molecule for the MgtC_Cter ACT domain, and its binding was tested by NMR. No chemical shift was observed upon Mg2+ addition (up to 20 mM) in MgtC_Cter (50 μM) or after EDTA treatment (5 mM), suggesting that MgtC_Cter does not bind Mg2+ and that the purified protein had no trace of metal cation attached to it (data not shown).
In conclusion, analysis at the sequence and structure levels argues for the presence of a folded ACT domain in M. tuberculosis MgtC that lacks an amino acid or small solute binding site.
Role of C-terminal domain in dimerization of M. tuberculosis MgtC.
The structural comparison of the M. tuberculosis MgtC ACT domain with the classical ACT domains also revealed a striking difference in dimerization ability. Indeed, the dimerization observed in most ACT domains, including NikR and SerA (Fig. 5A and C) is likely to be prevented for the M. tuberculosis MgtC ACT domain by several detrimental substitutions at the potential monomer-monomer interfaces (Fig. 5B and D). The positions buried by the dimerization of the ACT domain of NikR or of SerA are highlighted in the structural alignment (Fig. 3B). The small and often hydrophobic residues (mainly Gly, Thr, Val, Ile, and Leu) are replaced by large hydrophilic residues (e.g., Arg, His, and Gln) in M. tuberculosis MgtC. Furthermore, these putative interfaces are lined with residues that are highly variable in nature among the orthologous sequences of MgtC (Fig. 3A). This sequence-structure analysis is in line with the apparent monomeric nature of M. tuberculosis MgtC_Cter in solution as described above.
Fig 5.
Quaternary structure organization of ACT domains. (A) Dimeric structure of the ACT domain of NikR from Helicobacter pylori (blue ribbon) as deduced from its crystal structure (PDB 3LGH). Side chains participating in the dimerization are shown as sticks. (B) Solution structure of M. tuberculosis MgtC shown in the same orientation as the left monomer of NikR in panel A. Large side chains hindering the dimerization of MgtC are shown as sticks. (C) Dimeric structure of the ACT domains of SerA from Escherichia coli (pink ribbon) as deduced from the crystal structure (PDB 1PSD). The side chains involved in amino acid binding are shown as sticks. Note that mainly glycine residues are involved in the contacting helices (see alignment in Fig. 3B). (D) Solution structure of M. tuberculosis MgtC, shown in the same orientation as the left monomer of SerA in panel C. Residues hindering the dimerization of MgtC are shown as sticks. The figure was prepared using Pymol (http://www.pymol.org).
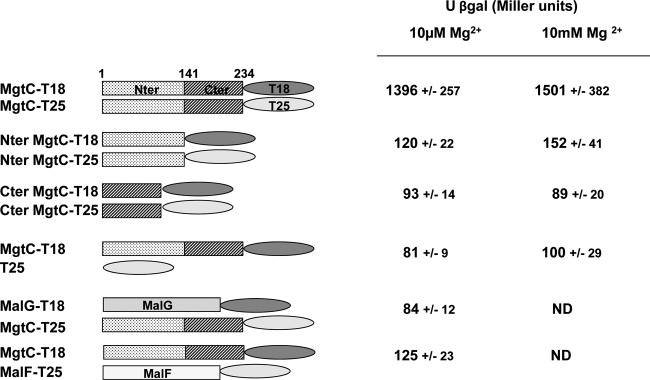
To further consider a potential role of MgtC_Cter in protein-protein interaction, we investigated whether this domain could be involved in a potential dimerization of the entire protein. To address this point, we used the bacterial two-hybrid (BACTH) system that has been validated for detecting interactions between inner-membrane proteins in living bacteria (18). We took advantage of our structural study to derive proper domain boundaries delimiting the transmembrane and cytoplasmic domains. Derivatives of pUT18 and pKNT25 were constructed to fuse the T18 or T25 fragment of adenylate cyclase to M. tuberculosis MgtC. Deletions were carried out on T18 and T25 derivatives to maintain only the N-terminal or C-terminal domain of the MgtC protein (see Materials and Methods) (Fig. 6). Plasmids encoding the T18 and T25 fusion proteins were introduced in an E. coli cya mutant (BTH101), and functional complementation was determined by measuring β-galactosidase activity from strains grown in medium containing 10 μM Mg2+ or 10 mM Mg2+ (Fig. 6). A significant level of β-galactosidase activity was observed when BTH101 was cotransformed with the MgtC-T18 and MgtC-T25 plasmids, indicating a dimerization of MgtC under these conditions. The level of β-galactosidase activity is similar in medium containing 10 μM Mg2+ or 10 mM Mg2+, suggesting that dimerization is not modulated by Mg2+. We have used the MalG and MalF proteins as specificity controls (18) to show that M. tuberculosis MgtC does not interact with any membrane protein (Fig. 6). A basal level of β-galactosidase activity, similar to that of the negative control (MgtC-T18/T25), was observed with the T18 and T25 proteins fused to the N-terminal part or the C-terminal part of M. tuberculosis MgtC, indicating that these domains alone do not dimerize (Fig. 6). In addition, no interaction was detected with T18 fused to the N-terminal part and T25 fused to the C-terminal part (data not shown). Taken together, these results indicate that the C-terminal domain of M. tuberculosis MgtC does not dimerize, which agrees with the biochemical analyses, but that this domain may promote the dimerization of the entire M. tuberculosis MgtC protein.
Fig 6.
Analysis of in vivo dimerization of M. tuberculosis MgtC using the BACHT system. The E. coli BTH101 strain was cotransformed with plasmids encoding MgtC-T18 and MgtC-T25 fusions, as well as derivatives that carry only the N-terminal or the C-terminal part of M. tuberculosis MgtC. The β-galactosidase activity from strains grown in NCE medium supplemented with 10 μM or 10 mM Mg2+ was measured. The two membrane proteins MalF and MalG of the maltose ABC transporter were used as controls (18). Under the same assay conditions, functional complementation between T25-MalF and T18-MalG yielded 3,950 ± 520 Miller units.
DISCUSSION
Despite its importance in the virulence of various bacterial pathogens, the precise function of the MgtC protein remains unknown. Genes encoding MgtC-like proteins are found in a limited number of eubacterial genomes, and phylogenetic analysis suggests that mgtC has been acquired by horizontal gene transfer repeatedly throughout bacterial evolution (4). MgtC-like proteins harbor a conserved hydrophobic N-terminal domain and a divergent soluble C-terminal domain (4). In the present study, we solved the first high-resolution structure of the cytoplasmic domain of an MgtC family member by determining the NMR structure of the C-terminal domain of the M. tuberculosis MgtC protein. This structure clearly indicates that M. tuberculosis MgtC_Cter is a monomeric ACT domain. Despite the low level of identity between C-terminal domains of MgtC proteins from other intracellular pathogens, our analysis indicates that they are predicted to adopt the same fold and conclusions described above for M. tuberculosis MgtC are likely to be suitable for other members of the family. In addition, the very few residues that are conserved in the MgtC family seem to have a structural role, suggesting a conservation constraint.
ACT domains are small modules (∼90 residue long) involved mainly in the binding of amino acids and in the regulation of various enzymatic activities related to amino acid metabolism (7, 13). Few ACT domains have been proposed to bind other small ligands, such as metal cations. The present structure indicates that M. tuberculosis MgtC harbors a folded ACT domain but does not harbor an amino acid binding site. It also lacks metal binding properties due to the absence of conserved residues adequately positioned for chelation (Met, Cys, Asp, or Glu). Indeed, the only conserved cysteine is buried, and the solvent-accessible glutamates are dispensable (30). In addition, domains with a βαββαβ fold, including ACT domains, have been involved in dimerization, and some modules appear to evolve in the protein interaction domain (1, 20). Our biochemical analysis and bacterial two-hybrid study supported a monomeric nature of the recombinant C-terminal domain, which contrasts with canonical ACT domains. Hence, the monomeric ACT domain of M. tuberculosis MgtC might have a regulatory role that would not bind amino acids or a small solute but rather would be involved in protein-protein interactions. Bacterial two-hybrid experiments indicate a possible dimerization of the M. tuberculosis MgtC protein that requires both the N-terminal and C-terminal domains, each domain by itself being insufficient for dimerization. The fact that the C-terminal domain somehow facilitates MgtC dimerization may rely on a conformation observed only in the complete protein.
Phenotypical analysis of mgtC mutants has linked MgtC to Mg2+ uptake in M. tuberculosis, as well as other intracellular bacterial pathogens (2). Our experimental data indicate that M. tuberculosis MgtC_Cter does not bind Mg2+. In addition, we have shown that the expression of M. tuberculosis MgtC is not significantly induced by Mg2+ deprivation. This result contrasts with the regulation of mgtC in S. Typhimurium and Y. pestis, which is highly induced upon Mg2+ deprivation (10, 41). The mgtC gene is linked to the mgtB gene, encoding a Mg2+ transporter, both in S. Typhimurium and in Y. pestis, and we propose that MgtC is regulated by Mg2+ in bacterial species where it is cotranscribed with a Mg2+ transporter but not in the other species, such as M. tuberculosis. These data further support the hypothesis that MgtC lacks an intrinsic function related to Mg2+ uptake but instead has an indirect regulatory role in Mg2+ uptake. MgtC might interact with other proteins, such as P-type ATPase cation transporters, and the C-terminal domain of MgtC might facilitate the interaction with such proteins. Further biochemical studies with the full-length protein would be required to identify potential partners for MgtC and better define the potential contribution of the C-terminal domain in protein-protein interactions.
ACKNOWLEDGMENTS
We thank Daniel Ladant (Paris, France) for providing plasmids and W. R. Jacobs for the generous gift of the M. tuberculosis mc27000 strain, which has been approved for use in biosafety level 2 by the Institutional Biosafety Committees of the Albert Einstein College of Medicine and the University of Montpellier 2.
This work was supported by Vaincre La Mucoviscidose (IC0902), and S.C.-K. was supported by ANR-06-MIME-027-01.
Footnotes
Published ahead of print 14 September 2012
REFERENCES
- 1. Agarwal S, et al. 2010. Structure and interactions of the C-terminal metal binding domain of Archaeoglobus fulgidus CopA. Proteins 78:2450–2458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Alix E, Blanc-Potard AB. 2007. MgtC: a key player in intramacrophage survival. Trends Microbiol. 15:252–256 [DOI] [PubMed] [Google Scholar]
- 3. Blanc-Potard AB, Groisman EA. 1997. The Salmonella selC locus contains a pathogenicity island mediating intramacrophage survival. EMBO J. 16:5376–5385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Blanc-Potard AB, Lafay B. 2003. MgtC as an horizontally-acquired virulence factor of intracellular bacterial pathogens: evidence from molecular phylogeny and comparative genomics. J. Mol. Evol. 57:479–486 [DOI] [PubMed] [Google Scholar]
- 5. Buchmeier N, et al. 2000. A parallel intraphagosomal survival strategy shared by Mycobacterium tuberculosis and Salmonella enterica. Mol. Microbiol. 35:1375–1382 [DOI] [PubMed] [Google Scholar]
- 6. Catherinot V, Labesse G. 2004. ViTO: tool for refinement of protein sequence-structure alignments. Bioinformatics 20:3694–3696 [DOI] [PubMed] [Google Scholar]
- 7. Chipman DM, Shaanan B. 2001. The ACT domain family. Curr. Opin. Struct. Biol. 11:694–700 [DOI] [PubMed] [Google Scholar]
- 8. Cléry A, Blatter M, Allain FH. 2008. RNA recognition motifs: boring? Not quite. Curr. Opin. Struct. Biol. 18:290–298 [DOI] [PubMed] [Google Scholar]
- 9. Cornilescu G, Delaglio F, Bax A. 1999. Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J. Biomol. NMR 13:289–302 [DOI] [PubMed] [Google Scholar]
- 10. Garcia Vescovi E, Soncini FC, Groisman EA. 1996. Mg2+ as an extracellular signal: environmental regulation of Salmonella virulence. Cell 84:165–174 [DOI] [PubMed] [Google Scholar]
- 11. Gouet P, Robert X, Courcelle E. 2003. ESPript/ENDscript: extracting and rendering sequence and 3D information from atomic structures of proteins. Nucleic Acids Res. 31:3320–3323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Grabenstein JP, Fukuto HS, Palmer LE, Bliska JB. 2006. Characterization of phagosome trafficking and identification of PhoP-regulated genes important for survival of Yersinia pestis in macrophages. Infect. Immun. 74:3727–3741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Grant GA. 2006. The ACT domain: a small molecule binding domain and its role as a common regulatory element. J. Biol. Chem. 281:33825–33829 [DOI] [PubMed] [Google Scholar]
- 14. Guntert P. 2004. Automated NMR structure calculation with CYANA. Methods Mol. Biol. 278:353–378 [DOI] [PubMed] [Google Scholar]
- 15. Günzel D, Kucharski LM, Kehres DG, Romero MF, Maguire ME. 2006. The MgtC virulence factor of Salmonella enterica serovar Typhimurium activates Na+,K+-ATPase. J. Bacteriol. 188:5586–5594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Holm L, Rosenström P. 2010. Dali server: conservation mapping in 3D. Nucleic Acids Res. 38:W545–W549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Karimova G, Pidoux J, Ullmann A, Ladant D. 1998. A bacterial two-hybrid system based on a reconstituted signal transduction pathway. Proc. Natl. Acad. Sci. U. S. A. 95:5752–5756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Karimova G, Dautin N, Ladant D. 2005. Interaction network among Escherichia coli membrane proteins involved in cell division as revealed by bacterial two-hybrid analysis. J. Bacteriol. 187:2233–2243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Koradi R, Billeter M, Wuthrich K. 1996. MOLMOL: a program for display and analysis of macromolecular structures. J. Mol. Graph. 14:51–55, 29–32 [DOI] [PubMed] [Google Scholar]
- 20. Kozlov G, et al. 2004. Structural similarity of YbeD protein from Escherichia coli to allosteric regulatory domains. J. Bacteriol. 186:8083–8088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Laskowski RA, Moss DS, Thornton JM. 1993. Main-chain bond lengths and bond angles in protein structures. J. Mol. Biol. 231:1049–1067 [DOI] [PubMed] [Google Scholar]
- 22. Lavigne JP, O'Callaghan D, Blanc-Potard AB. 2005. MgtC is required for Brucella suis intramacrophage growth: a potential mechanism of adaptation to low Mg2+ environment shared by Salmonella enterica and Mycobacterium tuberculosis. Infect. Immun. 73:3160–3163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lawley TD, et al. 2006. Genome-wide screen for Salmonella genes required for long-term systemic infection of the mouse. PLoS Pathog. 2:e11 doi:10.1371/journal.ppat.0020011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Maloney KE, Valvano MA. 2006. The mgtC gene of Burkholderia cenocepacia is required for growth under magnesium limitation conditions and intracellular survival in macrophages. Infect. Immun. 74:5477–5486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Miller JH. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 26. Moncrief MB, Maguire ME. 1998. Magnesium and the role of MgtC in growth of Salmonella typhimurium. Infect. Immun. 66:3802–3809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Nederveen, et al. 2005. RECOORD: a recalculated coordinate database of 500+ proteins from the PDB using restraints from the BioMagResBank. Proteins 59:662–672 [DOI] [PubMed] [Google Scholar]
- 28. Pons JL, Labesse G. 2009. @TOME-2: a new pipeline for comparative modeling of protein-ligand complexes. Nucleic Acids Res. 37:W485–W491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Pons JL, Malliavin TE, Delsuc MA. 1996. Gifa V 4: a complete package for NMR data set processing. J. Biomol. NMR 8:445–452 [DOI] [PubMed] [Google Scholar]
- 30. Rang C, et al. 2007. Dual role of the MgtC virulence factor in host and non-host environments. Mol. Microbiol. 63:605–623 [DOI] [PubMed] [Google Scholar]
- 31. Retamal P, Castillo-Ruiz M, Mora GC. 2009. Characterization of MgtC, a virulence factor of Salmonella enterica Serovar Typhi. PLoS One 4:e5551 doi:10.1371/journal.pone.0005551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sambandamurthy VK, et al. 2006. Mycobacterium tuberculosis DeltaRD1 DeltapanCD: a safe and limited replicating mutant strain that protects immunocompetent and immunocompromised mice against experimental tuberculosis. Vaccine 24:6309–6320 [DOI] [PubMed] [Google Scholar]
- 33. Sattler M, Schleucher J, Griesinger C. 1999. Heteronuclear multidimensional NMR experiments for the structure determination of proteins in solution employing pulsed field gradients. Prog. Nucl. Magn. Reson. Spectrosc. 34:93–158 [Google Scholar]
- 34. Schuller DJ, Grant GA, Banaszak LJ. 1995. The allosteric ligand site in the Vmax-type cooperative enzyme phosphoglycerate dehydrogenase. Nat. Struct. Biol. 2:69–76 [DOI] [PubMed] [Google Scholar]
- 35. Snavely MD, Miller CG, Maguire ME. 1991. The mgtB Mg2+ transport locus of Salmonella typhimurium encodes a P-type ATPase. J. Biol. Chem. 266:815–823 [PubMed] [Google Scholar]
- 36. Thompson JA, Liu M, Helaine S, Holden DW. 2011. Contribution of the PhoP/Q. regulon to survival and replication of Salmonella enterica serovar Typhimurium in macrophages. Microbiology 157:2084–2093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Veyron-Churlet R, Brust B, Kremer L, Blanc-Potard AB. 2011. Expression of OmpATb is dependent on small membrane proteins in Mycobacterium bovis BCG. Tuberculosis 91:544–548 [DOI] [PubMed] [Google Scholar]
- 38. Walters SB, et al. 2006. The Mycobacterium tuberculosis PhoPR two-component system regulates genes essential for virulence and complex lipid biosynthesis. Mol. Microbiol. 60:312–330 [DOI] [PubMed] [Google Scholar]
- 39. West AL, et al. 2010. Holo-ni(Ii)hpnikr is an asymmetric tetramer containing two different nickel-binding sites. J. Am. Chem. Soc. 132:14447–14456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ye Y, Godzik A. 2003. Flexible structure alignment by chaining aligned fragment pairs allowing twists. Bioinformatics 2:ii246–ii255 [DOI] [PubMed] [Google Scholar]
- 41. Zhou D, et al. 2006. Genome-wide transcriptional response of Yersinia pestis to stressful conditions simulating phagolysosomal environments. Microbes Infect. 8:2669–2678 [DOI] [PubMed] [Google Scholar]