Abstract
Enteropathogenic Escherichia coli employs a type III secretion system (T3SS) to translocate virulence effector proteins directly into enterocyte host cells, leading to diarrheal disease. The T3SS is encoded within the chromosomal locus of enterocyte effacement (LEE). The function of some of the LEE-encoded proteins remains unknown. Here we investigated the role of the Orf16 protein in T3SS biogenesis and function. An orf16 deletion mutant showed translocator and effector protein secretion profiles different from those of wild-type cells. The orf16 null strain produced T3S structures with abnormally long needles and filaments that caused weak hemolysis of red blood cells. Furthermore, the number of fully assembled T3SSs was also reduced in the orf16 mutant, indicating that Orf16, though not essential, is required for efficient T3SS assembly. Analysis of protein secretion revealed that Orf16 is a T3SS-secreted substrate and regulates the secretion of the inner rod component EscI. Both pulldown and yeast two-hybrid assays showed that Orf16 interacts with the C-terminal domain of an inner membrane component of the secretion apparatus, EscU; the inner rod protein EscI; the needle protein EscF; and the multieffector chaperone CesT. These results suggest that Orf16 regulates needle length and, along with EscU, participates in a substrate specificity switch from early substrates to translocators. Taken together, our results suggest that Orf16 acts as a molecular measuring device in a way similar to that of members of the Yersinia YscP and flagellar FliK protein family. Therefore, we propose that this protein be renamed EscP.
INTRODUCTION
The type III secretion system (T3SS) is a specialized macromolecular transport apparatus used by many pathogenic bacteria of plants and animals to translocate virulence proteins (termed effectors) into the host cell cytosol, in which they subvert multiple signaling pathways (43). This syringe-like protein secretion structure is also known as an “injectisome” (12). It is composed of around 20 proteins organized into three main structures, (i) a multiring basal complex embedded in the bacterial membranes that houses the export apparatus, (ii) a hollow extracellular filamentous structure that forms a conduit to the host cell, and (iii) a translocation pore in the host cell membrane formed by proteins known as translocators, which allow the effectors to be directly injected into eukaryotic host cells (12, 35, 50). Moreover, a T3SS is also used to assemble the bacterial flagellum, which is structurally related to the injectisome (28). Many proteins are conserved among both virulence and flagellar secretion assemblies, and it is proposed that these bacterial nanomachines evolved from a common ancestor (12, 28, 39).
The human pathogen enteropathogenic Escherichia coli (EPEC) is a major cause of severe infant diarrhea, producing significant morbidity and mortality in developing countries (78). EPEC belongs to the attaching and effacing (A/E) family of bacteria that colonize intestinal epithelial cells, creating a distinctive histopathological lesion known as an A/E lesion (11, 78). The genes involved in generating the A/E phenotype are contained within a chromosomal pathogenicity island (PAI) named the locus of enterocyte effacement (LEE), which encodes a T3SS (26, 62). EPEC uses this secretion system to direct the translocation of seven LEE-encoded effectors and many non-LEE-encoded (Nle) effectors (distributed in other PAIs around the chromosome) directly into enterocyte cells (16, 22). These effectors alter numerous cell functions and induce severe reorganization of host cytoskeletal proteins, leading to the formation of actin-rich, pedestal-like structures typical of the A/E lesion (16, 48).
The EPEC injectisome is a supermolecular complex spanning both bacterial membranes and extending an external needle structure and filament (14, 90). The EscC secretin forms the outer membrane (OM) ring, whereas EscD and the lipoprotein EscJ form the inner membrane (IM) rings (38, 82, 94, 114). The EscI protein polymerizes to make up the periplasmic inner rod of the T3SS basal complex (87). Additionally, integral and associated IM proteins, together with cytoplasmic components, constitute the export apparatus essential for type III secretion. Among these, five transmembrane proteins (EscR, EscS, EscT, EscU, and EscV) highly conserved among different-virulence T3SSs and with components of the flagellar export apparatus, are predicted to be localized in a patch of membrane within the IM ring (67, 73, 105). EscU, a member of the YscU/FlhB family of proteins, undergoes spontaneous autocleavage in an NPTH amino acid motif in its C-terminal cytosolic domain (EscUC), and it has been proposed to participate in a substrate specificity switch that regulates secretion hierarchy (17, 103, 115). Another fundamental component of the secretion apparatus is the ATPase EscN, a member of the YscN/FliI group of proteins, which energizes the secretion process and functions as a docking site for chaperone-effector complexes (3, 37, 100, 116). The extracellular needle is made by the polymerization of EscF subunits to an approximate length of 50 nm (91, 111). In addition, a long helical homopolymeric filament of the EspA protein extends the needle (49, 90). Upon contact with the host cell, the injectisome assembles a translocation pore formed by the proteins EspB and EspD in the enterocyte cell membrane (44).
The assembly of a functional T3SS is a sequential process that requires tight regulation (9, 12, 17). It initiates with a Sec-dependent stage in which OM and IM rings are assembled and membrane and associated cytosolic components of the export apparatus are incorporated (23, 24, 105). After this platform is formed, three distinct sets of proteins are secreted in an orderly manner: the early substrates (inner rod and needle subunits), then the middle substrates (translocators), and finally the late substrates (effectors) (9, 17, 54). The flagellar export apparatus also has different export substrate specificity states: early (rod/hook-type substrates) and late (filament-type substrates) (32). In both virulence and flagellar systems, hierarchical secretion is controlled by protein interactions within the so-called molecular switches. The length of the needle/hook is regulated by a switching substrate specificity mechanism (71). Proteins commonly named type III secretion substrate specificity switch (T3S4) proteins (1, 65) achieve the length control of these structures. They include YscP from Yersinia, Spa32 from Shigella, InvJ from Salmonella, and FliK from the flagellar system. Mutants with changes in the genes that encode these proteins exhibit abnormally long needles or hooks that can be up to 20 times as long as those observed in the wild-type (WT) strains; in addition, these mutants do not secrete translocators and effectors or flagellin (41, 45, 46, 52, 58, 95, 98, 110).
Many different mechanisms have been proposed to explain needle/hook length control (12, 27, 29, 45, 59, 60, 76, 93, 106, 109). Interestingly, extragenic suppressor mutations of fliK and yscP are localized within flhB and yscU (25, 41, 53, 110). Once the needle/hook has reached its full length, a protein interaction between the T3S4 protein and the component of the export apparatus regulates the first substrate specificity switch, which arrests needle/hook export while allowing the secretion of later substrates (7, 25, 31, 34, 41, 110).
Many of the EPEC LEE-encoded proteins have been characterized; however, the function of Orf16, encoded by the fifth gene of the LEE3 operon, has not been previously examined. In this study, we investigated the role of Orf16 in the biogenesis and function of the T3SS in EPEC. We show that Orf16 plays a critical role in T3SS assembly and regulation of protein secretion. Following the nomenclature used for Yersinia, we propose that this protein be renamed EscP, which is the name used here.
(This study was performed in partial fulfillment of the requirements for the Ph.D. degree in Biomedical Sciences of J.M.F at the Universidad Nacional Autónoma de México [UNAM].)
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The bacterial strains and plasmids used in the present study are described in Table 1. Bacterial strains were aerobically grown (with shaking at 250 rpm) at 37°C in either Luria-Bertani (LB) or Dulbecco's modified Eagle medium (DMEM; Gibco) supplemented with 1% LB and 4 mg/liter pyridoxal hydrochloride (Sigma). When required, the medium was supplemented with ampicillin at 100 μg/ml, streptomycin at 25 μg/ml, kanamycin at 50 μg/ml, chloramphenicol at 25 μg/ml, or tetracycline at 25 μg/ml.
Table 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s)a | Reference or source |
|---|---|---|
| E. coli | ||
| E2348/69 | WT EPEC O127:H6 strain; Smr | 55 |
| ΔescN mutant | E2348/69 carrying an in-frame deletion of escN; Smr | 38 |
| ΔescV mutant | E2348/69 carrying an in-frame deletion of escV; Smr | 38 |
| Δler (JPEP24) mutant | E2348/69 carrying an in-frame deletion of ler; Kmr Smr | 8 |
| ΔescP mutant | E2348/69 carrying an in-frame deletion of escP; Kmr Smr | This study |
| BL21(DE3)/pLysS | Used for overproduction of proteins from pET-based plasmids; Cmr | Novagen |
| XL1-Blue | Used for cloning; Tcr | Stratagene |
| Salmonella | ||
| JR501 | For converting plasmid to Salmonella compatibility | 85 |
| SJW1103 | WT for motility and chemotaxis | 113 |
| S. cerevisiae AH109 | Yeast reporter strain; MATa trp1-901 leu2-3 ura3-52 his3-200 gal4Δ gal80Δ LYS2::GAL1UAS-GAL1TATA-HIS3 GAL2UAS-GAL2TATA-ADE2 URA3::MEL1UAS-MEL1TATA-lacZ | Clontech |
| Plasmids | ||
| pKD46 | Red recombinase system plasmid under control of the araB promoter | 15 |
| pKD4 | Template plasmid containing the kanamycin resistance cassette for the Red recombinase system | 15 |
| PCR-Blunt-II TOPO | Cloning vector | Invitrogen |
| pTrc99AFF4 | Modified pTrc99A expression vector | 80 |
| pTOPO-2HA | pCR2.1-TOPO derivative carrying C. rodentium espG coding region fused to two HA epitopes at the C terminus | 21 |
| pMAL-c2x | Expression vector for production of recombinant proteins with an N-terminal MBP tag | New England Biolabs |
| pET19b | T7 expression vector for production of recombinant proteins with an N-terminal His10 tag | Novagen |
| pET23b | T7 expression vector for production of recombinant proteins with a C-terminal His6 tag | Novagen |
| pGADT7 | Y2H vector containing GAL4 activation domain and LEU2 nutritional marker | Clontech |
| pGBKT7 | Y2H vector containing GAL4 DNA binding domain and TRP1 nutritional marker | Clontech |
| pGADT7-T | Encodes a fusion of the GAL4 activation domain with murine p53 | Clontech |
| pGBKT7-53 | Encodes a fusion of the GAL4 DNA binding domain with murine p53 | Clontech |
| pGBKT7-Lam | Encodes a fusion of the GAL4 DNA binding domain with human lamin C | Clontech |
| pJRo16 | pCR-Blunt II-TOPO carrying escP | This study |
| pJTo16 | pTrc99AFF4 carrying escP | This study |
| pMTBitirHcT | pTrc99AFF4 carrying tir and cesT with a His10 tag sequence at the 5′ end | Unpublished |
| pJHo16 | pTOPO-2HA carrying escP with its native RBS | This study |
| pJHΔ11o16 | pTOPO-2HA carrying Δ1-11escP with its native RBS | This study |
| pJHeH | pTOPO-2HA carrying espH with its native RBS | This study |
| pJHtir | pTOPO-2HA carrying tir with its native RBS | This study |
| pJHmap | pTOPO-2HA carrying map with its native RBS | This study |
| pJHnC | pTOPO-2HA carrying nleC with its native RBS | This study |
| pJHnD | pTOPO-2HA carrying nleD with its native RBS | This study |
| pJHnH | pTOPO-2HA carrying nleH with its native RBS | This study |
| pJHnI | pTOPO-2HA carrying nleI with its native RBS | This study |
| pJHeI | pTOPO-2HA carrying escI with its native RBS | This study |
| pJLo16 | pMAL-c2X carrying escP | This study |
| pJEeUC | pET19b carrying escUC | This study |
| pJEeUCC | pET19b carrying escUCC | This study |
| pJEeF | pET19b carrying escF | This study |
| pJEeI | pET19b carrying escI | This study |
| pMEcT | pET19b carrying cesT | Unpublished |
| pJEo16 | pET19b carrying escP | This study |
| pMEsL | pET19b carrying sepL | Unpublished |
| pJE23bo16 | pET23b carrying escP | This study |
| pOGADo16 | pGADT7 carrying escP | This study |
| pOGBKeUC | pGBKT7 carrying escUC | This study |
| pOGBKeUCC | pGBKT7 carrying escUCC | This study |
| pOGBKeF | pGBKT7 carrying escF | This study |
| pNGBKeI | pGBKT7 carrying escI | This study |
| pMGBKcT | pGBKT7 carrying cesT | Unpublished |
Sm, streptomycin; Km, kanamycin; Tc, tetracycline; Cm, chloramphenicol.
Construction of escP deletion mutant and plasmids.
DNA manipulations were performed according to standard protocols. Restriction enzymes, T4 DNA ligase (New England BioLabs), and Taq DNA polymerase (Qiagen) were used according to the manufacturer's instructions. The oligonucleotides used in this work were synthesized by Sigma. DNA sequencing was performed by the sequencing facility at the Instituto de Fisiología Celular, UNAM.
Deletion of the escP gene from the WT EPEC chromosome was performed by the one-step mutagenesis method (15). The kanamycin resistance cassette was amplified with 70-mer oligonucleotides using plasmid pKD4 as a template. escP was eliminated and replaced with the resistance marker by recombination. The replacement was confirmed by PCR and sequencing.
For plasmid construction, the escP gene was PCR amplified from chromosomal DNA of WT EPEC using primers containing NdeI and BamHI restriction sites. The PCR product was cloned into pCR-BluntII-TOPO to generate plasmid pJRo16, sequenced, and subcloned into pTrc99AFF4 and pET19b expression vectors to generate plasmids pJTo16 and pJEo16. For the construction of a plasmid expressing EscP with a C-terminal histidine tag, the escP gene was PCR amplified from chromosomal DNA of WT EPEC using primers containing NdeI and XhoI restriction sites. The PCR product was cloned into pET23b, generating plasmid pJE23o16. For the construction of plasmids expressing proteins with a C-terminal double hemagglutinin (HA) epitope tag, the coding regions of EPEC espH, map, tir, nleC, nleD, nleI, nleH, escI, and escP containing their putative native ribosome binding sites (RBS) but without their stop codons were amplified by PCR and cloned as HindIII/XhoI fragments into pTOPO-2HA. To produce N-terminal maltose binding protein (MBP)-tagged EscP, escP was PCR amplified and cloned into the pMAL-c2X vector as a BamHI/HindIII fragment. The escUC, escUCC, cesT, escI, and escF genes were PCR amplified, double digested with NdeI and BamHI, and cloned into the pET19b expression vector, which allows the production of N-terminally His10-tagged proteins.
For construction of the Δ1-11escP allele encoding EscP without its first 11 amino acid (aa) residues but containing its putative native RBS, two independent PCRs flanking the region to be deleted were carried out. PCR A was performed using a forward oligonucleotide priming the beginning of orf15 and a reverse oligonucleotide priming the sequence located immediately upstream of the deletion start point and containing extra bases located immediately downstream of the deletion start point. PCR B was carried out using a forward oligonucleotide priming the sequence immediately downstream of the end of the deletion site and a reverse primer that was complementary to a sequence located in the middle of escQ. The overlapping PCR was performed with the same primers used for the escP amplification containing HindIII/XhoI restriction sites. The purified PCR product was cloned into pTOPO-2HA, generating plasmid pJHΔ11o16 (Table 1), and sequenced.
Protein secretion assays.
EPEC protein secretion assays were performed under shaking conditions. A 120-μl volume of an overnight (ON) EPEC culture (grown in LB with shaking) was inoculated into 6 ml prewarmed DMEM. Growth was continued at 37°C until the optical density at 600 nm (OD600) reached 0.6 to 0.8. Cultures were harvested by centrifugation (16,100 × g for 5 min), and the resulting pellets were resuspended in SDS-PAGE sample buffer normalized according to the OD600 value. The supernatant was carefully collected and precipitated with 10% trichloroacetic acid ON. Precipitated proteins were concentrated by centrifugation at 16,100 × g for 30 min. The pellet was air dried for 5 min and resuspended in SDS-PAGE sample buffer normalized according to the OD600 value, and 4 μl of saturated Tris was added to neutralize the sample. To evaluate the secretion of the recombinant proteins EscP-HA and EscI-HA, 8 ml of prewarmed DMEM was inoculated with 160 μl of ON cultures of WT and ΔescN and ΔescP mutant EPEC strains containing plasmid pJHo16 or pJHeI. Growth was continued at 37°C until the OD600 reached 0.6. Pellets and supernatants were collected and treated as described above. The resulting samples were subjected to SDS–15% PAGE and Western blot analysis.
Immunoblotting.
Immunoblotting was carried out using polyclonal anti-EspB, anti-EspA, anti-Tir, anti-EspC, anti-EspF, anti-EscF, anti-EscI, anti-EscJ, anti-EscUC, or anti-CesT antibodies, as well as anti-DnaK (Assay Designs), horseradish peroxidase (HRP)-conjugated anti-HA (Sigma), HRP-conjugated anti-His (Pierce), or anti-MBP (New England BioLabs) monoclonal antibodies. Secondary antibodies were HRP-conjugated goat anti-rabbit or anti-mouse (Santa Cruz Biotechnology). Immunodetection was performed with a SuperSignal West Pico (Pierce) or Immobilon Western kit (Millipore). For analysis of EscP-HA secretion, the SuperSignal Western Blot Enhancer kit (Pierce) was used before immunodetection. The relative intensities of the bands on Western blot assays were quantified with the Scion Image software (Scion Corporation). Polyclonal antibodies against EscF, EscI, and EscUC generated in this study were produced by subcutaneous immunization of female rabbits with recombinant His10-tagged purified proteins.
His-EscUC purification.
To overproduce His-EscUC, E. coli BL21(DE3)/pLysS (BDP) cells containing plasmid pJEeUC were grown in 250 ml of LB at 30°C until the OD600 was between 0.6 and 0.8; at that point, protein production was induced by the addition of 0.5 mM isopropyl-β-d-thiogalactopyranoside (IPTG). After 4 h of further incubation, cells were harvested by centrifugation (7,000 × g for 20 min, 4°C) and stored at −20°C. The cells were thawed, suspended in binding buffer (20 mM Tris-HCl, pH 8.0, 0.5 M NaCl) with 1 mM phenylmethylsulfonyl fluoride, and lysed by sonication. Cell lysates were centrifuged (38,000 × g for 20 min), and the cleared lysate was loaded onto a Ni-nitrilotriacetic acid agarose column pre-equilibrated with binding buffer. After extensive washing with binding buffer containing 30 mM and 60 mM imidazole, the protein was eluted with increasing concentrations of imidazole (100, 200, and 300 mM). Purified recombinant protein was dialyzed against TNED buffer (20 mM Tris-HCl, pH 7.5, 0.5 M NaCl, 0.1 mM EDTA, 0.2 mM dithiothreitol) containing 20% glycerol. The resulting samples were subjected to SDS–16.5% PAGE and Western blot analysis.
Pulldown assays.
To determine protein interactions between EscP and EscUC, EscUCC, EscF, EscI, CesT, SepL, and the Tir-His-CesT complex, log-phase cultures of BDP cells carrying recombinant plasmid pMAL-c2x, pJLo16, pJEescUC, pJEeUCC, pJEeF, pJEeI, pMEcT, pMEsL, or pMTBitirHcT were induced with 0.1, 0.3, 0.5, 0.5, 0.1, 0.3, 0.1, 0.1, and 0.5 mM IPTG, respectively, and then incubated at 30°C for 4 h. For affinity copurification, MBP- or MBP-EscP-containing cleared lysates were incubated for 2 h with His-EscUC, His-EscUCC, His-EscF, His-EscI, His-CesT, or Tir-His-CesT lysates at 4°C and bound to an amylose resin according to the standard purification procedure. After several washes with column buffer (50 mM Tris-Cl, pH 7.4, 200 mM NaCl, 1 mM EDTA), MBP or MBP-EscP and associated proteins were eluted with column buffer containing 10 mM maltose. Fractions of 300 μl were collected and subjected to SDS-PAGE and immunoblotting.
Y2H assays.
Yeast two-hybrid (Y2H) analysis was done using Matchmaker GAL4 Two-Hybrid System 3 (Clontech) according to the manufacturer's instructions. To generate plasmid pOGADo16, escP was cloned into NdeI/BamHI sites of pGADT7 (GAL4 activation domain). On the other hand, escUC, escUCC, escF, escI, and cesT were cloned into the NdeI/BamHI sites of pGBKT7 (GAL4 DNA binding domain) to produce pOGBKeUC, pOGBKeUCC, pOGBKeF, pNGBKeI, and pMGBKcT, respectively. The pGBKT7 and pGADT7 constructs were cotransformed into Saccharomyces cerevisiae AH109 cells using the lithium acetate transformation procedure in the Clontech yeast protocol handbook. Transformations were plated onto low-stringency minimal synthetic-dropout (SD) medium (lacking leucine and tryptophan) to select for the presence of both plasmids and incubated at 30°C. A single colony was inoculated into 2.5 ml of SD medium lacking leucine and tryptophan and grown ON at 30°C. Two-milliliter volumes of cell cultures were then pelleted, washed twice with 1 ml of water, and adjusted to an OD600 of 1.0 by dilution with water, except for cultures containing pMGBKcT, which were adjusted to an OD600 of 2.0. The samples were then 10-fold serially diluted, and 3 μl of each dilution was spotted onto medium-stringency SD medium (lacking leucine, tryptophan, and histidine) to select for interacting proteins. Plates were incubated at 30°C for 3 to 5 days, except for pMGBKcT, which was incubated for 7 to 9 days. Matchmaker GAL4 Two-Hybrid System 3 provides controls for protein interaction. As a positive control, we used plasmids pGADT7-T and pGBKT7-53, whereas the pGADT7-T and pGBKT7-Lam constructs were used as a negative control. To verify that constructs in pGBKT7 do not individually activate reporter gene expression, they were cotransformed with the pGADT7 vector alone.
Purification of EPEC injectisomes.
Samples were prepared essentially as described previously (51, 61), with some modifications. Cultures (20 ml) grown ON with shaking at 37°C in LB were used to inoculate 950 ml of DMEM. Cultures were grown with moderate shaking (100 rpm) at 37°C until the OD600 reached 0.8, and bacteria were collected by centrifugation and resuspended in 80 ml of sucrose solution (150 mM Tris-HCl, pH 8, 0.5 M sucrose). The suspension was stirred at 4°C, and 1 mg/ml lysozyme was slowly added. EDTA at pH 8 was then added to a final concentration of 2 mM, and the suspension was stirred at 4°C for 1 h. Cells were lysed with 0.3% lauryldimethylamine oxide (LDAO) before the addition of 5 mM MgSO4 and 100 mM NaCl. Cell debris removal was done by low-speed centrifugation, and needle complexes in the supernatant were pelleted by high-speed centrifugation (81,000 × g for 1 h, 4°C). The pellet was resuspended in buffer F (0.1% LDAO in 10 mM Tris-HCl [pH 8]–0.3 M NaCl–5 mM EDTA) and adjusted to a final concentration of 35% (wt/vol) CsCl. Samples were centrifuged for 12 h at 54,000 × g in a Beckman SW30 rotor. Fractions of 500 μl were collected from the gradient and diluted with 1 ml of buffer F previous to centrifugation in a Beckman TLA-100.4 rotor at 135,000 × g for 30 min at 4°C. The pellets containing the injectisomes were resuspended in 100 μl of buffer F and stored at 4°C.
Electron microscopy.
Samples were prepared as described previously (76). Injectisomes were negatively stained with 2% phosphotungstic acid (pH 4.5) on carbon-coated copper grids. Micrographs were recorded using a calibrated magnification of ×20,000 with a JEM-1011 transmission electron microscope (JEOL, Tokyo, Japan) operated at 100 kV. Needle lengths were measured by using the ImageJ software (scale set at 5.22 Å per pixel).
Hemolysis assay.
Quantification of hemolysis by EPEC strains was performed as described previously (44, 108), with some modifications (36). Briefly, 0.5 ml of an EPEC culture grown in DMEM to an OD600 of 0.4 was added to 0.5 ml of a 4% red blood cell (RBC)-DMEM solution and centrifuged at 2,500 × g for 1 min. After 4 h of static incubation at 37°C under a 5% CO2 atmosphere, the bacterium-RBC suspension was gently resuspended. Cells were centrifuged at 12,000 × g for 1 min, and the release of hemoglobin into the supernatant was measured at OD450. Assays were performed in triplicate.
Purification of EspA filaments.
Purification of extracellular EspA protein was done similarly to the method previously described (36). Cultures of WT and ΔescP and ΔescN mutant EPEC strains were grown in 150 ml of DMEM at 37°C with shaking to an OD600 of ∼0.7. Bacteria were harvested by centrifugation and resuspended in 20 ml of 20 mM Tris-HCl, pH 7.5. The filaments were sheared by vigorous vortexing for 4 min. Bacterial cells were pelleted at 38,000 × g for 20 min (twice), and the supernatant was further centrifuged at 160,000 × g for 1 h. The resulting pellets were resuspended in 20 mM Tris-HCl, pH 7.5, normalized to the OD600 of cultures (50 μl for an OD of 1), and analyzed by SDS-PAGE and immunoblotting. For membrane preparation, the bacterial cell pellets obtained after shearing were resuspended in 20 mM Tris-HCl, pH 7.5, and lysed by sonication. Cell lysates were centrifuged at 38,000 × g for 15 min, and the pellets were resuspended in SDS-PAGE sample buffer normalized according to the OD600 value.
Motility assays.
For analysis of multicopy effect on swimming, plasmid pJTo16 carrying escP was transformed into Salmonella enterica strain SJW1103. Fresh transformant cells were inoculated onto soft tryptone agar plates (0.25%) containing ampicillin and incubated at 30°C for 4 to 6 h with or without 1 mM IPTG.
RESULTS
EscP is a member of the YscP/FliK family of proteins.
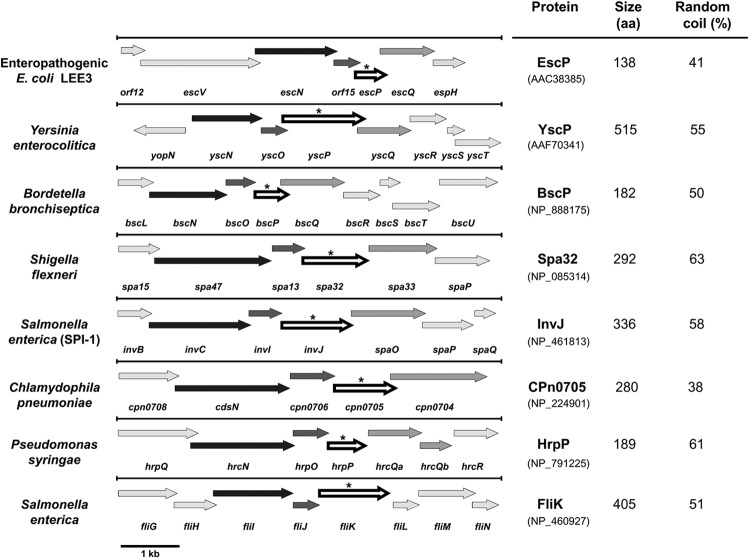
The escP gene (previously orf16) is encoded in the LEE3 operon within the LEE PAI. escP encodes a 138-amino-acid protein with a deduced molecular mass of 16.5 kDa. When subjected to bioinformatic analysis through PSI-BLAST searches, it failed to return any homolog (data not shown) (82). Nevertheless, a genomic context analysis showed that escP is syntenic to genes found in other well-characterized T3SS loci of bacterial pathogens of plants and animals and to the fliK gene of the flagellar T3SS (Fig. 1); i.e., it is always localized two genes downstream of the gene encoding the type III ATPase and between a small gene encoding a protein essential for T3S (30, 86) and the gene encoding the protein that forms the cytoplasmic ring or flagellar C ring (5, 33, 75).
Fig 1.
The LEE escP gene is syntenic to genes encoding members of the YscP/FliK family of proteins. Shown is a schematic representation of T3SS gene clusters showing the genomic context of escP and syntenic genes (arrows indicated by an asterisk), located two genes downstream of the gene encoding the ATPase (black arrows). These genetic maps were obtained from NCBI (http://www.ncbi.nlm.nih.gov/gene) and manually modified for presentation here. Genes are represented to scale (bar, 1 kb). The number of residues of the corresponding proteins (accession numbers are in parentheses) and the percentage of random coiling (calculated with SIMPA96) (http://npsa-pbil.ibcp.fr) are shown on the right.
The proteins encoded by the genes occupying a position equivalent to that of escP within the different type III gene clusters have a wide range of sizes (Fig. 1) (varying from 182 to 515 residues), and amino acid sequence alignment with EscP does not reveal significant similarity (see Fig. S1A in the supplemental material). However, protein sequence comparison using the most conserved C-terminal region, which includes the T3S4 domain (1, 65), showed moderate similarity to EscP (see Fig. S1B). In this regard, it is noteworthy that even the same protein (FliK or HrpP) in different species or pathovars is variable, the C terminus being the most conserved region (74, 109). Unlike some members of this group of proteins, EscP is not a proline-rich protein and it is the only one with a predicted basic isoelectric point; however, all of them possess a high percentage of random coil structure (Fig. 1), as previously suggested (82). In addition, many of these proteins have been shown to perform a similar function in the control of needle/hook length and regulation of protein secretion through its interaction with the soluble domain of a membrane protein of the export apparatus (7, 46, 53, 58, 95, 98, 110). In this study, we pursued the functional characterization of EscP from EPEC.
EscP affects the secretion of translocator and effector proteins differentially.
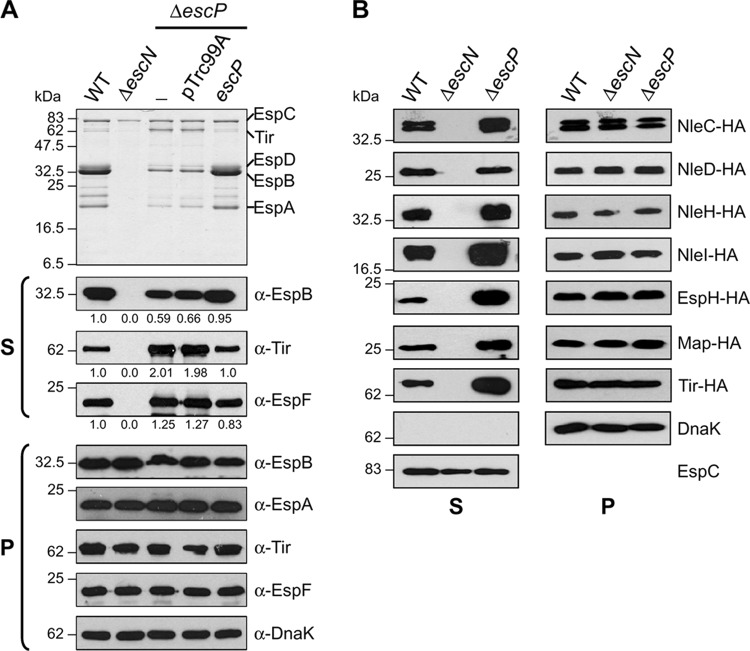
To assess the role of EscP in type III protein secretion, we generated an EPEC escP nonpolar deletion mutant (ΔescP) (Table 1). The escP-null strain was analyzed for secretion of the translocators EspA, EspB, and EspD, as well as LEE-encoded and Nle effectors, into culture supernatants by SDS-PAGE and immunoblotting. WT EPEC showed a typical DMEM-induced secretion profile of translocators and the LEE-encoded effectors Tir and EspF, while the ATPase-defective ΔescN mutant strain was unable to secrete type III proteins (EspC is a type V autotransported protein) (Fig. 2A). In contrast to WT EPEC levels of secretion, the escP mutant showed markedly reduced secretion of translocator proteins and increased secretion of Tir and EspF, as shown by Coomassie staining and/or Western blotting including densitometry (Fig. 2A, top and S panels). All of the proteins were stable and synthesized at similar levels by the different strains, as revealed by immunoblotting of whole-cell lysates (Fig. 2A, panel P). The chaperone DnaK was used as a loading control. Additionally, complementation of the escP deletion mutant was achieved by plasmid-produced EscP, confirming that the mutation is nonpolar. Notably, the oversecretion pattern of effectors was also completely restored to WT levels in the complemented mutant. Successful complementation was accomplished as well when producing the His-EscP, EscP-His, or EscP-HA-tagged protein (data not shown; see Fig. 5A), establishing that these recombinant proteins are fully functional for protein secretion.
Fig 2.
An escP mutant EPEC strain secretes reduced levels of translocators and increased levels of effectors. (A) Protein secretion profiles of WT EPEC and the ΔescN mutant strain, the ΔescP mutant strain (−), the ΔescP mutant strain with the empty vector (pTrc99A), and the ΔescP mutant strain expressing plasmid pJTo16 (escP), visualized by Coomassie brilliant blue-stained SDS–15% PAGE (upper panel). Immunodetection of secreted proteins (S) and whole-cell-lysates (P) was performed using anti-EspB, anti-EspA, anti-Tir, anti-EspF, and anti-DnaK antibodies. Densitometry analysis of secreted proteins in Western blot assays was performed using the Scion Image software (Scion Corporation) (B) Profiles of HA-tagged effector NleC, NleD, NleH, NleI, EspH, Map, and Tir secretion by WT and ΔescN and ΔescP mutant EPEC strains. Immunodetection of secreted proteins (S) and whole-cell lysates (P) was performed with anti-HA antibody. Anti-EspC and anti-DnaK antibodies were used as loading controls. The EspC protein is not secreted via the T3SS. Molecular masses of protein standards are indicated on the left of each panel.
Fig 5.
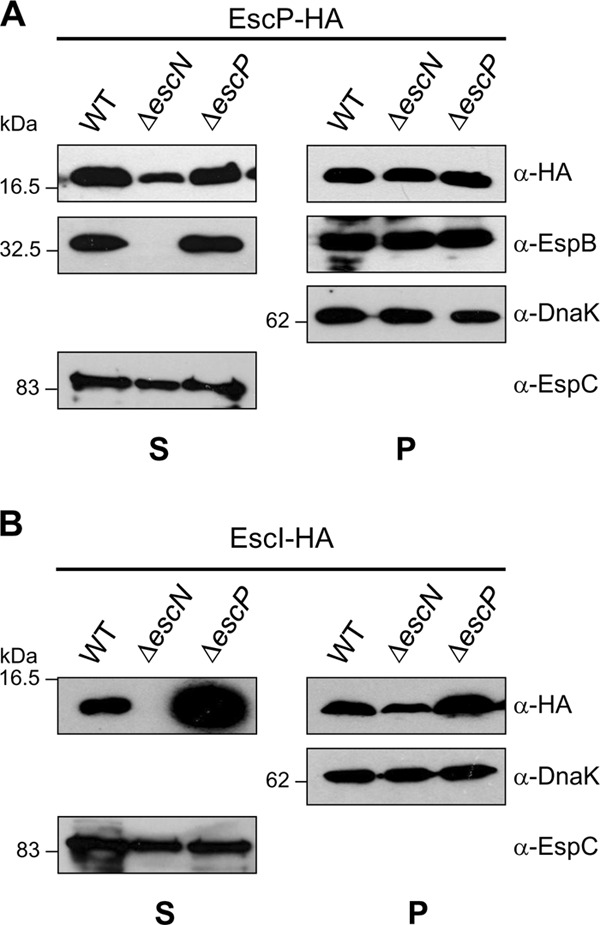
EscP is a type III-secreted substrate and regulates EscI secretion. Analysis of EscP-HA (A) and EscI-HA (B) protein secretion and production is shown. Immunodetection of secreted proteins (S) and whole-cell-lysates (P) of WT and ΔescN and ΔescP mutant EPEC strains overproducing EscP-HA or EscI-HA, separated by SDS–15% PAGE, was performed using anti-HA, anti-EspB, anti-DnaK, and anti-EspC antibodies. The EspC protein is not secreted via the T3SS. Molecular masses of protein standards are indicated on the left of each panel.
Likewise, to examine the in vitro secretion of other LEE-encoded and Nle effectors, the EPEC genes espH, map, tir, nleC, nleD, nleH, and nleI were cloned into plasmid pTOPO-2HA (Table 1), producing C-terminally double HA epitope-tagged proteins. Proteins secreted into culture supernatants from WT and ΔescN and ΔescP mutant EPEC strains were analyzed by immunoblotting using an anti-HA monoclonal antibody. Compared to those in the WT strain, higher (NleC, NleH, NleI, EspH, Map, and Tir) or equivalent levels (NleD) of effector protein secretion were observed in the escP mutant (Fig. 2B, panel S). A particularly enhanced secretion of effectors in the escP mutant background was detected for EspH, Tir, and NleI. It should be noted that an increased amount of Tir protein secretion was observed in the escP mutant, both when native tir was expressed from the chromosome and with recombinant tir from a plasmid, indicating that the observed hypersecretion is not due to a multicopy effect. Besides, proteins were expressed at similar levels in the three strains, as shown by immunoblotting of whole-cell extracts (Fig. 2B, panel P). DnaK was used as a loading control for whole-cell extracts, and EspC was used for supernatants (shown only for Tir). Neither effector nor translocator proteins were identified in supernatants of the escN mutant strain, indicating T3SS-dependent secretion.
EscP regulates injectisome needle length.
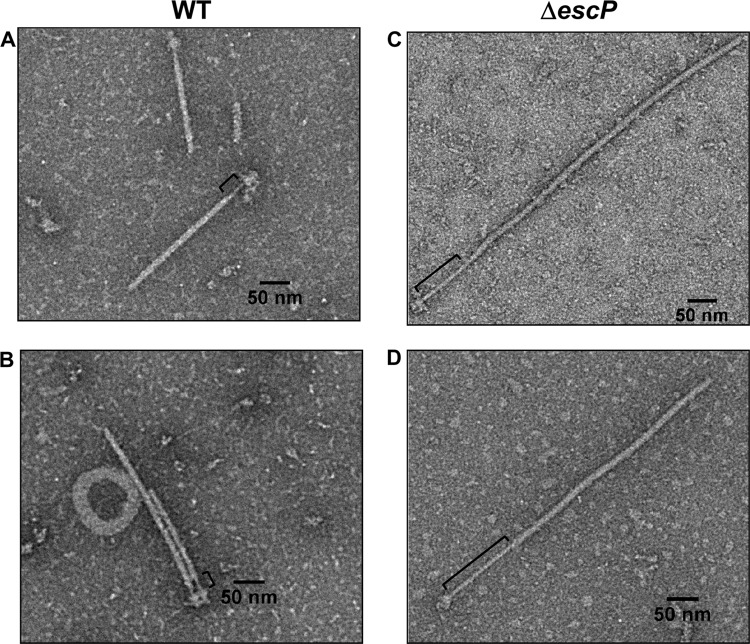
The ability of the escP mutant to secrete differential amounts of translocators and effectors suggested the existence of at least a partially functional T3SS. In a previous bioinformatic study, EscP was suggested as a candidate protein for needle length control (82). In this study, we sought to confirm this prediction experimentally by investigating its role in T3SS biogenesis. In order to determine if EscP participates in needle length regulation, as do other members of the YscP/FliK family of proteins, we purified injectisomes from WT and ΔescP mutant EPEC strains. The resulting samples were negatively stained and observed by transmission electron microscopy (TEM). As shown in Fig. 3, some of the purified injectisomes from the escP mutant possessed needles longer than those obtained from the WT strain. The average needle length of the WT strain was ∼23 nm (Fig. 3, left panels), whereas needles up to five times as long (127 nm) were seen in injectisomes purified from the ΔescP mutant strain (Fig. 3, right panels). Interestingly, the escP mutant strain was able to assemble EspA filament on top of a needle with deregulated length. Consistent with previous reports (79, 90), our TEM analysis showed that the needle width is ∼9 nm, smaller than that of the EspA filament, which is ∼12 nm. However, our measurements indicated that the needle length is shorter than those reported earlier (∼40 to 50 nm) (14, 91, 111). This discrepancy could be due to different methodologies and samples used for length determination (see Discussion). The presence of the T3SS in the CsCl density gradient centrifugation fractions used for TEM analysis was confirmed by immunoblotting with anti-EspA polyclonal antibodies (data not shown). Unexpectedly, a considerable reduction in the number of injectisomes was consistently observed by its purification from the escP mutant EPEC strain (see below).
Fig 3.
An escP mutant EPEC strain assembles abnormally long needle structures with filament. T3S complexes were purified from the WT (A and B) and ΔescP mutant (C and D) EPEC strains. Samples were negatively stained with 2% phosphotungstic acid and observed by TEM at a magnification of ×20,000. Brackets indicate the portion corresponding to the needle.
The escP mutant assembles fewer functional injectisomes.
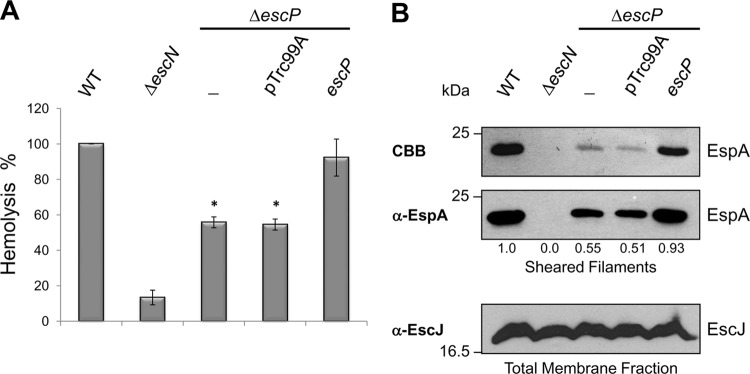
To gain functional evidence supporting the above-mentioned observation of a significant decrease in the number of purified injectisomes from the escP mutant than from the WT strain, we carried out T3SS-dependent RBC hemolysis experiments. EPEC-mediated hemoglobin release by osmotic lysis of erythrocyte cell membranes has been used to identify the formation of a functional type III translocon (91, 108). The WT and ΔescP and ΔescN mutant EPEC strains were incubated with RBCs, and the escP mutant displayed reduced hemolytic activity, ca. 55% of WT EPEC hemolysis (Fig. 4A). The secretion-deficient ΔescN mutant strain was used as a negative control. RBC hemolysis was restored to WT levels when EscP was produced from plasmid pJTo16 (Table 1). These results demonstrate the existence of functional injectisomes in the escP mutant that are capable of forming a translocation pore in RBC membranes. Nevertheless, the reduced hemolysis suggested that a smaller number of fully assembled injectisomes are formed in each cell. To corroborate this, we performed mechanical shearing of EspA filaments in the WT and ΔescP and ΔescN mutant strains. Consistent with our previous data, the results showed an ∼50% decrease in the amount of recovered EspA filament in the escP mutant compared to that in the WT strain (Fig. 4B), confirming that fewer mature injectisomes are assembled. EspA filamentation was restored when plasmid pJTo16 was introduced into the escP mutant (Fig. 4B). Besides, the amount of a T3SS IM ring component was estimated in the membrane fraction obtained from the same sheared cells by Western blot analysis. The results showed similar levels of EscJ protein in all of the strains (Fig. 4B, lower panel), suggesting that the number of needle complex basal structures is not affected in the escP mutant. Overall, these results indicate that EscP is required for complete and efficient biogenesis of the T3SS.
Fig 4.
An escP mutant EPEC strain shows reduced hemolytic activity and assembles fewer EspA filaments. (A) Hemolytic capabilities of WT EPEC strain, the ΔescN mutant strain, the ΔescP mutant strain (−), the ΔescP mutant strain with the empty vector (pTrc99A), and the ΔescP mutant strain expressing the plasmid pJTo16 (escP). Hemoglobin released into the supernatant was measured by determining OD450 after incubation of EPEC strains with human RBCs. Standard deviations of three independent experiments are shown. Asterisks indicate significant differences from WT EPEC. *, P < 0.0001, as determined by Student's t test. (B) EspA filament sheared from WT EPEC, the ΔescN mutant strain, the ΔescP mutant strain (−), the ΔescP mutant strain with the empty vector (pTrc99A), and the ΔescP mutant strain expressing plasmid pJTo16 (escP). EspA protein was visualized by Coomassie brilliant blue (CBB)-stained SDS–12.5% PAGE (upper panel) and by immunoblotting with anti-EspA antibody (middle panel). Densitometry analysis of EspA bands in the Western blot assay was performed with the Scion Image software (Scion Corporation). Membrane fractions of sheared bacterial cells were analyzed by immunoblotting with anti-EscJ antibody (lower panel). Molecular masses of protein standards are indicated on the left of each panel.
EscP is a weakly type III-secreted protein and is needed to regulate EscI secretion.
It has been demonstrated that YscP, Spa32, InvJ, and FliK are secreted through the T3SS (66, 83, 84, 95, 98) and that secretion is required for hook/needle length control (2, 58, 66, 109). To examine whether EscP is a type III-secreted substrate, plasmid pJHo16 (Table 1), producing a double-HA-tagged recombinant EscP protein (EscP-HA), was introduced into the WT and ΔescN and ΔescP mutant EPEC strains and its secretion was analyzed by immunoblotting. EscP-HA was detected in the supernatant from the WT and ΔescP mutant strains when the SuperSignal Western Blot Enhancer kit (Thermo Scientific) was used for detection (Fig. 5A). Unexpectedly, EscP-HA was consistently secreted, although in a smaller amount, from the type III-associated-ATPase-deficient ΔescN mutant, suggesting an injectisome-independent secretion mechanism (see Discussion). Since it has been reported that the flagellar type III ATPase is not indispensable for protein secretion (70), we evaluated EscP-HA secretion in the escV and ler T3S-deficient mutants in which injectisome-independent secretion was also observed (data not shown). It should be noted that none of the HA-tagged effectors or the early substrate EscI (87) was secreted in either of these mutant backgrounds under the same conditions (data not shown). Moreover, Western blot analysis showed that EspB secretion was restored to WT levels in the escP mutant when it was complemented with EscP-HA (Fig. 5A, panel S).
In addition, we evaluated the secretion of the inner rod component EscI by introducing plasmid pJHeI (Table 1) into the same strains. As shown in Fig. 5B, the escP mutant secreted much larger amounts of EscI-HA into the culture supernatant than did the WT strain. This result suggests that EscP regulates EscI secretion. Interestingly, the amount of EscI-HA detected in the escP mutant was somewhat higher than that in the WT strain (Fig. 5B, panel P), suggesting that EscP somehow contributes to the regulation of EscI protein levels. Furthermore, the amount of EscI-HA was slightly lower in the escN mutant background than that in the WT strain (Fig. 5B). Notably, both findings, i.e., increased production of the inner rod component in a T3S4 protein mutant background and decreased production in a type III secretion-deficient background, have been previously reported for the invJ and invA mutant strains of S. enterica (96). In agreement, the Yersinia YscI inner rod protein is not produced in a strain that lacks a functional T3SS (112).
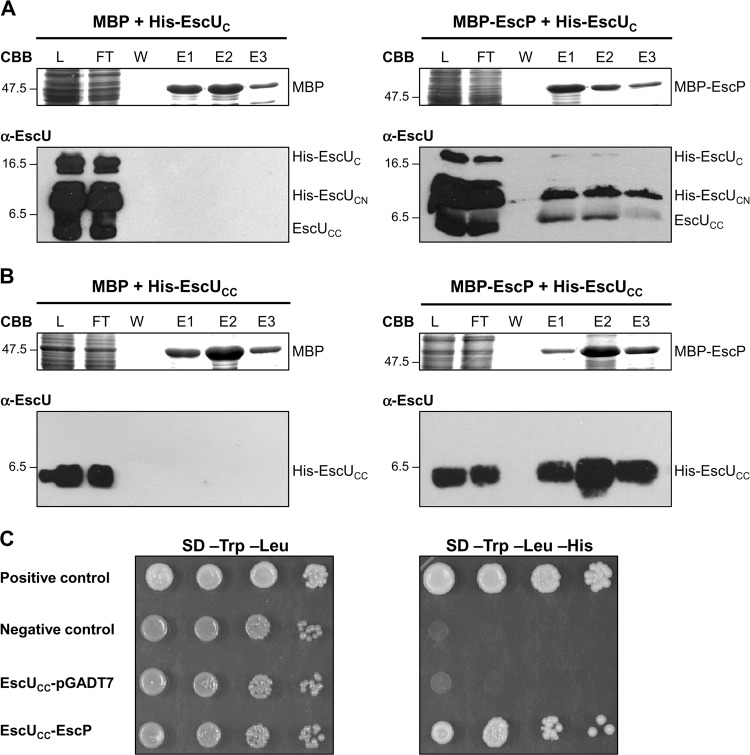
EscP interacts with EscUCC.
The YscU/FlhB family of proteins has been previously shown to participate in a secretion substrate-switching event together with members of the YscP/FliK protein family (7, 25, 34, 56, 57, 65, 68, 72). Therefore, we sought to assess the interaction between EscP and the conserved IM protein EscU. For this purpose, we first purified the soluble C-terminal domain of EscU as a His-tagged recombinant protein (His-EscUC) and generated polyclonal antibodies against this region of the protein. The autocleavage of His-EscUC at the NPTH site produced three versions of the C-terminal domain, i.e., a small amount of uncleaved His-EscUC and much larger amounts of His-EscUCN and EscUCC as cleaved forms, with predicted molecular masses of 17.1 kDa, 7.7 kDa, and 9.4 kDa, respectively, as visualized by Coomassie staining (see Fig. S2 in the supplemental material). In agreement with previous reports (6, 18, 31, 115), the two C-terminal subdomains remain tightly associated and so are copurified (see Fig. S2). To distinguish between the His-EscUCN and EscUCC cleaved products, we carried out a Western blot assay with an anti-His monoclonal antibody. From these results, we could determine that EscUCC behaved anomalously in SDS-PAGE since it ran below the 6.5-kDa molecular mass marker (see Fig. S2). This may be an effect of its predicted acidic pI (4.9), as has been previously noted for other acidic proteins (4), such as the needle component in EPEC (data not shown).
Next, to investigate whether there is an interaction between His-EscUc and an MBP recombinant version of EscP, we performed copurification assays and examined if MBP-EscP could retain His-EscUC on an amylose chromatography column. The soluble lysates of cultured BDP cells expressing plasmids pJLo16 and pJEeUC were mixed and loaded onto an amylose resin. After extensive washing, bound proteins were eluted with maltose and analyzed by SDS-PAGE. As a control, binding of His-EscUC to MBP was assayed in an identical manner. His-EscUC was copurified from the column with MBP-EscP but not with MBP alone (Fig. 6A). The identity of the EscU protein was confirmed by immunoblotting, and both His-EscUCN and EscUCC C-terminal fragments were detected (Fig. 6A). Further, in order to dissect this interaction, we constructed plasmid pJEeUCC encoding His-EscUCC. His-EscUCC was copurified from the column with MBP-EscP but not with MBP alone (Fig. 6B). These results indicate that the extreme C terminus (83 aa, beginning from the proline at the NPTH motif of the cytoplasmic C-terminal domain) is sufficient for the interaction with EscP. To confirm this interaction in vivo, we performed Y2H assays. EscUCC was fused to the GAL4 DNA binding domain, while EscP was fused to the GAL4 activation domain. Growth of S. cerevisiae containing both fusions on intermediate-stringency SD medium (lacking Trp, Leu, and His) confirmed the interaction between EscUCC and EscP (Fig. 6C). The EscUcc-GAL4 DNA binding domain cotransformed with the pGADT7 vector alone was used as a control to demonstrate that EscUCC fusion does not activate reporter gene expression (Fig. 6C).
Fig 6.
EscP interacts with the extreme C-terminal domain of the export apparatus component EscU. (A and B) Pulldown assays performed by affinity chromatography. The cleared lysate (L) containing MBP or MBP-EscP and His-EscUC or His-EscUCC was loaded onto amylose resin. MBP and MBP-EscP plus associated proteins were copurified as described in Materials and Methods. All fractions were analyzed by Coomassie brilliant blue (CBB)-stained SDS–16.5% PAGE and immunoblotted with anti-EscUC antibody. Flowthrough (FT), final wash (W), and elution (E1 to E3) fractions are shown. Molecular masses of protein standards (in kDa) are indicated on the left of each panel. (C) Y2H assay performed using S. cerevisiae strain AH109 cotransformed with plasmids pOGADo16 (GAL4AD-EscP) and pOGBKeUCC (GAL4DNABD-EscUCC) (Table 1). Cultures of the resulting S. cerevisiae colonies were spotted as 10-fold serial dilutions onto SD medium lacking Trp and Leu and onto SD medium lacking Trp, Leu, and His to select for interacting proteins. The S. cerevisiae strain cotransformed with pOGBKeUCC and the pGADT7 vector alone was used as an autoactivating control. The manufacturer (Clontech) provided positive and negative controls for protein interaction.
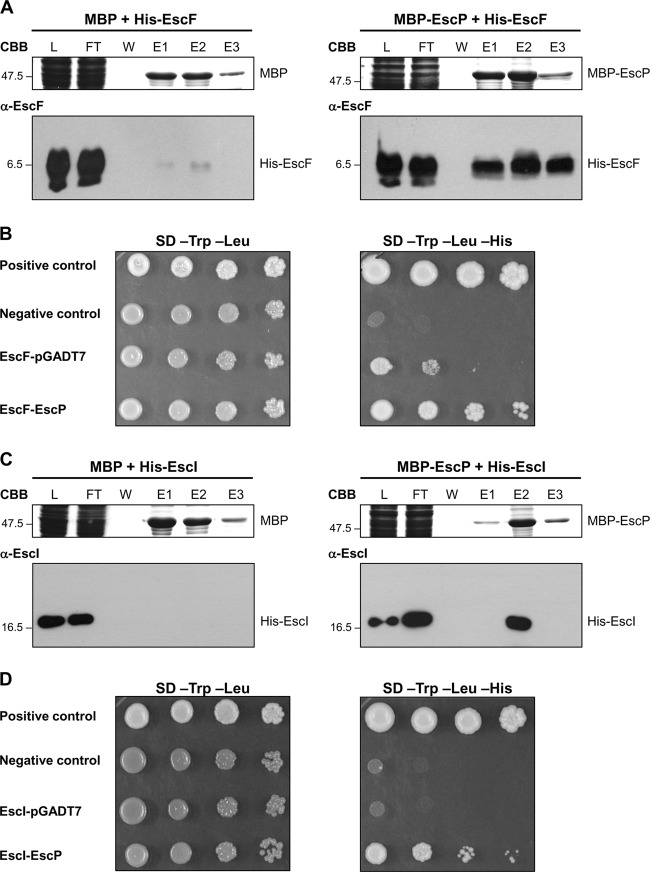
EscP interacts with the early substrates EscF and EscI.
We previously showed that EscP is secreted and regulates needle length. The precise mechanism by which proteins of the YscP/FliK family regulate needle/hook length has not been well established. For the flagellar system, it has been reported that the interaction of FliK with the hook protein FlgE contributes to the stable attachment of the N-terminal domain of FliK with the inner surface of the growing hook structure, allowing FliKC to trigger the switch of export specificity (69). However, this has not been demonstrated for any virulence T3SS. In order to determine whether EscP could interact with the needle component protein, we performed copurification assays as described above. The results showed that His-EscF was copurified from the column with MBP-EscP but not with MBP alone (Fig. 7A), indicating that EscP is capable of interacting with the needle component. This interaction was confirmed with an Y2H assay as previously described. Although EscF fused to the GAL4 DNA binding domain was capable of activating reporter gene expression on its own, the observed interaction with EscP fused to the GAL4 activation domain was consistently stronger (Fig. 7B).
Fig 7.
EscP interacts with the early substrates EscF and EscI. (A and C) Pulldown assays performed by affinity chromatography. The cleared lysate (L) containing MBP or MBP-EscP and His-EscF or His-EscI was loaded onto amylose resin. MBP and MBP-EscP plus associated proteins were copurified as described in Materials and Methods. All fractions were analyzed by Coomassie brilliant blue (CBB)-stained SDS–16.5% PAGE for His-EscF and SDS–15% PAGE for His-EscI and immunoblotted with anti-EscF or anti-EscI antibodies. Flowthrough (FT), final wash (W), and elution (E1 to E3) fractions are shown. Molecular masses of protein standards (in kDa) are indicated on the left of each panel. (B and D) Y2H assays were performed using S. cerevisiae strain AH109 cotransformed with plasmids pOGADo16 (GAL4AD-EscP) and pOGBKeF(GAL4DNABD-EscF) or pOGBKeI (GAL4DNABD-EscI) (Table 1). Cultures of the resulting S. cerevisiae colonies were spotted as 10-fold serial dilutions onto SD medium lacking Trp and Leu and onto SD medium lacking Trp, Leu, and His to select for interacting proteins. The S. cerevisiae strains cotransformed with pOGBKeF or pOGBKeI and the pGADT7 vector alone were used as autoactivating controls. The manufacturer (Clontech) provided positive and negative controls for protein interaction.
We found that EscP regulates EscI secretion (Fig. 5B). Therefore, we examined whether EscI directly interacts with EscP. His-EscI coeluted with MBP-EscP from the column but not with MBP alone (Fig. 7C), indicating that EscP binds to EscI. This interaction was also supported by an in vivo Y2H assay (Fig. 7D).
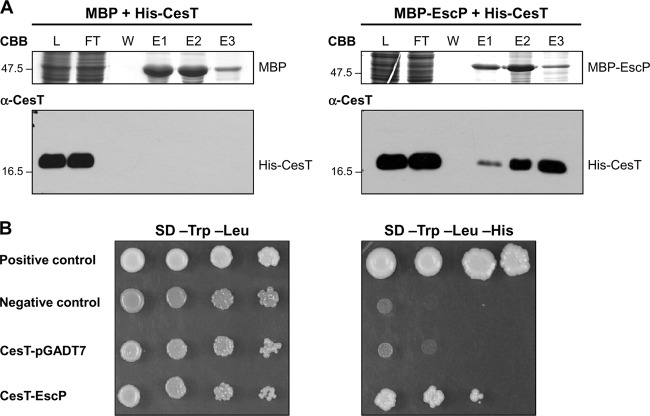
EscP interacts with the multicargo chaperone CesT.
We have shown an increased secretion of LEE-encoded and Nle effectors in the escP mutant strain (Fig. 2), which suggests that EscP is needed for regulation of effector secretion. In EPEC, the LEE-encoded type III chaperone CesT interacts with at least eight effectors (9, 13, 99, 100, 102) and with the type III ATPase EscN, serving to target effectors to the export apparatus (37). In order to determine whether EscP interacts with this chaperone, we performed copurification assays by MBP affinity chromatography. His-CesT was copurified from the column with MBP-EscP but not with MBP alone (Fig. 8A), indicating that EscP binds to CesT. This result is supported by a Y2H assay (Fig. 8B). As an additional negative control for our protein interaction experiments, it is worth mentioning that His-SepL, another LEE-encoded protein, was not copurified with MBP-EscP (data not shown).
Fig 8.
EscP interacts with the multicargo chaperone CesT. (A) Pulldown assay performed by affinity chromatography. The cleared lysate (L) containing MBP or MBP-EscP and His-CesT was loaded onto amylose resin. MBP and MBP-EscP plus associated proteins were copurified as described in Materials and Methods. All fractions were analyzed by Coomassie brilliant blue (CBB)-stained SDS–15% PAGE and immunoblotted with anti-CesT antibody. Flowthrough (FT), final wash (W), and elution (E1 to E3) fractions are shown. Molecular masses of protein standards (in kDa) are indicated on the left of each panel. (B) Y2H assay performed with S. cerevisiae strain AH109 cotransformed with plasmids pOGADo16 (GAL4AD-EscP) and pMGBKcT (GAL4DNABD-CesT) (Table 1). Cultures of the resulting S. cerevisiae colonies were spotted as 10-fold serial dilutions onto SD medium lacking Trp and Leu and onto SD medium lacking Trp, Leu, and His to select for interacting proteins. S. cerevisiae strain AH109 cotransformed with pMGBKcT and the pGADT7 vector alone was used as an autoactivating control. The manufacturer (Clontech) provided positive and negative controls for protein interaction.
DISCUSSION
T3SS assembly is strictly regulated and involves hierarchical protein secretion to guarantee an ordered progression. This sequential process requires the coordination of many cytosolic and membrane proteins to allow early, followed by middle, substrate secretion (translocators) and prevent late substrate secretion (effectors) while the injectisome is being assembled (9, 17, 80, 104). In the present study, we investigated the role of EscP (previously Orf16) in EPEC T3SS biogenesis and function and provide experimental evidence that it is involved in needle length control and efficient translocator secretion by establishing a substrate specificity switch with a conserved member of the export apparatus.
Bioinformatic analysis (Fig. 1; see Fig. S1 in the supplemental material) and the overall functional characterization of EscP led us to propose that it belongs to the YscP/FliK family of proteins. In agreement with our results, it has been proposed that they have diverged during evolution more than other T3SS proteins since members of this family do not share significant sequence similarity (45, 109). Nonetheless, despite different sizes and physicochemical parameters, these proteins are involved in length control and substrate specificity switching; hence, they are considered functional analogs (e.g., Spa32 function is interchangeable with InvJ and YscP function) (7, 98, 101).
It has been demonstrated that the EPEC switch proteins SepL and SepD form a complex that is essential for translocator protein secretion (19), although the precise molecular mechanism remains elusive. Here we found that escP deletion resulted in a considerable reduction in the amount of secreted translocators while the secretion of effectors was generally increased (Fig. 2). This suggests that EscP is involved in the regulation of protein secretion, although it is not absolutely required for T3SS assembly and secretion.
A similar phenotype, i.e., reduced secretion of translocators, has been reported for a C. rodentium orf16 mutant; however, in contrast to our results, effector secretion (represented by Tir) was reported to remain unaltered (21). EPEC EscP is 89% similar to its ortholog protein Orf16 in C. rodentium (see Fig. S3 in the supplemental material), so it is unlikely that these proteins have different functions. We believe that the discrepancy could be due to an incorrectly annotated orf16 gene in the C. rodentium LEE PAI (accession no. AF311901) (20). Protein sequence comparison of C. rodentium Orf16 (AAL06371) to its ortholog proteins in EPEC (AAC38385) and enterohemorrhagic E. coli (NP_312593) (see Fig. S3) indicates that an upstream TTG codon in the C. rodentium LEE3 operon could be the translation initiation codon. In agreement with this, the deletion of the first 11 aa residues of EscP (pJHΔ11o16) makes the protein unstable and hence incapable of complementing an escP mutant (data not shown). This result indicates that the EPEC escP/orf16 gene is correctly annotated in its LEE PAI (AF022236) (26). Deng et al. reported an Orf16 protein of 103 instead of 138 aa; therefore, its deletion mutant permits the expression of 40 aa from the N-terminal region (almost one-third of the protein) (21) that could somehow contribute to the prevention of effector secretion. In the present study, the enhanced secretion of LEE-encoded and Nle effectors by the escP mutant (Fig. 2) could be due to the novel interaction described here between EscP and the multicargo chaperone CesT (Fig. 8), which binds to multiple LEE-encoded and Nle effectors (99, 100). The EscP-CesT interaction could serve to prevent early effector secretion, so that translocator protein secretion is favored. Given the similar phenotypes, we can also hypothesize that EscP participates together with the SepD-SepL complex in promoting translocator secretion and detaining effector secretion. In addition, the existence of a translocation hierarchy among LEE-encoded effectors has been reported where the chaperones CesF and CesT play a critical role in regulating translocation efficiency (64). This could account for differences in secretion levels; e.g., NleD secretion is not affected in the escP mutant, suggesting that this effector is not subject to the same regulatory mechanism and probably does not depend on CesT. Interestingly, the T3S4 protein HpaC, which controls substrate specificity in the T3SS of Xanthomonas campestris pv. vesicatoria, forms a complex with the global T3S chaperone HpaB, which is proposed to regulate the translocation of only a subset of effectors. HpaC and HpaB are able to bind different type of substrates and the export apparatus component HrcV (10). In agreement, a CesT-Tir complex was copurified together with EscP in a pulldown assay (data not shown). Even so, it remains to be investigated how the EscP-CesT complex would be functioning in EPEC.
Injectisomes from animal pathogens possess a needle structure with a defined length. To characterize further the escP mutant phenotype, we purified injectisomes from WT and escP mutant EPEC strains and analyzed them by TEM. Previous measurements by TEM reported the EPEC needle length to be ca. 50 nm. However, these estimates were made with bacterium-attached injectisomes during RBC infection and considering the section of the EspA filament that was not gold labeled with EspA antiserum (91, 111). In a previous EPEC T3SS purification, a needle length of ca. 40 nm was reported (14). Our needle length measurements were achieved with purified injectisome samples by considering the distance between the OM ring and the initiation of the filament (the exact point where the diameter of the structure increases) by using a calibrated magnification scale. Our results showed a markedly shorter needle length estimate of ca. 23 nm. In accordance with this result, it has been proposed that needle length has evolved to match the dimensions of other bacterial or host cell surface structures (12, 77). Since EPEC possesses the filament atop the needle, a longer needle structure is probably not required.
When analyzing purified injectisomes from the ΔescP mutant, our results showed that it can produce longer needles than the WT EPEC strain (Fig. 3). Thus, the escP mutant strain is unable to arrest EscF secretion when the needle reaches its preset length. Different models of needle/hook length control have been proposed (2, 7, 27, 29, 45, 59, 60, 76, 93, 106, 112), some of which suggest that the T3S4 protein functions as a molecular ruler or measuring device that, upon secretion through the nascent structure, determines length. EscP is a much smaller protein than YscP or FliK (Fig. 1), and notably, there seems to be an approximate correlation between needle/hook length and the size of some of the length control proteins (EscP, 138 aa/∼23 nm; Spa32, 292 aa/∼45 nm; InvJ, 336 aa/∼50 nm; FliK, 409 aa/∼55 nm; YscP, 515 aa/∼60 nm) (41, 42, 45, 47, 97, 98), which would be in favor of the ruler/tape measure type of regulatory mechanism. By the same token, BscP is, like EscP, a very small protein (Fig. 1); this suggests a short needle structure, which agrees with the existence of the filament atop the needle in the T3SS of Bordetella bronchiseptica (63).
As aforementioned, a common feature of animal pathogen bacterial length control proteins is that they are secreted through their corresponding T3SS. In agreement, we found that EscP is a type III-secreted protein, although at very low levels (Fig. 5). According to recent length control models (7, 29), the EscP protein would be occasionally secreted during needle polymerization, so that only a few proteins will need to be secreted. Consistent with our finding of low-level EscP secretion, a recent proteomic analysis of the EPEC secretome failed to detect this protein (22). Interestingly, injectisome-independent secretion of EscP was consistently observed in different T3S-deficient mutant backgrounds (Fig. 5A) and was not observed under the same conditions for either effectors or another early substrate such as the inner rod protein EscI. For the time being, we can only speculate that EscP could also be secreted through the type III flagellar export system. In support of this hypothesis, EscP displays a considerable negative multicopy effect on the swimming of S. enterica (data not shown), suggesting that it could be recognized by the flagellar export apparatus. Nonetheless, further experimental evidence is needed to elucidate this injectisome-independent secretion mechanism.
It has been demonstrated in the flagellar T3SS that the interaction of FliK with the flagellar hook protein is required for efficient export specificity switching, suggesting a model in which FliK measures hook length inside the hook structure during its secretion process (69). In accordance with this, we found an interaction between EscP and the needle protein EscF (Fig. 7A and B), which is in favor of a mechanism that allows secreted EscP to detect needle assembly progression. As far as we know, besides the flagellum, this is the first report of such an interaction for any virulence system. In addition, we also found an interaction between EscP and the inner rod protein EscI (Fig. 7C and D). Since EscI is oversecreted in the escP mutant background (Fig. 5B), the interaction between these two proteins suggests that EscP regulates EscI secretion. An equivalent protein interaction between HpaC and HrpB2 has been previously reported in the plant pathogen Xanthomonas (57), which suggests the existence of regulatory mechanisms common to plant- and animal-pathogenic bacteria.
The secretion switching machinery consists, in addition to the length control protein, of an integral membrane component of the export apparatus that belongs to the YscU/FlhB family of proteins (9, 17, 32, 81). Several studies have reported the importance of the interaction among these proteins in mediating the substrate specificity switch (7, 25, 34, 56, 57, 65, 68, 72). In agreement with these observations, we demonstrated that EscP interacts with the membrane component EscU, specifically, with the C-terminal EscUCC domain (Fig. 6). This suggests that the EscP-EscUCC interaction is involved in the switching of secretion specificity from early substrates (EscI, EscF) to translocators (EspA, EspB, and EspD). It has been reported for the type III flagellar system that hook length control is intimately linked with filament assembly; i.e., the fliK mutant generates polyhooks but is unable to secrete filament-type substrates (110). Similarly, the invJ, spa32, and yscP mutants exhibit abnormally long needles and are unable to secrete late substrates (45, 52, 58, 98). Interestingly, we found that the injectisomes assembled by the EPEC escP mutant possess a filament, form a translocation pore (needed to display hemolytic activity), and are able to secrete effectors (Fig. 1, 3, and 4), indicating that the substrate switching can occur even in the absence of EscP, albeit with a low probability. Overall, these results suggest that EscP increases the flipping of the switch that otherwise occurs with low efficiency. In close agreement with our results, a recent study reported that a Δspa32 mutant can form translocation pores and hence possesses some hemolytic activity on RBCs, demonstrating that it autonomously switches substrate specificity and that this phenomenon is not firmly coupled to needle length control (92).
In this study, we also demonstrated that the escP mutant forms a smaller number of fully assembled injectisomes than the WT strain, although similar levels of the inner ring basal body component EscJ were observed in all strains (Fig. 4). Alternative models of needle length control state that assembly of the inner rod determines the size of the needle and is critical for substrate specificity switching (60, 112). It has been suggested that in Salmonella, InvJ stabilizes the conformation of the “socket” substructure at the base of the injectisome, which allows completion of the inner rod and results in firm anchoring of the needle (60). In the case of Yersinia spp., the inner rod protein YscI is hypersecreted in a yscP null strain and suppressor mutations of the yscP phenotype in YscUC reduce YscI secretion, suggesting that formation of the inner rod is needed for substrate switching (112). Recent work with Xanthomonas also showed that the inner rod protein HrpB2 is oversecreted in the hpaC mutant background (40, 89). According to these proposals and our finding of increased secretion of EscI in the escP null strain (Fig. 5), the assembly defect of injectisomes in the escP mutant could be the result of deficient formation of the inner rod. Since no detached long needles were observed during purification, it is probable that the inner rod is required for needle assembly in EPEC. However, in contrast to InvJ (60), our results and those of Shen et al. (92) showed that EscP and Spa32 are not indispensable for the formation of functional injectisomes (even though with abnormally long needles) (Fig. 1, 3, and 4), suggesting a mechanism different from that of Salmonella. Given that we did not observe long needles without filaments, we hypothesize that in the absence of EscP, (i) injectisomes are not formed because of a defect in inner rod assembly and (ii) some inner rod structures are spontaneously assembled, allowing needle anchoring and formation; the latter phenomenon somehow transmits a signal to the base that flips the switch to the secretion of translocators and effectors. Previous to the identification of a length control protein in this study, it was speculated that EscU could bypass the ruler regulating process (see Fig. S10 of reference 115). Here we demonstrated that although this is not the case, it is possible that the specific cleaved conformation of EscUC, which differs from that of Salmonella SpaSC (the latter having a longer C-terminal α-helix and being more similar to flagellar FlhBC) (115) could favor a length control protein-independent switching mechanism. In the WT scenario, EscP will be needed to regulate needle length and to switch substrate specificity at the right time during needle assembly. Ultimately, more studies will be required to understand the role of EscP in injectisome biogenesis. To our knowledge, this is the first report of a member of the YscP/FliK family affecting the number of fully assembled needle/filament macromolecular structures.
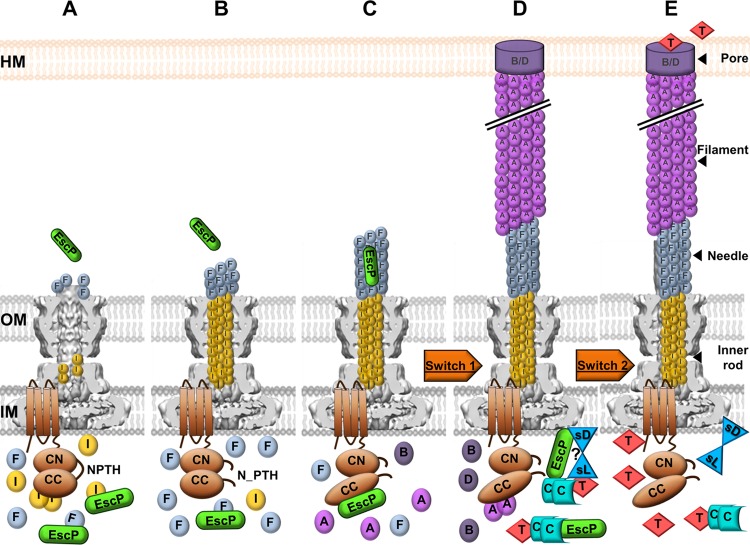
Taking our findings and previous studies together, we suggest the model of EscP function represented in Fig. 9. After the assembly of the OM and IM rings and export apparatus (23, 24, 105), early substrates EscI, EscF, and EscP are recognized and secreted. EscP interacts with both EscI and EscF, regulating their secretion. An EscI-EscU interaction has been reported, and it is independent of EscU autocleavage (87) (Fig. 9A). EscUC is autocleaved, and the two subdomains (EscUCC and EscUCN) remain tightly associated with each other. EscUC autocleavage creates an appropriate conformation essential for the interaction with other T3SS components (115). In addition, autocleavage might slow down the secretion rate of early substrates, as has been suggested in the flagellar T3SS (67). Inner rod assembly allows proper anchoring and assembly of the needle, and EscP is occasionally secreted while measuring needle length (Fig. 9B). When the needle length is within a certain range of its preset length, all interacting subdomains of EscP are able to make contact with the needle inner channel, causing a temporal blockade that results in a pause in the secretion process, which is needed to allow a productive EscUCC-EscP interaction. This interaction promotes a conformational change in EscUCC that flicks the specificity switch to permit translocator protein recognition and secretion (Fig. 9C). While the EspA filament is being assembled, EscP in complex with CesT-Tir, together with the SepL-SepD complex, prevents effector secretion until the translocation pore has been formed in the host cell membrane. It has been reported that the Tir-SepL interaction is critical in restraining the secretion of effector proteins in general, since Tir is required for the secretion of other effectors (99, 107). Here we suggest that EscP could have a similar effect via its interaction with the CesT-Tir chaperone-effector complex (Fig. 9D). Finally, after the translocation pore is formed, the SepD-SepL complex is altered, probably by detecting a change in calcium concentration and other environmental signals, controlling the switch from translocator to effector secretion (19) (Fig. 9E).
Fig 9.
Schematic representation of the EscP mechanism of needle length control and substrate specificity switching in EPEC. (A) Early substrates are secreted, and EscP interacts with both EscI (I) and EscF (F), regulating their secretion. The export apparatus component EscU is represented in the IM. (B) EscUC is autocleaved, and the two subdomains, EscUCC (CC) and EscUCN (CN), remain associated. The inner rod is assembled, allowing proper anchoring and assembly of the needle. EscP is occasionally secreted during needle polymerization. (C) EscP determines needle length within the needle inner channel. EscP interacts with EscUCC, promoting a conformational change that flicks the first specificity switch (Switch 1). (D) Translocator proteins EspA (A), EspB (B), and EspD (D) are secreted. EscP in complex with CesT (C)-Tir (T) prevents, together with the SepL (sL)-SepD (sD) complex, effector secretion until the translocation pore (B/D) has been formed in the host cell membrane (HM). (E) The SepD-SepL complex senses a signal and triggers the second switch (Switch 2) from translocator to effector secretion. The basal body background was adapted from reference 88.
The present study provides important input into this area of investigation, i.e., the existence of an EscU partner, EscP, in the regulation of substrate specificity switching in EPEC. Some of the open questions remaining from this study are the focus of our ongoing research.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Jonathan McMurry and Angel Andrade for critical reading of the manuscript. We thank Ángel Manjarrez-Hernández for kindly providing anti-EspC antibody, Eduardo Soto for bioinformatic analysis, and Onasis Vicente for two-hybrid plasmid constructs and preliminary experiments. We acknowledge Teresa Ballado, Javier de la Mora, and Juan Barbosa for excellent technical assistance.
This work was supported by grants from Dirección General de Asuntos del Personal Académico, UNAM (IN212911), and Consejo Nacional de Ciencia y Tecnología (CONACyT) (81847) to B.G.-P. Partial support was obtained from Fundación Miguel Alemán, A.C. This study was also supported by a fellowship from CONACyT (211365).
Footnotes
Published ahead of print 24 August 2012
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1. Agrain C, et al. 2005. Characterization of a type III secretion substrate specificity switch (T3S4) domain in YscP from Yersinia enterocolitica. Mol. Microbiol. 56:54–67. [DOI] [PubMed] [Google Scholar]
- 2. Agrain C, Sorg I, Paroz C, Cornelis GR. 2005. Secretion of YscP from Yersinia enterocolitica is essential to control the length of the injectisome needle but not to change the type III secretion substrate specificity. Mol. Microbiol. 57:1415–1427 [DOI] [PubMed] [Google Scholar]
- 3. Andrade A, Pardo JP, Espinosa N, Perez-Hernandez G, Gonzalez-Pedrajo B. 2007. Enzymatic characterization of the enteropathogenic Escherichia coli type III secretion ATPase EscN. Arch. Biochem. Biophys. 468:121–127 [DOI] [PubMed] [Google Scholar]
- 4. Armstrong DJ, Roman A. 1993. The anomalous electrophoretic behavior of the human papillomavirus type 16 E7 protein is due to the high content of acidic amino acid residues. Biochem. Biophys. Res. Commun. 192:1380–1387 [DOI] [PubMed] [Google Scholar]
- 5. Biemans-Oldehinkel E, Sal-Man N, Deng W, Foster LJ, Finlay BB. 2011. Quantitative proteomic analysis reveals formation of an EscL-EscQ-EscN type III complex in enteropathogenic Escherichia coli. J. Bacteriol. 193:5514–5519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Björnfot AC, Lavander M, Forsberg A, Wolf-Watz H. 2009. Autoproteolysis of YscU of Yersinia pseudotuberculosis is important for regulation of expression and secretion of Yop proteins. J. Bacteriol. 191:4259–4267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Botteaux A, Sani M, Kayath CA, Boekema EJ, Allaoui A. 2008. Spa32 interaction with the inner-membrane Spa40 component of the type III secretion system of Shigella flexneri is required for the control of the needle length by a molecular tape measure mechanism. Mol. Microbiol. 70:1515–1528 [DOI] [PubMed] [Google Scholar]
- 8. Bustamante VH, et al. 2011. PerC and GrlA independently regulate Ler expression in enteropathogenic Escherichia coli. Mol. Microbiol. 82:398–415 [DOI] [PubMed] [Google Scholar]
- 9. Büttner D. 2012. Protein export according to schedule: architecture, assembly, and regulation of type III secretion systems from plant- and animal-pathogenic bacteria. Microbiol. Mol. Biol. Rev. 76:262–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Büttner D, Lorenz C, Weber E, Bonas U. 2006. Targeting of two effector protein classes to the type III secretion system by a HpaC- and HpaB-dependent protein complex from Xanthomonas campestris pv. vesicatoria. Mol. Microbiol. 59:513–527 [DOI] [PubMed] [Google Scholar]
- 11. Chen HD, Frankel G. 2005. Enteropathogenic Escherichia coli: unravelling pathogenesis. FEMS Microbiol. Rev. 29:83–98 [DOI] [PubMed] [Google Scholar]
- 12. Cornelis GR. 2006. The type III secretion injectisome. Nat. Rev. Microbiol. 4:811–825 [DOI] [PubMed] [Google Scholar]
- 13. Creasey EA, et al. 2003. CesT is a bivalent enteropathogenic Escherichia coli chaperone required for translocation of both Tir and Map. Mol. Microbiol. 47:209–221 [DOI] [PubMed] [Google Scholar]
- 14. Daniell SJ, et al. 2001. The filamentous type III secretion translocon of enteropathogenic Escherichia coli. Cell. Microbiol. 3:865–871 [DOI] [PubMed] [Google Scholar]
- 15. Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Dean P, Kenny B. 2009. The effector repertoire of enteropathogenic E. coli: ganging up on the host cell. Curr. Opin. Microbiol. 12:101–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Deane JE, Abrusci P, Johnson S, Lea SM. 2010. Timing is everything: the regulation of type III secretion. Cell. Mol. Life Sci. 67:1065–1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Deane JE, et al. 2008. Crystal structure of Spa40, the specificity switch for the Shigella flexneri type III secretion system. Mol. Microbiol. 69:267–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Deng W, et al. 2005. Regulation of type III secretion hierarchy of translocators and effectors in attaching and effacing bacterial pathogens. Infect. Immun. 73:2135–2146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Deng W, Li Y, Vallance BA, Finlay BB. 2001. Locus of enterocyte effacement from Citrobacter rodentium: sequence analysis and evidence for horizontal transfer among attaching and effacing pathogens. Infect. Immun. 69:6323–6335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Deng W, et al. 2004. Dissecting virulence: systematic and functional analyses of a pathogenicity island. Proc. Natl. Acad. Sci. U. S. A. 101:3597–3602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Deng W, et al. 2012. Quantitative proteomic analysis of type III secretome of enteropathogenic Escherichia coli reveals an expanded effector repertoire for attaching/effacing bacterial pathogens. Mol. Cell. Proteomics 11:692–709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Diepold A, et al. 2010. Deciphering the assembly of the Yersinia type III secretion injectisome. EMBO J. 29:1928–1940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Diepold A, Wiesand U, Cornelis GR. 2011. The assembly of the export apparatus (YscR, S, T, U, V) of the Yersinia type III secretion apparatus occurs independently of other structural components and involves the formation of an YscV oligomer. Mol. Microbiol. 82:502–514 [DOI] [PubMed] [Google Scholar]
- 25. Edqvist PJ, et al. 2003. YscP and YscU regulate substrate specificity of the Yersinia type III secretion system. J. Bacteriol. 185:2259–2266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Elliott SJ, et al. 1998. The complete sequence of the locus of enterocyte effacement (LEE) from enteropathogenic Escherichia coli E2348/69. Mol. Microbiol. 28:1–4 [DOI] [PubMed] [Google Scholar]
- 27. Erhardt M, et al. 2010. The role of the FliK molecular ruler in hook-length control in Salmonella enterica. Mol. Microbiol. 75:1272–1284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Erhardt M, Namba K, Hughes KT. 2010. Bacterial nanomachines: the flagellum and type III injectisome. Cold Spring Harb. Perspect. Biol. 2(11):a000299 doi:10.1101/cshperspect.a000299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Erhardt M, Singer HM, Wee DH, Keener JP, Hughes KT. 2011. An infrequent molecular ruler controls flagellar hook length in Salmonella enterica. EMBO J. 30:2948–2961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Evans LD, Hughes C. 2009. Selective binding of virulence type III export chaperones by FliJ escort orthologues InvI and YscO. FEMS Microbiol. Lett. 293:292–297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ferris HU, et al. 2005. FlhB regulates ordered export of flagellar components via autocleavage mechanism. J. Biol. Chem. 280:41236–41242 [DOI] [PubMed] [Google Scholar]
- 32. Ferris HU, Minamino T. 2006. Flipping the switch: bringing order to flagellar assembly. Trends Microbiol. 14:519–526 [DOI] [PubMed] [Google Scholar]
- 33. Francis NR, Sosinsky GE, Thomas D, DeRosier DJ. 1994. Isolation, characterization and structure of bacterial flagellar motors containing the switch complex. J. Mol. Biol. 235:1261–1270 [DOI] [PubMed] [Google Scholar]
- 34. Fraser GM, et al. 2003. Substrate specificity of type III flagellar protein export in Salmonella is controlled by subdomain interactions in FlhB. Mol. Microbiol. 48:1043–1057 [DOI] [PubMed] [Google Scholar]
- 35. Galán JE, Wolf-Watz H. 2006. Protein delivery into eukaryotic cells by type III secretion machines. Nature 444:567–573 [DOI] [PubMed] [Google Scholar]
- 36. García-Gómez E, Espinosa N, de la Mora J, Dreyfus G, Gonzalez-Pedrajo B. 2011. The muramidase EtgA from enteropathogenic Escherichia coli is required for efficient type III secretion. Microbiology 157:1145–1160 [DOI] [PubMed] [Google Scholar]
- 37. Gauthier A, Finlay BB. 2003. Translocated intimin receptor and its chaperone interact with ATPase of the type III secretion apparatus of enteropathogenic Escherichia coli. J. Bacteriol. 185:6747–6755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Gauthier A, Puente JL, Finlay BB. 2003. Secretin of the enteropathogenic Escherichia coli type III secretion system requires components of the type III apparatus for assembly and localization. Infect. Immun. 71:3310–3319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Gophna U, Ron EZ, Graur D. 2003. Bacterial type III secretion systems are ancient and evolved by multiple horizontal-transfer events. Gene 312:151–163 [DOI] [PubMed] [Google Scholar]
- 40. Hartmann N, et al. 2012. Characterization of HrpB2 from Xanthomonas campestris pv. vesicatoria identifies protein regions that are essential for type III secretion pilus formation. Microbiology 158:1334–1349 [DOI] [PubMed] [Google Scholar]
- 41. Hirano T, Yamaguchi S, Oosawa K, Aizawa S. 1994. Roles of FliK and FlhB in determination of flagellar hook length in Salmonella typhimurium. J. Bacteriol. 176:5439–5449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hoiczyk E, Blobel G. 2001. Polymerization of a single protein of the pathogen Yersinia enterocolitica into needles punctures eukaryotic cells. Proc. Natl. Acad. Sci. U. S. A. 98:4669–4674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hueck CJ. 1998. Type III protein secretion systems in bacterial pathogens of animals and plants. Microbiol. Mol. Biol. Rev. 62:379–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ide T, et al. 2001. Characterization of translocation pores inserted into plasma membranes by type III-secreted Esp proteins of enteropathogenic Escherichia coli. Cell. Microbiol. 3:669–679 [DOI] [PubMed] [Google Scholar]
- 45. Journet L, Agrain C, Broz P, Cornelis GR. 2003. The needle length of bacterial injectisomes is determined by a molecular ruler. Science 302:1757–1760 [DOI] [PubMed] [Google Scholar]
- 46. Kawagishi I, Homma M, Williams AW, Macnab RM. 1996. Characterization of the flagellar hook length control protein FliK of Salmonella typhimurium and Escherichia coli. J. Bacteriol. 178:2954–2959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kimbrough TG, Miller SI. 2000. Contribution of Salmonella typhimurium type III secretion components to needle complex formation. Proc. Natl. Acad. Sci. U. S. A. 97:11008–11013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Knutton S, Baldwin T, Williams PH, McNeish AS. 1989. Actin accumulation at sites of bacterial adhesion to tissue culture cells: basis of a new diagnostic test for enteropathogenic and enterohemorrhagic Escherichia coli. Infect. Immun. 57:1290–1298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Knutton S, et al. 1998. A novel EspA-associated surface organelle of enteropathogenic Escherichia coli involved in protein translocation into epithelial cells. EMBO J. 17:2166–2176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Kosarewicz A, Konigsmaier L, Marlovits TC. 2012. The blueprint of the type-3 injectisome. Philos. Trans. R. Soc. Lond. B Biol. Sci. 367:1140–1154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kubori T, et al. 1998. Supramolecular structure of the Salmonella typhimurium type III protein secretion system. Science 280:602–605 [DOI] [PubMed] [Google Scholar]
- 52. Kubori T, Sukhan A, Aizawa SI, Galán JE. 2000. Molecular characterization and assembly of the needle complex of the Salmonella typhimurium type III protein secretion system. Proc. Natl. Acad. Sci. U. S. A. 97:10225–10230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Kutsukake K, Minamino T, Yokoseki T. 1994. Isolation and characterization of FliK-independent flagellation mutants from Salmonella typhimurium. J. Bacteriol. 176:7625–7629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Lara-Tejero M, Kato J, Wagner S, Liu X, Galán JE. 2011. A sorting platform determines the order of protein secretion in bacterial type III systems. Science 331:1188–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Levine MM, et al. 1978. Escherichia coli strains that cause diarrhoea but do not produce heat-labile or heat-stable enterotoxins and are non-invasive. Lancet i:1119–1122 [DOI] [PubMed] [Google Scholar]
- 56. Lorenz C, Buttner D. 2011. Secretion of early and late substrates of the type III secretion system from Xanthomonas is controlled by HpaC and the C-terminal domain of HrcU. Mol. Microbiol. 79:447–467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Lorenz C, et al. 2008. HpaC controls substrate specificity of the Xanthomonas type III secretion system. PLoS Pathog. 4:e1000094 doi:10.1371/journal.ppat.1000094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Magdalena J, et al. 2002. Spa32 regulates a switch in substrate specificity of the type III secreton of Shigella flexneri from needle components to Ipa proteins. J. Bacteriol. 184:3433–3441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Makishima S, Komoriya K, Yamaguchi S, Aizawa SI. 2001. Length of the flagellar hook and the capacity of the type III export apparatus. Science 291:2411–2413 [DOI] [PubMed] [Google Scholar]
- 60. Marlovits TC, et al. 2006. Assembly of the inner rod determines needle length in the type III secretion injectisome. Nature 441:637–640 [DOI] [PubMed] [Google Scholar]
- 61. Marlovits TC, et al. 2004. Structural insights into the assembly of the type III secretion needle complex. Science 306:1040–1042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. McDaniel TK, Kaper JB. 1997. A cloned pathogenicity island from enteropathogenic Escherichia coli confers the attaching and effacing phenotype on E. coli K-12. Mol. Microbiol. 23:399–407 [DOI] [PubMed] [Google Scholar]
- 63. Medhekar B, Shrivastava R, Mattoo S, Gingery M, Miller JF. 2009. Bordetella Bsp22 forms a filamentous type III secretion system tip complex and is immunoprotective in vitro and in vivo. Mol. Microbiol. 71:492–504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Mills E, Baruch K, Charpentier X, Kobi S, Rosenshine I. 2008. Real-time analysis of effector translocation by the type III secretion system of enteropathogenic Escherichia coli. Cell Host Microbe 3:104–113 [DOI] [PubMed] [Google Scholar]
- 65. Minamino T, Ferris HU, Moriya N, Kihara M, Namba K. 2006. Two parts of the T3S4 domain of the hook-length control protein FliK are essential for the substrate specificity switching of the flagellar type III export apparatus. J. Mol. Biol. 362:1148–1158 [DOI] [PubMed] [Google Scholar]
- 66. Minamino T, Gonzalez-Pedrajo B, Yamaguchi K, Aizawa SI, Macnab RM. 1999. FliK, the protein responsible for flagellar hook length control in Salmonella, is exported during hook assembly. Mol. Microbiol. 34:295–304 [DOI] [PubMed] [Google Scholar]
- 67. Minamino T, Imada K, Namba K. 2008. Mechanisms of type III protein export for bacterial flagellar assembly. Mol. Biosyst. 4:1105–1115 [DOI] [PubMed] [Google Scholar]
- 68. Minamino T, Macnab RM. 2000. Domain structure of Salmonella FlhB, a flagellar export component responsible for substrate specificity switching. J. Bacteriol. 182:4906–4914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Minamino T, Moriya N, Hirano T, Hughes KT, Namba K. 2009. Interaction of FliK with the bacterial flagellar hook is required for efficient export specificity switching. Mol. Microbiol. 74:239–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Minamino T, Namba K. 2008. Distinct roles of the FliI ATPase and proton motive force in bacterial flagellar protein export. Nature 451:485–488 [DOI] [PubMed] [Google Scholar]
- 71. Minamino T, Pugsley AP. 2005. Measure for measure in the control of type III secretion hook and needle length. Mol. Microbiol. 56:303–308 [DOI] [PubMed] [Google Scholar]
- 72. Minamino T, et al. 2004. Domain organization and function of Salmonella FliK, a flagellar hook-length control protein. J. Mol. Biol. 341:491–502 [DOI] [PubMed] [Google Scholar]
- 73. Moraes TF, Spreter T, Strynadka NC. 2008. Piecing together the type III injectisome of bacterial pathogens. Curr. Opin. Struct. Biol. 18:258–266 [DOI] [PubMed] [Google Scholar]
- 74. Morello JE, Collmer A. 2009. Pseudomonas syringae HrpP is a type III secretion substrate specificity switch domain protein that is translocated into plant cells but functions atypically for a substrate-switching protein. J. Bacteriol. 191:3120–3131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Morita-Ishihara T, et al. 2006. Shigella Spa33 is an essential C-ring component of type III secretion machinery. J. Biol. Chem. 281:599–607 [DOI] [PubMed] [Google Scholar]
- 76. Moriya N, Minamino T, Hughes KT, Macnab RM, Namba K. 2006. The type III flagellar export specificity switch is dependent on FliK ruler and a molecular clock. J. Mol. Biol. 359:466–477 [DOI] [PubMed] [Google Scholar]
- 77. Mota LJ, Journet L, Sorg I, Agrain C, Cornelis GR. 2005. Bacterial injectisomes: needle length does matter. Science 307:1278. [DOI] [PubMed] [Google Scholar]
- 78. Nataro JP, Kaper JB. 1998. Diarrheagenic Escherichia coli. Clin. Microbiol. Rev. 11:142–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Ogino T, et al. 2006. Assembly of the type III secretion apparatus of enteropathogenic Escherichia coli. J. Bacteriol. 188:2801–2811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Ohnishi K, Fan F, Schoenhals GJ, Kihara M, Macnab RM. 1997. The FliO, FliP, FliQ, and FliR proteins of Salmonella typhimurium: putative components for flagellar assembly. J. Bacteriol. 179:6092–6099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Osborne SE, Coombes BK. 2011. Expression and secretion hierarchy in the nonflagellar type III secretion system. Future Microbiol. 6:193–202 [DOI] [PubMed] [Google Scholar]
- 82. Pallen MJ, Beatson SA, Bailey CM. 2005. Bioinformatics analysis of the locus for enterocyte effacement provides novel insights into type-III secretion. BMC Microbiol. 5:9 doi:10.1186/1471-2180-5-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Payne PL, Straley SC. 1999. YscP of Yersinia pestis is a secreted component of the Yop secretion system. J. Bacteriol. 181:2852–2862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Rüssmann H, Kubori T, Sauer J, Galán JE. 2002. Molecular and functional analysis of the type III secretion signal of the Salmonella enterica InvJ protein. Mol. Microbiol. 46:769–779 [DOI] [PubMed] [Google Scholar]
- 85. Ryu J, Hartin RJ. 1990. Quick transformation in Salmonella typhimurium LT2. Biotechniques 8:43–45 [PubMed] [Google Scholar]
- 86. Sal-Man N, et al. 2012. EscA is a crucial component of the type III secretion system of enteropathogenic Escherichia coli. J. Bacteriol. 194:2819–2828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Sal-Man N, Deng W, Finlay BB. 2012. EscI: a crucial component of the type III secretion system forms the inner rod structure in enteropathogenic Escherichia coli. Biochem. J. 442:119–125 [DOI] [PubMed] [Google Scholar]
- 88. Schraidt O, Marlovits TC. 2011. Three-dimensional model of Salmonella's needle complex at subnanometer resolution. Science 331:1192–1195 [DOI] [PubMed] [Google Scholar]
- 89. Schulz S, Buttner D. 2011. Functional characterization of the type III secretion substrate specificity switch protein HpaC from Xanthomonas campestris pv. vesicatoria. Infect. Immun. 79:2998–3011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Sekiya K, et al. 2001. Supermolecular structure of the enteropathogenic Escherichia coli type III secretion system and its direct interaction with the EspA-sheath-like structure. Proc. Natl. Acad. Sci. U. S. A. 98:11638–11643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Shaw RK, Daniell S, Ebel F, Frankel G, Knutton S. 2001. EspA filament-mediated protein translocation into red blood cells. Cell. Microbiol. 3:213–222 [DOI] [PubMed] [Google Scholar]
- 92. Shen D-K, Moriya N, Martinez-Argudo I, Blocker AJ. 2012. Needle length control and the secretion substrate specificity switch are only loosely coupled in the type III secretion apparatus of Shigella. Microbiology 158:1884–1896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Shibata S, et al. 2007. FliK regulates flagellar hook length as an internal ruler. Mol. Microbiol. 64:1404–1415 [DOI] [PubMed] [Google Scholar]
- 94. Spreter T, et al. 2009. A conserved structural motif mediates formation of the periplasmic rings in the type III secretion system. Nat. Struct. Mol. Biol. 16:468–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Stainier I, et al. 2000. YscP, a Yersinia protein required for Yop secretion that is surface exposed, and released in low Ca2+. Mol. Microbiol. 37:1005–1018 [DOI] [PubMed] [Google Scholar]
- 96. Sukhan A, Kubori T, Galán JE. 2003. Synthesis and localization of the Salmonella SPI-1 type III secretion needle complex proteins PrgI and PrgJ. J. Bacteriol. 185:3480–3483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Tamano K, et al. 2000. Supramolecular structure of the Shigella type III secretion machinery: the needle part is changeable in length and essential for delivery of effectors. EMBO J. 19:3876–3887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Tamano K, Katayama E, Toyotome T, Sasakawa C. 2002. Shigella Spa32 is an essential secretory protein for functional type III secretion machinery and uniformity of its needle length. J. Bacteriol. 184:1244–1252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Thomas NA, Deng W, Baker N, Puente J, Finlay BB. 2007. Hierarchical delivery of an essential host colonization factor in enteropathogenic Escherichia coli. J. Biol. Chem. 282:29634–29645 [DOI] [PubMed] [Google Scholar]
- 100. Thomas NA, et al. 2005. CesT is a multi-effector chaperone and recruitment factor required for the efficient type III secretion of both LEE- and non-LEE-encoded effectors of enteropathogenic Escherichia coli. Mol. Microbiol. 57:1762–1779 [DOI] [PubMed] [Google Scholar]
- 101. Thomas NA, Finlay BB. 2004. Pathogens: bacterial needles ruled to length and specificity. Curr. Biol. 14:R192–R194 [DOI] [PubMed] [Google Scholar]
- 102. Thomas NA, Ma I, Prasad M, Rafuse C. 2012. Expanded roles for multicargo and class 1B effector chaperones in type III secretion. J. Bacteriol. 194:3767–3773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Thomassin JL, He X, Thomas NA. 2011. Role of EscU auto-cleavage in promoting type III effector translocation into host cells by enteropathogenic Escherichia coli. BMC Microbiol. 11:205 doi:10.1186/1471-2180-11-205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Tree JJ, Wolfson EB, Wang D, Roe AJ, Gally DL. 2009. Controlling injection: regulation of type III secretion in enterohaemorrhagic Escherichia coli. Trends Microbiol. 17:361–370 [DOI] [PubMed] [Google Scholar]
- 105. Wagner S, et al. 2010. Organization and coordinated assembly of the type III secretion export apparatus. Proc. Natl. Acad. Sci. U. S. A. 107:17745–17750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Wagner S, Stenta M, Metzger LC, Dal Peraro M, Cornelis GR. 2010. Length control of the injectisome needle requires only one molecule of Yop secretion protein P (YscP). Proc. Natl. Acad. Sci. U. S. A. 107:13860–13865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Wang D, Roe AJ, McAteer S, Shipston MJ, Gally DL. 2008. Hierarchal type III secretion of translocators and effectors from Escherichia coli O157:H7 requires the carboxy terminus of SepL that binds to Tir. Mol. Microbiol. 69:1499–1512 [DOI] [PubMed] [Google Scholar]
- 108. Warawa J, Finlay BB, Kenny B. 1999. Type III secretion-dependent hemolytic activity of enteropathogenic Escherichia coli. Infect. Immun. 67:5538–5540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Waters RC, O'Toole PW, Ryan KA. 2007. The FliK protein and flagellar hook-length control. Protein Sci. 16:769–780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Williams AW, et al. 1996. Mutations in fliK and flhB affecting flagellar hook and filament assembly in Salmonella typhimurium. J. Bacteriol. 178:2960–2970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Wilson RK, Shaw RK, Daniell S, Knutton S, Frankel G. 2001. Role of EscF, a putative needle complex protein, in the type III protein translocation system of enteropathogenic Escherichia coli. Cell. Microbiol. 3:753–762 [DOI] [PubMed] [Google Scholar]
- 112. Wood SE, Jin J, Lloyd SA. 2008. YscP and YscU switch the substrate specificity of the Yersinia type III secretion system by regulating export of the inner rod protein YscI. J. Bacteriol. 190:4252–4262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Yamaguchi S, Fujita H, Sugata K, Taira T, Iino T. 1984. Genetic analysis of H2, the structural gene for phase-2 flagellin in Salmonella. J. Gen. Microbiol. 130:255–265 [DOI] [PubMed] [Google Scholar]
- 114. Yip CK, et al. 2005. Structural characterization of the molecular platform for type III secretion system assembly. Nature 435:702–707 [DOI] [PubMed] [Google Scholar]
- 115. Zarivach R, et al. 2008. Structural analysis of the essential self-cleaving type III secretion proteins EscU and SpaS. Nature 453:124–127 [DOI] [PubMed] [Google Scholar]
- 116. Zarivach R, Vuckovic M, Deng W, Finlay BB, Strynadka NC. 2007. Structural analysis of a prototypical ATPase from the type III secretion system. Nat. Struct. Mol. Biol. 14:131–137 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.