Abstract
Spore formation in Bacillus subtilis is characterized by activation of RNA polymerase sigma factors, including the late-expressed σG. During spore formation an asymmetric division occurs, yielding the smaller prespore and the larger mother cell. At division, only 30% of the chromosome is in the prespore, and the rest is then translocated into the prespore. Following completion of engulfment of the prespore by the mother cell, σG is activated in the prespore. Here we tested the link between engulfment and σG activation by perturbing DNA translocation and replication, which are completed before engulfment. One approach was to have large DNA insertions in the chromosome; the second was to have an impaired DNA translocase; the third was to use a strain in which the site of termination of chromosome replication was relocated. Insertion of 2.3 Mb of Synechocystis DNA into the B. subtilis genome had the largest effect, delaying engulfment by at least 90 min. Chromosome translocation was also delayed and was completed shortly before the completion of engulfment. Despite the delay, σG became active only after the completion of engulfment. All results are consistent with a strong link between completion of engulfment and σG activation. They support a link between completion of chromosome translocation and completion of engulfment.
INTRODUCTION
Vegetative cells of Bacillus subtilis differentiate into highly resistant, dormant spores through a series of morphological changes. In response to starvation, the vegetative bacteria divide asymmetrically, yielding the smaller prespore (also called forespore) and the larger mother cell. The mother cell then engulfs the prespore and nurtures it as it develops into the mature spore. Ultimately, the mother cell lyses, releasing the mature spore. Changes in gene expression are coupled to the morphological changes (14, 22, 35). Of cardinal importance is the sequential, compartment-specific activation of four RNA polymerase sigma factors. The first of these, σF, is activated in the prespore after the asymmetric sporulation division. Then σE is activated in the mother cell. Following completion of engulfment, σG is activated in the prespore, and then σK is activated in the mother cell. Intercellular signals ensure that activation of succeeding sigma factors is dependent on the activity of the previous sigma factor in this sequence. The mechanisms for the activation of σF, σE, and σK are moderately well understood (14, 22, 29, 35). However, it is less clear how σG becomes activated. Here we focus on the coupling of σG activation to the completion of engulfment.
σG is encoded by sigG, also called spoIIIG (14, 22, 35), and becomes active upon the completion of engulfment (44). Prespore-specific transcription of sigG is initially directed by σF. Once active, σG directs transcription of its own structural gene in a positive feedback loop (25, 45). This mechanism provides a switch to rapid σG activity when needed during spore formation. However, activation of σG during vegetative growth is toxic, and premature activation during sporulation can block the process (28), so that this feedback loop must be tightly controlled. A number of approaches have been used to study σG activation, and they have identified a variety of controls, including those exercised by SpoIIAB, LonA, and CsfB. In general, these controls appear to prevent inappropriate activation of σG and thus to control the feedback loop. They do not signal activation at the correct time. Of the three proteins, CsfB has the most prominent role in that it prevents premature activation of σG in the prespore (5, 11, 26, 42). Transcription of csfB is initially directed by the prespore-specific factor σF (13, 42) and subsequently by σG, indicating complex regulatory circuits (42). Deletion of csfB results in a low level of σG activity in the prespore prior to the completion of engulfment but does not affect the large increase in σG activity that follows completion of engulfment (11, 26, 42). Thus, CsfB is inadequate to explain the postengulfment activation of σG. SpoIIAB is an anti-sigma factor with a key role in σF activation; its main role with respect to σG is thought to be to prevent inappropriate activation in the mother cell (8, 41). The protease LonA prevents activation of σG under nonsporulation conditions (40). A fourth protein, Fin, was recently shown to be necessary for an efficient transition from σF to σG activity in the prespore; in the absence of Fin, σG activity is substantially reduced (6). However, Fin does not affect the timing of σG activation upon completion of engulfment; rather, σG-directed fin transcription appears to enhance fin expression so as to ensure efficient σG activation (6). It appears to be an adjunct to the positive feedback loop of σG-directed sigG transcription in ensuring fast appearance of ample σG activity when it is needed.
A “feeding tube” channel, AA-AH · Q, has been shown to be necessary for σG to become active after the completion of engulfment. The components of the channel are encoded by eight genes expressed in the mother cell under σE control, spoIIIAA through spoIIIAH, and one gene expressed in the prespore under σF control, spoIIQ. The AA-AH · Q feeding tube provides a link between the mother cell and prespore, which is necessary for σG activation (4, 30) and for prespore integrity (16). No specific activator of σG has been shown to move through this channel. Rather, the channel is thought to be relatively nonspecific (4), needed after completion of engulfment for transport into the prespore of something (possibly ATP) required for σG activity. Notably, following completion of engulfment, it is also needed to obtain activity of σF and artificially expressed T7 RNA polymerase in the prespore. Both σF and T7 RNA polymerase are active before the completion of engulfment without the need for the feeding tube, whereas σG is not. Thus, although the feeding tube is necessary to ensure σG activity after engulfment, it is insufficient to explain why σG becomes active only after the completion of engulfment.
Here we tested the robustness of the link of σG activation to engulfment by perturbing engulfment. We took three approaches, based on the assumption that chromosome replication and chromosome translocation into the prespore are completed before engulfment is completed. The first approach, delaying replication and translocation, was to use derivatives of B. subtilis with 0.9 and 2.3 Mb of Synechocystis PCC6803 DNA inserted into the 4.2-Mb B. subtilis chromosome (24); the Synechocystis inserts lack rrn genes and appear to be largely silent (24). The second approach, affecting termination of replication and hence potentially the sequence of translocation events, was to use a strain in which a region containing the termination-associated TerI, TerII, and rtp loci had been relocated away from the normal replication termination site at 172°, resulting in termination of replication at 145° relative to the origin of replication at 0° (17, 19). The third approach, in which translocation was delayed, was to use a strain with a reduced SpoIIIE DNA translocase activity (3).
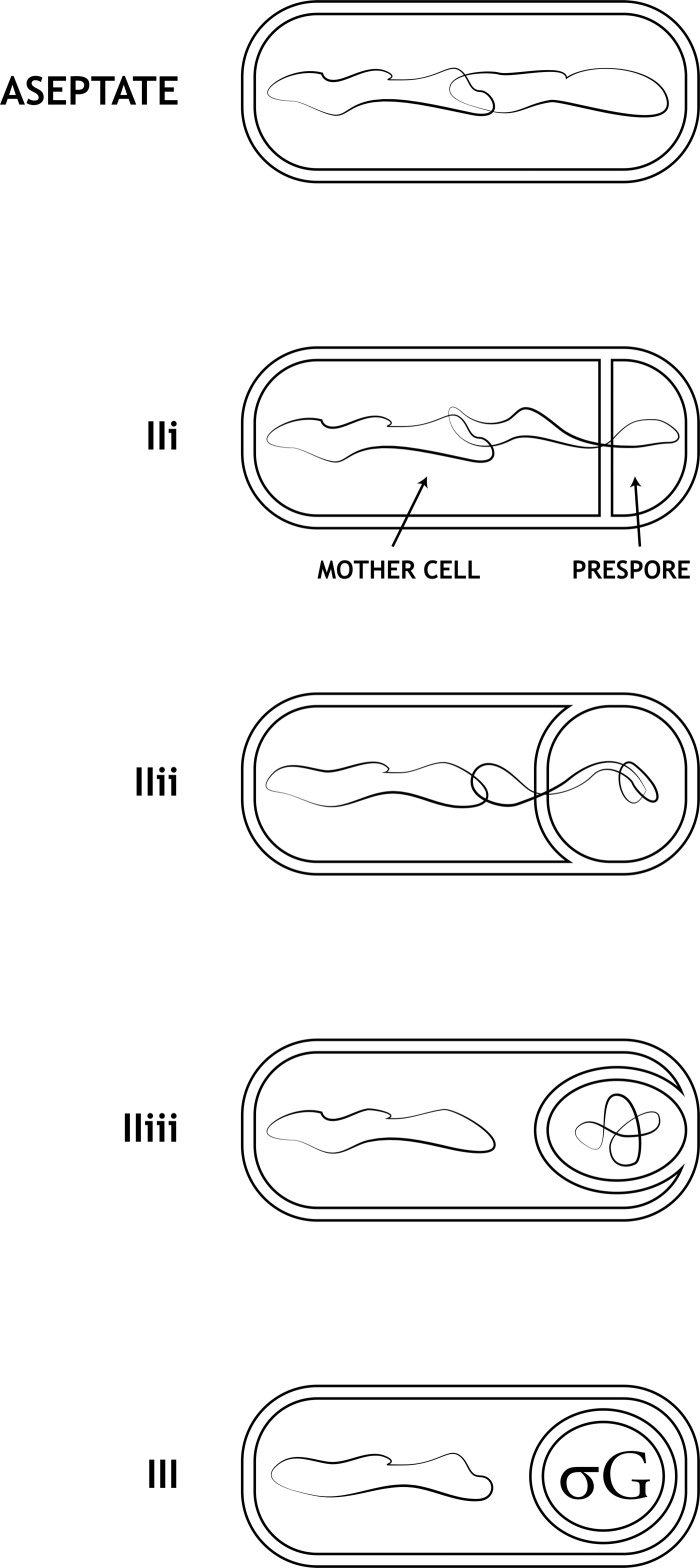
During the process of engulfment, the septal membrane undergoes a series of changes. When it is first formed, the septum is straight (designated stage IIi) (Fig. 1). The septum then becomes curved, bowing into the mother cell (stage IIii), as a result of the progressive loss of wall material starting from the center of the septal disk. The annulus of attachment of the septal membrane to the peripheral cell membrane then moves toward the cell pole (stage IIiii). Finally, the engulfing membranes fuse at the cell pole, releasing the completely engulfed prespore within the mother cell (stage III) (22, 23). The time from septation to completion of engulfment is not known precisely; the best estimate is about 70 min at 37°C (12, 32). Different studies indicate that chromosome replication is completed by stage IIi (2, 18, 21). However, in striking contrast to vegetative division, chromosome partitioning is not complete at the time of the sporulation division; approximately 70% of the chromosome destined for the prespore is still in the mother cell (47, 48). The DNA translocase SpoIIIE pumps the remaining chromosome into the prespore (1, 48). Completion of translocation of the chromosome into the prespore is estimated to take about 15 to 20 min (3, 27, 36) and occurs by stage IIiii (2). We found that insertion of 2.3 Mb of Synechocystis DNA into the B. subtilis chromosome delayed completion of translocation by at least 90 min, which nevertheless still preceded completion of engulfment and the activation of σG. Activation of σG also occurred after completion of engulfment in strains with impaired DNA translocase activity and with the site of termination of replication relocated. We infer that the completion of chromosome translocation into the prespore is a requirement for completion of engulfment, which is in turn required for activation of σG.
Fig 1.
Illustration of the stages of engulfment of the prespore by the mother cell.
MATERIALS AND METHODS
Media.
B. subtilis was grown at 37°C in modified Schaeffer's sporulation medium (MSSM) or on Schaeffer's sporulation agar (15, 33, 39). When required, the medium contained erythromycin at 1.5 μg/ml, neomycin at 3.5 μg/ml, chloramphenicol at 5 μg/ml, or spectinomycin at 100 μg/ml. Escherichia coli was grown on Luria Bertani lysogeny broth (LB) or LB agar, containing ampicillin at 100 μg/ml when required.
Strains.
The B. subtilis strains used are shown in Table 1. Strain SL14525 was constructed by introducing the PspoIIQ-gfp fusion into BEST7003 by double crossover from pGR6. pGR6 was constructed from pVK315 by replacing a PsspA-yfp fusion with PspoIIQ-gfp derived from pVK57 (kindly provided by Vasant Chary). To insert a σF-directed gfp reporter at various locations, the plasmid pGR14 was constructed by removing the erythromycin resistance gene and the region of homology to the 3′ end of thrC from pVK208 (kindly provided by Vasant Chary) and then inserting a spectinomycin resistance gene. The region of homology to the 5′ end of thrC was replaced with different regions of homology to generate a series of plasmids that could integrate by single crossover (Campbell mechanism) at various locations. A PCR-amplified fragment of ylnF replaced the thrC region in pGR14 to yield the plasmid pGR17. SL15634 was generated by Campbell integration of pGR17 into SU227. The thrC region in pGR14 was replaced with a PCR-amplified fragment of ynaE to yield pGR19, which was used to make SL15648. In pGR22 the thrC region was replaced by a region of amyE from the plasmid pVK2 (kindly provided by Vasant Chary); pGR22 was used to construct SL15660. Further details of strain construction are available on request. Plasmids were constructed and maintained in Escherichia coli DH5α.
Table 1.
B. subtilis strains used
| Strain | Relevant genotype or characteristics | Reference or source |
|---|---|---|
| BEST7003 | No Synechocystis insert | 24 |
| BEST8817 | 890-kb Synechocystis insert | 24 |
| BEST7527 | 2.3-Mb Synechocystis inserts | 24 |
| BR151 | Laboratory parental strain | F. E. Young |
| SL10969 | BR151 sspA::PsspA-gfp | Laboratory stock |
| SL14360 | BEST7003 sspA::PsspA-gfp | This study |
| SL14361 | BEST8817 sspA::PsspA-gfp | This study |
| SL14378 | BEST7527 sspA::PsspA-gfp | This study |
| SL14469 | SpoIIIE(D584A) sspA::PsspA-gfp | This study |
| SL14525 | BEST 7003 ppsB::PspoIIQ-gfp | This study |
| SL14539 | BEST 7527 ppsB::PspoIIQ-gfp | This study |
| SL14854 | BEST 7003 spoIIQ::PspoIIQ-gfp | This study |
| SL14856 | BEST 7527 spoIIQ::PspoIIQ-gfp | This study |
| SL15103 | SU227 amyE::PsspA-gfp | This study |
| SL15634 | SU227 PspoIIQ-gfp@ylnF | This study |
| SL15648 | SU227 PspoIIQ-gfp@ynaE | This study |
| SL15660 | SU227 PspoIIQ-gfp@amyE | This study |
| SL15671 | BEST 7003 amyE::PspoIIIG-gfp | This study |
| SL15673 | BEST 7527 amyE::PspoIIIG-gfp | This study |
| SpoIIIE(D584A) | Slow SpoIIIE DNA translocase | 3 |
| SU227 | Replication termination occurs at 145° | 7, 17,19 |
Spore formation.
Cultures were grown in MSSM in conical flasks where the medium occupied no more than 10% of the flask volume. The flasks were incubated at 37°C and shaken at 150 rpm on an orbital shaker. Growth was monitored by measuring the optical density at 600 nm. The end of exponential growth was defined as the start of spore formation. Cultures were analyzed by phase-contrast microscopy and by a heat killing assay (20 min at 80°C) to determine the extent of spore formation (9).
Fluorescence microscopy.
The conditions used for the growth and imaging of samples were similar to those described previously (2, 10). Samples (300 μl) were taken from cultures grown at 37°C in MSSM and mixed with FM4-64 to a final concentration of 1 ng/μl. The bacteria were allowed to settle for 10 min before being visualized with a Leica TCS SP5 confocal microscope. The assessment of engulfment was as described previously (2). The names of the intermediates in the process are essentially those suggested by Illing and Errington (23). At stage IIi, the sporulation septum is straight; at stage IIii, it bulges into the mother cell; at stage IIiii, the points of attachment of the septum to the peripheral membrane have moved toward the pole of the cell; at stage III, engulfment is complete, resulting in a detached prespore entirely within the mother cell. The engulfed prespore often moved away from the pole of the mother cell, suggestive of later stages of sporulation; however, subsequent stages could not be distinguished confidently, and all completely engulfed prespores are designated as having reached stage III. With the strains and conditions used here, FM4-64 efficiently stained completely engulfed prespores, although it does not do so under other conditions (43). Images were analyzed with a Leica TCS SP5 confocal system. Green fluorescent protein (GFP) emission was captured between 500 and 540 nm and FM4-64 emission between 600 and 794 nm; both fluorophores were excited at 488 nm.
Time lapse microscopy.
Bacteria were inoculated into 5 ml MSSM and grown overnight at 30°C, without shaking. Microscope slides were prepared essentially as described by Veening et al. (46). Agarose pads were prepared on the slides within 1.7- by 2.8-cm Gene Frames (Thermo Scientific) with 1.5% low-melting-point agarose in MSSM (50). Pads contained FM4-64 at a final concentration of 0.4 ng/ml. Pads were inoculated with 0.3 μl of early-exponential-phase culture, sealed with a 22- by 40-mm no. 1.5 cover glass (Fisher Scientific) and incubated for approximately 4 h at 30°C. Slides were then monitored with a TCS SP5 confocal microscope (Leica) contained within a temperature-controlled microscope environmental chamber which had been prewarmed to 30°C. For each slide, a field was chosen and focused upon, and the equipment was left to equilibrate for 2 h. After 2 h, the focus was adjusted, and bacteria were imaged at regular intervals using Leica time lapse software with autofocusing. GFP levels were quantified with the Leica TCS SP5 confocal system software. Derivatives of strain BEST7003 formed spores very poorly under these conditions and were not analyzed by time lapse microscopy.
Other methods.
The methods used to transform B. subtilis and to prepare plasmid and chromosomal DNA were essentially as described previously (33, 34).
RESULTS
Insertion of 2.3 Mb Synechocystis DNA into the B. subtilis chromosome delayed engulfment of the prespore by the mother cell.
B. subtilis strains harboring different Synechocystis DNA inserts (24) were tested for their ability to form spores in liquid media. Of those strains, BEST7527 was chosen for detailed study because it had the largest inserts of strains that formed substantial numbers of spores, although the extent varied from experiment to experiment. The BEST7527 derivative SL14378 formed 46% and 37% of the heat-resistant spores formed by the parental strain SL14360 with no inserts in the experiments described below. The inserts in BEST7527 are located origin distal to the ∼1.1 Mb of the B. subtilis chromosome that is ordinarily present in the prespore when it is first formed (49). There are a 908-kb insert in the right arm of the B. subtilis chromosome and two fragments totaling 1,387 kb in the left arm (Fig. 2) (24). The progression through the stages of engulfment for strains with (SL14378) and without (SL14360) the inserts is shown in Fig. 3.
Fig 2.
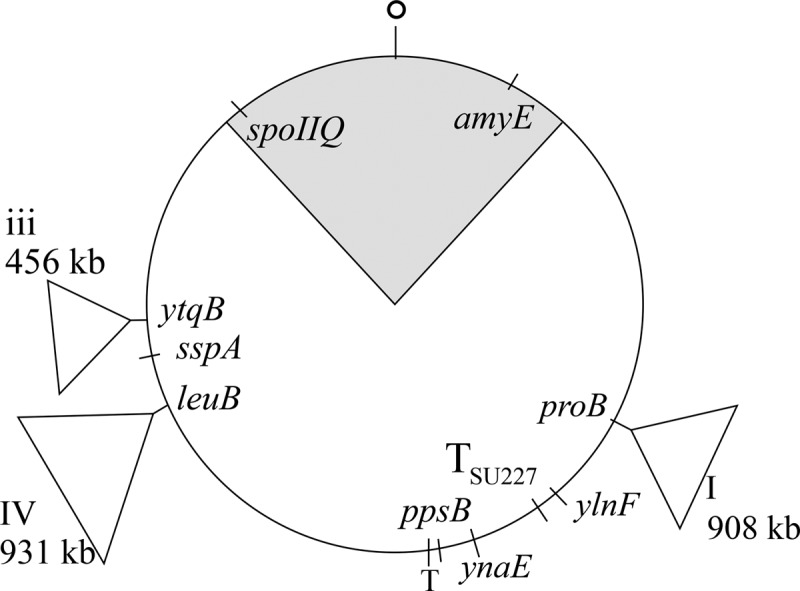
Schematic diagram of the B. subtilis chromosome showing the approximate location of Synechocystis DNA insertions in BEST7527 and of different genetic loci. BEST8817 had an 890-kb insert (not shown) in the position of insert iii of BEST7527. O, origin of replication; T, terminus; TSU227, terminus of replication in strain SU227. The shaded area indicates the portion of the chromosome that is in the prespore at the time of the sporulation division.
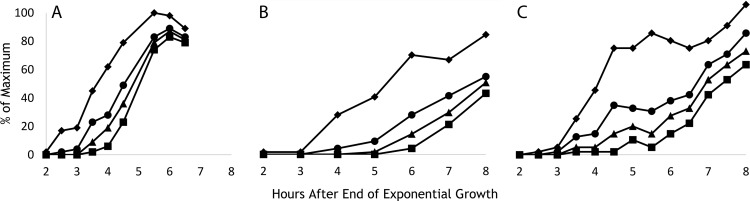
Fig 3.
Effect of insertion of Synechocystis DNA in B. subtilis on the progression of engulfment. The symbols indicate bacteria that had reached a given stage of engulfment (and may have progressed beyond it): diamonds, stage IIi; circles, stage IIii; triangles, stage IIiii; squares, stage III. Values are percentages of the maximum number that reached stage IIi. (A) Strain SL14360, which contains no Synechocystis DNA; for each point, at least 200 organisms were scored. (B and C) Independent experiments with strain SL14378, which contains 2.3 Mb Synechocystis DNA. (B) At least 100 organisms were scored for each point (except for 5 h, for which 89 were scored). (C) At least 200 organisms were scored for each point (except for 5 and 5.5 h, for which at least 140 organisms were scored).
For the parental strain with no insert, the sporulation division was first observed about 3 h after the end of exponential growth, and half the sporulating population had formed the septum by 4 h (stage IIi, strain SL14360) (Fig. 3A). Engulfment was completed (stage III) within 60 to 90 min of septation, similar to times reported previously for B. subtilis (12, 32). The strain containing the 2.3-Mb insert, SL14378, appeared to be slightly delayed in formation of the sporulation division septum compared to the parent strain. It was considerably delayed in the progression through engulfment stages IIii and IIiii, with half the sporulating population completing engulfment only about 3 h after they had formed the septum (Fig. 3B and C).
σG activity was detected only in bacteria that had completed engulfment, even when engulfment was delayed.
To test the effect on σG activation of a greatly enlarged chromosome and the resulting delay in engulfment, a transcriptional σG-directed PsspA-gfp fusion was used. The fusion was inserted at the sspA locus (Fig. 2). σG activity was detected in almost all prespores when engulfment had been completed. In contrast no expression was detected in any bacteria that had not completed engulfment (strain SL14378) (Table 2; examples of organisms at the different stages of engulfment are shown in Fig. 4A). The same result was obtained with the parental strain with no Synechocystis inserts (strain SL14360) (Table 2; Fig. 4B); namely, σG activity was detected only after the completion of engulfment. In contrast, σF-directed gene expression was detected soon after septation (strain SL14856, which has the Synechocystis inserts) (Table 2; Fig. 4C). Transcription from the sigG promoter was assessed with a transcriptional gfp fusion. It was detected well before the completion of engulfment (strain SL15673) (Table 2).
Table 2.
Effect on prespore-specific transcription of different perturbations of chromosome translocation and replication
| Strain | Perturbationa | σ tested | Promoterb | No. with GFP fluorescence/total at stage |
|||
|---|---|---|---|---|---|---|---|
| IIi | IIii | IIiii | III | ||||
| SL14360 | None | σG | PsspA | 0/263 | 0/87 | 0/59 | 432/439 |
| SL14361 | 0.9-Mb insert | σG | PsspA | 0/169 | 0/46 | 0/60 | 365/372 |
| SL14378 | 2.3-Mb insert | σG | PsspA | 0/564 | 0/212 | 0/153 | 505/523 |
| SL14539 | 2.3-Mb insert | σF | PspoIIQ(ter) | 0/445 | 0/227 | 14/218 | 261/261 |
| SL14856 | 2.3-Mb insert | σF | PspoIIQ(ori) | 15/196 | 124/147 | 261/262 | 806/806 |
| SL15673 | 2.3-Mb insert | σF/σG | PsigGc | 49/308 | 196/273 | 306/325 | 1,026/1,027 |
| SL15103 | Ter at 145° | σG | PsspA | 0/291 | 0/207 | 0/190 | 270/353 |
| SL14469 | Slow translocase | σG | PsspA | 0/94 | 0/37 | 0/36 | 177/178 |
Insertion of Synechocystis DNA, termination of replication (Ter) at 145°, or slow SpoIIIE (D584A) DNA translocase.
Promoter driving expression of gfp. ter and ori, promoter located near the terminus and origin.
Promoter of the structural gene for σG.
Fig 4.
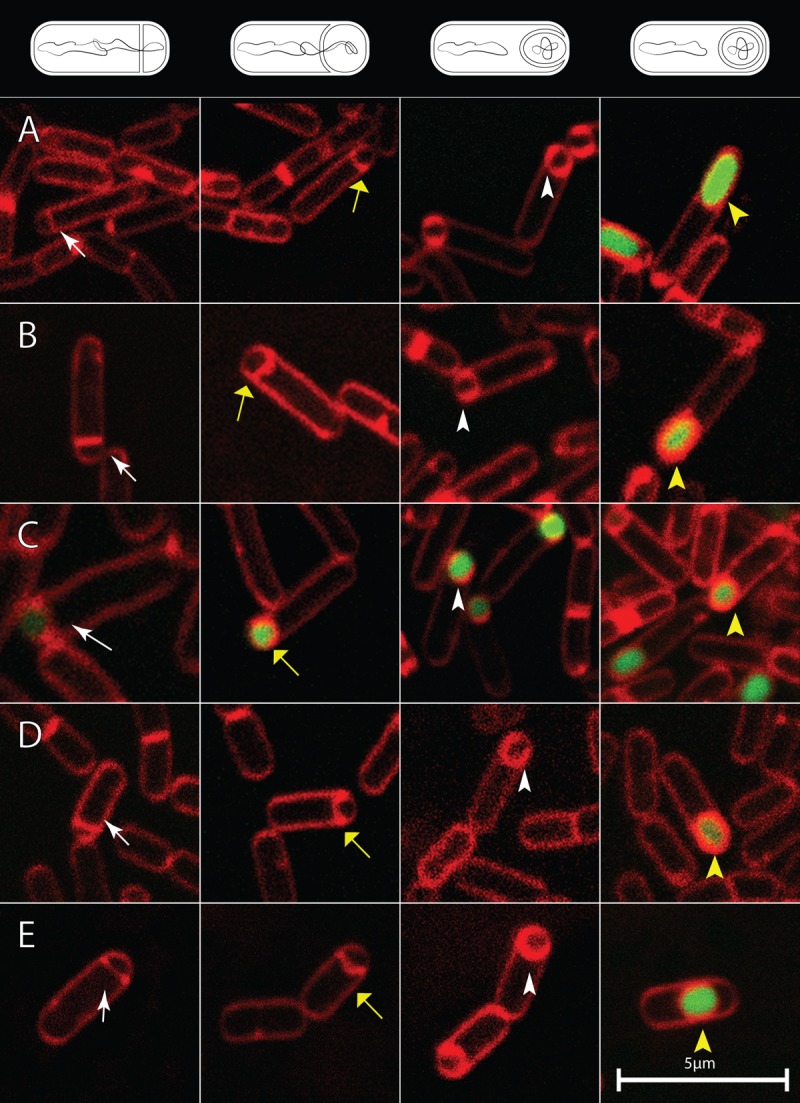
Examples of bacteria showing the activation of gfp transcription directed by σG or by σF at different stages of engulfment. White arrows, stage IIi; yellow arrows, stage IIii; white arrowheads, stage IIiii; yellow arrowheads, stage III. Diagrams of the different stages are shown at the top. Membranes were stained with FM4-64 (red). (A) Strain SL14378 with a 2.3-Mb Synechocystis insert and σG-directed PsspA-gfp; B, strain SL14360, σG-directed PsspA-gfp in BEST7003 background with no insert; C, strain SL14856 with a 2.3-Mb insert and origin-proximal σF-directed PspoIIQ-gfp; (D) strain SL15103 with relocated termination of replication at 145° and σG-directed PsspA-gfp; (E) strain SL14469 with slow DNA translocase and σG-directed PsspA-gfp. Bar, 5 μm (all images).
We also tested the effect of a large Synechocystis insert in just one arm of the B. subtilis chromosome. Strain SL14361 is derived from BEST8817 (24) and has 890 kb inserted in the left arm of the chromosome (in the position of insert iii) (Fig. 2). Such a large, asymmetrically located insertion might potentially disrupt the processes leading to σG activation. However, σG activity was again detected in almost all prespores that were completely engulfed but in no prespores that had not been completely engulfed (strain SL14631) (Table 2).
Completion of chromosome translocation into the prespore was delayed by the 2.3-Mb DNA insertion and occurred shortly before completion of engulfment.
Genes transcribed by RNA polymerase containing σF are expressed only in the prespore. Those located within about 1.1 Mb spanning the origin of replication of the circular 4.2-Mb chromosome are present in the prespore at the time of septation (49) and are expressed soon after septation (20). However, expression of genes located near the terminus requires their translocation into the prespore before they are expressed (3, 27, 49, 51). Expression of terminus-proximal σF-directed genes serves as an indicator of translocation of the terminus region into the prespore.
We used the σF-directed reporter PspoIIQ-gfp at origin-proximal and origin-distal locations to assess chromosome translocation in BEST7527 derivatives. In strain SL14856, the reporter was inserted at the origin-proximal spoIIQ locus (Fig. 2). As discussed above, expression was detected soon after septation (strain SL14856) (Table 2), with GFP fluorescence being observed in some bacteria at stage IIi and in most bacteria from stage IIii onwards. In contrast, when the reporter was inserted at ppsB, located close to the chromosome terminus (Fig. 2), no expression was detected at stages IIi or IIii (strain SL14539) (Table 2). GFP expression began to be detected only at stage IIiii, and by stage III most prespores displayed fluorescence. σF-directed genes located near the terminus are weakly expressed compared to the same gene at an origin-proximal location (27) (P. Xenopoulos and P. J. Piggot, unpublished results). Consequently, the number expressing at IIiii may be an underestimate. Nevertheless, it seems reasonable to infer that in the BEST7527 background, chromosome translocation was considerably delayed and was not completed until shortly before completion of engulfment.
σG activation remained coupled to completion of engulfment after relocation of the site for termination of replication and after impairment of DNA translocase function.
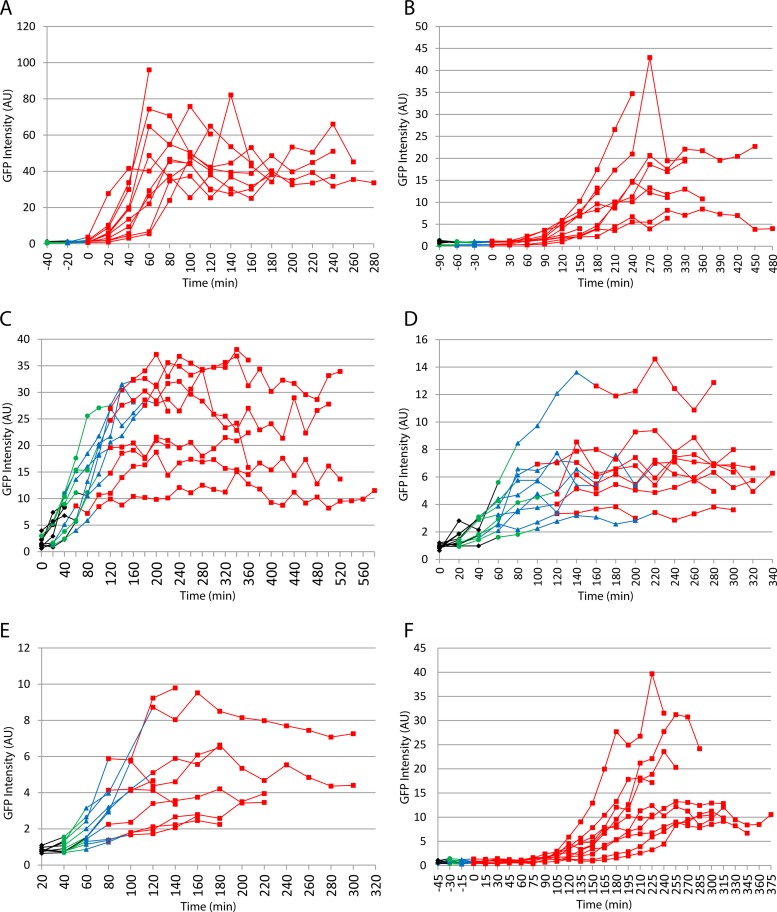
As described above, the robustness of the link(s) between translocation, engulfment, and σG activation was tested by examining the effects of large inserts of foreign DNA in the B. subtilis chromosome. We wished to test the effect of disrupting termination of replication, and hence potentially the sequence of translocation events, without large inserts of foreign DNA. For this purpose we used derivatives of a strain (SU227) in which termination of replication occurs at about 145° and not 172° as the result of relocation of key termination loci (17, 19). Sporulation efficiency was reduced to about 10%. Nevertheless, substantial numbers of sporulating organisms were present in batch cultures, and σG became active only after completion of engulfment (strain SL15103) (Table 2; examples of organisms at the different stages of engulfment are shown in Fig. 4D). To explore this relationship further, time lapse microscopy was performed to visualize σG activation in individual organisms forming spores on agarose pads. To assess σG activity, green fluorescence was measured in individual cells as they transitioned from septation to completion of engulfment. σG activity was detected in organisms that had completed engulfment but not in organisms that had not completed engulfment (strain SL15103) (Fig. 5B). The pattern was very similar to that of a derivative of our standard B. subtilis 168 strain, in which there had been no manipulation of the terminus region (strain SL10969) (Fig. 5A).
Fig 5.
Activation of σG- and σF-directed gene expression in individual cells analyzed by time lapse microscopy. Time traces of individual cells. (A) SL10969, standard strain, σG reporter; (B) Strain SL15103, termination of replication at 145° and σG reporter; (C) SL15660, termination of replication at 145° and σF reporter at 28°; (D) SL15634, termination of replication at 145° and σF reporter at 140°; (E) SL15648, termination of replication at 145° and σF reporter at 161°; (F) SL14469; slow DNA translocase and σG reporter. Bacteria were maintained at 30°C, and successive images were captured at the indicated times. Expression from gfp transcriptional fusions was observed as green fluorescence, which was measured in arbitrary units that are not comparable between strains. For strains expressing a σF-directed reporter, times are normalized (time zero) to the first image to show the sporulation septum (stage IIi). For strains expressing a σG-directed reporter, time traces of individual bacteria are normalized (time zero) to the first image showing completion of engulfment (stage III). The different stages of engulfment are indicated by different colors: black is stage IIi, green is stage IIii, blue is stage IIiii, and red is stage III.
The relocation of the terminus of replication did not uncouple σG activation from completion of engulfment. However, it remained possible that the relocation disturbed chromosome translocation and uncoupled that process from engulfment. To test for this possibility, time lapse studies were preformed using a σF-directed reporter, PspoIIQ-gfp, at different chromosomal locations. When located near the origin, at 28° (amyE), the reporter was expected to already be in the prespore at the time of septation. Indeed, GFP expression began soon after septum formation (strain SL15660) (Fig. 5C). Two locations were tested, 140° (ylnF) and 161° (ynaE), that flanked the replication termination site in SU227 (Fig. 2). In both cases (strains SL15634 and SL15648) (Fig. 5D and E, respectively), expression also began soon after septation and well before the completion of engulfment (although it was perhaps slightly delayed compared to expression at the 28° location). Thus, translocation into the prespore of these terminus-proximal loci preceded completion of engulfment.
We also tested σG activation in a B. subtilis strain in which translocation was delayed by about 30 min because of a mutation that reduced the efficiency of the DNA translocase SpoIIIE (3). Again, in batch cultures, σG activity was detected only after the completion of engulfment (strain SL14469) (Table 2; examples of organisms at the different stages of engulfment are shown in Fig. 4E). This conclusion was reinforced by time lapse microscopy of individual organisms forming spores on agarose pads (strain SL14469) (Fig. 5F). Interestingly, σG activity was first detected some time after completion of engulfment with SL14469, in contrast to strain SL10969 with wild-type spoIIIE, where σG activity was detected soon after completion of engulfment (Fig. 5A). The reason for the slow activation after engulfment in the strain with the inefficient translocase is not known. It was not apparent in batch culture (Table 2) and might result from the different temperatures used for batch cultures (37°C) and time lapse studies (30°C); this possibility was not tested.
DISCUSSION
During spore formation, σG becomes active after the completion of engulfment (44). Once activated, σG directs transcription of its own structural gene, sigG, in a positive feedback loop (25), so that there is the potential for strong, inappropriate activation. Premature activation of σG can be highly detrimental (28), and a series of controls have been identified that prevent its occurrence: CsfB prevents premature activation in the prespore (5, 11, 26, 42), SpoIIAB prevents activation in the mother cell (8, 41), and the protease LonA prevents activation under nonsporulating conditions (40). Mutations causing premature σG activation map to the genes encoding these regulators. Artificial overexpression of sigG can also uncouple σG activation from the completion of engulfment (16). Despite these findings, the nature of the link between activation and engulfment has remained enigmatic. Here we explored its robustness by testing the effect of perturbations of chromosome replication and translocation, without affecting the known σG regulators by mutation or overexpression. Our principal finding is that there is indeed a strong link between the completion of engulfment and activation of σG. In addition, the results are consistent with a link between the completion of chromosome translocation and the completion of engulfment.
The greatest effect on engulfment resulted from insertion of 2.3 Mb of Synechocystis DNA in the B. subtilis chromosome. The presence of the insert delayed completion of engulfment by at least 90 min. The results were consistent with chromosome translocation also being substantially delayed and with the delay in engulfment being a consequence of the delay in the completion of translocation. Even with such a long delay, activation of σG was detected only after engulfment had been completed, suggesting a robust linkage between the two processes.
In wild-type B. subtilis, it takes about 15 to 20 min to complete translocation of the origin-distal ∼3.1 Mb of the chromosome from the mother cell into the prespore (3, 27, 36). We do not know how much DNA is in the prespore when it is first formed in strains containing 2.3 Mb of Synechocystis DNA. However, the long time of translocation of the rest of the chromosome into the prespore seems disproportionate to the size of the additional DNA. A possible explanation is that the prolonged delay in translocation caused by the Synechocystis DNA results from the lack of directional skew in copies of the octamer GAGAAGGG, named SRS6, in the Synechocystis DNA (31, 38). In the B. subtilis chromosome, the orientation of SRS6 is highly skewed and directs the SpoIIIE translocase to the terminus (37).
Two other sets of experiments also tested the effects of perturbations of chromosome replication and translocation. In one, a SpoIIIE translocase was used in which translocation is delayed by about 30 min (3). Again, despite the delayed translocation, σG became active only after completion of engulfment (Table 2; Fig. 5F). In the other, a strain was used in which termination of replication occurred at 145° relative to the origin at 0° on the circular chromosome map, rather than at the normal position, 172° (17, 19). Relocating the terminus of replication had no effect on σG activation, which occurred only after the completion of engulfment (Table 2; Fig. 5B). It also appeared not to delay completion of translocation, as terminus-proximal σF-directed reporters were expressed before the completion of engulfment. Given the role of SRS6 octamers in directing the DNA translocase toward the terminus region (37), this result was somewhat surprising. However, there are just four SRS6 sequences between 145° and 172°, one pointing toward 172° and three away from it, so changing the terminus of replication within this region may not greatly affect translocation.
Together, the various experiments indicate that there is a robust link ensuring that σG becomes active only upon the completion of engulfment. They do not indicate what the link is. Nevertheless, the link is central to activation of σG. Probably the most dramatic changes upon the completion of engulfment are, first, that the prespore is no longer in direct contact with the external medium, being completely surrounded by the mother cell, and, second, that it is separated from the mother cell by two opposed membranes. The requirement for the AA-AH · Q channel (4, 30) emphasizes the effect of completing engulfment but does not provide a sufficient explanation for σG activation. The channel is not needed for σF or artificially expressed T7 RNA polymerase to be active in the prespore before engulfment completion, but it is needed for them to be active after the completion of engulfment. Hence, the idea that the channel is a feeding tube, necessary to provide some key metabolite (possibly ATP) (4). But before completion of engulfment, enough key metabolite was present in the prespore to ensure σF activity and T7 polymerase activity, so why is σG not active? Completion of engulfment presumably causes a general shock to the prespore. It could cause a conformational change in a regulatory protein that is associated with the engulfment machinery. It could reduce the uptake of micronutrients into the prespore. It could, at least transiently, disrupt energy generation in the prespore. Any of these effects could be sufficient to trip the autocatalytic loop to activate σG expression, overcoming the inhibitory effects of regulators such as CsfB.
ACKNOWLEDGMENTS
This work was supported by Public Health Service grants GM43577 (to P.J.P.) and T32AI07101 (to G.R.) from the National Institutes of Health.
We are very grateful to Gerry Wake, Liz Harry, David Rudner, and Vasant Chary for kindly providing strains. We thank Tina Buttaro, Vasant Chary, and Panos Xenopoulos for many helpful discussions.
Footnotes
Published ahead of print 14 September 2012
REFERENCES
- 1. Bath J, Wu LJ, Errington J, Wang JC. 2000. Role of Bacillus subtilis SpoIIIE in DNA transport across the mother cell-prespore division septum. Science 290:995–997 [DOI] [PubMed] [Google Scholar]
- 2. Bogush M, Xenopoulos P, Piggot PJ. 2007. Separation of chromosome termini during sporulation of Bacillus subtilis depends on SpoIIIE. J. Bacteriol. 189:3564–3572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Burton BM, Marquis KA, Sullivan NL, Rapoport TA, Rudner DZ. 2007. The ATPase SpoIIIE transports DNA across fused septal membranes during sporulation in Bacillus subtilis. Cell 131:1301–1312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Camp AH, Losick R. 2009. A feeding tube model for activation of a cell-specific transcription factor during sporulation in Bacillus subtilis. Genes Dev. 23:1014–1024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Camp AH, Losick R. 2008. A novel pathway of intercellular signalling in Bacillus subtilis involves a protein with similarity to a component of type III secretion channels. Mol. Microbiol. 69:402–417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Camp AH, Wang AF, Losick R. 2011. A small protein required for the switch from σF to σG during sporulation in Bacillus subtilis. J. Bacteriol. 193:116–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Carrigan CM, Pack RA, Smith MT, Wake RG. 1991. Normal terC-region of the Bacillus subtilis chromosome acts in a polar manner to arrest the clockwise replication fork. J. Mol. Biol. 222:197–207 [DOI] [PubMed] [Google Scholar]
- 8. Chary VK, Meloni M, Hilbert DW, Piggot PJ. 2005. Control of the expression and compartmentalization of σG activity during sporulation of Bacillus subtilis by regulators of σF and σE. J. Bacteriol. 187:6832–6840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chary VK, Piggot PJ. 2003. Postdivisional synthesis of the Sporosarcina ureae DNA translocase SpoIIIE either in the mother cell or in the prespore enables Bacillus subtilis to translocate DNA from the mother cell to the prespore. J. Bacteriol. 185:879–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chary VK, Xenopoulos P, Piggot PJ. 2006. Blocking chromosome translocation during sporulation of Bacillus subtilis can result in prespore-specific activation of σG that is independent of σE and of engulfment. J. Bacteriol. 188:7267–7273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chary VK, Xenopoulos P, Piggot PJ. 2007. Expression of the σF-directed csfB locus prevents premature appearance of σG activity during sporulation of Bacillus subtilis. J. Bacteriol. 189:8754–8757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dawes IW, Kay D, Mandelstam J. 1969. Sporulation in Bacillus subtilis. Establishment of a time scale for the morphological events. J. Gen. Microbiol. 56:171–179 [DOI] [PubMed] [Google Scholar]
- 13. Decatur AL, Losick R. 1996. Identification of additional genes under the control of the transcription factor σF of Bacillus subtilis. J. Bacteriol. 178:5039–5041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. de Hoon MJ, Eichenberger P, Vitkup D. 2010. Hierarchical evolution of the bacterial sporulation network. Curr. Biol. 20:R735–R745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. de Lencastre H, Piggot PJ. 1979. Identification of different sites of expression for spo loci by transformation of Bacillus subtilis. J. Gen. Microbiol. 114:377–389 [DOI] [PubMed] [Google Scholar]
- 16. Doan T, et al. 2009. Novel secretion apparatus maintains spore integrity and developmental gene expression in Bacillus subtilis. PLoS Genet. 5:e1000566 doi:10.1371/journal.pgen.1000566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Duggin IG, Wake RG, Bell SD, Hill TM. 2008. The replication fork trap and termination of chromosome replication. Mol. Microbiol. 70:1323–1333 [DOI] [PubMed] [Google Scholar]
- 18. Dunn G, Mandelstam J. 1977. Cell polarity in Bacillus subtilis: effect of growth conditions on spore positions in sister cells. J. Gen. Microbiol. 103:201–205 [DOI] [PubMed] [Google Scholar]
- 19. Franks AH, Wake RG. 1996. Replication fork arrest at relocated replication terminators on the Bacillus subtilis chromosome. J. Bacteriol. 178:4258–4265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Harry EJ, Pogliano K, Losick R. 1995. Use of immunofluorescence to visualize cell-specific gene expression during sporulation in Bacillus subtilis. J. Bacteriol. 177:3386–3393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hauser PM, Errington J. 1995. Characterization of cell cycle events during the onset of sporulation in Bacillus subtilis. J. Bacteriol. 177:3923–3931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hilbert DW, Piggot PJ. 2004. Compartmentalization of gene expression during Bacillus subtilis spore formation. Microbiol. Mol. Biol. Rev. 68:234–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Illing N, Errington J. 1991. Genetic regulation of morphogenesis in Bacillus subtilis: roles of σE and σF in prespore engulfment. J. Bacteriol. 173:3159–3169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Itaya M, Tsuge K, Koizumi M, Fujita K. 2005. Combining two genomes in one cell: stable cloning of the Synechocystis PCC6803 genome in the Bacillus subtilis 168 genome. Proc. Natl. Acad. Sci. U. S. A. 102:15971–15976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Karmazyn-Campelli C, Bonamy C, Savelli B, Stragier P. 1989. Tandem genes encoding sigma-factors for consecutive steps of development in Bacillus subtilis. Genes Dev. 3:150–157 [DOI] [PubMed] [Google Scholar]
- 26. Karmazyn-Campelli C, et al. 2008. How the early sporulation sigma factor σF delays the switch to late development in Bacillus subtilis. Mol. Microbiol. 67:1169–1180 [DOI] [PubMed] [Google Scholar]
- 27. Khvorova A, Chary VK, Hilbert DW, Piggot PJ. 2000. The chromosomal location of the Bacillus subtilis sporulation gene spoIIR is important for its function. J. Bacteriol. 182:4425–4429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kirchman PA, DeGrazia H, Kellner EM, Moran CP., Jr 1993. Forespore-specific disappearance of the sigma-factor antagonist SpoIIAB: implications for its role in determination of cell fate in Bacillus subtilis. Mol. Microbiol. 8:663–671 [DOI] [PubMed] [Google Scholar]
- 29. Kroos L, Zhang B, Ichikawa H, Yu YT. 1999. Control of sigma factor activity during Bacillus subtilis sporulation. Mol. Microbiol. 31:1285–1294 [DOI] [PubMed] [Google Scholar]
- 30. Meisner J, Wang X, Serrano M, Henriques AO, Moran CP., Jr 2008. A channel connecting the mother cell and forespore during bacterial endospore formation. Proc. Natl. Acad. Sci. U. S. A. 105:15100–15105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Mrazek J, Karlin S. 1998. Strand compositional asymmetry in bacterial and large viral genomes. Proc. Natl. Acad. Sci. U. S. A. 95:3720–3725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Partridge SR, Errington J. 1993. The importance of morphological events and intercellular interactions in the regulation of prespore-specific gene expression during sporulation in Bacillus subtilis. Mol. Microbiol. 8:945–955 [DOI] [PubMed] [Google Scholar]
- 33. Piggot PJ, Curtis CA. 1987. Analysis of the regulation of gene expression during Bacillus subtilis sporulation by manipulation of the copy number of spo-lacZ fusions. J. Bacteriol. 169:1260–1266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Piggot PJ, Curtis CA, de Lencastre H. 1984. Use of integrational plasmid vectors to demonstrate the polycistronic nature of a transcriptional unit (spoIIA) required for sporulation of Bacillus subtilis. J. Gen. Microbiol. 130:2123–2136 [DOI] [PubMed] [Google Scholar]
- 35. Piggot PJ, Losick R. 2002. Sporulation and intercompartmental regulation, p 483–518. In Sonenshein AL, Hoch JA, Losick R. (ed), Bacillus subtilis and its closest relatives: from genes to cells. American Society for Microbiology, Washington, DC [Google Scholar]
- 36. Pogliano J, et al. 1999. A vital stain for studying membrane dynamics in bacteria: a novel mechanism controlling septation during Bacillus subtilis sporulation. Mol. Microbiol. 31:1149–1159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ptacin JL, et al. 2008. Sequence-directed DNA export guides chromosome translocation during sporulation in Bacillus subtilis. Nat. Struct. Mol. Biol. 15:485–493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Salzberg SL, Salzberg AJ, Kerlavage AR, Tomb JF. 1998. Skewed oligomers and origins of replication. Gene 217:57–67 [DOI] [PubMed] [Google Scholar]
- 39. Schaeffer P, Millet J, Aubert JP. 1965. Catabolic repression of bacterial sporulation. Proc. Natl. Acad. Sci. U. S. A. 54:704–711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Schmidt R, Decatur AL, Rather PN, Moran CP, Jr, Losick R. 1994. Bacillus subtilis Lon protease prevents inappropriate transcription of genes under the control of the sporulation transcription factor σG. J. Bacteriol. 176:6528–6537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Serrano M, Neves A, Soares CM, Moran CP, Jr, Henriques AO. 2004. Role of the anti-sigma factor SpoIIAB in regulation of σG during Bacillus subtilis sporulation. J. Bacteriol. 186:4000–4013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Serrano M, et al. 2011. A negative feedback loop that limits the ectopic activation of a cell type-specific sporulation sigma factor of Bacillus subtilis. PLoS Genet. 7:e1002220 doi:10.1371/journal.pgen.1002220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sharp MD, Pogliano K. 1999. An in vivo membrane fusion assay implicates SpoIIIE in the final stages of engulfment during Bacillus subtilis sporulation. Proc. Natl. Acad. Sci. U. S. A. 96:14553–14558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Stragier P. 1989. Temporal and spatial control of gene expression during sporulation: from facts to speculations, p 243–254. In Smith I, Slepecky RA, Setlow P. (ed), Regulation of prokaryotic development. American Society for Microbiology, Washington, DC [Google Scholar]
- 45. Sun D, et al. 1991. Effect of chromosome location of Bacillus subtilis forespore genes on their spo gene dependence and transcription by σF: identification of features of good σF-dependent promoters. J. Bacteriol. 173:7867–7874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Veening JW, et al. 2008. Bet-hedging and epigenetic inheritance in bacterial cell development. Proc. Natl. Acad. Sci. U. S. A. 105:4393–4398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wu LJ, Errington J. 1994. Bacillus subtilis SpoIIIE protein required for DNA segregation during asymmetric cell division. Science 264:572–575 [DOI] [PubMed] [Google Scholar]
- 48. Wu LJ, Errington J. 1997. Septal localization of the SpoIIIE chromosome partitioning protein in Bacillus subtilis. EMBO 16:2161–2169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wu LJ, Errington J. 1998. Use of asymmetric cell division and spoIIIE mutants to probe chromosome orientation and organization in Bacillus subtilis. Mol. Microbiol. 27:777–786 [DOI] [PubMed] [Google Scholar]
- 50. Xenopoulos P, Piggot PJ. 2011. Regulation of growth of the mother cell and chromosome replication during sporulation of Bacillus subtilis. J. Bacteriol. 193:3117–3126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Zupancic ML, Tran H, Hofmeister AE. 2001. Chromosomal organization governs the timing of cell type-specific gene expression required for spore formation in Bacillus subtilis. Mol. Microbiol. 39:1471–1481 [DOI] [PubMed] [Google Scholar]