Abstract
Nitrosomonas europaea is a chemolithoautotroph that obtains energy by oxidizing ammonia in the presence of oxygen and fixes CO2 via the Benson-Calvin cycle. Despite its environmental and evolutionary importance, very little is known about the regulation and metabolism of glycogen, a source of carbon and energy storage. Here, we cloned and heterologously expressed the genes coding for two major putative enzymes of the glycogen synthetic pathway in N. europaea, ADP-glucose pyrophosphorylase and glycogen synthase. In other bacteria, ADP-glucose pyrophosphorylase catalyzes the regulatory step of the synthetic pathway and glycogen synthase elongates the polymer. In starch synthesis in plants, homologous enzymes play similar roles. We purified to homogeneity the recombinant ADP-glucose pyrophosphorylase from N. europaea and characterized its kinetic, regulatory, and oligomeric properties. The enzyme was allosterically activated by pyruvate, oxaloacetate, and phosphoenolpyruvate and inhibited by AMP. It had a broad thermal and pH stability and used different divalent metal ions as cofactors. Depending on the cofactor, the enzyme was able to accept different nucleotides and sugar phosphates as alternative substrates. However, characterization of the recombinant glycogen synthase showed that only ADP-Glc elongates the polysaccharide, indicating that ATP and glucose-1-phosphate are the physiological substrates of the ADP-glucose pyrophosphorylase. The distinctive properties with respect to selectivity for substrates and activators of the ADP-glucose pyrophosphorylase were in good agreement with the metabolic routes operating in N. europaea, indicating an evolutionary adaptation. These unique properties place the enzyme in a category of its own within the family, highlighting the unique regulation in these organisms.
INTRODUCTION
Synthesis of the main reserve polysaccharide in bacteria (glycogen) and plants (starch) occurs using ADP-glucose (ADP-Glc) as the glucosyl donor to elongate an α-1,4-glucosidic chain (5, 7, 8). The main regulatory step of this biosynthetic pathway in both bacteria and plants is catalyzed by ADP-glucose pyrophosphorylase (ADP-Glc PPase; EC 2.7.7.27) in the presence of a divalent metal ion: ATP + α-d-glucose-1-phosphate (Glc-1P) ↔ ADP-Glc + PPi (4). Although the in vitro reaction is freely reversible, in the cell it proceeds toward ADP-Glc synthesis, due to hydrolysis of PPi and use of the sugar nucleotide. To date, characterization of ADP-Glc PPases shows that key intermediates of the principal carbon assimilatory pathway in the organism allosterically regulate the enzyme. Regulatory properties of the enzyme ensure that maximal activity is reached under conditions of high carbon and metabolic energy contents in the cell (6, 7).
ADP-Glc PPases are clustered into nine different groups on the basis of their selectivity for allosteric regulators and structure, whereby their properties are correlated with the main carbon assimilation pathways of the respective organism (6, 7). For instance, in heterotrophic bacteria the enzyme is inhibited by AMP/ADP and principally activated by fructose-1,6-bisphosphate (Fru-1,6-bisP) or fructose-1,6-phosphate (Fru-6P) and pyruvate (Pyr). These metabolites are from key steps of either the Embden-Meyerhof-Parnas or the Entner-Doudoroff glycolytic route. Organisms performing oxygenic photosynthesis and fixing CO2 via the reductive pentose phosphate pathway or Benson-Calvin cycle (cyanobacteria, green algae, and higher plants) contain ADP-Glc PPase mainly activated by 3-phosphoglycerate (3P-glycerate; the first intermediate generated in carbon fixation) and inhibited by Pi (the substrate for ATP synthesis by photophosphorylation) (28). Recently, the enzyme from the Gram-positive bacterium Streptomyces coelicolor was characterized and revealed distinctive properties unlike those of other groups, which forced the launch of a new group among ADP-Glc PPases (3).
Nitrosomonas europaea is a betaproteobacterium that as a facultative chemolithoautotroph can grow either heterotrophically or autotrophically (2, 43). In the latter case, all of its energy and reducing power are derived from oxidation of ammonia to nitrite and fixation of atmospheric CO2 using the Benson-Calvin cycle (14). This microorganism has the ability to be used in bioremediation applications because it can metabolize chlorinated aliphatic hydrocarbons (30, 32). Additionally, it is moderately halotolerant and can grow in sewage disposal plants, freshwater habitats, and fertilized agricultural soils (32). Elucidation of genomes from ammonia-oxidizing bacteria over the last few years helped provide an understanding of the metabolism and physiology of these environmentally and biotechnologically significant microorganisms (2). However, knowledge regarding the functional genomics and operation and regulation of metabolic routes is limited due to a lack of biochemical studies to characterize their enzymes, especially from Nitrosomonas spp.
Herein, we report the molecular cloning of the genes coding for putative ADP-Glc PPase and glycogen synthase (GSase) in N. europaea. After heterologous expression of the genes, the recombinant enzymes were purified and characterized. GSase was specific for ADP-Glc, whereas ADP-Glc PPase exhibited a degree of promiscuity for substrates and divalent metal ions acting as essential cofactors. Results are analyzed in the context of the occurrence of glycogen accumulation in Nitrosomonas spp. and the relationship of this process with other major metabolic pathways operating in this bacterium. To the best of our knowledge, this is the first time that the enzymes involved in polysaccharide biosynthesis have been studied in chemolithoautotrophic organisms.
MATERIALS AND METHODS
Chemicals.
Inorganic pyrophosphatase, Glc-1P, ATP, ADP, AMP, oxaloacetate (OAA), Pyr, phosphoenolpyruvate (PEP), and oligonucleotides were purchased from Sigma-Aldrich (St. Louis, MO). All other reagents were of the highest quality available.
Cloning, expression, and purification.
The gene coding for a putative ADP-Glc PPase in N. europaea (locus tag, NE2030) was amplified by PCR in a reaction mixture containing 25 ng of N. europaea ATCC 19817 genomic DNA, 50 μM both primers (NeuAGPFoward [TACTTCCAATCCAATGCCGCAGCAATGAAAGTTCAACCAGCTGTCAGACG] and NeuAGPReverse [TTATCCACTTCCAAGTCGGATGTAGTGGATGCC]), 200 μM deoxynucleoside triphosphates, and 0.02 U/μl Phusion DNA polymerase (New England BioLabs). We used the procedure of initial denaturation for 30 s at 98°C, 30 cycles of denaturation at 98°C for 5 s, annealing at 50°C for 20 s, and extension at 72°C for 1 min, and finally, a 5-min extension at 72°C. The PCR product was gel purified, after running agarose gel electrophoresis, and cloned into the pMCSG9 vector as described previously (17, 37, 49). The final plasmid construct includes a maltose-binding protein (MBP) tag, a polyhistidine (10-histidine) tag, a tobacco etch virus (TEV) protease cleavage site, two extra alanine residues, and the open reading frame of the N. europaea ADP-Glc PPase gene. The plasmid was transformed into Escherichia coli BL21(DE3)/magic cells for protein expression. The magic plasmid is kanamycin resistant, codes for three rare-triplet tRNAs (AGG for Arg, AGA for Arg, and ATA for Ile), and is controlled by a T7 promoter and a kanamycin resistance gene (54).
Protein was expressed using the following procedure. A starter culture was inoculated, and cells were grown overnight at 37°C with shaking at 200 rpm in 3 ml LB supplemented with 100 μg/ml ampicillin and 50 μg/ml kanamycin. The overnight culture was diluted 1/100 in fresh medium and grown under identical conditions to exponential phase (optical density at 600 nm, 0.6) at 37°C. Recombinant protein expression was induced with 0.5 mM isopropyl-β-d-thiogalactopyranoside for 16 h at 16°C, and then cells were harvested by centrifugation. The bacterial pellet was resuspended in binding buffer C (20 mM Tris, pH 8.0, 200 mM NaCl, 5 mM β-mercaptoethanol, 10% [vol/vol] glycerol) and disrupted by sonication. The lysate was centrifuged (36,000 × g, 15 min) to remove cellular debris. The resulting crude extract was loaded onto a Ni2+-nitrilotriacetic acid (NTA) (GE Healthcare) resin column that had been equilibrated with binding buffer. The column was washed with 2 column volumes each of buffer C plus 20 mM and then 40 mM imidazole. Finally, the recombinant protein was eluted with buffer C plus 300 mM imidazole.
To remove the MBP and polyhistidine tag from the ADP-Glc PPase, a polyhistidine-tagged TEV protease was added (0.1 mg/ml plus 5 mM EDTA) to the eluted fraction of the fusion protein, and the mixture was incubated overnight at 4°C. The sample was precipitated with (NH4)2SO4 to 60% saturation, centrifuged (36,000 × g, 15 min) to obtain a precipitate that was resuspended in buffer C, and loaded in a Ni2+-NTA resin equilibrated with the same buffer. The flowthrough fraction was pooled and reequilibrated in buffer A (50 mM MOPS [morpholinepropanesulfonic acid], pH 8.0, 5% [wt/vol] sucrose, 1 mM MgCl2, 0.1 mM EDTA) using a BioGel fast-desalting P-6DG (Bio-Rad) column. The protein was further purified by ion-exchange chromatography using a Q-Resource (GE Healthcare) column and eluted with a linear gradient of buffer B (buffer A plus 1 M NaCl). Fractions with enzymatic activity were pooled, desalted in buffer A, concentrated using an ultrafiltration device (AmiconUltra-30; Millipore), and stored at −80°C. Under these conditions, the enzyme remained fully active for at least 3 months.
Enzyme activity assays.
ADP-Glc PPase activity was assayed in the physiological direction of ADP-Glc synthesis. The assays were performed at 37°C and pH 8.0 using a colorimetric method developed by Fusari et al. (22). The standard assay medium contained (unless otherwise indicated) 100 mM MOPS (pH 8.0), 1 mM ATP, 1 mM Glc-1P, 30 mM MgCl2, 0.0015 U/μl inorganic pyrophosphatase, and 0.2 mg/ml bovine serum albumin (BSA) plus enzyme in a total volume of 50 μl. The reaction was stopped with the addition of malachite green color reagent, and the absorbance was measured at 650 nm. One unit of enzymatic activity is defined as the amount of enzyme catalyzing the formation of 1 μmol PPi/min under the above-specified conditions.
Alternative metal ions for ADP-Glc PPase were studied by performing the reaction in the absence of inorganic pyrophosphatase and stopped with boiling water and 10 mM EDTA. Then, Mg2+ and inorganic pyrophosphatase were added to the samples, the reaction mixture was further incubated for 10 min at 37°C, and the reaction was stopped with malachite green as described above. This was necessary due to the essential requirement of Mg2+ for activity of inorganic pyrophosphatase.
Kinetic studies.
Enzyme activity was assayed using a saturating concentration of one substrate (and/or effector) and by varying the concentration of substrate or effector in question. Kinetic constants were determined by plotting experimental data as enzyme activity (U/mg) versus substrate (or effector) concentration (mM). Data were fit to the Hill equation using the Levenberg-Marquardt nonlinear least-squares algorithm provided by the computer program Origin (version 7.0), as described elsewhere (5). Hill plots were used to calculate the Hill coefficient (nH), maximal velocity (Vmax), and kinetic constants corresponding to the activator, substrate, or inhibitor concentration giving 50% of the maximal activation (A0.5), velocity (S0.5), or inhibition (I0.5). Values for all kinetic constants are the means of at least three sets of data, which were reproducible within ±10%.
Gel filtration.
The quaternary structure of the enzyme was determined using Superdex 200 resin (GE Healthcare) packed in a 20-cm-length Tricorn 5/200 column (GE Healthcare). The column was equilibrated with buffer G (25 mM Tris-HCl, pH 8.0, 100 mM NaCl). Approximately 50 μg of ADP-Glc PPase or molecular mass standards in a 50-μl volume was loaded onto the column using a flow rate of 0.2 ml/min. The column calibration was performed with a gel filtration high-molecular-mass kit (GE Healthcare) including protein standards thyroglobulin (669 kDa), ferritin (440 kDa), aldolase (158 kDa), conalbumin (75 kDa), and ovalbumin (44 kDa). The column void volume was determined using a dextran blue loading solution (Promega). The molecular mass of the ADP-Glc PPase protein was extrapolated from the standard semilog curve Kav versus log molecular mass. Kav is the parameter defined by the following equation: (Ve − Vo)/(Vc − Vo), where Ve is the elution volume, Vo is the void volume, and Vc is the column volume.
Protein methods.
Electrophoresis under denaturing conditions (SDS-PAGE) was performed in 15% gels as described by Laemmli (34), and Coomassie brilliant blue was used to stain protein bands. Protein concentration was measured using the Bradford method (10) with BSA as a standard.
Effect of pH and temperature on protein stability and activity.
To evaluate protein stability at different pHs, the purified N. europaea ADP-Glc PPase (∼8 μM) was incubated at 37°C for 10 min in 50 mM buffer at pH values ranging from 4.7 to 11.0. Temperature stability was assessed using purified enzyme incubated in 50 mM MOPS (pH 8.0) buffer for 10 min at temperatures ranging from 4 to 70°C. An aliquot of enzyme from each condition was used to measure activity under the standard assay conditions described. The buffer used for activity assays of the enzyme at various pH values was the same buffer used to test pH stability.
RESULTS
Cloning, expression, and purification.
Analysis of the reported genome from N. europaea (11) revealed genes involved in glycogen metabolism. Specifically, genes encoding glycogen synthase (glgA, NE2264), branching enzyme (glgB, NE2029), ADP-Glc PPase (glgC, NE2030), and glycogen phosphorylase (glgP, NE0466 and NE0074) were identified. To gain insight into the biochemical characteristics of this bacterium, we cloned the full-length glgC gene, since it is known that in other bacteria ADP-Glc PPase catalyzes the rate-limiting step for glycogen biosynthesis (6). The gene was amplified by PCR from genomic DNA, and its identity was confirmed by DNA sequencing. The N. europaea glgC gene is predicted to encode a 435-amino-acid protein with a molecular mass of 49.2 kDa and a theoretical pI of 5.82 (calculated with Vector NTI [version 10.0] software). The protein is similar in size to other ADP-Glc PPases, and it shares higher sequence identity with orthologs from heterotrophic bacteria (52.9%, 39.2%, and 39.4% to the enzyme from E. coli, Mycobacterium tuberculosis, and Agrobacterium tumefaciens, respectively) than with those from cyanobacteria (30.7% for Anabaena sp. strain PCC 7120) or from plants (31.0% for Solanum tuberosum, small subunit).
To elucidate the functional properties of the N. europaea ADP-Glc PPase, the protein was heterologously expressed in E. coli and purified using immobilized metal ion affinity chromatography followed by ion-exchange chromatography. The protein was determined to be greater than 90% pure from densitometry and was the major band at ∼49 kDa in SDS-PAGE (Fig. 1). The pure enzyme was active as an ADP-Glc PPase, exhibiting a specific activity of ∼20 U/mg for the synthesis of ADP-Glc when assayed under the standard conditions (see Materials and Methods). Moreover, the purified N. europaea enzyme migrated as a tetrameric protein (∼200 kDa) when analyzed by gel filtration chromatography (data not shown), in agreement with the structures reported for characterized ADP-Glc PPases from prokaryotes and eukaryotes (6, 7).
Fig 1.
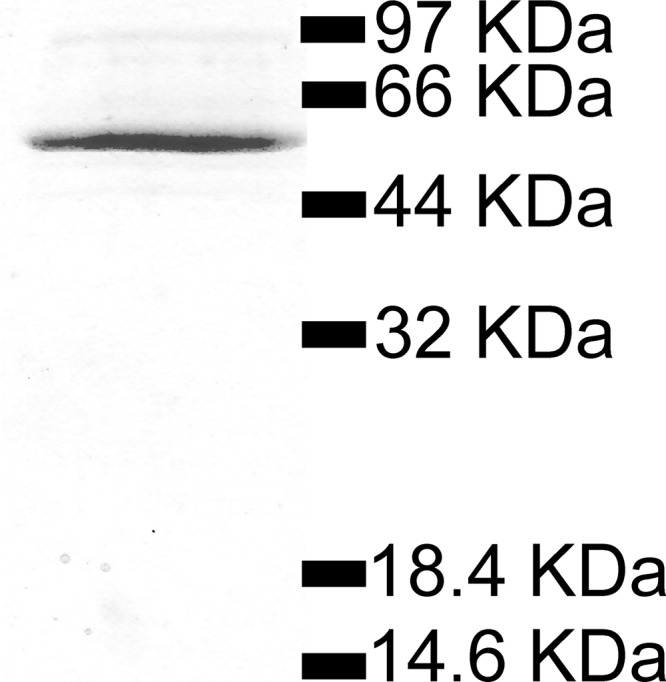
SDS-PAGE analysis of the purified N. europaea ADP-Glc PPase. Molecular masses correspond to those of standard proteins.
ADP-Glc PPase metal usage and kinetic parameters.
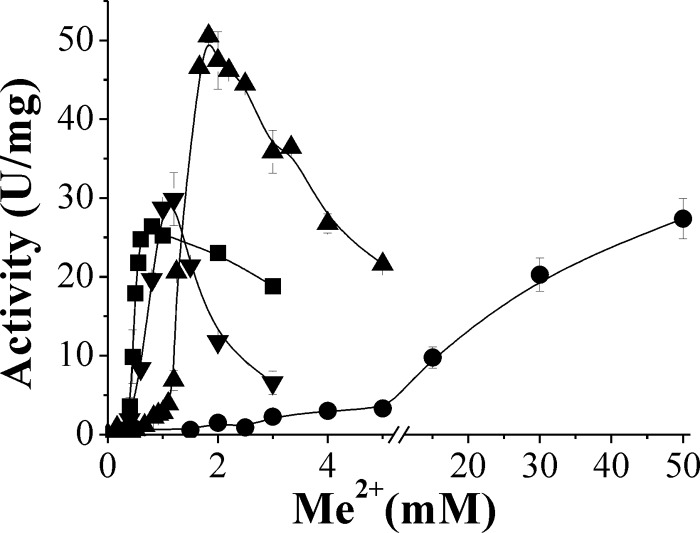
Metal ions are essential for charge compensation of the negatively charged sugar-phosphate backbone, they are instrumental for proper folding, and they are crucial cofactors for enzymes using nucleotides. Among divalent metal ions, Mg2+ is certainly the most abundant one, is freely available in any cell, and is involved in several physiological processes (20). ADP-Glc PPases from both plants and bacteria require a divalent metal ion as an essential cofactor (4–7). S0.5 values for Mg2+ are on the order of 1 to 10 mM. The enzymes from A. tumefaciens, E. coli, and Anabaena PCC 7120 have S0.5 values of 2.2 mM, 11.5 mM, and 1.5 mM, respectively (23, 27, 50). However, the N. europaea ADP-Glc PPase exhibited distinctive kinetic behavior with respect to saturation by Mg2+. In the direction of synthesis of ADP-Glc, this enzyme had a relatively low affinity for Mg2+ (S0.5, >40 mM), and it was practically impossible to reach saturation using this divalent metal (Fig. 2 and Table 1).
Fig 2.
Enzymatic activity of N. europaea ADP-Glc PPase versus concentrations of divalent metal ions (Me2+): Mg2+ (•), Mn2+ (▲), Co2+ (■), and Cd2+ (▼). ATP and Glc-1P concentrations were 1 mM, with the exception for assays with Cd2+, where the concentration of Glc-1P was 2 mM.
Table 1.
Kinetic parameters of N. europaea ADP-Glc PPase for Mg2+, Mn2+, Co2+, and Cd2+a
| Metal ion | Vmax (U/mg) | S0.5 (mM) | nH | kcat/S0.5 (mM−1 · s−1) |
|---|---|---|---|---|
| Mg2+ | >50 | >40 | ND | ∼4 |
| Mn2+ | 52 ± 2 | 1.37 ± 0.05 | >3 | 126 ± 7 |
| Co2+ | 26.45 ± 0.05 | 0.46 ± 0.02 | >3 | 192 ± 8 |
| Cd2+ | 34 ± 5 | 0.74 ± 0.06 | >3 | 150 ± 30 |
The reaction mixture had the standard composition except for Cd2+, in which case it contained 2 mM instead of 1 mM Glc-1P. Only points corresponding to the ascending activity (from zero to the maximal) in Fig. 2 were used for data fitting. ND, not determined.
We explored the possibility that the N. europaea ADP-Glc PPase might prefer an alternative divalent metal because of its poor affinity for Mg2+. Out of the metals tested, neither Ba2+ nor Cu2+ was effective, and only low levels of activity were observed with Zn2+, Ni2+, and Ca2+. Conversely, Mn2+, Co2+, and Cd2+ behaved as effective cofactors (Fig. 2), and their kinetic parameters are shown in Table 1. The enzyme reached higher Vmax values with Mg2+ or Mn2+ but exhibited a higher affinity toward Co2+ and Cd2+. The ratio kcat/S0.5 (analogous to catalytic efficiency [kcat/Km] for hyperbolic kinetics) of the recombinant enzyme was similar for Mn2+, Co2+, and Cd2+ but very low for Mg2+ (Table 1). A more detailed analysis, shown in Fig. 2, illustrates that all divalent metal ions inhibit the enzyme at concentrations higher than 1 mM (Cd2+ and Co2+) or 2 mM (Mn2+), with the exception of Mg2+. Our results also indicate that the enzyme achieves maximal activity at a 1:1 ratio of Me to ATP (where Me indicates the generic divalent metal ion) using Co2+ or Cd2+, while it seems to be necessary to have an excess of the cation using Mg2+ or Mn2+, particularly Mg2+. It is possible that in addition to a divalent metal ion complex with ATP (the actual substrate), a second Mg2+ or Mn2+ ion binds to the enzyme to effectively enhance its catalytic capacity.
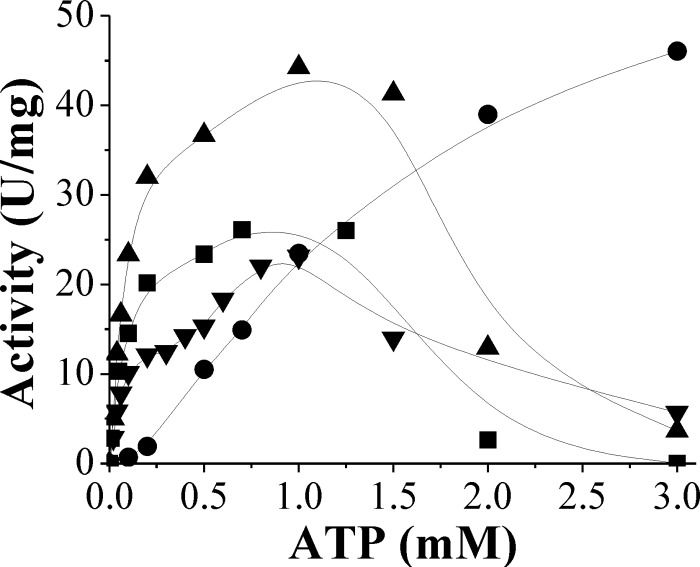
We determined the kinetic parameters of the purified N. europaea ADP-Glc PPase for the substrates ATP and Glc-1P and examined how they were affected by the different cofactors, Mg2+, Mn2+, Co2+, or Cd2+. The apparent affinity for Glc-1P was similar (S0.5, ∼0.1 mM) in the presence of either Mg2+, Mn2+, or Co2+ but about 5-fold lower in the presence of Cd2+ (Table 2). On the other hand, the kinetic parameters for ATP were more dependent upon the metal cofactor. Saturation plots for ATP at fixed concentrations of Co2+, Mn2+, or Cd2+ reached a maximal activity at between 25 and 40 U/mg, with S0.5 values of ∼0.1 to 0.3 mM and substrate inhibition beyond 1 to 2 mM (Fig. 3). Conversely, when assays were performed using 30 mM Mg2+, saturation plots were hyperbolic with estimated Vmax and S0.5 values of 50 U/mg and 1.4 mM, respectively. No substrate inhibition was observed up to 3 mM ATP (Fig. 3). These results agree with a model proposed by Segel (45) for enzymes in which the actual substrate is the Me-ATP complex and the free metal and/or nucleotide forms can act as inhibitors. This behavior is also consistent with the lag at the beginning of the metal saturation curves, where the Me-ATP complex-to-free ATP ratio is very low (Fig. 2). Absence of inhibition by ATP in the presence of Mg2+ can be explained because the nucleotide was never in excess relative to the cofactor, unlike the case of the other divalent metal ions. Remaining enzyme characterizations were performed using alternatively Mg2+ or Co2+. The former was used because of its relative abundance in nature (20, 31), and the latter was used because the enzyme exhibited the highest apparent affinity in the presence of this metal (Table 1).
Table 2.
Kinetic parameters of N. europaea ADP-Glc PPase for Glc-1P in the presence of different metal cofactorsa
| Glc-1P metal cofactor | Vmax (U/mg) | S0.5 (mM) | nH |
|---|---|---|---|
| Mg2+ | 18 ± 2 | 0.11 ± 0.02 | 1.1 ± 0.1 |
| Mn2+ | 40 ± 1 | 0.13 ± 0.01 | 1.32 ± 0.07 |
| Co2+ | 29.7 ± 0.2 | 0.13 ± 0.01 | 1.23 ± 0.03 |
| Cd2+ | 42 ± 1 | 0.72 ± 0.02 | 0.96 ± 0.01 |
Metal cofactor concentrations were 30 mM, 2 mM, 0.7 mM, and 1 mM for Mg2+, Mn2+, Co2+, and Cd2+, respectively.
Fig 3.
Saturation plots of N. europaea ADP-Glc PPase for ATP in the presence of 30 mM Mg2+ (•), 2 mM Mn2+ (▲), 0.7 mM Co2+ (■), or 1 mM Cd2+ (▼).
Temperature and pH dependence of ADP-Glc PPase stability and activity.
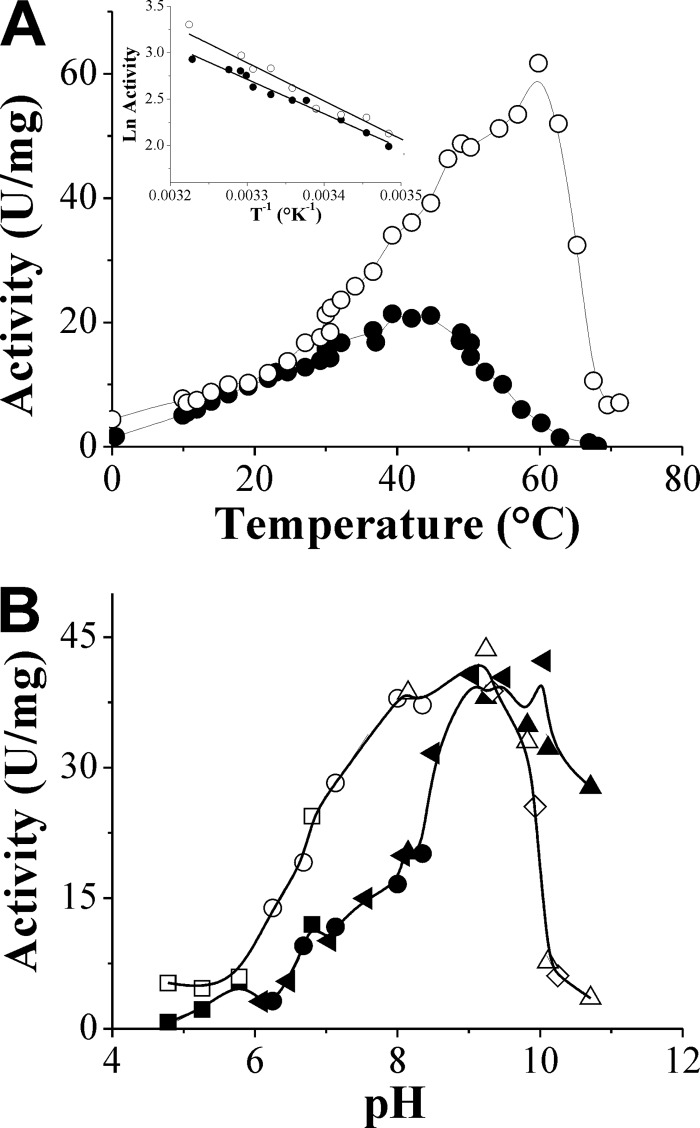
The activity and stability of N. europaea ADP-Glc PPase regarding its response to temperature and pH are shown in Fig. 4. The enzyme showed a remarkable thermal stability by remaining fully active at between 0 and 50°C for 15 min, 1 h, and 12 h at pH 8.0. It also remained stable for 30 min at 37°C over the broad pH range of 4.8 to 9.3. Optimal temperatures for enzyme activity were at 40 or 60°C, depending on whether the assays were performed using Mg2+ or Co2+ (Fig. 4A). The inset in Fig. 4A depicts Arrhenius plots (45), determined using either divalent cation over the temperature range where the enzyme was stable. The slopes of the lines of the Arrhenius plots were used to calculate the energy of activation (Ea) in the presence of Mg2+ or Co2+ as 31 ± 2 kJ mol−1 and 34 ± 2 kJ mol−1, respectively.
Fig 4.
Stability and enzymatic activity of N. europaea ADP-Glc PPase with temperature (A) and pH (B). Buffers used are morpholineethanesulfonic acid (■, □), bis-Tris-propane (◀), MOPS (•, ○), ethanolamine (▲, Δ), and N-cyclohexyl-3-aminopropanesulfonic acid (CAPS) (♢). Filled and empty symbols, the reaction mixture contained 30 mM Mg2+ or 1 mM Co2+, respectively. (Inset in panel A) Arrhenius plots of data in the main figure where the enzyme was stable. T, temperature.
The optimal pH for the recombinant N. europaea ADP-Glc PPase was 9.0 with either Mg2+ or Co2+, as shown in Fig. 4B. This optimal pH was the same as the one reported for the enzyme from Arthrobacter simplex (29) and somewhat different from the pH 7.5 found for the Chlorella vulgaris enzyme (36), where Mg2+ was used for the assays. Concerning the variations in the profiles of activity versus pH curves observed with Co2+ or Mg2+, results may be related to the distinct electronic characteristics determined for the metals and their particular group coordination preferences (16).
Promiscuity toward substrates of ADP-Glc PPase.
We investigated N. europaea ADP-Glc PPase substrate specificity for ATP and Glc-1P using either Co2+ or Mg2+ as essential cofactors. The enzyme was able to use other nucleoside triphosphates (NTPs) besides ATP as a substrate, with decreasing catalytic efficiencies being ATP > UTP ∼ CTP > dTTP > GTP (Table 3). These results indicate that there is a higher specificity between the two purines compared but a higher degree of promiscuity among the smaller pyrimidines. The catalytic efficiency for ATP was not dependent upon the divalent metal ion, but the promiscuity of the enzyme for other nucleotides was highly evident in the presence of Co2+, especially regarding UTP and CTP (Table 3). In assays performed with Mg2+, the catalytic efficiency with ATP was at least 2 orders of magnitude higher than that for the other NTPs. However, with Co2+ as the cofactor, catalytic efficiencies for ATP, UTP, or CTP were quite similar, with less than a 2-fold difference between them.
Table 3.
Kinetic parameters of N. europaea ADP-Glc PPase for alternative nucleotidesa
| Nucleotide | Mg2+ |
Co2+ |
||||||
|---|---|---|---|---|---|---|---|---|
| Vmax (U/mg) | S0.5 (mM) | nH | kcat/S0.5 (mM−1 · s−1) | Vmax (U/mg) | S0.5 (mM) | nH | kcat/S0.5 (mM−1 · s−1) | |
| ATP | 50 ± 9 | 1.38 ± 0.07 | 1.8 ± 0.1 | 120 ± 20 | 28.1 ± 0.8 | 0.09 ± 0.01 | 1.01 ± 0.01 | 1,000 ± 100 |
| UTP | 0.5 ± 0.2 | 2.0 ± 0.9 | 1.6 ± 0.5 | 0.8 ± 0.5 | 9 ± 2 | 0.54 ± 0.06 | 1.6 ± 0.2 | 60 ± 10 |
| CTP | 0.20 ± 0.08 | 1.0 ± 0.1 | 2.2 ± 0.4 | 0.7 ± 0.1 | 6.0 ± 0.3 | 0.21 ± 0.02 | 1.33 ± 0.07 | 90 ± 10 |
| dTTP | 0.05 ± 0.01 | 0.56 ± 0.09 | 3.8 ± 0.2 | 0.30 ± 0.06 | 0.26 ± 0.02 | 0.46 ± 0.05 | 2.0 ± 0.4 | 1.9 ± 0.2 |
| GTP | ∼0.4 | ∼2 | ND | ∼0.5 | 0.22 ± 0.03 | 0.58 ± 0.05 | 3.0 ± 0.9 | 1.2 ± 0.2 |
Mg2+ and Co2+ concentrations were 30 mM and 3 mM, respectively. ND, not determined.
We also investigated the substrate specificity for Glc-1P and observed that N. europaea ADP-Glc PPase could catalyze the reaction using mannose-1-phosphate (Man-1P) in place of Glc-1P. Vmax values for synthesis of ADP-Man were 20.7 ± 0.9 and 8 ± 1 U/mg using Co2+ and Mg2+, respectively. The apparent affinity for Man-1P was higher in the assays performed with Co2+ (S0.5 = 2.6 ± 0.2 mM) than in those performed with Mg2+ (S0.5 = 7 ± 1 mM). In contrast, no activity was detected with Man-1P (5 mM) and UTP (2 mM) in the presence of Mg2+ (30 mM), and a modest activity (∼0.15 U/mg) was measured with Co2+ (3 mM). The catalytic efficiency with Glc-1P is at least 1 order of magnitude higher than that with Man-1P (Table 2), and it is clear that Glc-1P is the preferred substrate.
Glycosyl donor for glycogen synthesis in N. europaea.
Since N. europaea ADP-Glc PPase was relatively promiscuous when ATP or UTP was used as the substrate, the question regarding which sugar-nucleotide (ADP-Glc and/or UDP-Glc) provides the glycosyl moiety necessary to elongate the glycogen molecule remained. To answer this, we explored the kinetic properties of the recombinant glycogen synthase from N. europaea (NeuGSase), which was produced and purified as described in the supplemental material. The NeuGSase was fully active with ADP-Glc but exhibited no activity with UDP-Glc. The specificity of the enzyme for ADP-Glc was not affected by divalent metal ions because similar results were obtained using Mg2+, Mn2+, or Co2+ in the assay medium. NeuGSase yielded hyperbolic saturation curves with a Vmax value of 50 U/mg using ADP-Glc, with S0.5 values of 0.14 mM (ADP-Glc) and 0.20 mg/ml (glycogen). The enzyme was found to be monomeric after size exclusion chromatography on Superdex 200, which is in agreement with the result found for glycogen synthase from E. coli (46).
Allosteric effectors for ADP-Glc PPase.
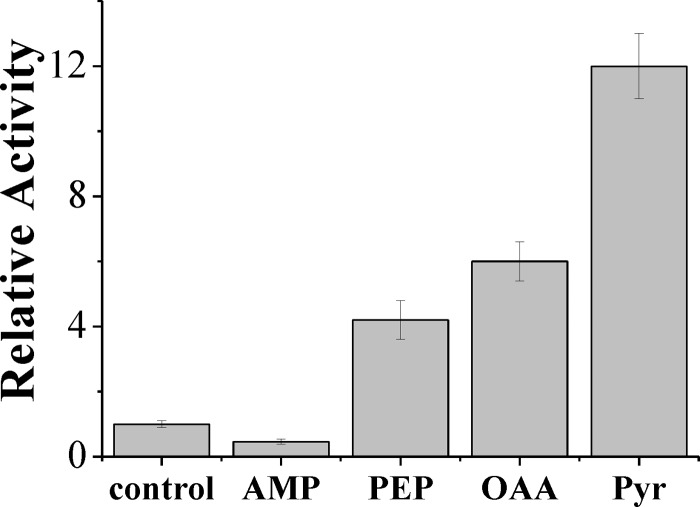
ADP-Glc PPase is finely regulated in bacteria and plants according to the main carbon metabolism occurring in the respective organism (4, 6, 7). It was therefore of interest to investigate the allosteric regulatory properties of the enzyme from N. europaea. To achieve this, we analyzed a wide variety of intermediates found in routes of carbon, nitrogen, and cellular energy metabolism. Specifically, we tested the following metabolites: NH4+, mannose-6-phosphate, glucose-6-phosphate, fructose-6-phosphate, fructose-1,6-bisphosphate, ribose-5-phosphate, NAD(P)+, NAD(P)H, serine, glutamine, asparagine, glutamate, aspartate, citrate, 3-phosphoglycerate, PEP, Pyr, OAA, 2-oxoglutarate, Pi, AMP, and ADP. Only AMP, PEP, OAA, and Pyr significantly modified the ADP-Glc PPase activity in the presence of Mg2+ (Fig. 5). AMP inhibited the enzyme by more than 50%, whereas PEP, OAA, and Pyr enhanced the activity by 4-, 6-, and 12-fold, respectively. Thus, in N. europaea glycogen synthesis could be finely modulated at the level of ADP-Glc PPase by the energy content in the cell. In this process, the enzyme is inhibited by a low energy signal (AMP) and activated by PEP, OAA, and Pyr, which comprise an important metabolic node in carbon/energy flux distribution in bacteria (43).
Fig 5.
Effect of metabolites on the activity of N. europaea ADP-Glc PPase. Assay conditions are the standard with 5 mM Mg2+. The metabolite concentration was 2 mM. control, enzymatic activity without effectors; bars, standard deviation of three independent assays.
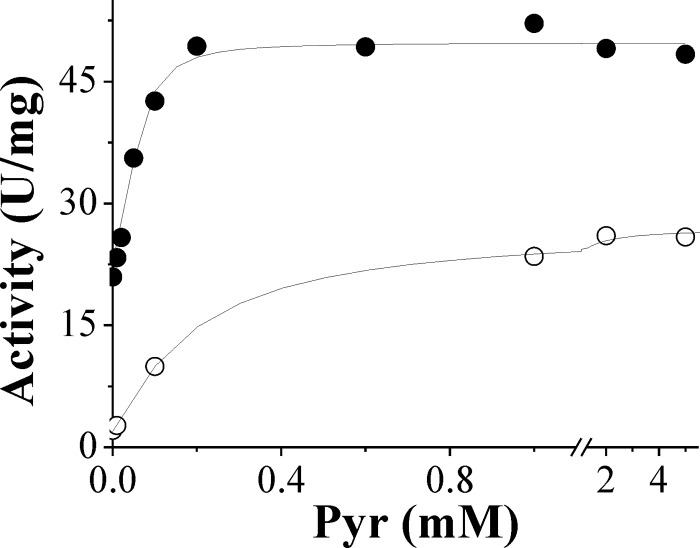
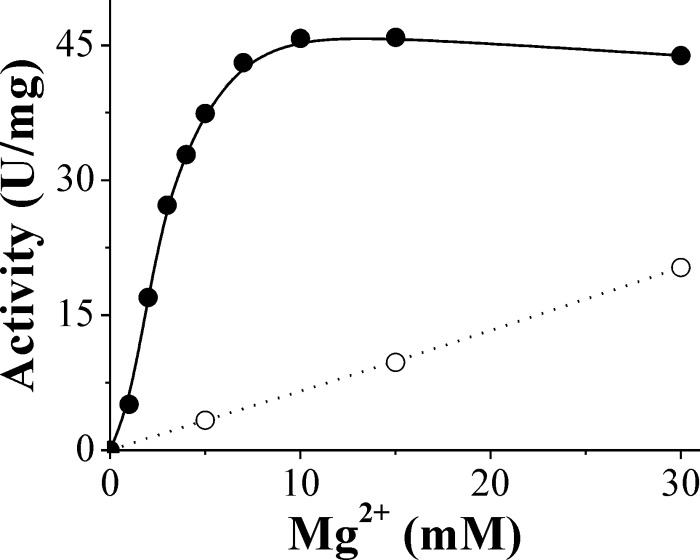
Pyruvate is the main activator of N. europaea ADP-Glc PPase, and its effect of activation is dependent upon Mg2+ concentrations. Saturation curves for Pyr show that the enzyme is activated 12- or 2.2-fold with A0.5 values for Pyr of 0.14 mM and 0.054 mM in the presence of either 5 mM and 30 mM Mg2+, respectively (Fig. 6). The divalent metal ion concentration effect on activation of the enzyme with Pyr was further investigated. It seems that the purpose of the activator is to enhance the enzyme's affinity for Mg2+. In the presence of 5 mM Pyr, the enzyme activity reached saturation at 10 mM Mg2+, with an S0.5 of 2.6 ± 0.1 mM (Fig. 7). In the absence of Pyr, more than 5-fold higher levels of Mg2+ were required to reach saturation (Fig. 2 and Table 1). Consequently, in the presence of Pyr, the catalytic efficiency of the enzyme using Mg2+ (Vmax/S0.5, 75 mM−1 · s−1) was about 20-fold higher than that in the absence of the allosteric activator (∼4 mM−1 · s−1). The effect of Pyr was specific for Mg2+, and its affinity was not significantly increased in the presence of other divalent cations (Mn2+, Co2+, and Cd2+) (data not shown). This synergy indicates a specific positive allosteric interaction between Pyr and Mg2+. Their respective affinities are reciprocally increased by their joint presence.
Fig 6.
Activation of N. europaea ADP-Glc PPase by Pyr in the presence of 5 mM (○) or 30 mM (•) Mg2+.
Fig 7.
Enzymatic activity of N. europaea ADP-Glc PPase against Mg2+ determined in the absence (○) or in the presence (•) of 5 mM Pyr.
N. europaea ADP-Glc PPase showed no significant difference in kinetic parameters for Glc-1P in the presence of Pyr (data not shown); however, kinetics for ATP or UTP were affected (Table 4). This alteration of kinetic parameters depended on whether Mg2+ or Co2+ was used as the essential cofactor. The main effect of Pyr was to enhance the catalytic efficiency of the enzyme for ATP with Mg2+. Vmax increased 1.5-fold, and the enzyme affinity toward ATP and Mg2+ increased 3-fold (Table 4). Alternatively, there was almost no effect of Pyr on catalytic efficiency using UTP with Mg2+ because the Vmax increased 2.8-fold but the affinity decreased by a similar level. Kinetic parameters for Pyr in the presence of ATP or UTP with Co2+ showed no change in catalytic efficiency for ATP and decreased efficiency by 2-fold for UTP (Table 4).
Table 4.
Kinetic parameters of N. europaea ADP-Glc PPase for ATP and UTP in presence of Pyra
| Nucleotide | Metal cofactor | Vmax (U/mg) | Km (mM) | kcat/Km (mM−1 · s−1) |
|---|---|---|---|---|
| ATP | Mg2+ | 73 ± 2 (1.5 ± 0.3) | 0.44 ± 0.03 (0.32 ± 0.03) | 550 ± 40 (4.6 ± 0.8) |
| Co2+ | 25 ± 1 (0.89 ± 0.04) | 0.10 ± 0.01 (1.1 ± 0.2) | 800 ± 70 (0.8 ± 0.1) | |
| UTP | Mg2+ | 1.4 ± 0.4 (3 ± 1) | 5 ± 2 (3 ± 1) | 1.0 ± 0.4 (1.2 ± 0.9) |
| Co2+ | 11.5 ± 0.5 (1.3 ± 0.3) | 1.3 ± 0.1 (2.4 ± 0.3) | 30 ± 3 (0.5 ± 0.1) |
The concentration of Mg2+ was 15 mM in both ATP and UTP assays, while that of Co2+ was 1 and 3 mM, respectively, when ATP and UTP were the substrates. The numbers in parentheses denote the ratio between the respective parameter determined in the presence and in the absence of 5 mM Pyr.
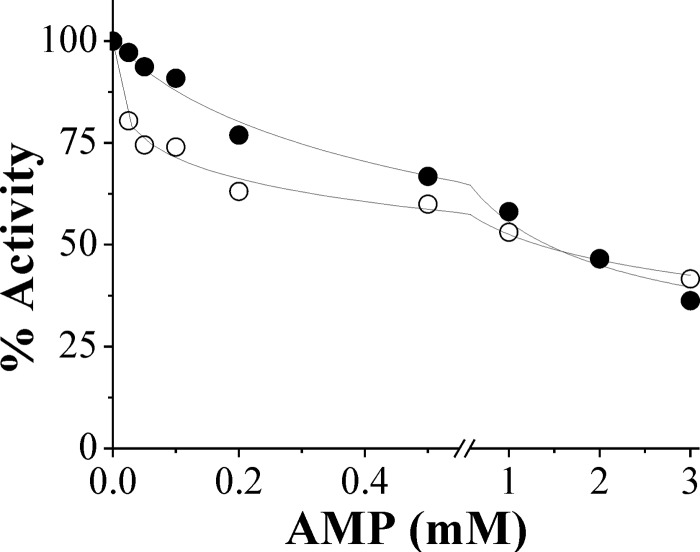
The effect of AMP on the activity of the N. europaea ADP-Glc PPase assayed with ATP and Mg2+ in the absence or in the presence of saturating concentrations of Pyr was also determined (Fig. 8). AMP produced a concentration-dependent inhibition of the enzyme, but it was of lesser magnitude when Pyr was present, although under either condition the inhibitory effect was only partial, with the maximal decrease in enzyme activity being about 60% (Fig. 8). Additionally, the presence of AMP produced almost no change in S0.5 for ATP (data not shown), a finding that implies that the interaction of AMP with the enzyme is partial noncompetitive with respect to the substrate (45). AMP has also been reported to be the main inhibitor of the ADP-Glc PPase from E. coli, but for this enzyme, the inhibitory effect is evident only in the presence of the allosteric activator fructose-1,6-bisphosphate (6), which is different from the effect of the N. europaea enzyme.
Fig 8.
Inhibition of N. europaea ADP-Glc PPase by AMP, expressed as percent activity, determined in the absence (○) or in the presence (•) of 5 mM Pyr.
DISCUSSION
At least three variants for glycogen metabolism can be distinguished in prokaryotes; namely, the GlgC-GlgA, the GlgE, and the Rv3032 pathways (recently reviewed by Chandra et al. [12]). The occurrence and regulation of the classical GlgC-GlgA pathway have been studied and are relatively well understood (4, 6, 7, 23, 27). This pathway involves ADP-Glc PPase and a glycogen synthase specific for ADP-Glc. It is widely present in Gram-positive and Gram-negative species, including cyanobacteria (and also resembles the pathway for starch metabolism in plants), but it seems to be absent in most archaebacteria (12). A second glycogen pathway has recently been identified (GlgE route) and is related to trehalose, which is a storage disaccharide in certain bacteria (18). GlgE is a maltosyltransferase that uses maltose-1-phosphate (derived from trehalose) to elongate a linear α-1,4-glucan chain. The GlgE pathway is particularly relevant in actinomycetes having high G+C content, such as streptomycetes, mycobacteria, and related corynebacteria (12, 18). The third route for polyglucan biosynthesis (Rv3032) was characterized in mycobacteria and is associated with fatty acid metabolism. This pathway generates methylglucose lipopolysaccharide through a GlgA paralog that uses UDP-Glc as well as ADP-Glc (12). All these routes for glycogen metabolism are associated with viability and important physiological functions (e.g., pathogenicity in mycobacteria) of the microorganism.
An analysis of the N. europaea genome (2, 11) shows the genes related to the classical GlgC-GlgA pathway, specifically, those coding for glycogen synthase (GlgA), ADP-Glc PPase (GlgC), α-1,4-alpha-glucan branching enzyme (GlgB), and glycogen phosphorylase (GlgP). Unlike other bacteria, these genes are not arranged in one operon, and the putative gene coding for a glycogen debranching enzyme (glgX) seems to be absent in N. europaea (11). Hitherto, it was concluded that the GlgC-GlgA pathway is present only for glycogen synthesis in ammonia-oxidizing lithoautotrophic bacteria. Many transcriptomic reports (14, 24, 38, 39, 41, 52) support the occurrence of the glycogen pathway in N. europaea, which is reinforced by reports showing the accumulation of the polysaccharide in Nitrosomonas eutropha (9). However, the actual role of the specific genes coding for putative enzymes for glycogen biosynthesis has not been confirmed by biochemical studies, limiting the understanding of the real incidence and regulation of the pathway. Our results reported in the present work contribute to solving such a limitation. After recombinant production in a highly purified form, we determined key characteristics of the two main enzymes involved in linear α-1,4-glucan elongation in N. europaea. We found that the specificity of glycogen synthase for ADP-Glc (typical of a GlgA) and the distinctive kinetic and regulatory properties exhibited by ADP-Glc PPase (in agreement with GlgC) support the in vivo action of glycogen biosynthesis in the microorganism.
A phylogenetic analysis of the ADP-Glc PPase and glycogen synthase gene products (GlgC and GlgA, respectively), which are the ones that determine the specificity for the nucleotides used in the synthetic pathway and its regulation, revealed that N. europaea forms belong to a different class than the most studied of these enzymes. The phylogenetic branches for proteobacterial ADP-Glc PPases are very complex. There seem to be at least four different clusters or groups (see Fig. S1 in the supplemental material). We cannot discard the possibility of horizontal transfers, but the fact that many bacteria have more than one gene falling into different (paralog) groups suggests the possibility of early duplications, some of which were kept and some of which were lost (see Table S1 in the supplemental material). Cluster 1 has representatives from the alpha-, beta-, and gammaproteobacteria, cluster 2 has beta- and gammaproteobacteria, cluster 3 has delta- and gammaproteobacteria, and cluster 4 has beta-, gamma-, and deltaproteobacteria. There are two deltaproteobacterial genes that fall outside these clusters that are more similar to cyanobacterial ADP-Glc PPases (see Table S1 in the supplemental material). Cluster 1 comprises some ADP-Glc PPases that have been biochemically studied, such as the ones from E. coli (gammaproteobacteria), Agrobacterium tumefaciens, Rhodospirillum rubrum, and Rhodobacter sphaeroides (alphaproteobacteria). The ADP-Glc PPase from N. europaea falls into cluster 2, which is a phylogenetic branch that is largely unexplored. Together with N. europaea, all the other ammonia-oxidizing betaproteobacteria are present here (see Fig. S1 in the supplemental material). Interestingly, a similar pattern is observed with the glycogen synthase, in which proteobacterial forms are divided into three clusters, with some bacteria having genes located in more than one (see Table S2 in the supplemental material). Most studied glycogen synthases are in cluster 1, and the one from N. europaea is in cluster 2 (see Fig. S2 in the supplemental material). It will be very interesting to determine whether enzymes from the other clusters maintain similar biochemical roles as the ones from cluster 1 and the one from cluster 2 studied in this paper. In addition, future genetic, biochemical, and phylogenetic studies for other enzymes of glycogen metabolism will be very important to understand whether the properties of this pathway are conserved or not.
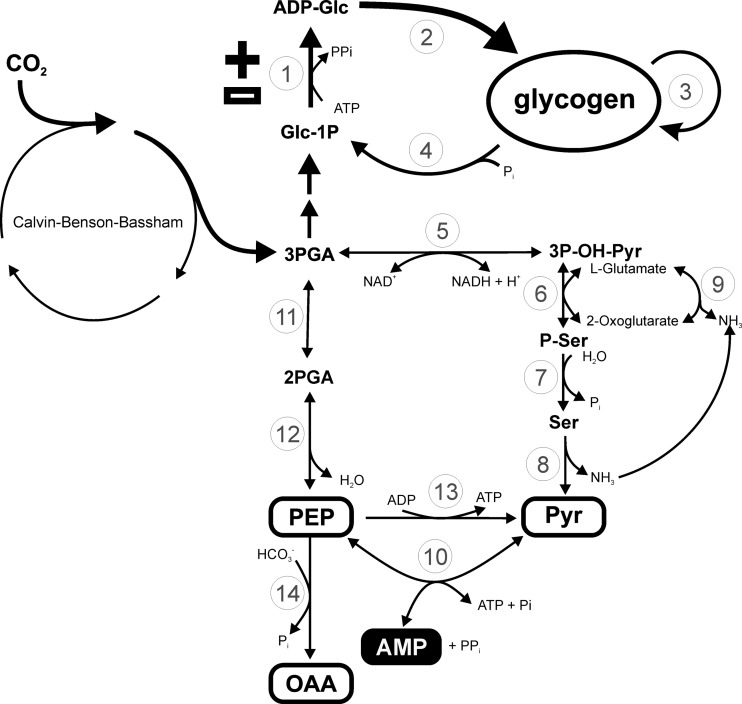
It is important to place the analysis of the properties determined for recombinant N. europaea ADP-Glc PPase into the context of the bacterium metabolism. For this reason, Fig. 9 was constructed from the genomic information available (2, 11). As described, the Benson-Calvin cycle is a major operative path that gives the microorganism the autotrophy to assimilate inorganic CO2. The first product of carbon fixation, 3-phosphoglycerate, as well as fructose-6-phosphate, is a key intermediate connecting the cycle to other metabolic pathways. Thus, the flux of assimilated carbon can be directed toward (i) glycolysis and generation of energy (ATP), (ii) the route of keto and amino acids associated with nitrogen metabolism, and (iii) storage of polysaccharides, mainly glycogen. In this context, the regulatory properties determined for N. europaea ADP-Glc PPase support a modulation of glycogen synthesis coordinated with the carbon and energy availability in the bacterium.
Fig 9.
Scheme for carbon flux in Nitrosomonas europaea deduced from information available in the genome (11). The pathway from carbon dioxide fixation to glycogen accumulation is indicated in bold. White and black boxes, metabolites behaving as allosteric activators (+) and inhibitors (−) of ADP-Glc PPase, respectively. Enzymes are numbered as follows: 1, ADP-Glc pyrophosphorylase (EC 2.7.7.27, NE2030); 2, glycogen synthase (EC 2.4.1.21, NE2264); 3, branching enzyme (EC 2.4.1.18, NE2029); 4, glycogen phosphorylase (EC 2.4.1.1, NE0074, NE0466); 5, phosphoglycerate dehydrogenase (EC 1.1.1.95, NE0334, NE1688); 6, phosphoserine transaminase (EC 2.6.1.52, NE0333); 7, phosphoserine phosphatase (EC 3.1.3.3, NE0439); 8, l-serine ammonia lyase (EC 4.3.1.17, NE0380); 9, glutamate dehydrogenase (EC 1.4.1.4, NE1616); 10, pyruvate, water dikinase (EC 2.7.9.2, NE2359, NE2366); 11, phosphoglycerate mutase (EC 5.4.2.1, NE0178, NE1780); 12, phosphopyruvate hydratase (EC 4.2.1.11, NE1044); 13, pyruvate kinase (EC 2.7.1.40, NE0325); 14, phosphoenolpyruvate carboxylase (EC 4.1.1.31, NE0589). PGA, phosphoglyceric acid.
On one hand, synthesis of ADP-Glc in N. europaea would be strictly linked to high energy contents since the enzyme uses ATP as a substrate and also because it is sensitive to AMP as a main allosteric inhibitor. In addition, activation of ADP-Glc PPase by Pyr, PEP, and OAA supports high glycogen synthesis when carbon is in relative excess, since increased levels of these metabolites oversatisfy the demand for many metabolic pathways in the cell (Fig. 9). In fact, regulation by Pyr, PEP, and OAA is of remarkable importance because they are part of a central metabolic enclave of carbon and energy distribution. This is in agreement with several studies performed in bacteria (Fig. 9) (43), especially in lithoautotrophic microorganisms having an incomplete tricarboxylic acid cycle (actually performing a Krebs horseshoe; see reference 53). In addition, the conversion of PEP into OAA, via carboxylation by anaplerotic PEP carboxylase, is an important alternative to bypass the Benson-Calvin cycle to assimilate CO2 in the economic chemolithoautotrophic lifestyle (11). Under the latter metabolic alternative, ADP-Glc PPase and glycogen biosynthesis could also be effectively regulated (Fig. 9).
The kinetic properties determined for the N. europaea ADP-Glc PPase are distinct with respect to those known for the enzyme from other sources. For pyrophosphorylases, including ADP-Glc PPases, it has been hypothesized that the divalent metal ion is involved in stabilization of negative charges of the phosphate backbone in the nucleotide and also in coordinating the nucleophilic attack between the substrates (33, 47, 48). This family of enzymes has a degree of promiscuity toward the divalent metal ion as an essential cofactor, where Mg2+, Mn2+, or Co2+ is effective at fulfilling such a requirement (6, 7, 33, 47, 48). A similar situation using metallic cofactors was found for the N. europaea ADP-Glc PPase, except that it exhibited a remarkably low affinity toward Mg2+, and the use of other cations (mainly Co2+) enhanced its promiscuity for other nucleotides (mainly UTP and CTP) and mannose-1-phosphate as the substrates. This is different from all previous reports on the high specificity of ADP-Glc PPase toward ATP and Glc-1P as the substrates (6, 7). The distinctive properties for the recombinant ADP-Glc PPase determined in this study should be analyzed by considering that the enzyme heterologously expressed in E. coli may differ slightly from that expressed in N. europaea; however, it is unlikely that the qualitative properties would change. No posttranslational modification for these bacterial ADP-Glc PPases has been described.
The allosteric activator Pyr markedly altered the N. europaea ADP-Glc PPase low affinity for Mg2+ and the degree of promiscuity for substrates. It mainly reduced the S0.5 for Mg2+ and increased the use of ATP (largely in combination with the divalent metal ion), making the enzyme more specific toward synthesizing ADP-Glc. This is a distinctive characteristic among activators of ADP-Glc PPases, as they mostly affect Vmax, and some increase the affinity for ATP, as is the case of fructose-1,6-bisphosphate activation of the enzyme from E. coli (5, 6). In addition, Pyr is the main activator in N. europaea and is remarkably different from the activator of ADP-Glc PPase from autotrophic (although photosynthetic) related cyanobacteria, where 3-phosphoglycerate is the most important allosteric activator (13, 27). Despite this major difference, the selectivity for activator is still in good agreement with the general view that glycogen synthesis regulation is related to main metabolic routes (6, 7). That is because the relevance of carbon fixation in cyanobacteria is different from that in N. europaea. In cyanobacteria, the Benson-Calvin cycle (producing 3-phosphoglycerate as a first product) is a central metabolic pathway related to obligate photosynthetic characteristics. Conversely, carbon fixation in N. europaea could have less relevance or at least be secondary to carbon distribution, which is a functional aspect of facultative chemolithoautotrophy (2, 44). For instance, under anoxic conditions, N. europaea and N. eutropha could grow heterotrophically using different low-molecular-mass organic compounds as the carbon source (44). Although the ability of N. europaea to use reduced carbon compounds as a sole energy source is very limited, the microorganism can grow under mixotrophic conditions, reducing the energy cost for CO2 fixation (15, 25, 44). An early work performed by Rao and Nicholas (42) showed that serine is a main metabolite identified after radioactive CO2 exposure of N. europaea, highlighting the relevance of the 3-phosphohydroxypyruvate branch in the metabolism of the bacterium (Fig. 9). Interestingly, it has been demonstrated in N. europaea growing anaerobically that Pyr and nitrite can serve as electron donor and acceptor, respectively. This reinforces the central role of the keto acid in the energy metabolism of the bacterium (1, 26).
The physiological relevance for using different divalent metal ions by N. europaea ADP-Glc PPase should be carefully considered due to the diversity of the relative abundance of the cations in natural environments of living microorganisms (19). Many genes putatively coding for specific metal ion transporters have been identified in ammonia-oxidizing chemolithoautotrophic bacteria (11). Results reported herein open the possibility that N. europaea ADP-Glc PPase could alternatively use Mg2+, Mn2+, or Cd2+ in vivo as an essential cofactor. Consequently, the enzyme could produce ADP-Glc for glycogen synthesis or provide other sugar-nucleotides for different carbohydrate pathways. However, the intracellular levels of divalent cations (35 mM Mg2+, 0.16 mM Mn2+, 6.6 μM Cd2+, and < 0.6 μM Co2+) determined in the extremophile Thermus thermophilus (31) suggest that Mg2+ would be the cation physiologically serving as an effective cofactor (particularly in the presence of Pyr) for N. europaea ADP-Glc PPase mainly involved in the synthesis of ADP-Glc. Still, the promiscuity exhibited by the enzyme has significance for biotechnological purposes. Thus, the capacity of the enzyme to synthesize different sugar-nucleotides (e.g., ADP-Man) may be very useful for in vitro production of libraries for glycorandomized strategies currently of interest for pharmacological uses (8, 21, 35). In this respect, the effect of the divalent metal ion on enzyme promiscuity could be considered a key tool to expand components in sugar-nucleotide libraries. On the other hand, the promiscuity of N. europaea ADP-Glc PPase toward different metal cations could be a valuable tool to perform spectroscopic studies to determine the structural domains in the enzyme related to the binding of the metallic cofactors and possible interactions between them.
An analysis of the overall characteristics of ADP-Glc PPase from N. europaea compared with those known for the enzyme in other microorganisms is important to understand their functional evolution. The known regulatory properties of ADP-Glc PPases from different bacteria (mostly Gram-negative bacteria) have been reviewed, and the PPases have been classified into nine different groups (6, 7). Recently, it was found that the enzyme from the Gram-positive bacterium Streptomyces coelicolor represents a new type due to its different regulatory properties (3). In the stated classification, oxygenic photoautotrophic microorganisms (e.g., cyanobacteria) with a Benson-Calvin cycle that fixes CO2 possess an enzyme included in class VIII. The first product of the cycle, 3-phosphoglycerate, is the major activator (27). Interestingly, the properties of N. europaea ADP-Glc PPase exclude it from class VIII, but they are close to those reported for the enzymes included in class VI. This class is found in members of the genus Rhodospirillum (R. rubrum, R. fulvum, R. molischianum, and R. tenue), which are anaerobic bacteria capable of growth heterotrophically (mainly using Pyr) in the dark or autotrophically by performing anoxygenic photosynthesis with the tricarboxylic acid cycle (including operating in the reverse, reductive direction) as the central metabolism (51). The ADP-Glc PPase from Rhodospirillum spp. was characterized as being specifically activated by Pyr (it increases the affinity of the enzyme for Mg2+ and ATP and is inhibited by ADP or AMP) (40).
In the present work, we have contributed to a better understanding of glycogen biosynthesis in N. europaea and other bacteria with related trophic characteristics and how it is placed in the context of the metabolism of different prokaryotes. We found an ADP-Glc PPase with distinctive regulatory properties that fit the metabolic needs of N. europaea and most likely other chemolithoautotrophic bacteria. Future structural studies that elucidate the mechanism for the unique regulatory properties of ADP-Glc PPase will contribute to understanding how the structure of this enzyme has evolved. This knowledge will be instrumental in learning about the metabolic evolution of this group of autotrophic bacteria, which are very important in the fields of both evolution and ecology.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grants from CONICET (PIP 2519), ANPCyT (PICT'08 1754), and UNL (CAID Orientados y Redes) (to A.A.I.) and the National Science Foundation (grant MCB 1024945 to M.A.B.). M.M. is a fellow from CONICET. A.A.I. is principal investigator from CONICET, and he was recipient of a John Simon Guggenheim Memorial Foundation Fellowship.
We kindly thank the Midwest Center for Structural Genomics for providing the pMCSG9 vector for cloning.
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1. Abeliovich A, Vonshak A. 1992. Anaerobic metabolism of Nitrosomonas europaea. Arch. Microbiol. 158:267–270 [Google Scholar]
- 2. Arp DJ, Chain PS, Klotz MG. 2007. The impact of genome analyses on our understanding of ammonia-oxidizing bacteria. Annu. Rev. Microbiol. 61:503–528 [DOI] [PubMed] [Google Scholar]
- 3. Asencion Diez MD, Peiru S, Demonte AM, Gramajo H, Iglesias AA. 2012. Characterization of recombinant UDP- and ADP-glucose pyrophosphorylases and glycogen synthase to elucidate glucose-1-phosphate partitioning into oligo- and polysaccharides in Streptomyces coelicolor. J. Bacteriol. 194:1485–1493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ball SG, Morell MK. 2003. From bacterial glycogen to starch: understanding the biogenesis of the plant starch granule. Annu. Rev. Plant Biol. 54:207–233 [DOI] [PubMed] [Google Scholar]
- 5. Ballicora MA, et al. 2007. Identification of regions critically affecting kinetics and allosteric regulation of the Escherichia coli ADP-glucose pyrophosphorylase by modeling and pentapeptide-scanning mutagenesis. J. Bacteriol. 189:5325–5333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ballicora MA, Iglesias AA, Preiss J. 2003. ADP-glucose pyrophosphorylase, a regulatory enzyme for bacterial glycogen synthesis. Microbiol. Mol. Biol. Rev. 67:213–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ballicora MA, Iglesias AA, Preiss J. 2004. ADP-glucose pyrophosphorylase: a regulatory enzyme for plant starch synthesis. Photosynth. Res. 79:1–24 [DOI] [PubMed] [Google Scholar]
- 8. Barton WA, et al. 2001. Structure, mechanism and engineering of a nucleotidylyltransferase as a first step toward glycorandomization. Nat. Struct. Biol. 8:545–551 [DOI] [PubMed] [Google Scholar]
- 9. Bock E, Wagner M. 2006. Oxidation of inorganic nitrogen compounds as an energy source, p 457–495 In Dworkin M, Falkow S, Rosenberg E, Schleifer KH, Stackebrandt E. (ed), The prokaryotes. A handbook on the biology of bacteria: ecophysiology and biochemistry, 3rd ed, vol 2 Springer Science, New York, NY [Google Scholar]
- 10. Bradford MM. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248–254 [DOI] [PubMed] [Google Scholar]
- 11. Chain P, et al. 2003. Complete genome sequence of the ammonia-oxidizing bacterium and obligate chemolithoautotroph Nitrosomonas europaea. J. Bacteriol. 185:2759–2773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chandra G, Chater KF, Bornemann S. 2011. Unexpected and widespread connections between bacterial glycogen and trehalose metabolism. Microbiology 157:1565–1572 [DOI] [PubMed] [Google Scholar]
- 13. Charng YY, Iglesias AA, Preiss J. 1994. Structure-function relationships of cyanobacterial ADP-glucose pyrophosphorylase. Site-directed mutagenesis and chemical modification of the activator-binding sites of ADP-glucose pyrophosphorylase from Anabaena PCC 7120. J. Biol. Chem. 269:24107–24113 [PubMed] [Google Scholar]
- 14. Cho CM, et al. 2006. Transcriptome of a Nitrosomonas europaea mutant with a disrupted nitrite reductase gene (nirK). Appl. Environ. Microbiol. 72:4450–4454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Clark C, Schmidt EL. 1966. Effect of mixed culture on Nitrosomonas europaea simulated by uptake and utilization of pyruvate. J. Bacteriol. 91:367–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Dokmanic I, Sikic M, Tomic S. 2008. Metals in proteins: correlation between the metal-ion type, coordination number and the amino-acid residues involved in the coordination. Acta Crystallogr. D Biol. Crystallogr. 64:257–263 [DOI] [PubMed] [Google Scholar]
- 17. Donnelly MI, et al. 2006. An expression vector tailored for large-scale, high-throughput purification of recombinant proteins. Protein Expr. Purif. 47:446–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Elbein AD, Pastuszak I, Tackett AJ, Wilson T, Pan YT. 2010. Last step in the conversion of trehalose to glycogen: a mycobacterial enzyme that transfers maltose from maltose 1-phosphate to glycogen. J. Biol. Chem. 285:9803–9812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Fraústo da Silva J, Williams R. 2001. The biological chemistry of the elements: the inorganic chemistry of life. Oxford University Press, New York, NY [Google Scholar]
- 20. Freisinger E, Sigel RKO. 2007. From nucleotides to ribozymes—a comparison of their metal ion binding properties. Coord. Chem. Rev. 251:1834–1851 [Google Scholar]
- 21. Fu X, et al. 2003. Antibiotic optimization via in vitro glycorandomization. Nat. Biotechnol. 21:1467–1469 [DOI] [PubMed] [Google Scholar]
- 22. Fusari C, Demonte AM, Figueroa CM, Aleanzi M, Iglesias AA. 2006. A colorimetric method for the assay of ADP-glucose pyrophosphorylase. Anal. Biochem. 352:145–147 [DOI] [PubMed] [Google Scholar]
- 23. Govons S, Gentner N, Greenberg E, Preiss J. 1973. Biosynthesis of bacterial glycogen. XI. Kinetic characterization of an altered adenosine diphosphate-glucose synthase from a “glycogen-excess” mutant of Escherichia coli B. J. Biol. Chem. 248:1731–1740 [PubMed] [Google Scholar]
- 24. Gvakharia BO, et al. 2007. Global transcriptional response of Nitrosomonas europaea to chloroform and chloromethane. Appl. Environ. Microbiol. 73:3440–3445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hommes NG, Sayavedra-Soto LA, Arp DJ. 2003. Chemolithoorganotrophic growth of Nitrosomonas europaea on fructose. J. Bacteriol. 185:6809–6814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hyman MR, Arp DJ. 1995. Effects of ammonia on the de novo synthesis of polypeptides in cells of Nitrosomonas europaea denied ammonia as an energy source. J. Bacteriol. 177:4974–4979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Iglesias AA, Kakefuda G, Preiss J. 1991. Regulatory and structural properties of the cyanobacterial ADPglucose pyrophosphorylases. Plant Physiol. 97:1187–1195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Iglesias AA, Podestá FE. 2005. Photosynthate formation and partitioning in crop plants, p 525–545 In Pessarakli M. (ed), Handbook of photosynthesis, 2nd ed CRC Press, Taylor and Francis Group, Boca Raton, FL [Google Scholar]
- 29. Kawai H, Kaneko M, Maejima K, Kato I, Yamasaki M. 1985. Preparation of ADP-glucose with an enzyme from Arthrobacter simplex. Agric. Biol. Chem. 49:2905–2911 [Google Scholar]
- 30. Keener WK, Arp DJ. 1994. Transformations of aromatic compounds by Nitrosomonas europaea. Appl. Environ. Microbiol. 60:1914–1920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kondo N, et al. 2008. Insights into different dependence of dNTP triphosphohydrolase on metal ion species from intracellular ion concentrations in Thermus thermophilus. Extremophiles 12:217–223 [DOI] [PubMed] [Google Scholar]
- 32. Koops H-P, Pommerening-Röser A. 2001. Distribution and ecophysiology of the nitrifying bacteria emphasizing cultured species. FEMS Microbiol. Ecol. 37:1–9 [Google Scholar]
- 33. Koropatkin NM, Cleland WW, Holden HM. 2005. Kinetic and structural analysis of alpha-d-glucose-1-phosphate cytidylyltransferase from Salmonella typhi. J. Biol. Chem. 280:10774–10780 [DOI] [PubMed] [Google Scholar]
- 34. Laemmli UK. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685 [DOI] [PubMed] [Google Scholar]
- 35. Mizanur RM, Zea CJ, Pohl NL. 2004. Unusually broad substrate tolerance of a heat-stable archaeal sugar nucleotidyltransferase for the synthesis of sugar nucleotides. J. Am. Chem. Soc. 126:15993–15998 [DOI] [PubMed] [Google Scholar]
- 36. Nakamura Y, Imamura M. 1985. Regulation of ADP-glucose pyrophosphorylase from Chlorella vulgaris. Plant Physiol. 78:601–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Nallamsetty S, Waugh DS. 2007. A generic protocol for the expression and purification of recombinant proteins in Escherichia coli using a combinatorial His6-maltose binding protein fusion tag. Nat. Protoc. 2:383–391 [DOI] [PubMed] [Google Scholar]
- 38. Park S, Ely RL. 2008. Candidate stress genes of Nitrosomonas europaea for monitoring inhibition of nitrification by heavy metals. Appl. Environ. Microbiol. 74:5475–5482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Park S, Ely RL. 2008. Genome-wide transcriptional responses of Nitrosomonas europaea to zinc. Arch. Microbiol. 189:541–548 [DOI] [PubMed] [Google Scholar]
- 40. Preiss J, Greenberg E. 1981. Biosynthesis of bacterial glycogen: activator specificity of the adenosine diphosphate glucose pyrophosphorylases from the genus Rhodospirillum. J. Bacteriol. 147:711–719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Radniecki TS, Dolan ME, Semprini L. 2008. Physiological and transcriptional responses of Nitrosomonas europaea to toluene and benzene inhibition. Environ. Sci. Technol. 42:4093–4098 [DOI] [PubMed] [Google Scholar]
- 42. Rao PS, Nicholas DJ. 1966. Studies on the incorporation of CO2 by cells and cell-free extracts of Nitrosomonas europaea. Biochim. Biophys. Acta 124:221–232 [DOI] [PubMed] [Google Scholar]
- 43. Sauer U, Eikmanns BJ. 2005. The PEP-pyruvate-oxaloacetate node as the switch point for carbon flux distribution in bacteria. FEMS Microbiol. Rev. 29:765–794 [DOI] [PubMed] [Google Scholar]
- 44. Schmidt I. 2009. Chemoorganoheterotrophic growth of Nitrosomonas europaea and Nitrosomonas eutropha. Curr. Microbiol. 59:130–138 [DOI] [PubMed] [Google Scholar]
- 45. Segel IH. 1993. Enzyme kinetics: behavior and analysis of rapid equilibrium and steady-state enzyme systems. Wiley Interscience, New York, NY [Google Scholar]
- 46. Sheng F, Jia X, Yep A, Preiss J, Geiger JH. 2009. The crystal structures of the open and catalytically competent closed conformation of Escherichia coli glycogen synthase. J. Biol. Chem. 284:17796–17807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Sivaraman J, Sauve V, Matte A, Cygler M. 2002. Crystal structure of Escherichia coli glucose-1-phosphate thymidylyltransferase (RffH) complexed with dTTP and Mg2+. J. Biol. Chem. 277:44214–44219 [DOI] [PubMed] [Google Scholar]
- 48. Thoden JB, Holden HM. 2007. Active site geometry of glucose-1-phosphate uridylyltransferase. Protein Sci. 16:1379–1388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Tropea JE, Cherry S, Nallamsetty S, Bignon C, Waugh DS. 2007. A generic method for the production of recombinant proteins in Escherichia coli using a dual hexahistidine-maltose-binding protein affinity tag. Methods Mol. Biol. 363:1–19 [DOI] [PubMed] [Google Scholar]
- 50. Uttaro AD, Ugalde RA, Preiss J, Iglesias AA. 1998. Cloning and expression of the glgC gene from Agrobacterium tumefaciens: purification and characterization of the ADPglucose synthetase. Arch. Biochem. Biophys. 357:13–21 [DOI] [PubMed] [Google Scholar]
- 51. Wang X, Modak HV, Tabita FR. 1993. Photolithoautotrophic growth and control of CO2 fixation in Rhodobacter sphaeroides and Rhodospirillum rubrum in the absence of ribulose bisphosphate carboxylase-oxygenase. J. Bacteriol. 175:7109–7114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wei X, et al. 2006. Transcript profiles of Nitrosomonas europaea during growth and upon deprivation of ammonia and carbonate. FEMS Microbiol. Lett. 257:76–83 [DOI] [PubMed] [Google Scholar]
- 53. Wood AP, Aurikko JP, Kelly DP. 2004. A challenge for 21st century molecular biology and biochemistry: what are the causes of obligate autotrophy and methanotrophy? FEMS Microbiol. Rev. 28:335–352 [DOI] [PubMed] [Google Scholar]
- 54. Wu N, Christendat D, Dharamsi A, Pai EF. 2000. Purification, crystallization and preliminary X-ray study of orotidine 5′-monophosphate decarboxylase. Acta Crystallogr. D Biol. Crystallogr. 56:912–914 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.