Abstract
Among the 57 genes that encode ribosomal proteins in the genome of Bacillus subtilis, a Gram-positive bacterium, 50 genes were targeted by systematic inactivation. Individual deletion mutants of 16 ribosomal proteins (L1, L9, L15, L22, L23, L28, L29, L32, L33.1, L33.2, L34, L35, L36, S6, S20, and S21) were obtained successfully. In conjunction with previous reports, 22 ribosomal proteins have been shown to be nonessential in B. subtilis, at least for cell proliferation. Although several mutants that harbored a deletion of a ribosomal protein gene did not show any significant differences in any of the phenotypes that were tested, various mutants showed a reduced growth rate and reduced levels of 70S ribosomes compared with the wild type. In addition, severe defects in the sporulation frequency of the ΔrplA (L1) mutant and the motility of the ΔrpsU (S21) mutant were observed. These data provide the first evidence in B. subtilis that L1 and S21 are required for the progression of cellular differentiation.
INTRODUCTION
The eubacterial ribosome (70S) is a complex macromolecule that is composed of a small (30S) subunit and a large (50S) subunit. The small subunit is comprised of the 16S rRNA and more than 20 proteins, whereas the large subunit is comprised of the 23S and 5S rRNAs and more than 30 proteins. Each ribosome has three binding sites for tRNA, namely, the A-site, which accepts the aminoacylated tRNA, the P-site, which binds the tRNA with the nascent peptide chain, and the E-site, which binds the deacylated tRNA before it leaves the ribosome (46, 47). The small subunit associates with the mRNA and the anticodon stem-loop of the bound tRNA and participates in ensuring the fidelity of translation by checking for correct pairing between the codon and anticodon (25, 40, 45, 60, 63). The large subunit associates with the acceptor arms of the tRNA and catalyzes the formation of a peptide bond between the amino acid attached to the tRNA in the A-site and the nascent peptide chain bound to the tRNA in the P-site (5, 38). The molecular mechanisms of translation have been elucidated in detail by the convergence of various approaches, including crystal structure analysis (20, 46, 47, 54, 62).
Most ribosomal proteins in eubacteria are highly conserved (42), and it has been proposed that several ribosomal proteins play essential roles in translation, as well as ribosome assembly (13, 17, 18, 32, 64). However, the detailed roles of most of the ribosomal proteins in cell proliferation, as well as the progression of cellular differentiation, have not been investigated fully. Mutation of the genes that encode the ribosomal proteins is one of the most effective ways to obtain further information about their function. In Escherichia coli, which is the best-characterized Gram-negative bacterium, several mutants in which specific ribosomal proteins are absent from the ribosome have been isolated (15, 16) and characterized (21, 31, 37, 51, 52). More recently, E. coli has been subjected to a systematic deletion of the genes encoding ribosomal proteins (49). Taken together with results that were obtained previously from a genome-wide analysis of gene inactivation (4), the results revealed that 22 of the 54 E. coli genes for ribosomal proteins could be deleted individually (49).
The Gram-positive bacterium Bacillus subtilis has been the subject of intensive scientific study because of its ability to sporulate, as well as to incorporate external DNA readily. Sequencing of the genome of the B. subtilis strain 168 was completed in 1997 (28). Systematic gene inactivation enabled us to clarify that, among 4,101 genes in the genome of B. subtilis, 271 genes are essential for cell proliferation at 37°C in LB medium (27). However, the 52 ribosomal protein genes that are present in the B. subtilis genome were excluded from the targets of the study on systematic gene inactivation because of their conservation among eubacteria. Despite the assumed importance of the ribosomal proteins, it was reported that in a B. subtilis mutant that harbored a mutation in rplK, which encodes ribosomal protein L11, L11 was absent from the ribosome (59). L11 is one of the ribosomal proteins that have been well investigated, because mutations in the rplK gene often result in thiostrepton resistance (9, 39, 59). The N-terminal domain of L11, which interacts with the elongation factor G and the antibiotic thiostrepton, is implicated in the termination of translation (8, 56). L11 is also required for the stringent response through the regulation of RelA activity, which synthesizes the signaling molecules GDP 3′-diphosphate (ppGpp) and GTP 3′-diphosphate (pppGpp), generally referred to as (p)ppGpp (11, 59, 61). Furthermore, L11 is involved in the activation of σB, which regulates the general stress regulon of B. subtilis (65). Thus, it is most likely that, under adverse growth conditions, L11 contributes to cell survival by transmitting various signals that occur in the ribosome during translation to these regulatory proteins. These findings raised the question of whether the 52 ribosomal proteins in B. subtilis are actually all required for cell growth and, as is the case for L11, whether any others act as mediators that transmit signals from the ribosome to certain regulatory factors, and vice versa.
In the present study, we attempted to obtain a series of genetic deletion mutants that covered 50 ribosomal protein genes in the B. subtilis genome. The ribosomal protein homologues (Ctc, YpfD, YtiA, and YhzA), as well as the ribosomal proteins L11 and L31, the deletion mutants of which have been analyzed previously (34, 36, 50, 55), and S14, which is known to be essential (36), were not included among the targets of the study. Furthermore, the effects of the absence of each ribosomal protein on cell proliferation, as well as on 70S ribosome formation, sporulation, and motility, were examined to reveal phenotypes of deletions in individual ribosomal proteins in the Gram-positive, endospore-forming bacterium B. subtilis.
MATERIALS AND METHODS
Media and culture conditions.
LB (44), LB agar, and 2× Schaeffer's sporulation medium supplemented with 0.1% glucose (2× SG) (29) were used. The culture conditions and media for preparation of competent cells have been described previously (3). When required, 5 μg ml−1 chloramphenicol or 5 μg ml−1 kanamycin was added to the media. Min-CH medium (43) is Spizizen's minimal glucose medium supplemented with 0.05% Amicase (Sigma).
Gene deletion experiments.
All of the B. subtilis strains used in the study were isogenic with B. subtilis strain 168 trpC2. The ribosomal protein genes were replaced with a chloramphenicol acetyltransferase (cat) gene from pCBB31 (24). When the target gene was considered to be a monocistronic unit or was located at the end of the operon, a part of the open reading frame (ORF) was replaced with an amplified version of the cat gene that included its own promoter, which was obtained by PCR using the primers catF and catpt1R. In contrast, when the transcript of the target gene included downstream gene(s), a part of the ORF was replaced by an amplified version of the cat gene that lacked any promoter or Rho-independent terminator sequence (catΔpt1) (35), which was amplified with primers catpt1F and catpt1R. For the disruption, upstream and downstream regions of each target gene, at least 600 bp in length, were amplified by PCR using the appropriate primers (rpxXuF and rpxXuR for the upstream region, rpxXdF and rpxXdR for the downstream region). (For all primers used in the study, see Table S1 in the supplemental material.) The two resultant fragments and the cat (or catΔpt1) fragment were then ligated and amplified by PCR using the appropriate primers (rpxXuF and rpxXdR; see Table S1). The resultant amplified DNA fragment contained regions homologous to the genomic regions that flanked the target gene and, to disrupt the gene, was used to transform strain 168 at a final concentration of 5 μg ml−1, which was considered to be a saturating DNA concentration (2). The transformants that harbored a deletion mutation of the target ribosomal protein gene as a result of a double-crossover event were selected for resistance to chloramphenicol at 30°C, 37°C, and 47°C. The development of competence was monitored by analysis of Trp+ transformation using chromosomal DNA from strain 168W; in each analysis, the activity was higher than 105 Trp+ transformants ml−1. Each assessment of gene disruption was performed more than twice. Correct disruption was verified by PCR and two-dimensional (2-D) electrophoresis.
The ΔfliE mutant was constructed by replacing the fliE ORF with the kanamycin resistance gene of pET41b (Novagen), which was amplified by PCR using the primers fliEuRKmF and fliEdFKmR. The upstream and downstream regions of the fliE gene were amplified by PCR using the primers fliEuF and fliEuRKmF for the upstream region and fliEdFKmR and fliEdR for the downstream region (see Table S1 in the supplemental material). The two resulting fragments and the kanamycin resistance gene were then ligated and amplified by PCR using fliEuF and fliEdR. The fliE gene was then disrupted using the resulting product. Correct disruption was verified by PCR.
Sucrose density gradient sedimentation analysis.
Bacillus subtilis cells were grown in LB medium at 37°C with shaking to early exponential phase (optical density at 600 nm [OD600] of ∼0.2) and harvested. The sucrose density gradient sedimentation analysis was performed as described previously (35). Briefly, cells were disrupted by passage through a French pressure cell, and cell debris was removed by centrifugation. Aliquots of extract were layered onto 10 to 40% sucrose density gradients and centrifuged at 4°C for 17.5 h at 65,000 × g (Hitachi P40ST rotor; 10 A260 units per tube). Samples were collected with a piston gradient fractionator (BioComP), and absorbance profiles were monitored at 254 nm (A254) using a Bio-mini UV monitor (ATTO, Japan).
Preparation of crude ribosomes.
Cells were grown in LB medium at 37°C and were harvested in early exponential phase (OD600 of ∼0.2). Crude ribosomes were obtained as described previously (34). Radical free and highly reducing (RFHR) 2-D gel electrophoresis (57) was performed essentially in accordance with the published procedures (34).
Assay for sporulation.
Bacillus subtilis cells were grown in 2× SG medium for 24 h at 37°C with shaking. Heat-resistant spores were counted by heating the cells at 80°C for 10 min, plating them on LB agar plates, and then incubating them at 37°C for 24 h. Spores were observed by phase-contrast microscopy (Olympus BX50).
Microscopic imaging.
Cells were grown in LB medium at 37°C with shaking to the exponential phase, and 500 μl of the culture was centrifuged at 12,000 × g for 1 min. The cell pellet was resuspended in 20 μl of the culture supernatant containing FM4-64 (10 μg ml−1, Invitrogen) and DAPI (4′,6-diamidino-2-phenylindole; 5 μg ml−1) (Wako Pure Chemical Industries). The cell suspension was mounted on microscope slides covered with a thin film of agarose (1.0%) in distilled water, and phase-contrast and fluorescence images were obtained with a SenSys-1401E air-cooled charge-coupled device (CCD) camera (Roper Scientific) attached to an Olympus BX50 microscope equipped with a 100× UPlanApo objective.
Motility assay.
The motility of the ribosomal protein deletion strains was assayed on plates that contained 1% Bacto Tryptone (Difco) and 0.5% NaCl, which were solidified by the addition of 0.4% Bacto agar (Difco). An aliquot of a liquid culture (1 μl), which had been grown in LB medium at 37°C to early exponential phase (OD600 of ∼0.2), was spotted onto the center of the plate. After incubation at 37°C for 24 h, the presence of swarm colonies was observed.
RESULTS
Systematic inactivation of genes encoding ribosomal proteins in the B. subtilis genome.
Bacillus subtilis has the ability to take up exogenous DNA with high efficiency. When cells are grown in the appropriate minimal medium, a transformation efficiency as high as 106 transformants ml−1 can be obtained (2). We employed this procedure to obtain a set of mutants that each harbored a deletion mutation of a gene that encoded a ribosomal protein. Among the 57 genes that were annotated to encode ribosomal proteins in the genome of B. subtilis, 50 genes were chosen as targets. Six genes, which encoded Ctc, YpfD, YtiA, YhzA, L11, and L31, were not included in the present study, because successful attempts to obtain deletion mutants of these genes had already been reported (34, 36, 50, 55, 59). The rpsN gene encoding S14 was also excluded, because its essentiality has already been established (36).
In B. subtilis, most of the genes that encode ribosomal proteins are located in the large S10-spc-α gene cluster, which contains 25 ribosomal protein genes (30). In the case of genes in this cluster, the target genes were replaced with an amplified version of the chloramphenicol resistance gene (cat) that lacked any promoter or Rho-independent terminator sequence (catΔpt1), to eliminate possible polar effects on downstream genes within the same operon. In these mutants, the cat gene was cotranscribed with the other genes that constitute the cluster, as a result of the gene replacement. In contrast, when the target gene was considered to be transcribed as a monocistronic message or was located at the end of the operon, the ORF of the target gene was replaced by an amplified version of the cat gene that included its own promoter. In either case, the DNA fragment that contained the cat (or catΔpt1) gene was ligated with at least 600 bp of the upstream and downstream regions of each target gene as described in Materials and Methods. The fragments obtained were used to transform B. subtilis strain 168 (trpC2). The transformants were selected for chloramphenicol resistance at three different temperatures (30°C, 37°C, and 47°C), because of the possibility that ribosomal protein deletion mutants will show high/low-temperature-sensitive phenotypes. In each experiment, to confirm that the recipient cells were competent to take up exogenous DNA, cells were exposed to chromosomal DNA extracted from strain 168W Trp+ at a final concentration of 5 μg ml−1, which was considered to give the maximum number of transformants (2), and conversion to the Trp+ phenotype was monitored. Each assessment of gene disruption was performed more than twice.
Using this approach, it was possible to obtain deletion mutants for 16 genes that encoded ribosomal proteins (Table 1). Among these, three genes encoded ribosomal proteins found in the 30S subunit (S6, S20, and S21), and 13 genes encoded proteins in the 50S subunit (L1, L9, L15, L22, L23, L28, L29, L32, L33.1, L33.2, L34, L35, and L36). Disruption of the target genes was confirmed by PCR (data not shown). In addition, crude ribosomes were prepared from the obtained mutants, as well as from three mutants (the ΔrpmE, ΔytiA, and ΔyhzA strains) that were isolated previously (34, 36), and RFHR 2-D gel electrophoresis of the crude ribosomes was performed to confirm the absence of the products of the deleted genes. As shown in Fig. 1 (see Fig. S1 in the supplemental material), the products of the deleted genes were not detected in the 2-D gels of crude ribosomes prepared from the mutants. However, it should be noted that L9, L33.2, and L36 could not be detected by the 2-D gel analysis of high-salt-washed ribosomes even from wild-type cells (34). Therefore, we did not expect to see any differences between the 2-D gel patterns of the wild-type strain and the ΔrplI (L9), ΔrpmGB (L33.2), and ΔrpmJ (L36) mutants. These results, in combination with previous reports (34, 36, 50, 55, 59), revealed that it is possible to delete 22 of the 57 ribosomal proteins, including ribosomal protein homologues, that are found in B. subtilis (Table 1).
Table 1.
Summary of disruption of ribosomal protein genes
| Gene | Protein | Transcriptional unit | Replacementa | Gene disruption in B. subtilis | Essentiality in E. colib |
|---|---|---|---|---|---|
| rplA | L1 | rplK-rplA | catΔpt1 | Yes | Nonessential |
| rplB | L2 | S10-spc-α | catΔpt1 | No | Essential |
| rplC | L3 | S10-spc-α | catΔpt1 | No | Essential |
| rplD | L4 | S10-spc-α | catΔpt1 | No | Essential |
| rplE | L5 | S10-spc-α | catΔpt1 | No | Essential |
| rplF | L6 | S10-spc-α | catΔpt1 | No | Essential |
| rplI | L9 | yybS-yybT-rplI | cat | Yes | Nonessential |
| rplJ | L10 | rplJ-rplL | cat | No | Essential |
| rplK | L11 | rplK-rplA | Yesc | Nonessential | |
| rplL | L7/L12 | rplJ-rplL | cat | No | Essential |
| rplM | L13 | rplM-rpsI | catΔpt1 | No | Essential |
| rplN | L14 | S10-spc-α | catΔpt1 | No | Essential |
| rplO | L15 | S10-spc-α | catΔpt1 | Yes | Nonessential |
| rplP | L16 | S10-spc-α | catΔpt1 | No | Essential |
| rplQ | L17 | S10-spc-α | cat | No | Essential |
| rplR | L18 | S10-spc-α | catΔpt1 | No | Essential |
| rplS | L19 | rplS | cat | No | Essential |
| rplT | L20 | infC-rpmI-rplT | cat | No | Essential |
| rplU | L21 | rplU-ysxB-rpmA | cat | No | Nonessential |
| rplV | L22 | S10-spc-α | catΔpt1 | Yes | Essential |
| rplW | L23 | S10-spc-α | catΔpt1 | Yes | Essential |
| rplX | L24 | S10-spc-α | catΔpt1 | No | Nonessential |
| ctc | L25 homologue | ctc | Yesc | Nonessential | |
| rpmA | L27 | rplU-ysxB-rpmA | cat | No | Nonessential |
| rpmB | L28 | rpmB | cat | Yes | Essential |
| rpmC | L29 | S10-spc-α | catΔpt1 | Yes | Nonessential |
| rpmD | L30 | S10-spc-α | catΔpt1 | No | Nonessential |
| rpmE | L31 | rpmE | cat | Yesc | Nonessential |
| ytiA | L31 homologue | ytiA | cat | Yesc | — |
| rpmF | L32 | rpmF | cat | Yes | Nonessential |
| rpmGA | L33.1 | rpmGA | cat | Yesc | Nonessential |
| rpmGB | L33.2 | rpmGB | cat | Yesc | — |
| rpmH | L34 | rpmH | cat | Yes | Nonessential |
| rpmI | L35 | infC-rpmI-rplT | catΔpt1 | Yes | Nonessential |
| rpmJ | L36 | S10-spc-α | catΔpt1 | Yes | Nonessential |
| ypfD | S1 homologue | ypfD-cmk | Yesc | Essential | |
| rpsB | S2 | rpsB | cat | No | Essential |
| rpsC | S3 | S10-spc-α | catΔpt1 | No | Essential |
| rpsD | S4 | rpsD | cat | No | Essential |
| rpsE | S5 | S10-spc-α | catΔpt1 | No | Essential |
| rpsF | S6 | rpsF-ssb-rpsR | catΔpt1 | Yes | Nonessential |
| rpsG | S7 | rpsL-rpsG-fus | catΔpt1 | No | Essential |
| rpsH | S8 | S10-spc-α | catΔpt1 | No | Essential |
| rpsI | S9 | rplM-rpsI | cat | No | Nonessential |
| rpsJ | S10 | S10-spc-α | catΔpt1 | No | Essential |
| rpsK | S11 | S10-spc-α | catΔpt1 | No | Essential |
| rpsL | S12 | rpsL-rpsG-fus | catΔpt1 | No | Essential |
| rpsM | S13 | S10-spc-α | catΔpt1 | No | Essential |
| rpsN | S14 | S10-spc-α | catΔpt1 | No | Essential |
| yhzA | S14 homologue | yhzA | cat | Yesc | — |
| rpsO | S15 | rpsO | cat | No | Nonessential |
| rpsP | S16 | rpsP | catΔpt1 | No | Essential |
| rpsQ | S17 | S10-spc-α | catΔpt1 | No | Nonessential |
| rpsR | S18 | rpsF-ssb-rpsR | cat | No | Essential |
| rpsS | S19 | S10-spc-α | catΔpt1 | No | Essential |
| rpsT | S20 | rpsT | cat | Yes | Nonessential |
| rpsU | S21 | rpsU-yqeT | catΔpt1 | Yes | Nonessential |
Fig 1.
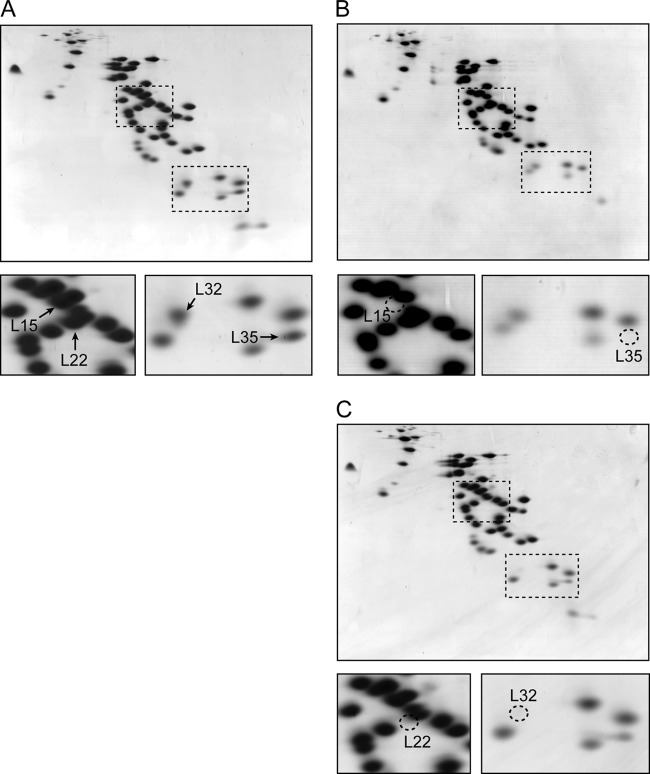
RFHR 2-D gel electrophoresis of ribosomal proteins prepared from deletion mutants of the ribosomal protein genes. Ribosomal proteins (750 μg) were prepared from cells in the early exponential phase (OD600 of ∼0.2) of the wild type (wt) (A), ΔrplO (L15) mutant (B), or ΔrplV (L22) mutant (C), grown in LB medium at 37°C and were used for RFHR two-dimensional gel electrophoresis as described in Materials and Methods. The areas of the two-dimensional gels that contained the spots of the L15 and L22 or L32 and L35 proteins were extracted from the gel images. Arrows indicate each ribosomal protein spot (A). Circles with dotted lines indicate protein spots that have disappeared (B and C). The deletion of rplO and rplV was confirmed by the disappearance of spots that correspond to L15 and L22, respectively. Significant reductions in the amount of L35 and L32 proteins were observed in the ribosomes prepared from the ΔrplO (L15) and ΔrplV (L22) mutants, respectively.
Characterization of the deletion mutants.
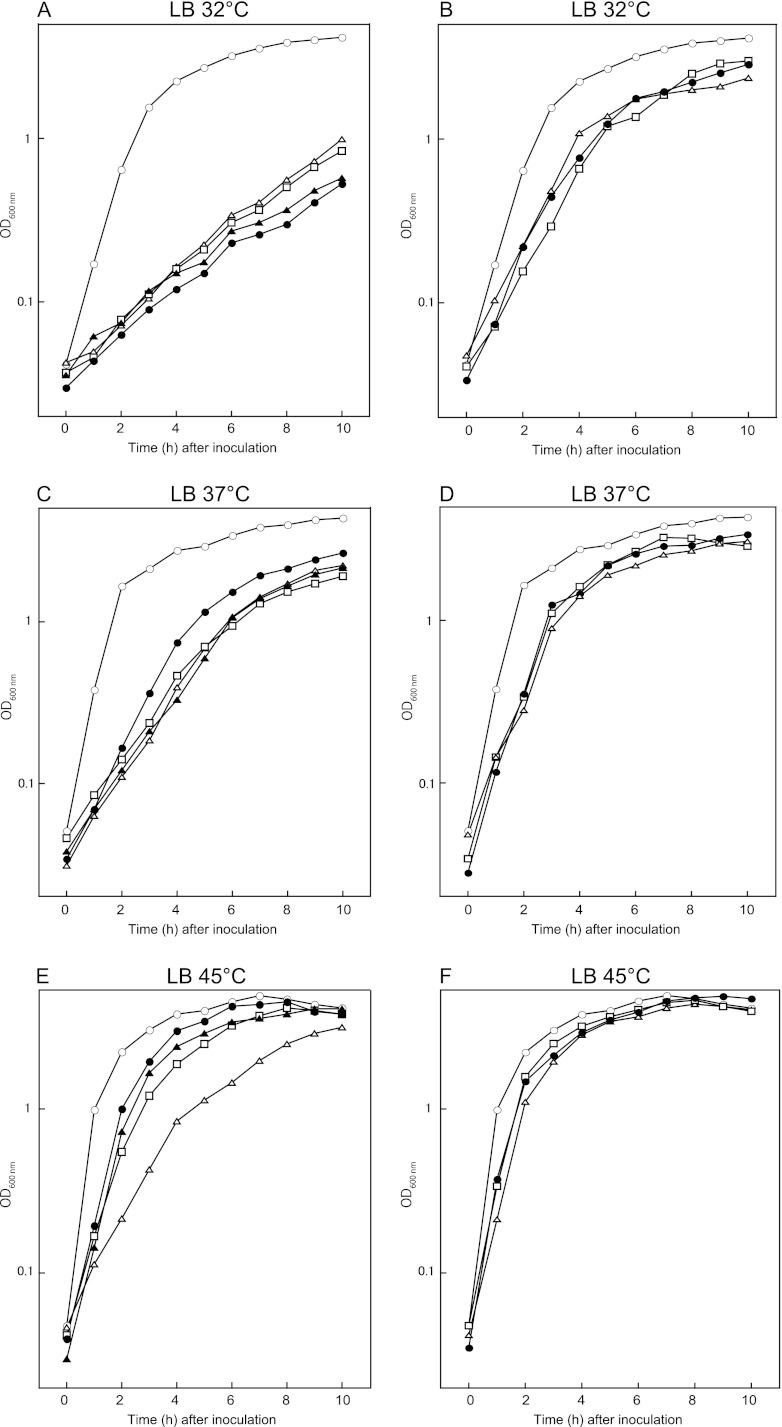
We attempted to characterize the phenotypes of the deletion mutants obtained above, together with those of the ΔrpmE (L31), ΔytiA, and ΔyhzA mutants, which were constructed previously in our laboratory (1, 34, 36). Among the 19 mutants tested in the present study, seven mutants, which harbored deletions of the genes that encoded ribosomal proteins L1, L22, L23, L34, L36, S6, and S21, respectively, showed slow growth phenotypes in LB medium compared with the wild type (Fig. 2 and Table 2). In particular, all of these mutants grew more slowly than the wild type at the lowest temperature (32°C) (Fig. 2A and B and Table 2). In contrast, except for the mutant lacking L1 (ΔrplA), which showed severe growth defects at all temperatures tested, the growth rate of these mutants was largely restored at 45°C (Fig. 2E and F and Table 2). Interestingly, the growth of the ΔrplV (L22), ΔrplW (L23), and ΔrpmH (L34) mutants, which was extremely slow at 32°C, was markedly restored at the higher temperature. The three ribosomal proteins encoded by these genes might have a role in ribosomal assembly at low temperatures. The other 12 mutants did not show severe growth defects in LB medium at 37°C, even though some of the mutants (e.g., the ΔrpmE mutant, expressing L31) showed a slight decrease in growth rate (see Fig. S3 in the supplemental material). These results indicated that these gene products are dispensable for the growth in LB medium at 37°C.
Fig 2.
Growth characteristics of the deletion mutants. Cells were grown in LB medium at 32°C (A and B), 37°C (C and D), or 45°C (E and F), and the optical density at 600 nm (OD600) was measured. Symbols in panels A, C, and E: ○, wild type; △, ΔrplA (L1) mutant; □, ΔrplV (L22) mutant; ●, ΔrplW (L23) mutant; ▲, ΔrpmH (L34) mutant. Symbols in panels B, D, and F: ○, wild type; △, ΔrpmJ (L36) mutant; □, ΔrpsF (S6) mutant; ●, ΔrpsU (S21) mutant. Results that are representative of three independent experiments are shown.
Table 2.
Doubling times of ribosomal protein gene deletion strains at various temperatures
| Strain | Doubling time (min) of strain ata: |
||
|---|---|---|---|
| 32°C | 37°C | 45°C | |
| Wild type | 31.4 ± 0.9 | 19.9 ± 0.6 | 14.4 ± 0.7 |
| ΔrplA (L1) mutant | 122 ± 13 | 66.3 ± 4.3 | 52.1 ± 3.6 |
| ΔrplV (L22) mutant | 107 ± 9.8 | 78.4 ± 2.6 | 36.7 ± 0.5 |
| ΔrplW (L23) mutant | 111 ± 2.1 | 58.0 ± 3.8 | 32.7 ± 0.6 |
| ΔrpmH (L34) mutant | 111 ± 12 | 70.5 ± 1.4 | 31.8 ± 0.5 |
| ΔrpmJ (L36) mutant | 58.3 ± 6.5 | 46.2 ± 1.2 | 27.0 ± 1.3 |
| ΔrpsF (S6) mutant | 62.1 ± 0.4 | 36.1 ± 0.1 | 24.8 ± 1.2 |
| ΔrpsU (S21) mutant | 45.6 ± 2.1 | 34.4 ± 1.1 | 23.4 ± 1.0 |
Means of three independent experiments ± standard deviations are shown.
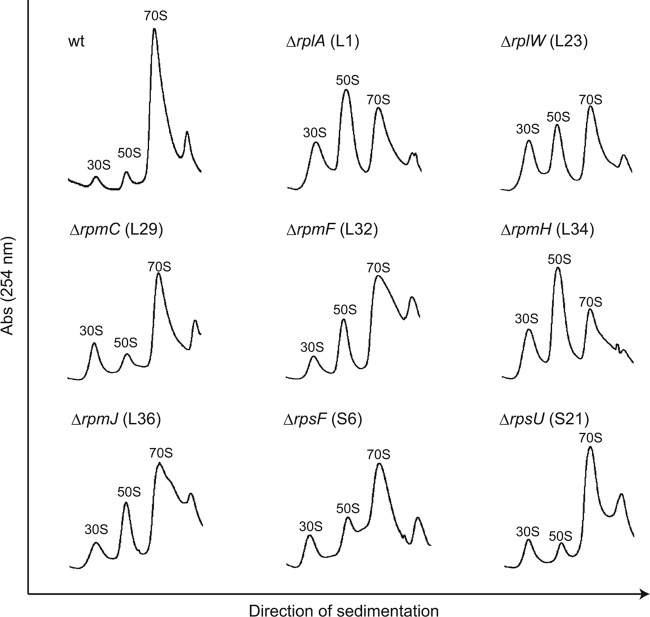
Next, we assumed that, in the deletion mutants, the absence of each individual ribosomal protein might cause a defect in the formation of 50S or 30S subunits, and thus formation of the 70S ribosome might be impaired. To test for this, the formation of the 70S ribosome was monitored by sucrose density gradient sedimentation analysis (Fig. 3; see Fig. S3 in the supplemental material). Ribosomes from eight of the mutants (L1, L23, L29, L32, L34, L36, S6, and S21) showed notably unusual profiles compared with that of the wild type (Fig. 3). In particular, there were extremely high levels of 30S and 50S subunits in the ΔrplA (L1) and ΔrpmH (L34) mutants; the areas of the peaks that corresponded to the 50S subunit were larger in these mutants than that of the 70S ribosome. The growth of almost all of these mutants was inhibited significantly at 37°C (Fig. 2 and Table 2), and it is likely that defects in the assembly of the 70S ribosome affected their growth. However, the ΔrplV mutant (L22) showed a severe growth defect (Fig. 2 and Table 2), even though a sufficient amount of the 70S ribosome was detected (see Fig. S3). This result strongly suggests that L22 is not involved in the assembly of the 50S subunit or formation of the 70S ribosome, but rather is required for the efficient functioning of the 70S ribosomal complex.
Fig 3.
Ribosome sedimentation profiles for the deletion mutant strains. Crude cell extracts were sedimented through a 10 to 40% sucrose gradient as described in Materials and Methods. The 30S, 50S, and 70S peaks are indicated in each profile. Abs, absorbance.
The process of assembly of 30S and 50S ribosomal subunits in E. coli was investigated previously by reconstituting the subunits from purified rRNA and ribosomal proteins in vitro (23, 33). Given that the in vitro reconstitution of ribosomal subunits indicated that some ribosomal proteins are necessary for the binding of other ribosomal proteins to the ribosome, it was plausible that changes in the constitution of 70S ribosomes might occur in these mutants. If this was true, it should be possible to monitor these differences through 2-D gel analyses of the 70S ribosome. In fact, in 2-D gels of ribosomes prepared from the ΔrplO (L15) and ΔrplV (L22) mutants, significant reductions in L35 and L32, respectively, were detected (Fig. 1). However, 2-D gels of ribosomes prepared from the other mutants used in the study did not show any significant changes, except for the disappearance of the target proteins whose genes were deleted (see Fig. S1 in the supplemental material). These observations suggested strongly that the in vivo assembly of ribosomal proteins into the ribosome might differ, at least in part, from assembly in vitro.
Next, we studied the effects of the absence of each ribosomal protein on spore formation. Interestingly, although nearly all of the deletion mutant strains showed sporulation frequencies that were almost identical to that of the wild type, the sporulation frequencies of the ΔrplA (L1) and ΔrplV (L22) mutant strains were reduced markedly (Table 3). In particular, the sporulation frequency of the ΔrplA (L1) mutant was less than 0.01%. It was assumed that this marked reduction in spore formation might be due to a combination of this mutant's severe growth defect (doubling time in LB at 37°C of 66.3 ± 4.3 min) and its abnormal profile for 70S formation. In contrast, although the ΔrpmH (L34) mutant also showed a severe growth defect (doubling time in LB at 37°C of 70.5 ± 1.4 min) and abnormal accumulation of ribosomal subunits (Fig. 2 and Fig. 3), the sporulation frequency of the ΔrpmH mutant was almost the same as that of the wild type (Table 3). The sporulation defect of the ΔrplA (L1) mutant was also confirmed by phase-contrast microscopy. Whereas refractile spores were observed with wild-type B. subtilis and the ΔrpmH (L34) mutant grown in 2× SG medium at 37°C for 24 h, virtually no spores were detected with the ΔrplA (L1) mutant (data not shown). Thus, it is unlikely that the sporulation defect of the ΔrplA mutant can be attributed to slow growth.
Table 3.
Sporulation of ribosomal protein gene deletion strains
| Strain | Sporulation (CFU ml−1)a |
Frequency (%)a | |
|---|---|---|---|
| Total | Spores | ||
| Wild type | 5.6 × 108 | 4.8 × 108 | 84 ± 3.9 |
| ΔrplA (L1) mutant | 1.5 × 108 | 1.5 × 102 | (1.1 ± 1.3) × 10−4 |
| ΔrplV (L22) mutant | 3.8 × 108 | 5.5 × 106 | 1.6 ± 0.5 |
| ΔrpmH (L34) mutant | 6.2 × 108 | 5.1 × 108 | 84 ± 7.4 |
Means of three independent experiments (± standard deviation for percent sporulation frequency) are shown.
It was observed that some mutants, such as the ΔrpsF (S6) and ΔrpsU (S21) strains, formed heteromorphic colonies that included hard and/or predominant clumps (data not shown). This observation prompted us to monitor the cell morphology and the motility in the mutants tested in this study. Microscopic observation revealed that the ΔrpsF (S6) and ΔrpsU (S21) cells were significantly more filamentous than the wild-type cells in LB medium at 37°C, even though apparently normal septa were observed in each of the mutants (see Fig. S4 in the supplemental material). These results suggest that a late stage of cell division is impaired in the absence of proteins S6 and S21. Next, to investigate cell motility in these mutants, aliquots of cultures that had been grown to the exponential phase in LB medium at 37°C were spotted onto soft agar plates, and the presence of swarm colonies was observed after 24 h at 37°C. As shown in Fig. 4, swarm colonies of the ΔrpsU (S21) strain (with a doubling time in LB medium at 37°C of 34.4 ± 1.1 min) were barely detected, whereas the ΔrpsF (S6) strain, whose growth rate is same as that of the ΔrpsU (S21) strain (with a doubling time in LB medium at 37°C of 36.1 ± 0.1 min), showed a small swarm circle compared with the wild type. These results suggest that S21 is necessary for cell motility.
Fig 4.
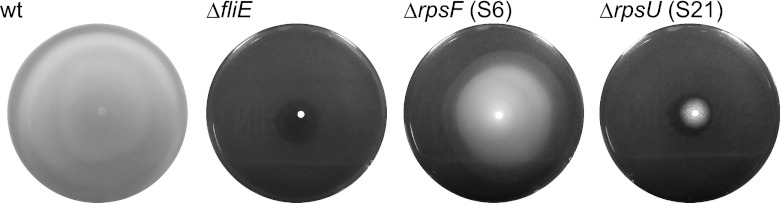
Disruption of rpsU resulted in a nonmotile phenotype. Motility plates showing the behavior of wild-type and ΔfliE, ΔrpsF, and ΔrpsU mutant cells, after 16 h at 37°C.
DISCUSSION
In the present study, we constructed a set of deletion mutants of ribosomal proteins in the Gram-positive bacterium B. subtilis, which has high spontaneous transformation activity. We then characterized the fundamental properties of these mutants, including their involvement in cell growth, ribosome assembly, and cell development. Among the 53 genes that encode ribosomal proteins and the four homologues of ribosomal protein genes that are found in the genome of B. subtilis, we were able to obtain 16 mutants that each harbored a deletion mutation of a gene that encoded a ribosomal protein (L1, L9, L15, L22, L23, L28, L29, L32, L33.1, L33.2, L34, L35, L36, S6, S20, and S21) (Table 1). Given that deletion mutants for the genes that encode L11, L25 (Ctc), L31, YtiA, S1 (YpfD), and YhzA have already been isolated (34, 36, 50, 55, 59), the results revealed that 22 ribosomal proteins are not individually essential for growth under the conditions tested in the study. It was surprising that the ribosomal proteins L1, L15, L22, L23, and L29, which are conserved among all three domains of life (42), could be deleted. Many of the deletion mutants that were obtained in the present and previous studies (1, 19, 34, 36) were deletions of genes that encoded the smaller ribosomal proteins, with molecular masses of less than 10 kDa (L28, L29, L31, L32, L33.1, L33.2, L34, L35, L36, YtiA, S20, and S21). It has been suggested that the sizes of ribosomal proteins have increased during evolution to complement the function of the rRNA, which acted originally as a ribozyme (7). This leads us to suggest that the smaller ribosomal proteins may have been incorporated into the ribosome more recently than the larger proteins and that the role of these small, nonessential proteins is to enhance the activity of the ribosome beyond that simply of protein translation. Indeed, under conditions of zinc deficiency, the zinc-containing ribosomal protein L31 (7.3 kDa) is replaced on the ribosome by YtiA (9.4 kDa), which enables L31 to be degraded to provide the essential element zinc to the cell (1, 19, 34).
Whereas several mutants showed a defect in the formation of the 70S ribosome, ribosomes from almost all of the deletion mutants, except for the L15 and L22 mutants, did not lack any ribosomal proteins apart from the target protein (Fig. 1; see Fig. S1 in the supplemental material). Although this result was consistent with the observation that the composition of the ribosome did not change markedly in any ribosomal protein deletion mutants of E. coli (49), it was not consistent with the results of in vitro reconstitution of the E. coli ribosome (23, 33). Therefore, it is possible that assembly of the ribosome in vitro might differ from that in vivo in some aspects as reported previously (53). At present, we cannot exclude the possibility that immature 50S and 30S subunits, formed as the result of the absence of a particular ribosomal protein, might not be detected readily by our 2-D gel analyses, and this might explain the difference in the in vitro and in vivo results. Indeed, we reported previously that depletion of S14 causes the accumulation of immature 30S subunits in which S2 and S3 are decreased markedly (36). This result was in good agreement with previous reports based on in vitro studies (23, 33). Further investigation to clarify the composition of ribosomal proteins in the 50S and 30S subunits that accumulate abnormally in deletion mutants should provide more detailed information about the in vivo pathway of ribosome assembly.
The amount of L32 protein was decreased significantly in ribosomes from the ΔrplV (L22) mutant (Fig. 1). Thus, it is likely that the absence of both L22 and L32 is responsible for the severe effect on cell proliferation, as well as the reduction of sporulation frequency, in the ΔrplV mutant (Fig. 2 and Table 3). A previous study has reported that L22 is located close to L32 in the 50S ribosome of Thermus thermophilus (62). Cross-linking of L22 to L32 in E. coli has also been reported (58). In contrast, L22 protein could be detected at an appreciable level in the 2-D gel of ribosomal proteins prepared from the ΔrpmF (L32) strain (see Fig. S1 in the supplemental material). Taken together, these results suggested that L22 is required for binding of L32 to the ribosome, and thus that a lack of L22 causes structural alteration of the 50S subunit. Similarly, although the relationship between L15 and L35 had not been elucidated previously, a significant reduction of L35 in ribosomes of the ΔrplO (L15) mutant was observed (Fig. 1), whereas the amount of L15 in ribosomes from the ΔrpmI (L35) mutant was almost the same as that in wild-type ribosomes (see Fig. S1). Thus, it is most likely that L15 is required for binding of L35 to the ribosome.
The ΔrplA (L1) and ΔrplV (L22) mutants showed a reduction in sporulation frequency (Table 3). A previous study had shown that inactivation of ctc, which encodes a homologue of L25, causes a temperature-sensitive sporulation phenotype in B. subtilis (55). In addition, the sporulation frequency of an rpmGB::pMutinT3-rpmGB mutant, in which transcription of the rpmGB (L33.2) gene is under the control of an isopropyl-β-d-thiogalactoside (IPTG)-regulated promoter, decreased slightly at 47°C (41). However, the involvement of these ribosomal proteins in sporulation remains unclear.
The other observation that suggests a relationship between ribosomal proteins and cell development is the loss of motility of the S21 deletion mutant strain (Fig. 4). The expression of the genes that are involved in the motility of B. subtilis cells is directed mainly by σD, a sigma factor that activates the expression of the genes that encode the components of the flagellar hook and motor, and the flagellar filament protein (48). The activity of σD is controlled by an anti-sigma factor, FlgM, which binds to σD and inhibits σD-dependent gene expression (6, 10). In addition, SwrA activates the fla-che operon, which includes flagellum genes, genes involved in chemotaxis, and the gene for the σD, while SwrB is required for maximal σD-dependent gene expression (26). It is likely that S21 is involved in the sufficient expression of these factors via protein translation. Further research using the S21 mutant described herein should help elucidate the details of this relationship between the ribosomal protein and cell motility.
In the present study, we showed that 22 out of 57 genes that encode ribosomal proteins can be deleted. Among them, the genes that encode L22, L23, and L28 have been reported to be essential for cell proliferation in E. coli (Table 1) (4, 49). Although an L28 mutant, VT423, in which the altered L28 was unable to bind to the ribosome, had been isolated (14), L22 and L23 defective mutations were shown to be lethal in E. coli (49). Our strategy to obtain the deletion mutants was based on the replacement of the target gene with a chloramphenicol resistance gene, as described in Materials and Methods. During the course of the study, we carried out transformation experiments under various temperature conditions. Hence, these procedures differed from those used in the study of Shoji et al. (49), in which the essential nature of the genes was confirmed by gene complementation experiments. It is plausible that it might be possible to introduce deletions of the genes that encode L22 and L23 into E. coli at high temperature. Comparison of the functions of individual ribosomal proteins between B. subtilis and E. coli using two sets of ribosomal protein deletion mutants should help to elucidate the evolutionary processes that each ribosomal protein has undergone.
Although many of the strains that harbored a deletion mutation in a ribosomal protein exhibited various defective phenotypes with regard to cell proliferation, 70S formation, spore development, and cell motility, several mutants did not show any significant phenotype. It is possible that the phenotypes of the deletion mutants are masked by the vast number of ribosomes in the cell, because the B. subtilis genome contains 10 rRNA (rrn) operons. We have already constructed mutants that harbor only a single rrn operon (either rrnA, -B, -D, -E, -I, -J, or -O) in their genome and have confirmed a reduction in the number of ribosomes in these mutants (35). Novel phenotypes that have not been observed in the present study might appear if the deletion mutations of the ribosomal proteins are introduced into the strains that harbor a single rrn operon. It is known that some ribosomal proteins regulate the expression of their own genes. For example, in B. subtilis, the expression of the infC-rpmI-rplT operon, which encodes the translation factor IF3 and the ribosomal proteins L35 and L20, is regulated by L20 and the expression of rpsD, which encodes S4, is autoregulated (12, 22). However, the function of ribosomal proteins in cell development has not been elucidated. Further investigations, including genome-wide approaches such as a transcriptomic or proteomic analyses, are needed to clarify whether ribosomal proteins are involved in the adjustment to adverse environmental conditions.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to C. R. Harwood for helpful discussion and critical reading of the manuscript. We thank Naofumi Nomura, Rikinori Murayama, Kazuma Furuya, Mai Honda, Masato Katano, and Sawako Yoshida for constructing the mutants.
This work was supported in part by Grants-in-Aid for Scientific Research (C) (F.K.) and the Strategic Research Foundation Grant-Aided Project for Private Universities (S1201003) (F.K. and Y.S.) from the Ministry of Education, Culture, Sports, Science, and Technology of Japan. In addition, this work was partly supported by a research grant from the Institute for Fermentation, Osaka, to F.K.
Footnotes
Published ahead of print 21 September 2012
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1. Akanuma G, Nanamiya H, Natori Y, Nomura N, Kawamura F. 2006. Liberation of zinc-containing L31 (RpmE) from ribosomes by its paralogous gene product, YtiA, in Bacillus subtilis. J. Bacteriol. 188:2715–2720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Anagnostopoulos C, Spizizen J. 1961. Requirements for transformation in Bacillus subtilis. J. Bacteriol. 81:741–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ashikaga S, Nanamiya H, Ohashi Y, Kawamura F. 2000. Natural genetic competence in Bacillus subtilis natto OK2. J. Bacteriol. 182:2411–2415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Baba T, et al. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2:2006.0080 doi:10.1038/msb4100050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ban N, Nissen P, Hansen J, Moore PB, Steitz TA. 2000. The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. Science 289:905–920 [DOI] [PubMed] [Google Scholar]
- 6. Bertero MG, Gonzales B, Tarricone C, Ceciliani F, Galizzi A. 1999. Overproduction and characterization of the Bacillus subtilis anti-sigma factor FlgM. J. Biol. Chem. 274:12103–12107 [DOI] [PubMed] [Google Scholar]
- 7. Bokov K, Steinberg SV. 2009. A hierarchical model for evolution of 23S ribosomal RNA. Nature 457:977–980 [DOI] [PubMed] [Google Scholar]
- 8. Bowen WS, Van Dyke N, Murgola EJ, Lodmell JS, Hill WE. 2005. Interaction of thiostrepton and elongation factor-G with the ribosomal protein L11-binding domain. J. Biol. Chem. 28:2934–2943 [DOI] [PubMed] [Google Scholar]
- 9. Cameron DM, Thompson J, Gregory ST, March PE, Dahlberg AE. 2004. Thiostrepton-resistant mutants of Thermus thermophilus. Nucleic Acids Res. 32:3220–3227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Caramori T, BarillÀ D, Nessi C, Sacchi L, Galizzi A. 1996. Role of FlgM in σD-dependent gene expression in Bacillus subtilis. J. Bacteriol. 178:3113–3118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cashel M, Gentry DR, Hernandez VJ, Vinella D. 1996. The stringent response, p 1458–1496 In Neidhardt FC, et al. (ed), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, DC [Google Scholar]
- 12. Choonee N, Even S, Zig L, Putzer H. 2007. Ribosomal protein L20 controls expression of the Bacillus subtilis infC operon via a transcription attenuation mechanism. Nucleic Acids Res. 35:1578–1588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cukras AR, Southworth DR, Brunelle JL, Culver GM, Green R. 2003. Ribosomal proteins S12 and S13 function as control elements for translocation of the mRNA:tRNA complex. Mol. Cell 12:321–328 [DOI] [PubMed] [Google Scholar]
- 14. Dabbs ER. 1978. Mutational alterations in 50 proteins of the Escherichia coli ribosome. Mol. Gen. Genet. 165:73–78 [DOI] [PubMed] [Google Scholar]
- 15. Dabbs ER. 1979. Selection for Escherichia coli mutants with proteins missing from the ribosome. J. Bacteriol. 140:734–737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Dabbs ER. 1991. Mutants lacking individual ribosomal proteins as a tool to investigate ribosomal properties. Biochimie 73:639–645 [DOI] [PubMed] [Google Scholar]
- 17. Diaconu M, et al. 2005. Structural basis for the function of the ribosomal L7/12 stalk in factor binding and GTPase activation. Cell 121:991–1004 [DOI] [PubMed] [Google Scholar]
- 18. Fei J, Kosuri P, MacDougall DD, Gonzalez RL. 2008. Coupling of ribosomal L1 stalk and tRNA dynamics during translation elongation. Mol. Cell 30:348–359 [DOI] [PubMed] [Google Scholar]
- 19. Gabriel SE, Helmann JD. 2009. Contributions of Zur-controlled ribosomal proteins to growth under zinc starvation conditions. J. Bacteriol. 191:6116–6122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gao YG, et al. 2009. The structure of the ribosome with elongation factor G trapped in the posttranslocational state. Science 326:694–699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Götz F, Dabbs ER, Gualerzi CO. 1990. Escherichia coli 30S mutants lacking protein S20 are defective in translation initiation. Biochim. Biophys. Acta 1050:93–97 [DOI] [PubMed] [Google Scholar]
- 22. Grundy FJ, Henkin TM. 1991. The rpsD gene, encoding ribosomal protein S4, is autogenously regulated in Bacillus subtilis. J. Bacteriol. 173:4595–4602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Herold M, Nierhaus KH. 1987. Incorporation of six additional proteins to complete the assembly map of the 50 S subunit from Escherichia coli ribosomes. J. Biol. Chem. 262:8826–8833 [PubMed] [Google Scholar]
- 24. Imamura D, et al. 2004. spoIVH (ykvV), a requisite cortex formation gene, is expressed in both sporulating compartments of Bacillus subtilis. J. Bacteriol. 186:5450–5459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jenner L, Demeshkina N, Yusupova G, Yusupov M. 2010. Structural rearrangements of the ribosome at the tRNA proofreading step. Nat. Struct. Mol. Biol. 17:1072–1078 [DOI] [PubMed] [Google Scholar]
- 26. Kearns DB, Losick R. 2005. Cell population heterogeneity during growth of Bacillus subtilis. Genes Dev. 19:3083–3094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kobayashi K, et al. 2003. Essential Bacillus subtilis genes. Proc. Natl. Acad. Sci. U. S. A. 100:4678–4683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kunst F, et al. 1997. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature 390:249–256 [DOI] [PubMed] [Google Scholar]
- 29. Leighton TJ, Doi RH. 1971. The stability of messenger ribonucleic acid during sporulation in Bacillus subtilis. J. Biol. Chem. 246:3189–3195 [PubMed] [Google Scholar]
- 30. Li X, Lindahl L, Sha Y, Zengel JM. 1997. Analysis of the Bacillus subtilis S10 ribosomal protein gene cluster identifies two promoters that may be responsible for transcription of the entire 15-kilobase S10-spc-alpha cluster. J. Bacteriol. 179:7046–7054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lotti M, Dabbs ER, Hasenbank R, Stöffler-Meilicke M, Stöffler G. 1983. Characterisation of a mutant from Escherichia coli lacking protein L15 and localisation of protein L15 by immuno-electron microscopy. Mol. Gen. Genet. 192:295–300 [DOI] [PubMed] [Google Scholar]
- 32. Maguire BA, Beniaminov AD, Ramu H, Mankin AS, Zimmermann RA. 2005. A protein component at the heart of an RNA machine: the importance of protein L27 for the function of the bacterial ribosome. Mol. Cell 20:427–435 [DOI] [PubMed] [Google Scholar]
- 33. Mizushima S, Nomura M. 1970. Assembly mapping of 30S ribosomal proteins from E. coli. Nature 226:1214–1218 [DOI] [PubMed] [Google Scholar]
- 34. Nanamiya H, et al. 2004. Zinc is a key factor in controlling alternation of two types of L31 protein in the Bacillus subtilis ribosome. Mol. Microbiol. 52:273–283 [DOI] [PubMed] [Google Scholar]
- 35. Nanamiya H, et al. 2010. Bacillus subtilis mutants harbouring a single copy of the rRNA operon exhibit sever defects in growth and sporulation. Microbiology 156:2944–2952 [DOI] [PubMed] [Google Scholar]
- 36. Natori Y, et al. 2007. A fail-safe system for the ribosome under zinc-limiting conditions in Bacillus subtilis. Mol. Microbiol. 63:294–307 [DOI] [PubMed] [Google Scholar]
- 37. Nishi K, Dabbs ER, Schnier J. 1985. DNA sequence and complementation analysis of a mutation in the rplX gene from Escherichia coli leading to loss of ribosomal protein L24. J. Bacteriol. 163:890–894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Nissen P, Hansen J, Ban N, Moore PB, Steitz TA. 2000. The structural basis of ribosome activity in peptide bond synthesis. Science 289:920–930 [DOI] [PubMed] [Google Scholar]
- 39. Ochi K, Zhang D, Kawamoto S, Hesketh A. 1997. Molecular and functional analysis of the ribosomal L11 and S12 protein genes (rplK and rpsL) of Streptomyces coelicolor A3(2). Mol. Gen. Genet. 256:488–498 [DOI] [PubMed] [Google Scholar]
- 40. Ogle JM, et al. 2001. Recognition of cognate transfer RNA by the 30S ribosomal subunit. Science 292:897–902 [DOI] [PubMed] [Google Scholar]
- 41. Ohashi Y, et al. 2003. Expression profiling of translation-associated genes in sporulating Bacillus subtilis and consequence of sporulation by gene inactivation. Biosci. Biotechnol. Biochem. 67:2245–2253 [DOI] [PubMed] [Google Scholar]
- 42. Roberts E, Sethi A, Montoya J, Woese CR, Luthey-Schulten Z. 2008. Molecular signatures of ribosomal evolution. Proc. Natl. Acad. Sci. U. S. A. 105:13953–13958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Rutberg L. 1969. Mapping of a temperate bacteriophage active on Bacillus subtilis. J. Virol. 3:38–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 45. Schluenzen F, et al. 2000. Structure of functionally activated small ribosomal subunit at 3.3 Å resolution. Cell 102:615–623 [DOI] [PubMed] [Google Scholar]
- 46. Schmeing TM, Ramakrishnan V. 2009. What recent ribosome structures have revealed about the mechanism of translation. Nature 461:1234–1242 [DOI] [PubMed] [Google Scholar]
- 47. Schmeing TM, et al. 2009. The crystal structure of the ribosome bound to EF-Tu and aminoacyl-tRNA. Science 326:688–694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Serizawa M, et al. 2004. Systematic analysis of SigD-regulated genes in Bacillus subtilis by DNA microarray and Northern blotting analyses. Gene 329:125–136 [DOI] [PubMed] [Google Scholar]
- 49. Shoji S, Dambacher CM, Shajani Z, Williamson JR, Schultz PG. 2011. Systematic chromosomal deletion of bacterial ribosomal protein genes. J. Mol. Biol. 413:751–761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Sorokin A, Serror P, Pujic P, Azevedo V, Ehrlich SD. 1995. The Bacillus subtilis chromosome region encoding homologues of the Escherichia coli mssA and rpsA gene products. Microbiology 141:311–339 [DOI] [PubMed] [Google Scholar]
- 51. Stöffler G, Noah M, Stöffler-Meilicke M, Dabbs ER. 1984. The localization of protein L19 on the surface of 50 S subunits of Escherichia coli aided by the use of mutants lacking protein L19. J. Biol. Chem. 259:4521–4526 [PubMed] [Google Scholar]
- 52. Stöffler-Meilicke M, Dabbs ER, Albrecht-Ehrlich R, Stöffler G. 1985. A mutant from Escherichia coli which lacks ribosomal proteins S17 and L29 used to localize these two proteins on the ribosomal surface. Eur. J. Biochem. 150:485–490 [DOI] [PubMed] [Google Scholar]
- 53. Sykes MT, Shajani Z, Sperling E, Beck AH, Williamson JR. 2010. Quantitative proteomic analysis of ribosome assembly and turnover in vivo. J. Mol. Biol. 403:331–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Takyar S, Hickerson RP, Noller HF. 2005. mRNA helicase activity of the ribosome. Cell 120:49–58 [DOI] [PubMed] [Google Scholar]
- 55. Truitt CL, Weaver EA, Haldenwang WG. 1988. Effects on growth and sporulation of inactivation of a Bacillus subtilis gene (ctc) transcribed in vitro by minor vegetative cell RNA polymerases (E-σ37, E-σ32). Mol. Gen. Genet. 212:166–171 [DOI] [PubMed] [Google Scholar]
- 56. Van Dyke N, Xu W, Murgola EJ. 2002. Limitation of ribosomal protein L11 availability in vivo affects translation termination. J. Mol. Biol. 31:329–339 [DOI] [PubMed] [Google Scholar]
- 57. Wada A. 1986. Analysis of Escherichia coli ribosomal proteins by an improved two dimensional gel electrophoresis. I. detection of four new proteins. J. Biochem. 100:1583–1594 [DOI] [PubMed] [Google Scholar]
- 58. Walleczek J, Redl B, Stöffler-Meilicke M, Stöffler G. 1989. Protein-protein cross-linking of the 50 S ribosomal subunit of Escherichia coli using 2-iminothiolane. Identification of cross-links by immunoblotting techniques. J. Biol. Chem. 264:4231–4237 [PubMed] [Google Scholar]
- 59. Wienen B, et al. 1979. Ribosomal protein alterations in thiostrepton- and micrococcin-resistant mutants of Bacillus subtilis. J. Biol. Chem. 254:8031–8041 [PubMed] [Google Scholar]
- 60. Wimberly BT, et al. 2000. Structure of the 30S ribosomal subunit. Nature 407:327–339 [DOI] [PubMed] [Google Scholar]
- 61. Yang X, Ishiguro EE. 2001. Involvement of the N terminus of ribosomal protein L11 in regulation of the RelA protein of Escherichia coli. J. Bacteriol. 183:6532–6537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Yusupov MM, et al. 2001. Crystal structure of the ribosome at 5.5 Å resolution. Science 292:883–896 [DOI] [PubMed] [Google Scholar]
- 63. Yusupova GZ, Yusupov MM, Cate JH, Noller HF. 2001. The path of messenger RNA through the ribosome. Cell 106:233–241 [DOI] [PubMed] [Google Scholar]
- 64. Zaher HS, Green R. 2010. Hyperaccurate and error-prone ribosomes exploit distinct mechanisms during tRNA selection. Mol. Cell 39:110–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Zhang S, Scott JM, Haldenwang WG. 2001. Loss of ribosomal protein L11 blocks stress activation of the Bacillus subtilis transcription factor σB. J. Bacteriol. 183:2316–2321 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.