Abstract
The twin arginine translocation (Tat) pathway exports folded proteins from the cytoplasm to the periplasm of bacteria. The targeting of the exported proteins to the Tat pathway relies on a specific amino-terminal signal sequence, which is cleaved after exportation. In the phytopathogen Dickeya dadantii, the pectin lyase homologue PnlH is exported by the Tat pathway without cleavage of its signal sequence, which anchors PnlH into the outer membrane. In proteobacteria, the vast majority of outer membrane proteins consists of β-barrel proteins and lipoproteins. Thus, PnlH represents a new kind of outer membrane protein. In Escherichia coli, periplasmic chaperones SurA, Skp, and DegP work together with the β-barrel assembly machinery (Bam) to target and insert β-barrel proteins into the outer membrane. In this work, we showed that SurA is required for an efficient targeting of PnlH to the outer membrane. Moreover, we were able to detect an in vitro interaction between SurA and the PnlH signal sequence. Since the PnlH signal sequence contains a highly hydrophobic region, we propose that SurA protects it from the hydrophobic periplasm during targeting of PnlH to the outer membrane. We also studied the nature of the information carried by the PnlH signal sequence responsible for its targeting to the outer membrane after exportation by the Tat system.
INTRODUCTION
Proteobacteria are divided into four compartments: the cytoplasm surrounded by the inner membrane (IM), which along with the outer membrane (OM) delimitates the periplasm. In order to maintain this organization, proteobacteria have evolved different processes allowing the targeting of molecules to the appropriate compartment. OM biogenesis may be the most challenging process. Indeed, OM components, synthesized in the cytoplasm, have to cross the inner membrane and the periplasm before being inserted into the OM (38). The large majority of OM proteins are either lipoproteins, anchored in the OM by an amino-terminal lipid moiety, or outer membrane integral proteins (OMPs) forming a transmembrane β barrel in the OM. Like periplasmic or some secreted proteins, OM proteins are synthesized in the cytoplasm as precursors containing an amino-terminal signal sequence specifically targeting them to either the Sec translocon or the twin arginine translocation (Tat) pathway. The bulk mass of OM proteins is exported to the periplasm in an unfolded state by the Sec translocon. However, a few proteins are folded in the cytoplasm and then targeted to the Tat translocon by a particular signal sequence containing two arginine residues in their N-terminal part (11, 32).
After exportation, OMP precursors are maturated by the cleavage of their signal sequence by the signal peptidase LepB (34). Due to their propensity to form a β barrel, OMPs are prone to aggregation and have to be kept in a folding competent state during the transport from the exit of the Sec translocon to the OM. This process relies on three periplasmic proteins: (i) SurA, a chaperone that also has two peptidyl-prolyl isomerase domains and that is the main OMP-targeting factor in the periplasm of Escherichia coli (5, 39, 48); (ii) DegP, a multimeric protein that has both a chaperone and a protease activity depending on its oligomerization state (49); and (iii) Skp, a chaperone that has been described to bind to unfolded OMPs to protect them from aggregation (54). In E. coli, Skp and DegP form a rescue pathway that deals with OMPs falling off the SurA main pathway (39, 48). However, whichever periplasmic chaperone is involved in OMP transport, the last step of OMP biogenesis, folding and insertion into the OM, is catalyzed by the β-barrel assembly machinery (Bam) complex (56). This complex of five OM proteins consists of a widely conserved OMP from the Omp85 family, BamA, and four lipoproteins, BamB, BamC, BamD, and BamE (47). Interestingly, the Bam complex has been shown to catalyze in vitro OMP folding and insertion into proteoliposomes in the presence of SurA, reinforcing the idea of a SurA main pathway (13). Since OMPs are not fully folded before insertion into the OM, the signal allowing their targeting to the OM should reside in the linear sequence of the protein. Consistent with this idea, the periplasmic chaperone SurA has been widely described to exhibit a preferential affinity for Ar-X-Ar sequences, where Ar is an aromatic amino acid, which are more common in OMPs than in soluble proteins because of the presence of an aromatic girdle in β barrels (15, 45). Furthermore, it has been suggested that the E. coli BamA could recognize OMPs via the 12 carboxy-terminal residues and notably via a carboxy-terminal phenylalanine (40). However, this signal is not the only one allowing the interaction between BamA and OMPs, and it is not conserved among all bacteria (4, 40).
Lipoprotein maturation relies on a signal peptidase (LspA) and on two enzymes involved in lipoprotein acylation (Lgt, Lnt) (14). Together, these three enzymes are responsible for signal sequence cleavage and addition of three acyl chains to the lipoprotein amino-terminal cysteine residue, resulting in the insertion of the lipoprotein lipid moiety within the IM. Lipoproteins lacking an IM retention signal (or Lol avoidance signal) are then extracted from the IM by the ABC transporter LolCDE and transferred to the chaperone LolA to form a water-soluble complex (24, 26, 58). The exact nature of the Lol avoidance signal varies from one organism to another but often relies on the nature of the residues following the amino-terminal cysteine. In E. coli, the presence of an aspartate residue at position +2 seems to constitute the strongest LolCDE avoidance signal (50). Moreover, the presence of three acyl chains on the amino-terminal cysteine is indispensable for interaction with LolCDE and therefore for transfer to LolA (9). Eventually, lipoproteins are transferred in an energy-independent manner from LolA to the OM lipoprotein LolB that is thought to ensure insertion of the lipoprotein lipid moiety into the OM (25, 55).
Recent work in the gammaproteobacterial phytopathogen Dickeya dadantii has shown the existence of a third kind of OM protein. The pectin lyase homologue, PnlH, is anchored in the OM by an uncleaved Tat signal sequence (8). Fusion of the PnlH Tat signal sequence to various proteins allows targeting of these fusion proteins to the outer membrane. This indicates that, contrary to other OM proteins, all the information required for PnlH targeting to the OM relies in its signal sequence. Moreover, PnlH does not share any similarity (transmembrane β barrel or amino-terminal acylation) with OMPs or lipoproteins. Taken together, these data suggest the existence of a new pathway for the targeting of proteins to the OM of D. dadantii. Since the expression of PnlH in E. coli results in a correct insertion of PnlH into the OM, this pathway should be conserved in other bacteria. The aim of this work was to identify proteins involved in the targeting of this third kind of OM protein. To this end, we analyzed the impact of mutations in the OMP-targeting pathway on the targeting of PnlH to the OM of D. dadantii. Surprisingly, we showed that SurA is involved in the targeting of PnlH to the OM. We also studied the nature of the information carried by the PnlH signal sequence.
MATERIALS AND METHODS
Bacterial strains, plasmids, and oligonucleotides.
The bacterial strains and plasmids used in this study are described in Table 1. Oligonucleotides are listed in Table S1 in the supplemental material. D. dadantii and E. coli were grown at 30°C and 37°C, respectively, in LB medium. When required, the following antibiotics were added at the indicated concentrations: ampicillin, 100 μg/ml; kanamycin, 25 μg/ml; and chloramphenicol, 25 μg/ml and 5 μg/ml for E. coli and D. dadantii, respectively.
Table 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant characteristics | Source or reference |
|---|---|---|
| Strains | ||
| D. dadantii 3937 derivatives | ||
| A350 | rafR ganB | 17 |
| A5275 | rafR ganB bamE::Kanr | This study |
| A5276 | rafR ganB bamC::Kanr | This study |
| A5281 | rafR ganB bamB::Kanr | This study |
| A5476 | rafR ganB degP::Kanr | This study |
| A5477 | rafR ganB skp::uidA Kanr | This study |
| A5478 | rafR ganB surA::Kanr | This study |
| Escherichia coli | ||
| NM522 | F′ proAB lacIq Δ(lacZ)M15 Δ(lac-proAB) thi hsd-5 | Stratagene |
| SR1458 | F− araD139 Δ(argF-lac)U169 ϕ(PdegP-lacZ) | 36 |
| Plasmids | ||
| pGEM-T | Ampr; allows the AT cloning of a PCR product | Promega |
| pBluescript | pUC ori Ampr | Stratagene |
| pACT3 | pACYC ori Cmr | 23 |
| pL247 | Ampr uidA::Kanr cassette | Laboratory collection |
| pHP45ΩKan | pMB1 ori Ampr Kanr cassette | 7 |
| pGEX-6P3 | pBR322 ori Ampr, codes for GST | GE Healthcare |
| pGEM-T surA | Contains the DNA fragment amplified with SurA+ and SurA− | This study |
| pGEM-T surA::Kanr | pHP45ΩKan Kanr cassette in the HindIII site of pGEM-T surA | This study |
| pGEM-T skp | Contains the DNA fragment amplified with Skp+ and Skp− | This study |
| pGEM-T skp::Kanr | pL247 uidA::Kanr cassette in the PstI site of pGEM-T skpΔPstI | This study |
| pGEM-T degP | Contains the DNA fragment amplified with DegP+ and DegP− | This study |
| pGEM-T degP::Kanr | pHP45ΩKan Kanr cassette in the MunI site of pGEM-T degP | This study |
| pGEM-T bamB | Contains the DNA fragment amplified with BamB+ and BamB− | This study |
| pGEM-T bamB::kanr | pHP45ΩKan Kanr cassette in the BclI site of pGEM-T bamB | This study |
| pGEM-T bamC | Contains the DNA fragment amplified with BamC+ and BamC− | This study |
| pGEM-T bamC::kanr | pHP45ΩKan Kanr cassette in the BclI site of pGEM-T bamC | This study |
| pGEM-T bamE | Contains the DNA fragment amplified with BamE+ and BamE− | This study |
| pGEM-T bamE::kanr | pHP45ΩKan Kanr cassette between BamHI sites of pGEM-T bamE | This study |
| pGEM-T pnlH | Contains the DNA fragment amplified with PnlHHis+ and PnlHHis− | This study |
| pBSpnlH | Insertion of pGEM-T pnlH ApaI SacI DNA fragment containing pnlH CDS in pBS | This study |
| pPnlH | pACT3 containing the gene of PnlH with a C-terminal His tag between SacI and KpnI sites | This study |
| PnlH-PemA | pBAD18 containing a fusion of the PnlH signal sequence with the mature form of PemA | 8 |
| pPnlHssPemA | pACT3 containing a fusion of the PnlH signal sequence with the mature form of PemA | This study |
| pD16A | pPnlH containing a D16A substitution in PnlH | This study |
| pL30SL31T | pPnlH containing L30SL31T substitutions in PnlH | This study |
| pP32L | pPnlH containing a P32L substitution in PnlH | This study |
| pF34A | pPnlH containing a F34A substitution in PnlH | This study |
| pP35A | pPnlH containing a P35A substitution in PnlH | This study |
| pP32LP35A | pPnlH containing P32LP35A substitutions in PnlH | This study |
| pY36A | pPnlH containing a Y36A substitution in PnlH | This study |
| pQ41A | pPnlH containing a Q41A substitution in PnlH | This study |
| pPnlHΔ28-41 | pPnlH containing a deletion of nucleotides 81 to 123 | This study |
| pSurA | pGEM-T containing SurA with a C-terminal Strep tag under the control of the lac promoter | This study |
| pXss20 | pGEX-6P3 containing of a fusion of GST with the first 41 amino acids of PnlH | This study |
To construct strains A5275, A5276, A5281, A5476, A5477, and A5478, DNA fragments containing the genes bamE, bamC, bamB, degP, skp, and surA, respectively, were amplified with the corresponding primers. The resulting DNA fragments were inserted into the pGEM-T plasmid. The Kanr cassette was extracted from pHP45ΩKan and inserted into the different coding sequences (CDSs), except for skp, for which the uidA::Kanr cassette of pL247 was prepared and inserted into its CDS. All the resulting constructs were inserted into the D. dadantii A350 chromosome by recombination in low-phosphate medium (41). Recombinations were checked by PCR. Before phenotype analysis, all mutations were transduced into a fresh A350 background (37).
To construct plasmid pPnlH, a DNA fragment containing pnlH was amplified with the oligonucleotides PnlHHis+ and PnlHHis−. The resulting DNA was subcloned into pBluescript (pBS) between the SacI and ApaI sites. The resulting plasmid (pBSpnlH) was digested by SacI and KpnI, and the DNA fragment containing pnlH was inserted into pACT3. To construct plasmids carrying one or more substitutions in the PnlH-6His signal sequence, site-directed mutagenesis with appropriate primers (see Table S1 in the supplemental material) was performed. To construct pPnlHssPemA, a DNA fragment containing the CDS of the PnlH signal sequence fused to the mature form of PemA (PnlHssPemA) was amplified from the PnlH-PemA plasmid (8) with PnlHssPemA+ and PnlHssPemA−. The PCR product was digested by SacI and KpnI and then inserted into pACT3. To construct pPnlHΔ28-41, site-directed mutagenesis with primers Δ28-41+ and Δ28-41− and pPnlH as the template was performed to add an extra HindIII site at nucleotide 123 of the pnlH CDS. The resulting DNA was digested by HindIII and recircularized to remove the DNA fragment between the two HindIII sites. To construct pSurA, the SurAStrep− primer was designed to add a Strep tag at the carboxy terminus of SurA, and a DNA fragment containing surA was amplified with SurAStrep+ and SurAStrep−. The PCR product was inserted into pGEM-T. Insertions allowing expression of SurA with the carboxy-terminal Strep tag under the control of the pGEM-T lac promoter were selected. To construct pXss20, the sequence of the PnlH signal sequence was amplified with Xss20+ and Xss20−. The PCR product was inserted in frame into pGEX-6P3 between the BamHI and SalI sites downstream of the glutathione S-transferase (GST)-coding sequence. All constructions were checked by sequencing.
Drug sensitivity assay.
The sensitivity to EDTA, SDS, and rifampin of different mutant strains was determined by disk diffusion assay. A 0.1-ml sample from an LB overnight culture was mixed with 3 ml of molten LB top agar and poured over an LB agar plate. Three 6-mm filter paper disks (Dominique Dutscher) were put on the top of molten agar, and 7.5 μl of EDTA (0.5 M), SDS (10%, wt/wt), or rifampin (20 mg/ml) was added onto a disk. The diameters of the growth inhibition zones were measured after 16 h.
Cell fractionation.
To prepare the periplasm, cells were grown overnight at 30°C in LB medium complemented with the appropriate antibiotics. Cells were harvested by centrifugation for 7 min at 5,000 × g, and the periplasm was prepared by lysozyme treatment and osmotic shock as previously described (2). Whole-cell fractions from steady-state cultures or exponential-phase cultures were obtained by concentrating cells up to an optical density at 600 nm (OD600) of 7.5. To prepare membranes, cells from exponential-phase cultures were harvested by centrifugation for 7 min at 5,000 × g, concentrated up to an OD600 of 7.5 in TE (10 mM Tris-HCl, pH 8.0, 1 mM EDTA), 1 mM phenylmethylsulfonyl fluoride (PMSF), and broken with a French press. Debris was removed by centrifugation for 20 min at 7,000 × g, and the supernatant was ultracentrifuged for 1 h at 100,000 × g. The supernatant and the pellet were taken as the soluble fraction and membrane fraction, respectively. When necessary, membranes were separated by centrifugation for 48 h at 200,000 × g on a 38 to 65% sucrose flotation gradient as previously described (46). Fractions were collected from the bottom of the tube and precipitated with ethanol.
To analyze the composition of the different fractions, proteins were separated by SDS-PAGE and transferred onto a PVDF membrane (Millipore). PnlH, KdgR, PemA, BlaM, and TolA were detected by immunodetection. Anti-PnlH, anti-KdgR, anti-PemA, anti-BlaM, and anti-TolA antibodies were used at dilutions of 1/3,000, 1/5,000, 1/5,000, 1/3,000, and 1/10,000, respectively. The position of the inner membrane in the sucrose gradient was detected by NADH oxidase activity assay (31) or immunoblotting with TolA antibodies; the position of porins was detected by staining the immunoblot membrane with Coomassie blue.
In vitro interactions with SurA-Strep.
To prepare PnlH-6His inclusion bodies, NM522/pPnlH cells were grown at 30°C. At an OD600 of 0.6, isopropyl-β-d-thiogalactopyranoside (IPTG) was added at a final concentration of 1 mM and cells were grown for an extra 2 h. Cells were harvested by centrifugation for 7 min at 5,000 × g, concentrated up to an OD600 of 7.5 in TE, 1 mM PMSF, and broken with a French press. The lysate was centrifuged for 20 min at 7,000 × g, and PnlH inclusion bodies were harvested in the pellet. To get pure GST and derivatives, NM522/pGEX-6P3 or NM522/pXSS20 cells were grown at 30°C. At an OD600 of 0.5 mM, IPTG was added at a final concentration of 1 mM and cells were grown for an extra 2 h. Cells were harvested by centrifugation for 7 min at 5,000 × g, concentrated up to an OD600 of 7.5 in TE, 1 mM PMSF, and broken with a French press. Triton X-100 was added at a final concentration of 1% (vol/vol). The lysate was centrifuged for 20 min at 7,000 × g to removed debris, and the supernatant was loaded onto Protino glutathione agarose 4B (Macherey-Nagel) resin previously equilibrated with TES (50 mM Tris-HCl, pH 7.4, 1 mM EDTA, 100 mM NaCl), 1% (vol/vol) Triton X-100. The resin was then washed 3 times with TES, 1% (vol/vol) Triton X-100. GST and derivatives were eluted with 2× Laemmli buffer. For overlay experiments, samples containing proteins of interest were run on an SDS-polyacrylamide gel and transferred onto a PVDF membrane. The membrane was saturated with 4% gelatin in phosphate-buffered saline (PBS; 81 mM Na2HPO4, 19 mM NaH2PO4, 150 mM NaCl, pH 7.4). As after all following steps, the membrane was washed 3 times for 5 min each time with PBS containing 1% gelatin (PBS-G). After incubation for 45 min at room temperature with 8.5 ml of PBS-G complemented with 1.5 ml of periplasm containing or not SurA-Strep, the membrane was washed and finally incubated for 45 min with streptavidin conjugated with horseradish peroxidase (HRP; Pierce) at a dilution of 1/30,000 in PBS-G. After the membrane was washed, an Immobilon Western ECL kit (Millipore) was used to reveal the membrane. In order to compare two conditions of overlay, samples were loaded twice on a gel, the resulting membrane was cut in two, and both parts were treated in parallel.
β-Galactosidase assays.
Cells from overnight cultures were diluted to an OD600 of approximately 0.02 and grown at 33°C to an OD600 of 0.5 in LB medium supplemented with the appropriate antibiotic and 1 mM IPTG. Growth was stopped by adding toluene, and β-galactosidase activity was assayed on permeabilized cells as previously described (28). Three independent replications were averaged to obtain the indicated values.
RESULTS
Analysis of D. dadantii mutants in the OMP-targeting pathway.
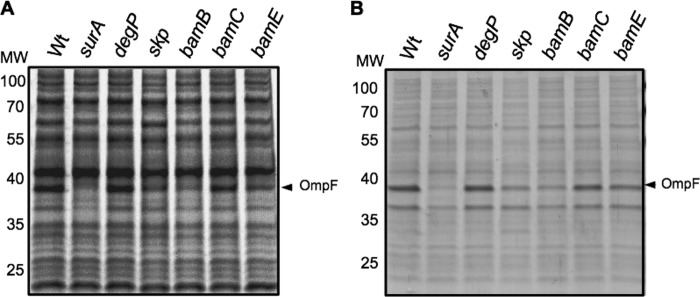
In E. coli, the OMP-targeting pathway mainly relies on the three periplasmic chaperones SurA, Skp, and DegP and on the OM complex BamABCDE. The D. dadantii genome contains homologues of E. coli surA, skp, degP, bamA, bamB, bamC, bamD, and bamE encoded by the loci ADN00090, ADM97288, ADM99598, ADM97287, ADM99516, ADM97479, ADM99647, and ADM97110, respectively. Notably, all these loci were found in a genetic context similar to that in E. coli. Since bamA and bamD mutations are lethal in E. coli, these genes were excluded from our analysis (22, 51). D. dadantii surA, skp, degP, bamB, bamC, and bamE mutant strains were constructed by insertion of a kanamycin resistance cassette. Mutant strains did not exhibit obvious growth defects, indicating that, as in E. coli, none of these genes is essential in D. dadantii. To check the effect of these mutations on OMP biogenesis, an analysis of the protein profiles of whole-cell and membrane fractions of the corresponding strains was performed (Fig. 1). Compared to the wild-type strain, the surA mutant exhibited a strong decrease in the quantity of the major porin OmpF in both whole-cell and membrane fractions. This decrease was also observed to a lower extent for the skp and bamB mutants. This indicates that the OMP-targeting pathway is at least partially affected in these three mutants. Accordingly, surA, skp, and bamB mutants also presented an increased sensitivity to SDS, EDTA, and rifampin (Table 2), indicating that the integrity of their outer membrane is affected (43). The bamE mutant presented a weak but reproducible decrease in the quantity of OmpF and an increased sensitivity to EDTA and, to a lower extent, to rifampin, indicating an alteration of its OM biogenesis. Taken together, these data suggest that in D. dadantii, as in E. coli, SurA is the major periplasmic factor for OMP targeting. Moreover, the relative importance of each component of the Bam complex in OM biogenesis seems to be conserved between D. dadantii and E. coli.
Fig 1.
Porin depletion in D. dadantii mutants with mutations in the OMP-targeting pathway. Protein profiles of whole cells of D. dadantii wild-type (Wt) strain A350 and derivative strains (indicated at the top) carrying the chromosomal mutation (A) and protein profiles of membrane fractions of the same strains (B). Equivalent amounts of cell materials from exponential-phase cultures of the different strains were separated by SDS-PAGE, followed by Coomassie blue staining. The arrowhead indicates the D. dadantii major porin OmpF. MW, molecular weight (in thousands).
Table 2.
Growth inhibition of D. dadantii mutants for the OMP targeting pathwaya
| Strain | Characteristic or mutated gene | Inhibition zone diam (mm) |
||
|---|---|---|---|---|
| SDS (10%) | EDTA (0.5 M) | Rifampin (20 mg/ml) | ||
| A350 | Wild type | 11 ± 0 | 19 ± 2 | 16 ± 1 |
| A5275 | bamE | 14 ± 1 | 29 ± 1 | 21 ± 1 |
| A5276 | bamC | 13 ± 1 | 20 ± 1 | 17 ± 1 |
| A5281 | bamB | 19 ± 1 | 32 ± 1 | 22 ± 0 |
| A5476 | degP | 12 ± 1 | 24 ± 2 | 16 ± 1 |
| A5477 | skp | 18 ± 1 | 30 ± 1 | 25 ± 1 |
| A5478 | surA | 17 ± 0 | 31 ± 1 | 22 ± 1 |
Sensitivity to SDS (10%), EDTA (0.5 M), and rifampin (20 mg/ml) was determined by measuring the growth inhibition zone on LB agar plates topped with filter paper disks containing these compounds, as described in Materials and Methods. The values indicated are the means of 3 experiments.
Efficient targeting of PnlH to the OM depends on SurA but not on Skp and DegP.
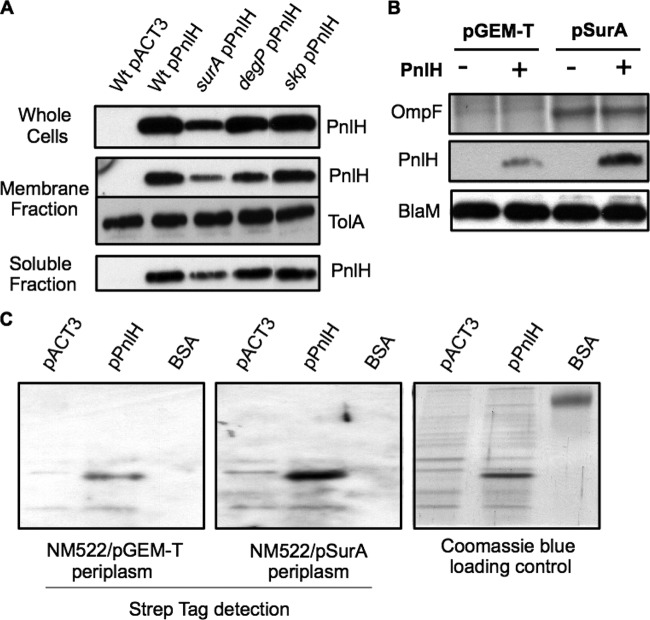
To determine if the periplasmic chaperones SurA, Skp, and DegP could be involved in PnlH targeting to the OM, we checked PnlH localization in corresponding D. dadantii mutant strains. Since PnlH is not expressed in strain A350 and derivatives under the conditions used in this study (8), we expressed PnlH with a carboxy-terminal 6His tag (PnlH-6His) from a pACT3 plasmid (pPnlH). Additionally, expression of PnlH-6His from this plasmid allowed us to avoid any possible feedback regulatory effect due to the activation of the σE stress response, as described for the E. coli surA mutant (42). Cell and membrane separation experiments showed that PnlH-6His is mostly located in the OM, when expressed in the wild-type D. dadantii A350 strain (see Fig. S1 in the supplemental material). To analyze PnlH-6His localization in the different genetic backgrounds, appropriate strains were transformed with the pPnlH plasmid. The amount of PnlH-6His in whole-cell fractions of skp and degP mutants was equivalent to that for the wild-type strain but was strongly decreased for the surA mutant (Fig. 2A). Accordingly, the same diminution was observed in the membrane and soluble fractions of the last mutant (Fig. 2A). This phenomenon was not due to a global protein synthesis/stability decrease, since the level of the IM protein TolA was identical in the membrane fraction of all the tested strains (Fig. 2A). To determine if the small amount of PnlH-6His present in the surA mutant membrane fraction was in the IM or the OM, we performed membrane separation on a flotation sucrose gradient. However, we were not able to properly separate the IM from the OM of the surA mutant. This observation is consistent with the decreased OM density described for an E. coli surA mutant strain (48). Since surA is part of an operon, we tested whether the decrease in PnlH-6His level in the surA mutant was due to a polar effect. When SurA carrying a carboxy-terminal Strep tag (SurA-Strep) was expressed from plasmid pSurA, wild-type quantities of OmpF and PnlH-6His were restored in the surA mutant (Fig. 2B). These results indicate that the effect of the surA mutation on OmpF and PnlH stability is not due to a polar effect.
Fig 2.
Role of periplasmic chaperones SurA, Skp, and DegP in the targeting of PnlH-6His to the OM of D. dadantii. (A) Immunodetection of PnlH in whole-cell, membrane, and soluble fractions of D. dadantii wild-type strain and derivative strains carrying a chromosomal mutation (indicated at the top) and either empty pACT3 vector or pPnlH. Equivalent amounts of cell material from exponential-phase cultures of the different strains were loaded in each well. Immunodetection of the inner membrane protein TolA was used as a loading control. (B) Complementation of a surA mutant carrying either empty pACT3 vector or pPnlH with either empty pGEM-T vector or pSurA. Equivalent amounts of whole cells of steady-state cultures were analyzed. Proteins were separated on an SDS-polyacrylamide gel and transferred onto a PVDF membrane for immunodetection of PnlH and BlaM (loading control). OmpF was detected by SDS-PAGE, followed by Coomassie blue staining. (C) The in vitro interaction between PnlH-6His and SurA-Strep was assayed by overlay. BSA and cell lysates of NM522 carrying either empty pACT3 vector or pPnlH were separated by SDS-PAGE and transferred onto a PVDF membrane. The membrane was next incubated with the periplasm of NM552 carrying either empty pGEM-T vector or pSurA. SurA-Strep bound to proteins on the membrane was detected with streptavidin conjugated with HRP. The amounts of protein loaded on the gel were estimated by Coomassie blue staining.
Our data suggest that SurA could be involved directly or indirectly in the stabilization of PnlH in the periplasm. Overlay experiments have previously been used to demonstrate the interaction of SurA with the passenger domain of the E. coli autotransporter EspP (44). We performed such an experiment to assess whether PnlH can interact directly with SurA in vitro. To this end, PnlH-6His was immobilized on a PVDF membrane and used as a matrix to detect a possible interaction with SurA-Strep. The membrane was incubated with either the periplasm of an E. coli strain expressing SurA-Strep or the periplasm of an E. coli control strain. The use of streptavidin conjugated with HRP allowed us to detect SurA-Strep bound on the immobilized PnlH-6His (Fig. 2C). No interaction between SurA-Strep and the control, bovine serum albumin (BSA), could be detected in this way, indicating that SurA-Strep specifically binds to PnlH-6His. Taken together, these data indicate that SurA is able to bind to PnlH in vitro. Moreover, since a surA mutation leads to a strong decrease in PnlH stability, it is likely that SurA is involved in the targeting of PnlH to the OM by preventing its periplasmic degradation.
SurA is able to interact with the PnlH signal sequence.
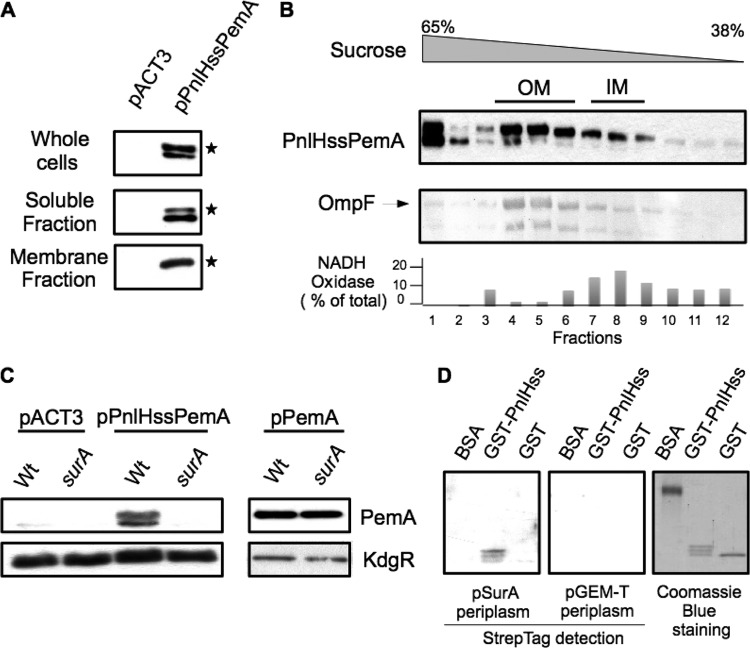
SurA is thought to ensure a specific targeting of OMPs by binding to Ar-X-Ar motifs, which are particularly enriched in OMPs (15). To ensure a specific targeting of PnlH to the OM, SurA should also recognize and bind a specific signal within this protein. Since a fusion of the PnlH signal sequence to the mature form of the D. dadantii pectin methylesterase PemA (PnlHssPemA) is sufficient to target it to the OM of D. dadantii (8), we hypothesized that the PnlH signal sequence could contain the SurA recognition site. To test this hypothesis, we examined whether PnlHssPemA is dependent on SurA for its targeting to the OM. First, we checked the expression and the localization of PnlHssPemA in the wild-type strain A350 carrying the pPnlHssPemA plasmid. Antibody directed against PemA detected two bands in the cells producing this fusion (Fig. 3A). After cell fractionation, the upper band (indicated by a star) was mainly found in the membranes. The smallest one was found in the soluble fraction and could be a degradation product. Membrane separation on a sucrose gradient showed that the upper band was associated with the OM, while the lower band remained at the bottom of the gradient (Fig. 3B). Contrary to the wild-type strain, no PnlHssPemA was detected in the surA mutant (Fig. 3C), suggesting that its stability was affected in the absence of SurA. Since a surA mutation has no effect on the amount of the native PemA expressed from pACT3 (Fig. 3C), we concluded that the PnlH signal sequence is responsible for the degradation of PnlHssPemA in the surA mutant and that it could be the part of PnlH that interacts with SurA. Since SurA interacts in vitro with full-length PnlH in an overlay experiment, we verified if SurA could bind the PnlH signal sequence. For this purpose, the first 41 amino acids of PnlH were fused to the carboxy-terminal end of the GST (GST-PnlHss). When GST-PnlHss was produced in NM522, it was mostly found in inclusion bodies, indicating a high propensity of the PnlH signal sequence to aggregate. Purification of GST-PnlHss on glutathione agarose generated three forms of the protein (Fig. 3D, right). Apparently, the upper band corresponds to the full-length GST-PnlHss and the smaller bands correspond to degradation products. Overlay experiments showed interactions between SurA-Strep and the different forms of GST-PnlHss (Fig. 3D). Since a larger amount of SurA-Strep bound to the full-length GST-PnlHss and since SurA-Strep did not bind to GST or BSA, we concluded that the interaction between SurA-Strep and GST-PnlHss was due to the PnlH signal sequence. Altogether, these data suggest that SurA is able to recognize and bind the PnlH signal sequence to protect PnlH from degradation in the periplasm.
Fig 3.
Role of the PnlH signal sequence in the interaction with SurA. (A) Cell fractionation of D. dadantii A350 strains carrying either empty pACT3 vector or pPnlHssPemA. PnlHssPemA was detected with antibodies against PemA. Equivalent amounts of cell materials from exponential-phase cultures of the different strains were loaded in each well. (B) The membrane fraction from strain A350 carrying pPnlHssPemA was separated by flotation sucrose gradient centrifugation and analyzed by immunoblotting with PemA antibodies or stained with Coomassie blue to detect the major porin, OmpF, which reflects the position of the outer membrane. NADH oxidase activity indicates the position of the inner membrane. (C) Immunodetection of PemA in whole cells of D. dadantii wild-type strain A350 and a surA mutant carrying either empty pACT3 vector or pPnlHssPemA. Equivalent amounts of cell material from steady-state cultures were loaded in each well. The cytoplasmic protein KdgR was used as a loading control. (D) The in vitro interaction between the PnlH signal sequence and SurA-Strep was assayed by overlay. Purified BSA, GST, and GST fused to the PnlH signal sequence (GST-PnlHss) were run on an SDS-polyacrylamide gel and transferred onto a PVDF membrane. The membrane was next incubated with the periplasm of NM552 carrying either empty pGEM-T vector or pSurA. SurA-Strep bound to proteins on the membrane was detected with streptavidin conjugated with HRP. The amounts of protein loaded on the gel were estimated by Coomassie blue staining.
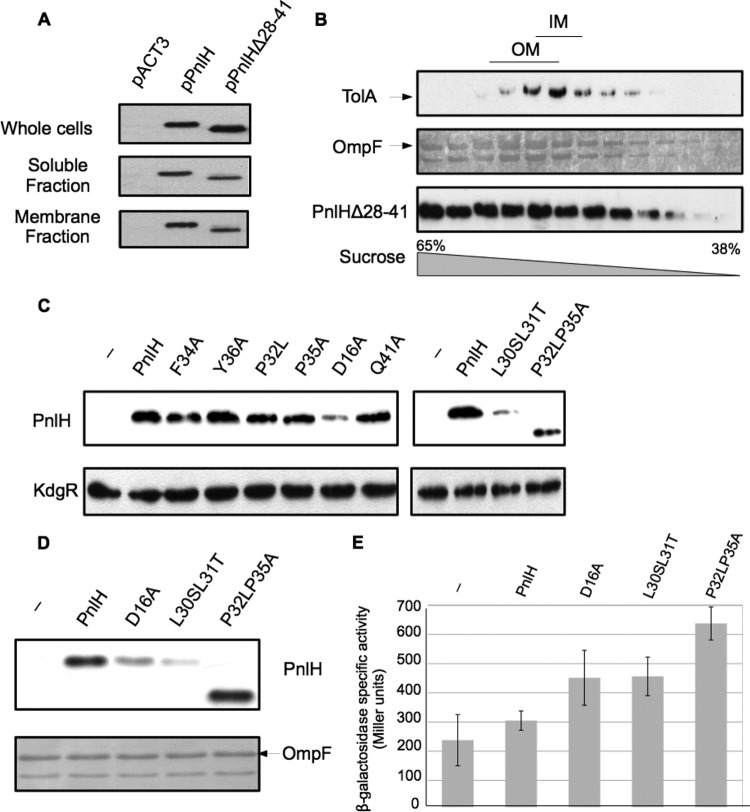
The secondary structure of the PnlH signal sequence could be an important factor for the targeting of PnlH to the OM.
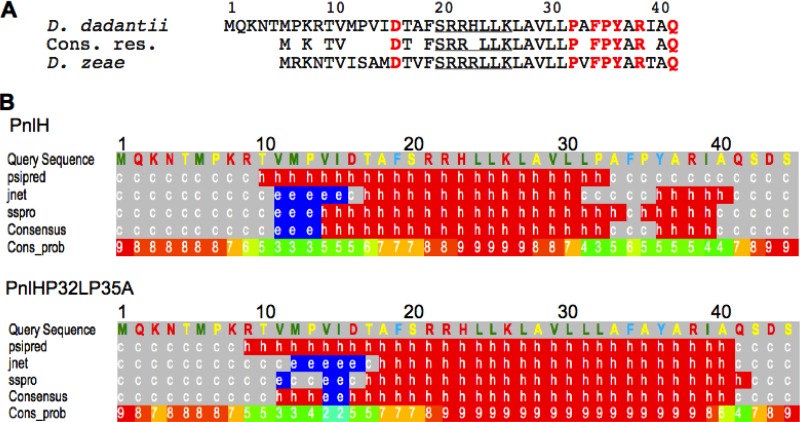
If it is clear that the PnlH signal sequence is responsible for the targeting of PnlH to the OM, the exact nature of the information carried by this signal sequence remains unknown. The only known homologue of the D. dadantii PnlH can be found in D. zeae, and alignment of the two signal sequences reveals that besides the Tat consensus, the carboxy-terminal part of the signal sequence is the most conserved (Fig. 4A). To check whether this part of the signal sequence could carry a signal for the targeting of PnlH, we constructed a deletion of the region between residues 28 and 41 (PnlHΔ28-41). When PnlHΔ28-41 was expressed in D. dadantii wild-type strain A350, its distribution between the soluble and the membrane fractions was similar to that for wild-type PnlH (Fig. 5A), and sucrose gradient separation failed to properly separate the membranes. The protein seemed to be present in both membranes (Fig. 5B). This observation could indicate that the region of the PnlH signal sequence contained between residues 28 and 41 carries a signal necessary to the proper targeting of PnlH to the OM. To identify such a putative signal, we realized amino acid substitutions in this region of the PnlH signal (Fig. 4A). Additionally, we replaced aspartate 16, which is the only negatively charged amino acid in the PnlH signal sequence. When PnlHD16A, PnlHP32L, PnlHF34A, PnlHP35A, PnlHY36A, and PnlHQ41A were expressed in strain A350, PnlHD16A was the only one whose amount was significantly decreased in comparison with that of the wild-type PnlH (Fig. 5C, left). This decrease was also observed in the membrane fraction prepared from strain A350 expressing PnlHD16A (Fig. 5D). This phenomenon could be linked either to a periplasmic instability of PnlHD16A or to its incapacity to be exported or inserted in the OM, leading to degradation. Accumulation of misfolded or aggregated proteins in the periplasm leads to the activation of periplasmic stress responses that increase the expression of many periplasmic factors, such as DegP, that help to release the stress (18). Since the stress response is highly compartmentalized (29), we tested whether PnlHD16A could induce a periplasmic stress. A positive result would mean that PnlHD16A is still exported to the periplasm. For this purpose, we expressed PnlHD16A in E. coli strain SR1458, carrying a chromosomal fusion between the degP promoter and lacZ. Since PnlHD16A induced the degP-lacZ fusion approximately 1.5-fold more than the wild-type PnlH (Fig. 5E), we concluded that PnlHD16A is able to induce a periplasmic stress response and is therefore exported to the periplasm.
Fig 4.
In silico analysis of the PnlH signal sequence. (A) Amino acid sequence alignment of PnlH from D. dadantii (upper line, GenBank accession number YP_003883450.1) and D. zeae (lower line, GenBank accession number YP_003005603.1). Conserved residues (Cons. res.) are indicated in the middle line. The Tat consensus is underlined. Residues that were substituted are in red bold. (B) Secondary structure prediction of the first 41 residues of PnlH and PnlHP32LP235A by the Phyre protein fold recognition software (available at www.sbg.bio.ic.ac.uk/∼phyre/). Letters indicate the type of secondary structure predicted at the corresponding amino acid site (h, alpha helix; c, coil; e, β sheet). Numbers indicate the reliability of the prediction on a scale of from 0 (low) to 9 (high).
Fig 5.
Study of the PnlH signal sequence by directed mutagenesis. (A) Cell fractionation of D. dadantii A350 strains carrying either empty pACT3 vector, pPnlH, or pPnlHΔ28-41. PnlH and PnlHΔ28-41 were detected with antibodies against PnlH. Equivalent amounts of cell materials from exponential-phase cultures of the different strains were loaded in each well. (B) The membrane fraction from strain A350 carrying pPnlHΔ28-41 was separated by flotation sucrose gradient centrifugation and analyzed by immunoblotting with PemA antibodies or stained with Coomassie blue to detect the major porin OmpF, which reflects the position of the outer membrane. Immunoblotting with TolA antibodies indicates the position of the inner membrane. (C) Immunodetection of PnlH in whole-cell lysates of D. dadantii A350 strains carrying either empty pACT3 vector (−) or pPnlH (PnlH) or pPnlH derivatives carrying one or two substitutions in the PnlH signal sequence. Equivalent amounts of cell material from steady-state cultures were loaded in each well. Immunoblotting with KdgR antibodies was used as a loading control. (D) Membrane fraction of A350 strains carrying either empty pACT3 vector (−), pPnlH (PnlH), pD16A (D16A), pL30SL31T (L30SL31T), or pP32LP35A (P32LP35A). PnlH and derivatives were detected with antibodies against PnlH. Equivalent amounts of membrane fractions from exponential-phase cultures were loaded in each well. OmpF was visualized by the coloration of the PVDF membrane with Coomassie blue and served as a loading control. (E) Induction of periplasmic stress in E. coli SR1458 expressing PnlH and derivatives. E. coli SR1458 carrying either empty pACT3 vector (−), pPnlH (PnlH), pD16A (D16A), pL30SL31T (L30SL31T), or pP32LP35A (P32LP35A) was grown at 33°C in LB medium supplemented with chloramphenicol and 1 mM IPTG. The value for β-galactosidase specific activity is the average of three independent determinations.
None of the single substitutions that we realized in the region between residues 28 and 41 presented a defect in its stability. Therefore, we searched for a signal that could not rely on only one amino acid. To identify this signal, we used the Phyre protein fold recognition software to predict an eventual secondary structure in the PnlH signal sequence. Phyre establishes a consensus between PsiPred, jnet, and sspro secondary structure prediction algorithms and is thus considered reliable structure prediction software (19). According to the Phyre consensus prediction, the most probable secondary structure that could be present in the PnlH signal sequence is a 19-residue-long alpha helix (residues 14 to 32) that contains both the Tat consensus motif (residues 20 to 26) and a region rich in hydrophobic residues (residues 24 to 31) (Fig. 4B). Interestingly, the region between residues 28 and 41 contains two prolines, which are good alpha-helix breakers and which could be involved in the introduction of a coiled region after the predicted alpha helix. To test the importance of these two prolines, we constructed the double substitution P32L P35A (PnlHP32LP35A). The predicted alpha helix in PnlHP32LP35A was significantly longer than the one in the wild-type PnlH (Fig. 4B), suggesting that prolines 32 and 35 could determine the length of the alpha helix in the PnlH signal sequence. When PnlHP32LP35A was expressed in strain A350, only a protein migrating faster than the wild-type PnlH and present in a smaller quantity could be detected in whole cells (Fig. 5C, left). After cell fractionation, this protein was found to be present in the membrane fraction (Fig. 5D), and when membranes were separated on a sucrose gradient, it was mostly found in the OM (see Fig. S2A in the supplemental material). Interestingly, when PnlHP32LP35 was expressed in E. coli, it was present in two forms, one migrating as the wild-type PnlH and one migrating faster (see Fig. S2B in the supplemental material), indicating that the protein that is associated with the OM in the D. dadantii A350 strain is a degradation product of PnlHP32LP35A. Since we were able to detect the fast-migrating form of PnlHP32LP35A through its carboxy-terminal His tag (see Fig. S2C in the supplemental material), we concluded that this extremity was intact and that the degradation should have occurred at the amino-terminal part of the protein. Expression of PnlHP32LP35A in E. coli strain SR1458 provoked a strong increase in the expression of the degP-lacZ fusion (approximately 2-fold) (Fig. 5E), indicating that PnlHP32LP35A is able to induce a periplasmic stress response and is therefore exported to the periplasm. Altogether, these data suggest that the double substitution P32L P35A strongly affects the stability of the PnlH signal sequence without impairing its targeting to the OM.
The predicted alpha helix of the PnlH signal sequence is rich in hydrophobic residues (residues 24 to 31). To decrease the global hydrophobicity of the predicted alpha helix, leucines 30 and 31were replaced by serine and threonine (PnlHL30SL31T). When PnlHL30SL31T was expressed in the D. dadantii A350 strain, it was barely detectable in either the whole-cell (Fig. 5C) or the membrane (Fig. 5D) fraction. Moreover, expression of PnlHL30SL31T in E. coli strain SR1458 provoked a significant increase in β-galactosidase activity (approximately 1.5-fold) (Fig. 5E), indicating that PnlHL30SL31T is able to induce a periplasmic stress response. These data suggest that the reduction of the hydrophobicity in the predicted alpha helix of the PnlH signal sequence significantly impairs the stability of the protein and could alter its targeting to the OM.
bamB and bamE null mutations affect the stability of PnlH in D. dadantii.
By interacting with SurA, PnlH seems to follow the main pathway used by OMPs. Therefore, PnlH could also depend on a functional Bam system to be inserted into the membrane. To assess this question, we studied the targeting of PnlH to the membranes of D. dadantii lacking different components of the Bam system. In comparison to wild-type strain A350, a somewhat smaller quantity of PnlH was detected in the cells of the bamB and bamE strains (Fig. 6A, top). This difference became more evident in the membrane fraction prepared from these strains (Fig. 6A, middle). Since similar quantities of the IM protein TolA were detected in all these strains (Fig. 6A, bottom), the observed diminution of PnlH quantity was specific. However, separation of the bamB and bamE mutant membranes on a sucrose gradient showed that PnlH-6His is still efficiently associated with the OM of these mutants (Fig. 6B). Moreover, the level of PnlH-6His-targeting defect in the three mutants was proportional to the level of OMP-targeting defect observed in the same mutants (Fig. 1). Taken together, these data show that bamB and bamE null mutations slightly affect the stability of PnlH without clearly affecting its targeting to the OM.
Fig 6.
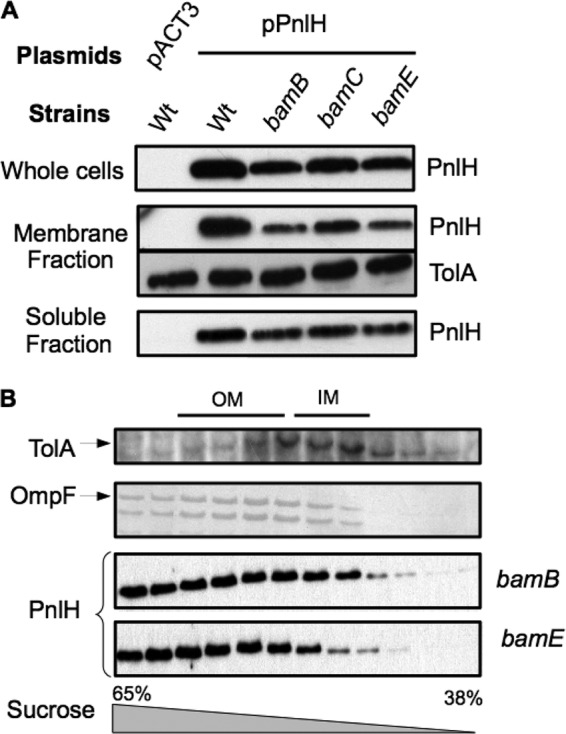
Effect of bamB, bamC, and bamE mutations on the targeting of PnlH-6His to the OM of D. dadantii. (A) Immunodetection of PnlH in whole-cell, membrane, and soluble fractions of D. dadantii wild-type strain A350 and derivative strains carrying chromosomal mutations (indicated at the top) and either empty pACT3 vector or plasmid pPnlH. Equivalent amounts of cell materials from exponential-phase cultures of the different strains were loaded in each well. Immunodetection of the inner membrane protein TolA was used as a loading control. (B) The membrane fractions from the bamB and bamE mutant strains carrying pPnlH were separated by flotation sucrose gradient centrifugation and analyzed by immunoblotting with PnlH antibodies or stained with Coomassie blue to detect the major porins, which reflect the position of the outer membrane. Immunoblotting with TolA antibodies indicates the position of the inner membrane. For clarity, porins and TolA profiles are presented only for the bamB mutant, but the profiles were equivalent for the bamE mutant.
DISCUSSION
The large majority of proteobacteria outer membrane proteins are either lipoproteins or β-barrel proteins (OMPs), and two distinct specific pathways ensure their targeting to the OM. Lipoproteins are sorted and targeted by the Lol pathway, while OMP targeting relies on the combined action of three periplasmic chaperones, SurA, Skp, and DegP, together with the Bam complex (30, 38). In this work, we have studied the targeting of PnlH, a D. dadantii atypical outer membrane protein which is anchored in the OM by an uncleaved Tat signal sequence (8). Since PnlH is not acylated at its amino-terminal extremity, it is unlikely that it directly interacts with the Lol system (8, 9). Thus, we focused our study on the possible role of the OMP-targeting pathway in the targeting of PnlH to the OM.
After exportation by the Tat system, PnlH is released in a folded state in the periplasm. Since the pectin lyase portion of PnlH exhibits 43% sequence identity with a Pseudomonas marginalis pectin lyase, PNL, that has been described to be a soluble protein (33), it should be soluble in the periplasm. However, the PnlH signal sequence is highly hydrophobic, and its fusion to a soluble protein (GST) makes it nonsoluble. Thus, the PnlH signal sequence should be buried in the course of the periplasmic transit. Our data show that a D. dadantii surA mutant exhibits a strong PnlH stability defect. The same defect was observed for a fusion of the PnlH signal sequence to the mature form of the pectin methylesterase PemA but not for the native PemA, indicating that the role of SurA in the stability of PnlH is linked to the signal sequence. Since OMP-targeting defects, such as the one provoked by the absence of SurA, lead to a strong activation of the periplasmic stress response in E. coli (42, 53, 10), the PnlH stability defect observed in the D. dadantii surA mutant could be an indirect effect due to the activation of the periplasmic stress response by mistargeted OMPs. However, we favor a direct role of SurA on the stability of PnlH because (i) a skp mutant also presents a OMP-targeting defect that should activate a periplasmic stress response (38) but does not exhibit any effect on PnlH stability and (ii) SurA binds to the Tat signal sequence of PnlH in an overlay experiment. Therefore, a possible role for SurA could be to hide the hydrophobic PnlH signal sequence and to prevent the periplasmic degradation of PnlH. SurA has been shown to be involved in the transport of the passenger domain of the IcsA autotransporter in the periplasm of Shigella flexneri (35). Moreover, direct binding of SurA on the unfolded passenger domain of the E. coli autotransporter EspP has been demonstrated (44). Altogether our results and these observations indicate that SurA specificity and chaperone activity are not restricted only to unfolded β-barrel proteins. SurA specificity for OMPs has been linked to a strong preference for aromatic amino acid-rich sequences containing the Ar-X-Ar motif that is particularly frequent in OMPs (1, 15). Such a motif is present in the PnlH signal sequence (phenylalanine 34 and tyrosine 36). Surprisingly, single substitution of each of these aromatic residues did not obviously affect PnlH targeting, indicating that an Ar-X-Ar motif on the PnlH signal sequence could be dispensable for the interaction with SurA. Consistent with this observation, it has been shown that SurA can strongly bind peptides that do not fit the Ar-X-Ar consensus by adapting its tertiary and quaternary structure (57). Moreover, SurA can fold to form a nonspecific conduit for unfolded polypeptide, as can the domain III of the trigger factor, a general cytoplasmic chaperone (21, 57).
SurA has been described to ensure the delivery of OMPs to the Bam system by interacting with BamA (3, 47, 52). Therefore, it is possible that SurA delivers PnlH to the OM, but how the PnlH signal sequence is inserted in the OM remains unknown. Our data show that bamB and bamE null mutations slightly affect the stability of PnlH. However, the PnlH insertion in the OM is not disrupted in the corresponding mutants, and the effect of these null mutations on the level of PnlH is proportional to the effect on the level of the β-barrel protein OmpF. This suggests that the role of BamB and BamE on the stability of PnlH could be indirect, provoked by the activation of a periplasmic stress response (38) or the misinsertion of a protein involved in the targeting of PnlH. However, it is worth noting that a model of spontaneous insertion in the OM has been proposed for some bacterial secretins and for mitochondrial signal-anchored proteins (12, 16, 27). Therefore, further work is necessary to determine how the PnlH signal sequence is inserted in the OM and if this process requires proteins other than those of the Bam complex.
Since the fusion of the PnlH signal sequence to PemA is able to target it to the OM, this signal sequence should carry all the information necessary to the targeting of PnlH. The first step of PnlH targeting is exportation through the Tat system, and the PnlH signal sequence thus has the same organization as other Tat signal sequences with a polar amino-terminal part, a Tat consensus, a hydrophobic region, and a polar carboxy-terminal part (32). In this work, we tried to identify the information that ensures the targeting of PnlH to the OM after exportation by the Tat system, and the difficulty was sorting between information allowing the targeting to the Tat system (32) and information allowing the transit in the periplasm and the insertion in the OM. We showed that the substitution PnlHD16A strongly impairs the stability of PnlH in D. dadantii without blocking its exportation, as shown by the activation of a periplasmic stress by PnlHD16A in E. coli. Since negatively charged residues are not a compulsory characteristic of the amino-terminal part of the Tat signal sequence (32) and since aspartate 16 is conserved in D. zeae, it is possible that it constitutes a specific signal for the targeting of PnlH to the OM. We could also identify a region of the PnlH signal sequence contained between residues 28 and 41 that could be specifically involved in the targeting of PnlH to the OM after exportation since PnlH with deletion of this region is present in both the IM and the OM of D. dadantii. Consistent with this experimental observation, this part of the signal sequence is well conserved in the D. zeae PnlH. However, single amino acid substitutions in this part of the protein did not lead to an obvious targeting defect, indicating that the signal may not rely on a single amino acid. Interestingly, structure prediction of the PnlH signal sequence suggests the existence of an alpha helix between residues 14 and 32, followed by a coil region formed by prolines 32 and 35. Substitution of these two prolines (PnlHP32LP35A) leads to a degradation of the amino-terminal part of the PnlH signal sequence but does not impair the targeting of PnlH to the OM of D. dadantii. Since PnlHP32LP35A induces a strong periplasmic stress in E. coli, it is likely that the coil region and thus the length of the predicted alpha helix are important factors for the stability of the PnlH signal sequence in the periplasm but not for its insertion in the OM. The PnlH signal sequence-predicted alpha helix contains a highly hydrophobic region between residues 24 and 31. Replacement of leucines 30 and 31 by serine and threonine (PnlHL30SL31T), respectively, led to a strong decrease in the PnlH stability in D. dadantii and activation of a periplasmic stress in E. coli. This could indicate that the decrease of the hydrophobicity of the predicted alpha helix of PnlHL30SL31T is not sufficient to impair the exportation but could impair the targeting to the OM after exportation. Since the PnlH signal sequence anchors PnlH in the OM, it is tempting to think that its predicted hydrophobic alpha helix is responsible for the anchoring of PnlH in the OM. The presence of a hydrophobic alpha helix in an OM protein seems surprising, since the presence of such a structure in membrane proteins constitutes a strong IM retention signal. However, it has been shown that the lipoproteins of the outer membrane auxiliary family PelC of Pseudomonas aeruginosa and Wza of E. coli are anchored in the OM by a carboxy-terminal amphipathic alpha helix (6, 20), indicating that alpha helices can be inserted in the OM. Another surprising point is the presence of a cluster of hydrophobic residues in the PnlH signal sequence. Indeed, β-barrel proteins do not have such clusters in their primary sequences, but their folding in the OM forms a hydrophobic surface allowing the insertion into the OM (38). Similarly, the oligomerization of Wza forms in the OM a helical barrel that exposes the hydrophobic residues of the carboxy-terminal amphipathic alpha helices to the membrane, while polar residues are localized in the central cavity of the barrel (6). Such an oligomerization has not been reported for PnlH. Thus, the presence and the importance of the hydrophobic cluster in the PnlH signal sequence could be attributed to the requirement for a hydrophobic surface to anchor proteins in the OM.
The pnlH is located in a 13-gene cluster, bordered by tRNA genes, that has been acquired by lateral transfer (8). This cluster is not present in most sequenced Dickeya strains. No gene that could encode a dedicated targeting system is present in this cluster. OM Tat-anchored proteins, such as PnlH, do not seem to be widespread, and they probably have to rely on existing targeting systems. As shown by this study, SurA could be versatile enough to ensure the targeting of noncanonical OM proteins. This could, at least in part, explain how newly acquired OM proteins can be targeted to the OM to ensure their functions. Such versatility in an OM-targeting pathway could, in short, ensure the acquisition of new functions by lateral gene transfer.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by a BQR from the INSA de Lyon. Arnaud Rondelet was supported by Cluster 10: Infectiologie of the French Region Rhône Alpes.
We thank J. M. Betton for providing us the SR1458 strain. We also thank Vladimir Shevchik for his precious help and carefully reading of the manuscript and Geraldine Effantin for technical support.
Footnotes
Published ahead of print 7 September 2012
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1. Alcock FH, et al. 2008. Conserved substrate binding by chaperones in the bacterial periplasm and the mitochondrial intermembrane space. Biochem. J. 409:377–387 [DOI] [PubMed] [Google Scholar]
- 2. Allen JWA, Tomlinson EJ, Hong L, Ferguson SJ. 2002. The Escherichia coli cytochrome c maturation (Ccm) system does not detectably attach heme to single cysteine variants of an apocytochrome c. J. Biol. Chem. 277:33559–33563 [DOI] [PubMed] [Google Scholar]
- 3. Bennion D, Charlson ES, Coon E, Misra R. 2010. Dissection of β-barrel outer membrane protein assembly pathways through characterizing BamA POTRA 1 mutants of Escherichia coli. Mol. Microbiol. 77:1153–1171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. de Cock H, Struyvé M, Kleerebezem M, van der Krift T, Tommassen J. 1997. Role of the carboxy-terminal phenylalanine in the biogenesis of outer membrane protein PhoE of Escherichia coli K-12. J. Mol. Biol. 269:473–478 [DOI] [PubMed] [Google Scholar]
- 5. Denoncin K, Schwalm J, Vertommen D, Silhavy TJ, Collet J-F. 2012. Dissecting the Escherichia coli periplasmic chaperone network using differential proteomics. Proteomics 12:1391–1401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dong C, et al. 2006. Wza the translocon for E. coli capsular polysaccharides defines a new class of membrane protein. Nature 444:226–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Fellay R, Frey J, Krisch H. 1987. Interposon mutagenesis of soil and water bacteria: a family of DNA fragments designed for in vitro insertional mutagenesis of Gram-negative bacteria. Gene 52:147–154 [DOI] [PubMed] [Google Scholar]
- 8. Ferrandez Y, Condemine G. 2008. Novel mechanism of outer membrane targeting of proteins in Gram-negative bacteria. Mol. Microbiol. 69:1349–1357 [DOI] [PubMed] [Google Scholar]
- 9. Fukuda A, et al. 2002. Aminoacylation of the N-terminal cysteine is essential for Lol-dependent release of lipoproteins from membranes but does not depend on lipoprotein sorting signals. J. Biol. Chem. 277:43512–43518 [DOI] [PubMed] [Google Scholar]
- 10. Gerken H, Leiser OP, Bennion D, Misra R. 2010. Involvement and necessity of the Cpx regulon in the event of aberrant β-barrel outer membrane protein assembly. Mol. Microbiol. 75:1033–1046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gralnick JA, Vali H, Lies DP, Newman DK. 2006. Extracellular respiration of dimethyl sulfoxide by Shewanella oneidensis strain MR-1. Proc. Natl. Acad. Sci. U. S. A. 103:4669–4674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Guilvout I, et al. 2008. In vitro multimerization and membrane insertion of bacterial outer membrane secretin PulD. J. Mol. Biol. 382:13–23 [DOI] [PubMed] [Google Scholar]
- 13. Hagan CL, Kim S, Kahne D. 2010. Reconstitution of outer membrane protein assembly from purified components. Science 328:890–892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hayashi S, Wu HC. 1990. Lipoproteins in bacteria. J. Bioenerg. Biomembr. 22:451–471 [DOI] [PubMed] [Google Scholar]
- 15. Hennecke G, Nolte J, Volkmer-Engert R, Schneider-Mergener J, Behrens S. 2005. The periplasmic chaperone SurA exploits two features characteristic of integral outer membrane proteins for selective substrate recognition. J. Biol. Chem. 280:23540–23548 [DOI] [PubMed] [Google Scholar]
- 16. Hoang HH, et al. 2011. Outer membrane targeting of Pseudomonas aeruginosa proteins shows variable dependence on the components of Bam and Lol machineries. mBio 2:e00246–11 doi:10.1128/mBio.00246-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hugouvieux-Cotte-Pattat N, Charaoui-Boukerzaza S. 2009. Catabolism of raffinose, sucrose, and melibiose in Erwinia chrysanthemi 3937. J. Bacteriol. 191:6960–6967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hunke S, Betton J-M. 2003. Temperature effect on inclusion body formation and stress response in the periplasm of Escherichia coli. Mol. Microbiol. 50:1579–1589 [DOI] [PubMed] [Google Scholar]
- 19. Kelley LA, Sternberg MJE. 2009. Protein structure prediction on the Web: a case study using the Phyre server. Nat. Protoc. 4:363–371 [DOI] [PubMed] [Google Scholar]
- 20. Kowalska K, et al. 2010. The C-terminal amphipathic α-helix of Pseudomonas aeruginosa PelC outer membrane protein is required for its function. Biochimie 92:33–40 [DOI] [PubMed] [Google Scholar]
- 21. Ludlam AV, Moore BA, Xu Z. 2004. The crystal structure of ribosomal chaperone trigger factor from Vibrio cholerae. Proc. Natl. Acad. Sci. U. S. A. 101:13436–13441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Malinverni JC, et al. 2006. YfiO stabilizes the YaeT complex and is essential for outer membrane protein assembly in Escherichia coli. Mol. Microbiol. 61:151–164 [DOI] [PubMed] [Google Scholar]
- 23. Maneewannakul S, Maneewannakul K, Ippen-Ihler K. 1994. The pKSM710 vector cassette provides tightly regulated lac and T7lac promoters and strategies for manipulating N-terminal protein sequences. Plasmid 31:300–307 [DOI] [PubMed] [Google Scholar]
- 24. Masuda K, Matsuyama S, Tokuda H. 2002. Elucidation of the function of lipoprotein-sorting signals that determine membrane localization. Proc. Natl. Acad. Sci. U. S. A. 99:7390–7395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Matsuyama S, Yokota N, Tokuda H. 1997. A novel outer membrane lipoprotein, LolB (HemM), involved in the LolA (p20)-dependent localization of lipoproteins to the outer membrane of Escherichia coli. EMBO J. 16:6947–6955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Matsuyama S, Tajima T, Tokuda H. 1995. A novel periplasmic carrier protein involved in the sorting and transport of Escherichia coli lipoproteins destined for the outer membrane. EMBO J. 14:3365–3372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Merklinger E, et al. 2012. Membrane integration of a mitochondrial signal-anchored protein does not require additional proteinaceous factors. Biochem. J. 442:381–389 [DOI] [PubMed] [Google Scholar]
- 28. Miller JH. 1992. A short course in bacterial genetics: a laboratory manual and handbook for Escherichia coli and related bacteria. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 29. Miot M, Betton J-M. 2004. Protein quality control in the bacterial periplasm. Microb. Cell Fact. 3:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Okuda S, Tokuda H. 2011. Lipoprotein sorting in bacteria. Annu. Rev. Microbiol. 65:239–259 [DOI] [PubMed] [Google Scholar]
- 31. Osborn MJ, Gander JE, Parisi E, Carson J. 1972. Mechanism of assembly of the outer membrane of Salmonella typhimurium. Isolation and characterization of cytoplasmic and outer membrane. J. Biol. Chem. 247:3962–3972 [PubMed] [Google Scholar]
- 32. Palmer T, Berks BC. 2012. The twin-arginine translocation (Tat) protein export pathway. Nat. Rev. Microbiol. 10:483–496 [DOI] [PubMed] [Google Scholar]
- 33. Papi R, Kyriakidis D. 2003. Overexpression of the pectin lyase gene of Pseudomonas marginalis in Escherichia coli and purification of the active enzyme. Biotechnol. Appl. Biochem. 37:187–194 [DOI] [PubMed] [Google Scholar]
- 34. Pugsley AP. 1993. The complete general secretory pathway in gram-negative bacteria. Microbiol. Rev. 57:50–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Purdy GE, Fisher CR, Payne SM. 2007. IcsA surface presentation in Shigella flexneri requires the periplasmic chaperones DegP, Skp, and SurA. J. Bacteriol. 189:5566–5573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Raina S, Missiakas D, Georgopoulos C. 1995. The rpoE gene encoding the sigma E (sigma 24) heat shock sigma factor of Escherichia coli. EMBO J. 14:1043–1055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Resibois A, Colet M, Faelen M, Schoonejans E, Toussaint A. 1984. phiEC2, a new generalized transducing phage of Erwinia chrysanthemi. Virology 137:102–112 [DOI] [PubMed] [Google Scholar]
- 38. Ricci DP, Silhavy TJ. 2012. The Bam machine: a molecular cooper. Biochim. Biophys. Acta 1818:1067–1084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Rizzitello AE, Harper JR, Silhavy TJ. 2001. Genetic evidence for parallel pathways of chaperone activity in the periplasm of Escherichia coli. J. Bacteriol. 183:6794–6800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Robert V, et al. 2006. Assembly factor Omp85 recognizes its outer membrane protein substrates by a species-specific C-terminal motif. PLoS Biol. 4:e377 doi:10.1371/journal.pbio.0040377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Roeder DL, Collmer A. 1985. Marker-exchange mutagenesis of a pectate lyase isozyme gene in Erwinia chrysanthemi. J. Bacteriol. 164:51–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Rouviere PE, Gross CA. 1996. SurA, a periplasmic protein with peptidyl-prolyl isomerase activity, participates in the assembly of outer membrane porins. Genes Dev. 10:3170–3182 [DOI] [PubMed] [Google Scholar]
- 43. Ruiz N, Falcone B, Kahne D, Silhavy TJ. 2005. Chemical conditionality: a genetic strategy to probe organelle assembly. Cell 121:307–317 [DOI] [PubMed] [Google Scholar]
- 44. Ruiz-Perez F, et al. 2009. Roles of periplasmic chaperone proteins in the biogenesis of serine protease autotransporters of Enterobacteriaceae. J. Bacteriol. 191:6571–6583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Schulz GE. 2000. beta-Barrel membrane proteins. Curr. Opin. Struct. Biol. 10:443–447 [DOI] [PubMed] [Google Scholar]
- 46. Shevchik VE, Condemine G, Hugouvieux-Cotte-Pattat N, Robert-Baudouy J. 1996. Characterization of pectin methylesterase B, an outer membrane lipoprotein of Erwinia chrysanthemi 3937. Mol. Microbiol. 19:455–466 [DOI] [PubMed] [Google Scholar]
- 47. Sklar JG, et al. 2007. Lipoprotein SmpA is a component of the YaeT complex that assembles outer membrane proteins in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 104:6400–6405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Sklar JG, Wu T, Kahne D, Silhavy TJ. 2007. Defining the roles of the periplasmic chaperones SurA, Skp, and DegP in Escherichia coli. Genes Dev. 21:2473–2484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Subrini O, Betton J-M. 2009. Assemblies of DegP underlie its dual chaperone and protease function. FEMS Microbiol. Lett. 296:143–148 [DOI] [PubMed] [Google Scholar]
- 50. Terada M, Kuroda T, Matsuyama SI, Tokuda H. 2001. Lipoprotein sorting signals evaluated as the LolA-dependent release of lipoproteins from the cytoplasmic membrane of Escherichia coli. J. Biol. Chem. 276:47690–47694 [DOI] [PubMed] [Google Scholar]
- 51. Voulhoux R, Bos MP, Geurtsen J, Mols M, Tommassen J. 2003. Role of a highly conserved bacterial protein in outer membrane protein assembly. Science 299:262–265 [DOI] [PubMed] [Google Scholar]
- 52. Vuong P, Bennion D, Mantei J, Frost D, Misra R. 2008. Analysis of YfgL and YaeT interactions through bioinformatics, mutagenesis, and biochemistry. J. Bacteriol. 190:1507–1517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Walsh NP, Alba BM, Bose B, Gross CA, Sauer RT. 2003. OMP peptide signals initiate the envelope-stress response by activating DegS protease via relief of inhibition mediated by its PDZ domain. Cell 113:61–71 [DOI] [PubMed] [Google Scholar]
- 54. Walton TA, Sandoval CM, Fowler CA, Pardi A, Sousa MC. 2009. The cavity-chaperone Skp protects its substrate from aggregation but allows independent folding of substrate domains. Proc. Natl. Acad. Sci. U. S. A. 106:1772–1777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Watanabe S, Oguchi Y, Yokota N, Tokuda H. 2007. Large-scale preparation of the homogeneous LolA lipoprotein complex and efficient in vitro transfer of lipoproteins to the outer membrane in a LolB-dependent manner. Protein Sci. 16:2741–2749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Wu T, et al. 2005. Identification of a multicomponent complex required for outer membrane biogenesis in Escherichia coli. Cell 121:235–245 [DOI] [PubMed] [Google Scholar]
- 57. Xu X, Wang S, Hu Y- X, McKay DB. 2007. The periplasmic bacterial molecular chaperone SurA adapts its structure to bind peptides in different conformations to assert a sequence preference for aromatic residues. J. Mol. Biol. 373:367–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Yakushi T, Masuda K, Narita S, Matsuyama S, Tokuda H. 2000. A new ABC transporter mediating the detachment of lipid-modified proteins from membranes. Nat. Cell Biol. 2:212–218 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.