Abstract
A novel anti-varicella-zoster virus compound, a derivative of pyrazolo[1,5-c]1,3,5-triazin-4-one (coded as 35B2), was identified from a library of 9,600 random compounds. This compound inhibited both acyclovir (ACV)-resistant and -sensitive strains. In a plaque reduction assay under conditions in which the 50% effective concentration of ACV against the vaccine Oka strain (V-Oka) in human fibroblasts was 4.25 μM, the 50% effective concentration of 35B2 was 0.75 μM. The selective index of the compound was more than 200. Treatment with 35B2 inhibited neither immediate-early gene expression nor viral DNA synthesis. Twenty-four virus clones resistant to 35B2 were isolated, all of which had a mutation(s) in the amino acid sequence of open reading frame 40 (ORF40), which encodes the major capsid protein (MCP). Most of the mutations were located in the regions corresponding to the “floor” domain of the MCP of herpes simplex virus 1. Treatment with 35B2 changed the localization of MCP in the fibroblasts infected with V-Oka but not in the fibroblasts infected with the resistant clones, although it did not affect steady-state levels of MCP. Overexpression of the scaffold proteins restored the normal MCP localization in the 35B2-treated infected cells. The compound did not inhibit the scaffold protein-mediated translocation of MCP from the cytoplasm to the nucleus. Electron microscopic analysis demonstrated the lack of capsid formation in the 35B2-treated infected cells. These data indicate the feasibility of developing a new class of antivirals that target the herpesvirus MCPs and inhibit normal capsid formation by a mechanism that differs from those of the known protease and encapsidation inhibitors. Further biochemical studies are required to clarify the precise antiviral mechanism.
INTRODUCTION
Varicella-zoster virus (VZV) causes varicella upon primary infection and herpes zoster upon reactivation. Both conditions may be associated with severe complications (2). A recent study demonstrated that viral loads after zoster correlated with severity of postherpetic neuralgia (PHN) (33). Antiviral drugs, including acyclovir (ACV), valacyclovir, and famciclovir, have been widely used for the treatment of VZV-associated diseases (2, 45). However, the number of approved drugs for treating VZV diseases is limited, and all of them act by inhibiting viral DNA replication. The development of resistant strains, as well as the ineffectiveness of available antivirals for preventing complications, have driven a search for alternative compounds (1, 15, 26, 38). New anti-VZV compounds have been under investigation, including the bicyclic nucleoside analog FV100 (27), cidofovir derivatives (60), nonnucleoside DNA polymerase inhibitors (17, 40), helicase-primase inhibitors (7), encapsidation inhibitors (55), and cyclin-dependent kinase inhibitors (39, 50).
Plaque reduction assays are commonly used for screening and evaluation of novel antiviral compounds. Such assays are laborious and time-consuming due to the slow growth of VZV in tissue culture. Several modified high-throughput assays have been described that are less inconvenient than the plaque reduction assays for detecting infectious VZV in clinical specimens and screening anti-VZV compounds (9, 14, 31). In addition, biochemical assays have been developed for the characterization of ACV-resistant strains (36, 48). Previously, we established reporter cell lines for varicella-zoster virus (VZV), human cytomegalovirus (HCMV), and human herpesvirus 8 (HHV-8) and demonstrated their utility for the evaluation of antiviral compounds and the detection of drug-resistant strains (12, 22, 57).
In this study, we used the VZV reporter cell line to screen 9,600 compounds for anti-VZV activity, identified several candidates, and found that one of the compounds targeted the VZV major capsid protein (MCP) encoded by open reading frame 40 (ORF40) and blocked capsid formation. Given that MCP is structurally conserved among the Herpesviridae, precise analyses of this novel mode of antiviral action may offer possibilities for the development of a pan-herpesvirus inhibitor.
MATERIALS AND METHODS
Cells and viruses.
HEL (human embryonic lung fibroblast, a gift of I. Kosugi), MeWo (a human melanoma cell line) (16), MV9G (a VZV reporter cell line established from MeWo cells) (57), and U4C (an HCMV reporter cell line established from U373MG cells) (12) cells were grown in Dulbecco's modified Eagle medium (DMEM) (Gibco-BRL) supplemented with 10% fetal bovine serum (FBS) (HyClone), 100 U/ml penicillin, and 100 μg/ml streptomycin (Gibco-BRL). Guinea pig lung (GPL) fibroblasts (ATCC CCL-158) were grown in F-12 medium with the same supplements. VZV vaccine Oka strain (V-Oka), thymidine kinase (TK)-deficient strains V-Oka-YSR (47) and Kanno (43), laboratory strains CaQu (43), Ellen (5), and MSP (52), and some clinical isolates (49), were used for the evaluation of the anti-VZV compounds. Herpes simplex virus 1 (HSV-1) (F), HCMV (Towne), and HHV-6B (Z29) were also used for the evaluation. Molt-3 cells were cultured and infected with HHV-6B (Z29) as described previously (21). U4C was used for the evaluation of anti-HCMV activity.
Screening of anti-VZV compounds.
A random library of 9,600 chemical compounds (Maybridge) dissolved in dimethyl sulfoxide (DMSO) were diluted to 80 μM with medium in 96-well 0.5-ml assay blocks (Costar); 50 μl of the diluted compounds (final concentration, 20 μM) and ∼2,000 MeWo cells infected with the VZV V-Oka strain (cytopathic effect, ∼50%) in 100 μl of medium were added to MeWo cells (5 × 104 cells/well in 50 μl of medium) in 96-well plates. Eighty compounds were examined per plate along with 4 control wells. In the control wells, ∼2,000 uninfected MeWo cells or ∼1,000, ∼500, or ∼250 MeWo cells infected with VZV were added in the absence of chemical compounds. After 44 h of incubation, MV9G VZV reporter cells in 25 to 40 μl of medium (5 × 104 cells/well) were overlaid and cultured for an additional 24 h. Firefly luciferase activity was measured as chemiluminescence (Dual-Glo luciferase assay system; Promega) with a luminometer (JNR AB2300; ATTO, Japan). Five to ten 96-well plates were tested in each experiment. Compounds for which luciferase levels were less than that for the control of ∼250 infected MeWo cells were evaluated further for anti-VZV activities and for cytotoxicity.
Plaque reduction and focus reduction assays.
Semiconfluent cells in wells of 12-well plates were inoculated with 60 to 300 PFU of cell-free VZV (V-Oka). After a 2-h incubation at 37°C, the inocula were replaced with media containing 2- to 4-fold serial dilutions of compounds, 1% methylcellulose, and 2% FBS. After 6 days the media were removed, and the cells were stained with 2% crystal violet in 10% formalin and then washed with water. Plaques were counted under a dissecting microscope. The assay was performed in triplicate. The 50% effective concentrations (EC50s) were calculated by linear regression from plots of the logs of drug concentrations against percentage reductions in plaque numbers at each antiviral concentration compared to the plaque number for the no-drug control. Cell viabilities were measured by a commercial kit (CellTiter-Glo, Promega) as described previously (12).
The focus reduction assay was performed as follows. Semiconfluent cells in wells of 24-well plates were inoculated with 40 to 200 VZV-infected cells per well. After 2 h at 37°C, the inocula were replaced with media containing chemical compounds and then were cultured for 3 days. Cells were then rinsed with phosphate-buffered saline (PBS), fixed with 3.7% formalin, permeabilized with 0.5% Triton X-100, and reacted with a 1:500 dilution of anti-VZV IE62 (ORF62) monoclonal antibody (Mab8616, Millipore) at 37°C for 1 h and then with horseradish peroxidase-conjugated anti-mouse immunoglobulin G antibody (Histofine simple stain; MAX PO, Nichirei, Japan) at 37°C for 1 h. VZV-positive foci were visualized with 3,3′-diaminobenzidine (DAB) substrate (Roche).
Quantitative PCR assays.
Nuclei of cells infected with cell-free VZV were prepared as follows: infected cells were washed with PBS, treated extensively with trypsin, and collected by centrifugation. After incubation of the cells in an NP-40 buffer (10 mM Tris-HCl [pH 7.4], 150 mM NaCl, 1.5 mM MgCl2, 0.1% Nonidet P-40 [NP-40]) at room temperature for 10 min, nuclei were collected by centrifugation and washed with the same buffer. DNA samples were prepared from VZV-infected cells or from nuclei of VZV-infected cells by using a commercial kit (QIAamp DNA minikit; Qiagen). Primers, 5′-CCTCCGTATCGGGACTTCAA and 5′-TGACCGTCCTCGCATACGTA, and a probe, 5′-FAM-TTGGCGAAGAGCTAAC-MGB, were used for a quantitative PCR (qPCR) assay for the VZV ORF62 gene. The assay thermal conditions were 50°C for 2 min and 95°C for 10 min, with cycles of 95°C for 30 s and 60°C for 30 s. Dilutions of a plasmid bearing the entire ORF62 gene of VZV (V-Oka) were used as quantitative standards. The qPCR assay for the human albumin gene was previously described (30).
Total RNA samples were prepared by using a commercial kit (RNeasy Plus minikit; Qiagen). Quantitative reverse-transcription PCR (RT-qPCR) was done by using a commercial kit (one-step RT-PCR master mix; Applied Biosciences) with the primers and probes that target the ORF62 (8) and glucose-6-phosphate dehydrogenase (12) mRNAs.
Isolation and sequence analysis of virus clones resistant to 35B2.
MeWo and GPL cells were infected with VZV (V-Oka) in 96-well plates and cultured in the presence of 4 μM 35B2 until an obvious cytopathic effect (CPE) was observed. Cells in the well that contained the CPE-positive cells were plated into wells of a 48-well plate in the presence of 4 μM 35B2 per well. Once the cells showed a CPE again, the infected cells were cocultured with uninfected cells in the presence of 4 μM or higher concentrations of 35B2 and then expanded to a T25 flask.
DNA fragments covering each ORF of the resistant clones were amplified by PCR and purified by a commercial kit (Qiaex II; Qiagen). A commercial kit (BigDye Terminator cycle sequencing kit, version 3.1; PE Applied Biosystems) was used for sequencing reactions, which were analyzed with an ABI Prism 7500 system (PE Applied Biosystems).
Expression and detection of ORF40.
A fragment covering the full-length VZV (V-Oka) ORF40 gene was amplified by PCR and cloned into phCMV2 (Genlantis, San Diego, CA) to generate an N-terminal hemagglutinin (HA) epitope-tagged protein (phCMV-ORF40). Similarly amplified fragments covering the full-length ORF33 and ORF33.5 genes were cloned into pcDNA3 (Invitrogen) and pCMV-Tag2 (Stratagene) to express untagged and N-terminally FLAG-tagged proteins, respectively. As a control of FLAG-tagged nuclear protein, HHV-8 ORF50 transactivator protein was expressed (22).
A DNA fragment containing the ORF40 amino acid positions 309 to 929 was cloned into a glutathione S-transferase (GST) fusion vector (pGEX-6P-1; GE Healthcare), resulting in pGEX6-ORF40. The GST-ORF40 fusion protein was expressed in BL21 cells (Invitrogen) after addition of IPTG (isopropyl-β-d-thiogalactopyranoside), and then partially purified as insoluble inclusion bodies. Rabbits were immunized with the fusion protein (Japan Biotest Corporation, Tokyo, Japan). An immunofluorescent assay (IFA) was done as previously described (24). The anti-GST-ORF40 rabbit serum, anti-FLAG tag (M2; Sigma), and anti-HA tag (TANA2; Medical & Biological Laboratories, Nagoya, Japan) monoclonal antibodies were used as primary antibodies, and fluorescein isothiocyanate (FITC)-conjugated goat anti-rabbit IgG F(ab′)2 (Sigma) and Alexa Fluor 594-conjugated anti-mouse IgG F(ab′)2 (Invitrogen) fragments were used as secondary antibodies for IFA. 4′,6-Diamidino-2-phenylindole (DAPI) was used for counterstaining, if necessary. Confocal images were captured with an LSM5 Pascal digital confocal microscope (Zeiss) and analyzed by using its LSM Image Browser software.
293T cells transfected with plasmids were harvested after a 44-h incubation period and solubilized in an immunoprecipitation (IP) buffer (20 mM Tris-HCl [pH 8.0], 0.15 M NaCl, 0.5% NP-40, 0.2% Triton X-100). After sonication and centrifugation, the supernatants were incubated with rabbit anti-GST-ORF40 antibody along with protein G Sepharose beads (GE Healthcare) for 2 h at 4°C. The beads were washed with the IP buffer 4 times, and the bound proteins were dissociated by incubation in the SDS loading buffer. The eluates were analyzed by immunoblotting using the anti-FLAG tag antibody.
Transmission electron microscopy.
Pellets of cells infected with VZV (V-Oka) were prefixed with 2.5% glutaraldehyde in 0.1 M phosphate buffer (pH 7.4) for 2 h at room temperature, postfixed in 1% osmium tetroxide, and embedded in Epon. Ultrathin sections were stained with uranyl acetate and lead citrate and then examined with a transmission electron microscope (model H-7650; Hitachi Ltd., Japan) at 80 kV.
RESULTS
Identification of novel anti-VZV compounds with a reporter cell assay.
To identify anti-VZV compounds, MeWo cells were infected with VZV (V-Oka) in the presence of chemical compounds (final concentration, 20 μM) and then cocultured with the MV9G VZV reporter cells, which express luciferase in a dose-dependent manner upon VZV infection. In this study we used mainly V-Oka, since only V-Oka can infect guinea pig fibroblast cells and the transfection efficiency of guinea pig fibroblast cells is better than that of human fibroblast cells. Four hundred compounds that decreased luciferase activity were identified from the 9,600 compounds in the reporter assay. Approximately 350 compounds were excluded from further study because they were cytotoxic at a 20 μM or 50 μM concentration. Seven compounds were selected for further study based on their strong anti-VZV activities in the reporter cell assay. Their anti-VZV activities were confirmed by immunostaining of VZV-infected cells that were cultured in the presence of each compound for 3 days (data not shown). One compound, 2-[(2,6-dichlorophenyl)methylthio]-3H-pyrazolo[1,5-c]1,3,5-triazin-4-one (35B2) (Fig. 1A), was selected for further analysis.
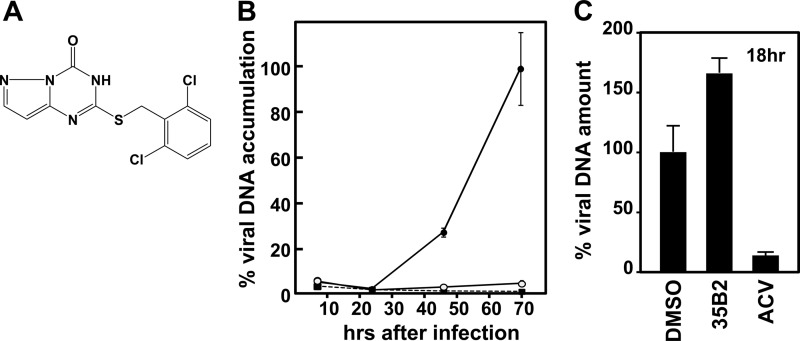
Fig 1.
(A) Structure of 35B2. (B) Confirmation of antiviral activity of 35B2. HEL cells were plated in wells of 24-well plates, infected with 2,000 PFU of cell-free VZV (V-Oka), cultured in the presence of DMSO (closed circles), 20 μM 35B2 (open circles), and 36 μM (8 μg/ml) ACV (closed squares), and then harvested at the indicated time points. VZV copy numbers per cell were obtained by real-time PCR assays for the VZV IE62 gene and the human albumin gene. The relative amounts of viral DNA under each of the conditions are shown by using the copy numbers per cell obtained from the infected cells treated with DMSO at 70 h after infection as a 100% control. The experiments were done in triplicate, and the means of the relative amounts and the standard deviations are shown in the graph. (C) HEL cells were plated in the wells of 12-well plates, infected with 6,000 PFU of cell-free VZV (V-Oka), and cultured in the presence of DMSO, 20 μM 35B2, and 20 μg/ml ACV. At 18 h after infection, nuclei were prepared from the infected cells. VZV copy numbers per cell were obtained as described above, and the relative amounts of viral DNA are shown by using the copy numbers per cell obtained from the infected cells treated with DMSO as a 100% control.
Antiviral activity of 35B2.
In addition to the immunostaining experiment, we confirmed the antiviral activity of 35B2 by measuring VZV DNA amounts in cultures after a multiple round of infection (Fig. 1B).
EC50s of 35B2 against VZV (V-Oka) determined by the plaque reduction assay in GPL and HEL cells were less than 1 μM, which were significantly lower than those for ACV (Table 1). Since 50% cytotoxic concentrations (CC50s) were ∼160 μM, 35B2 had selective indexes (SIs) of >200 in HEL and GPL cells. EC50s of 35B2 against several laboratory and clinical strains were determined by the focus reduction assay using cell-associated viruses as inocula, since most low-passage-number VZV strains are cell associated. It is well known that EC50s obtained by using cell-associated viruses as inocula are higher than those obtained by using cell-free viruses (44, 57). Growth of ACV-resistant strains V-Oka-YSR and Kanno as well as other ACV-sensitive strains and clinical isolates was inhibited by the 35B2 treatment at similar concentrations (Table 2). The compound only weakly inhibited growth of HSV-1 and had no effect on HHV-6B or HCMV (data not shown).
Table 1.
EC50s determined by plaque reduction assay
| Strain | HEL cells |
GPL cells |
||||
|---|---|---|---|---|---|---|
| EC50 (μM) | CC50 (μM) | SI | EC50 (μM) | CC50 (μM) | SI | |
| 35B2 | 0.75 ± 0.18 | 162.1 ± 1.3 | 216 | 0.47 ± 0.06 | 152.5 ± 5.9 | 324 |
| ACV | 4.25 ± 0.48 | NDa | >25 | ND | ||
ND, not determined.
Table 2.
EC50s of 35B2 against several strains
| Strain | ACV phenotypea | EC50 (μM)b |
|---|---|---|
| V-Oka | S | 5.0 ± 0.2 |
| V-Oka-YSR | R | 4.6 ± 0.2 |
| Kanno | R | 4.0 ± 0.0 |
| CaQu | S | 4.4 ± 0.2 |
| Ellen | S | 4.1 ± 0.4 |
| MSP | S | 4.3 ± 0.1 |
| 79 | ND | 4.3 ± 0.2 |
| 132 | ND | 3.4 ± 0.3 |
| 429 | ND | 4.0 ± 0.3 |
S, ACV sensitive; R, ACV resistant; ND, not determined.
EC50s were determined by the focus reduction assay.
Phase of infection inhibited by 35B2.
To elucidate the phase of infection inhibited by 35B2, we examined whether the compound inhibited viral DNA synthesis in a single cycle of infection. Since cell-free VZV stocks usually contain a great excess of defective but DNA-positive particles, to minimize carryover from the defective particles attached to cells, nuclei of infected cells were prepared for the quantification of replicating viral DNA. As shown in Fig. 1C, 35B2 did not inhibit viral DNA synthesis, while ACV did, suggesting that 35B2 targets a step after viral DNA synthesis or a step that is independent from viral DNA synthesis.
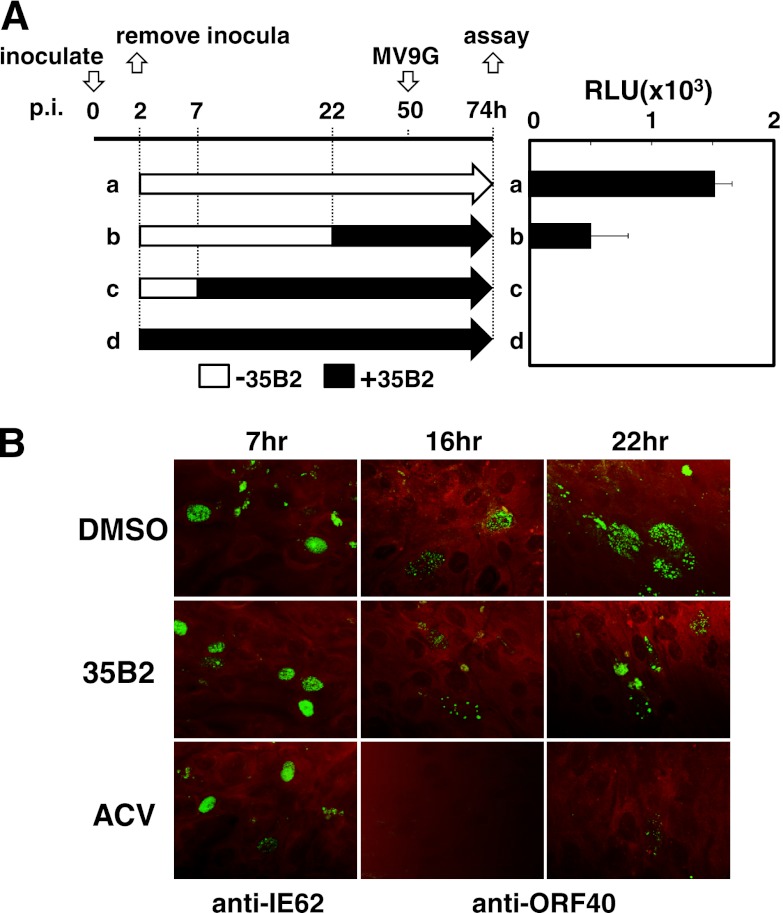
Next, the effect of “time of addition” of 35B2 on virus growth was analyzed. Addition of 35B2 to the cultures at 2 and 7 h but not at 22 h after inoculation of cell-free virus had an inhibitory effect (Fig. 2A). In addition, ORF62 mRNA and IE62 (ORF62) protein were detected by RT-qPCR and by IFA with monoclonal antibody against IE62, respectively, although there were slight decreases of the expression levels (Fig. 2B; data not shown). MCP encoded by ORF40, a late gene product, was not detectable at 12 h after infection of HEL cells, became detectable at 16 h, and accumulated at 22 h (Fig. 2B; data not shown). At 22 h after infection, a part of the IE62-positive cells formed foci containing 5 to 10 IE62-positive cells along with an MCP-positive cell, while most IE62-positive cells appeared to be single at 18 h after infection (data not shown), suggesting that one cycle of infection takes 18 to 22 h. A single cycle of VZV infection in this experiment seemed to be longer than what was previously reported (34), probably due to infection with cell-free virus in this experiment instead of infection with cell-associated virus in the reported study. As expected, ACV inhibited accumulation of MCP. In contrast to ACV, 35B2 did not inhibit expression of MCP. Thus, 35B2 inhibits the early or late phase but not the immediate-early phase of infection.
Fig 2.
(A) “Time of addition” experiment. MeWo cells were inoculated with cell-free VZV (V-Oka) at the time point 0 h. The inocula were removed at 2 h, and fresh media without (a, b, and c) and with (d) 35B2 were added to the cells. 35B2 was added to the cells at the time points 7 h (c) and 22 h (b). At 50 h, MV9G VZV reporter cells were added and cultured for an additional 24 h. Luciferase activities were expressed in terms of relative light units (RLU). (B) HEL cells were infected with cell-free VZV (V-Oka). Two hours later, the inocula were removed. The cells were cultured in the presence of DMSO, 20 μM 35B2, and 20 μg/ml ACV for the indicated durations and fixed with acetone. The antibodies used for IFA are indicated. Evans blue was used for counterstaining.
Characterization of 35B2-resistant virus clones.
Four independent resistant clones were isolated from MeWo cells infected with VZV (V-Oka) and 20 from V-Oka-infected GPL cells. EC50s for inhibition of focus formation were 2- to >20-fold greater than for V-Oka (Table 3).
Table 3.
EC50/focus reduction assay and alterations in ORF40 of 35B2-resistant clones
| Clone | EC50/FRA (μM)a | Amino acid change(s) in: |
||||
|---|---|---|---|---|---|---|
| ORF40 | ORF62 | ORF0 | ORF1 | ORF31 | ||
| V-Oka | 1.8 ± 0.2 | |||||
| V-Oka-YSR | 1.1 ± 0.5 | |||||
| R1 | 37.6 ± 4.1 | L196F | H275R | S197G | L52P | S88A |
| R2 | 4.2 ± 0.2 | F1110S | V733A, Q1215R | — | — | — |
| R3 | 3.5 ± 0.5 | V80A, A1078T | —b | — | — | — |
| R4 | 28.8 ± 1.6 | F1093S | Several | I106V | — | — |
| G01 | 14.0 ± 2.8 | T12I | — | NDc | ND | ND |
| G02 | 26.3 ± 3.8 | T192A | — | ND | ND | ND |
| G03 | 32.8 ± 4.5 | C93R | — | ND | ND | ND |
| G04 | 9.4 ± 1.3 | F1110S | — | ND | ND | ND |
| G08 | 6.0 ± 0.7 | V184F | — | ND | ND | ND |
| G09 | 17.3 ± 1.3 | F79L | — | ND | ND | ND |
| G11 | 16.4 ± 1.5 | V1094F | — | ND | ND | ND |
| G15 | 18.5 ± 1.6 | V1094F | — | ND | ND | ND |
| G17 | 25.2 ± 0.7 | V343I | — | ND | ND | ND |
| G18 | 25.0 ± 0.3 | F96C | — | ND | ND | ND |
| G10 | 10.8 ± 0.2 | F1110S | ND | ND | ND | ND |
| G13 | ND | F1110S | ND | ND | ND | ND |
| G16 | ND | C93R | ND | ND | ND | ND |
| G28 | >40 | L196F | ND | ND | ND | ND |
| G29 | 31.5 ± 0.6 | T192A | ND | ND | ND | ND |
| G31 | >40 | F79I | ND | ND | ND | ND |
| G32 | 6.1 ± 0.3 | Q962L | ND | ND | ND | ND |
| G36 | ND | C93R | ND | ND | ND | ND |
| G40 | 22.2 ± 1.2 | V343I | ND | ND | ND | ND |
| G41 | ND | F1110S | ND | ND | ND | ND |
EC50s were obtained by the focus reduction assay (FRA).
—, no alteration.
ND, not determined.
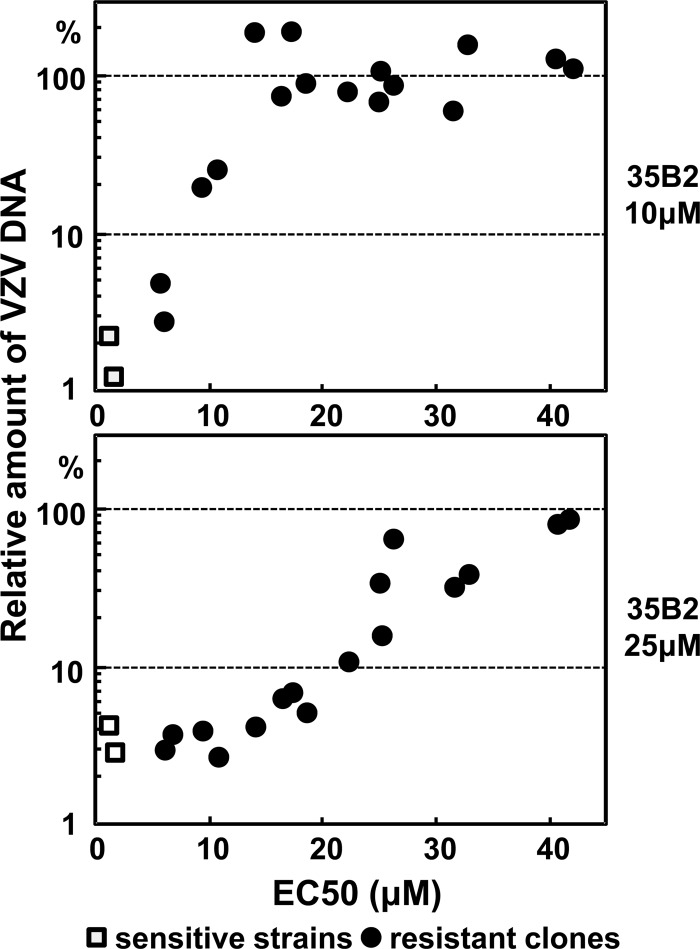
Resistance of the clones was also analyzed by comparison of the amounts of VZV DNA in GPL cells that were infected with cell-associated viruses and cultured in the absence or in the presence of 35B2 for 72 h. In contrast to the original V-Oka and ACV-resistant V-Oka derivative V-Oka-YSR, most 35B2-resistant clones had no inhibition of the accumulation of viral DNA in the cells cultured in the presence of 10 μM 35B2, and some clones had no inhibition even in the presence of 25 μM 35B2. There was a good correlation between the amounts of VZV DNA after a multiple round of infection and the EC50 values obtained by the focus reduction assay (Fig. 3). In the absence of 35B2, the resistant clones and the original V-Oka grew similarly (data not shown).
Fig 3.
Comparison of the EC50 values (Table 3) with the relative VZV DNA amounts in cells cultured in the presence of 10 μM (top panel) or 25 μM (bottom panel) 35B2 after multiple rounds of infection.
Sequence analyses of the resistant clones.
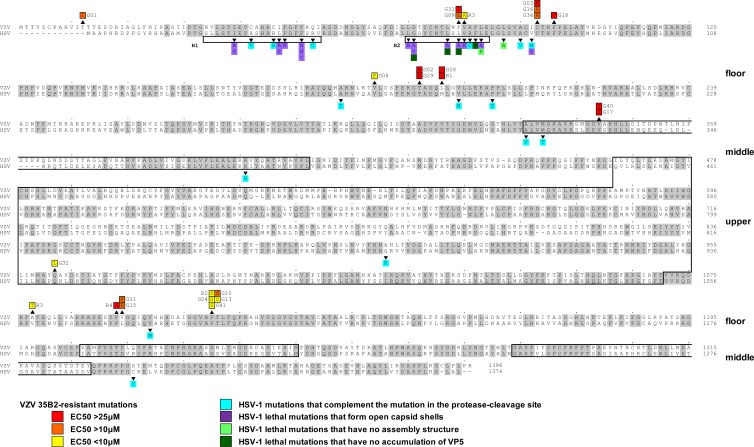
Nucleotide sequences were determined for all 69 ORFs of R1, one of the strongly resistant clones. Nine positions differed from the parental V-Oka sequence, five of which were silent mutations. The four coding mutations altered the amino acid sequences of ORFs 0, 1, 31, 40, and 62. A single mutation changed amino acid sequences of both ORFs 0 and 1, since these ORFs overlap in V-Oka (23). To see which of the mutated genes is responsible for the resistant phenotype, sequences of the 5 mutated ORFs were determined for three additional resistant clones, R2, R3, and R4 (Table 3). All three of the clones had at least one amino acid sequence alteration in ORF40, while two were in ORF62 and one in ORF0. Since it appeared most likely that amino acid changes in ORF40 or ORF62 contributed to the resistance, sequences of ORF40 and ORF62 were determined for 10 additional clones. None of these clones had a mutation in ORF62 but all had a single mutation in ORF40 (Table 3). The ORF40 mutations were clustered; for example, F1110S was in 5 clones, C93R was in 3 clones, and L196F, T192A, V343I, and V1094F were in 2 clones. Interestingly, 21 of the 24 resistant clones contained mutations in ORF40 amino acid sequences that corresponded to the floor domain of HSV-1 MCP (VP-5) (Fig. 4).
Fig 4.
Comparison of amino acid sequences between HSV-1 VP5 (UL19) and VZV MCP (ORF40) and locations of mutations identified in 35B2-resistant clones. Amino acid sequences identical in both HSV-1 VP5 and VZV MCP are highlighted in gray. Numbers at the ends of the sequences indicate the positions in the amino acid sequences. Mutations identified in 35B2-resistant clones are indicated above the VZV MCP sequences. The mutations in the clones for which EC50s were <10 μM, 10 to 25 μM, and >25 μM are highlighted in yellow, orange, and red, respectively. Shown beneath the HSV-1 VP5 sequence and highlighted in blue, purple, light green, and dark green, respectively, are the previously characterized HSV-1 VP5 mutations that complemented a virus defective in protease cleavage of the scaffold proteins (10, 58) and the lethal mutations that formed open capsid shells, those that formed no capsids, and those that did not accumulate VP5 in infected cells (20, 56). The HSV-1 VP5 helices, H1 and H2, and the previously predicted VP5 upper and middle domains (3, 4) are boxed.
No effect of 35B2 on scaffold protein-mediated translocation of MCP.
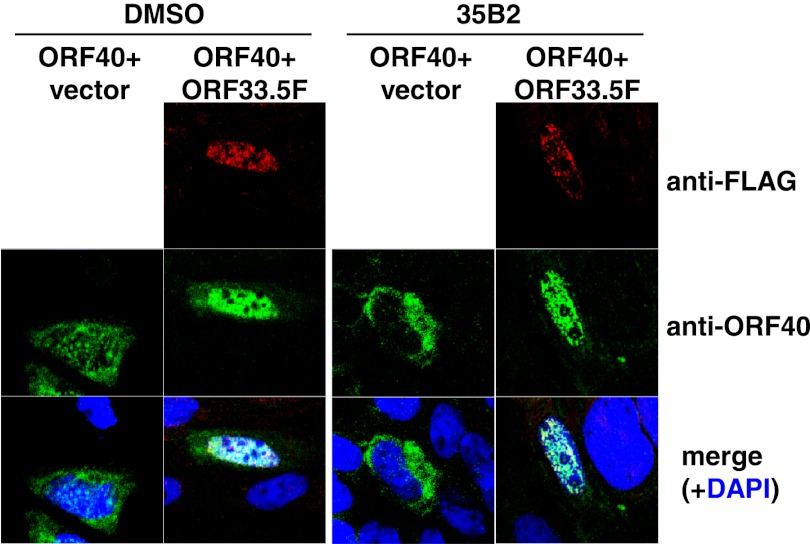
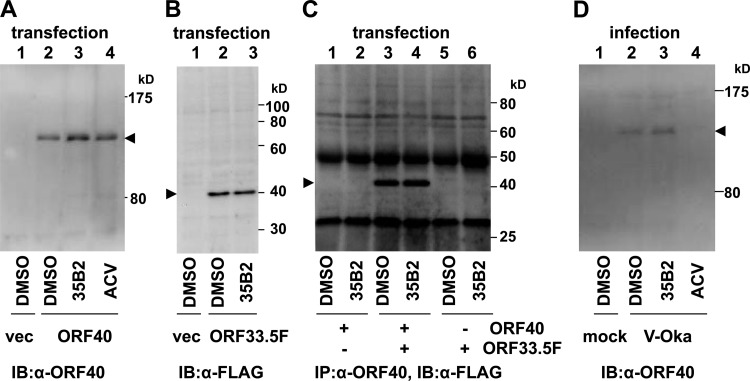
As previously reported (6), expression of ORF40 in the absence of other VZV gene products resulted in the cytoplasmic localization of MCP, and coexpression of ORF40 with ORF33.5 (VZV scaffold protein lacking the protease domain) resulted in translocation of MCP from the cytoplasm to the nucleus (Fig. 5). 35B2 treatment did not affect MCP localization under either condition (Fig. 5). An immunoblotting analysis demonstrated that steady-state levels of MCP in cells transfected with the ORF40 construct and those with the ORF33.5 construct were not apparently affected by 35B2 (Fig. 6A and B). The effect of 35B2 on the interaction between MCP and the scaffold proteins was analyzed by using extracts from 293T cells transfected with the ORF40 and/or FLAG-tagged ORF33.5 constructs. FLAG-tagged ORF33.5 was immunoprecipitated by anti-GST-ORF40 antibody only from the extract of the cells cotransfected with the ORF40 and ORF33.5 constructs, confirming the interaction between MCP and scaffold proteins. In this assay, 35B2 did not affect that interaction (Fig. 6C). 35B2 did not decrease the levels of MCP in VZV-infected cells (Fig. 6D), in addition to the transfected cells, and caused no apparent delay of the MCP expression (Fig. 2B).
Fig 5.
No effect of 35B2 on the scaffold protein-mediated translocation of the VZV MCP. GPL cells were plated in media containing DMSO (left) or 20 μM 35B2 (right), transfected with phCMV-ORF40 along with pCMV-ORF33.5F expressing FLAG-tagged ORF33.5 (ORF40+ORF33.5) or with an empty vector (ORF40+vector), cultured for 2 days, and then fixed with acetone. FLAG-tagged scaffold protein (ORF33.5F) and MCP (ORF40) were visualized by IFA using the anti-FLAG tag monoclonal antibody and the rabbit anti-GST-ORF40 polyclonal antibody as primary antibodies, Alexa Fluor 594-conjugated anti-mouse IgG Fab′ and FLAG-conjugated anti-rabbit IgG Fab′ as secondary antibodies, and DAPI for counterstaining. Alexa Fluor 594, FITC, and DAPI signals are shown in red, green, and blue, respectively.
Fig 6.
(A) No effect of 35B2 on steady-state levels of MCP in ORF40-transfected cells. 293T cells were transfected with empty vector (vec) (lane 1) or with phCMV-ORF40 (lanes 2 to 4), cultured in the presence of DMSO (lanes 1 and 2), 20 μM 35B2 (lane 3), or 20 μg/ml ACV (lane 4), and harvested at 44 h after transfection. MCP was detected by immunoblotting using rabbit anti-GST-ORF40 antibody and peroxidase-conjugated anti-rabbit IgG. (B) No effect of 35B2 on steady-state levels of scaffold protein in transfected cells. 293T cells were transfected with empty vector (lane 1) or with pCMV-ORF33.5F (lanes 2 and 3), cultured in the presence of DMSO (lanes 1 and 2) or 20 μM 35B2 (lane 3), and harvested at 44 h after transfection. Scaffold protein was detected by immunoblotting using anti-FLAG tag antibody and peroxidase-conjugated anti-mouse IgG. (C) No effect of 35B2 on the interaction between MCP and scaffold proteins in transfected cells. 293T cells were transfected with phCMV-ORF40 (lanes 1 and 2), with phCMV-ORF40 and pCMV-ORF33.5F (lanes 3 and 4), or with pCMV-ORF33.5F (lanes 5 and 6), cultured in the presence of DMSO (lanes 1, 3, and 5) or 35B2 (lanes 2, 4, and 6) and harvested at 44 h after transfection. The cell extracts were reacted with anti-GST-ORF40 antibody and protein G. Samples after immunoprecipitation (IP) were analyzed by immunoblotting (IB) using the anti-FLAG antibody. (D) No effect of 35B2 on steady-state levels of MCP in infected cells. HEL cells infected with V-Oka were harvested at 22 h after infection. MCP was detected as described for panel A.
Effect of 35B2 on MCP localization.
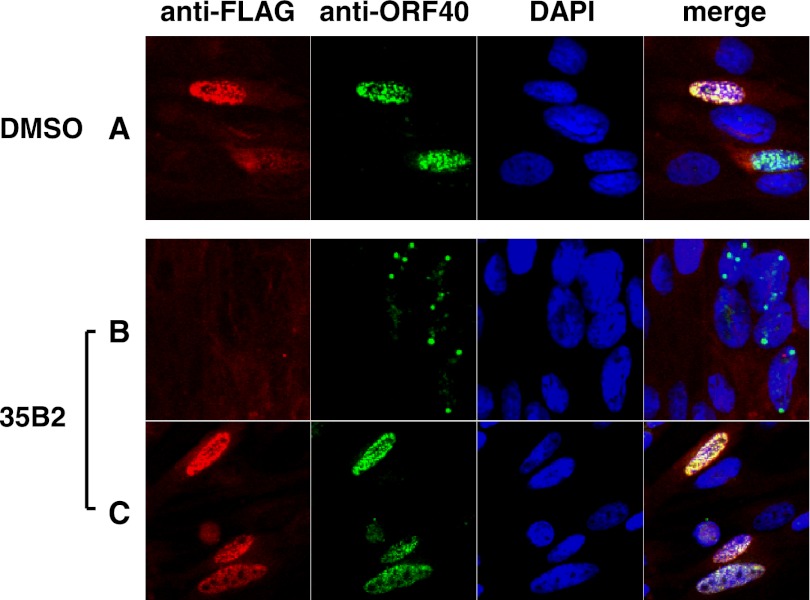
In GPL cells infected with cell-free V-Oka, treatment with 35B2 changed MCP from being widely distributed throughout much of the nucleus to being localized in a small number (<20 per cell) of prominent discrete nuclear speckles (Fig. 7A and B). Similar results were obtained in V-Oka-infected HEL cells (data not shown). Such speckles were also present in the vicinity of the nuclear membrane in GPL and HEL cells that were infected with cell-associated V-Oka and cultured in the presence of 35B2 (Fig. 7D; data not shown). However, 35B2 did not change the localization of MCP in cells infected with the 35B2-resistant clones R1 and G18 (Fig. 7F and H); thus, the absence of speckles correlated with resistance.
Fig 7.
Effect of 35B2 on localization of the VZV MCP in infected cells. (A and B) GPL cells were infected with cell-free virus (V-Oka), cultured in the presence of DMSO (A) or 20 μM 35B2 (B) and fixed 26 h after infection. (C through H) GPL cells were infected with the cell-associated virus V-Oka (C and D) and 35B2-resistant clones R1 (E and F) and G18 (G and H), cultured in the presence of DMSO (C, E, and G) or 20 μM 35B2 (D, F, and H), and fixed 44 h after infection. The cells were reacted with rabbit anti-GST-ORF40 and then with FITC-conjugated anti-rabbit IgG and counterstained with DAPI. The top panels show only FITC signals (green), and the bottom panels show both FITC (green) and DAPI (blue) signals.
Overexpression of the VZV scaffold protein encoded by ORF33 by transient transfection restored the normal MCP localization in the 35B2-treated infected GPL cells (Fig. 8C). Similar results were obtained in ORF33.5-transfected 35B2-treated GPL cells (data not shown). In these cells, scaffold proteins and MCP colocalized as widely distributed signals in the nuclei, similar to those in the transfected and DMSO-treated cells (Fig. 8A). In contrast, MCP exhibited perinuclear localization with discrete speckles in the cells lacking the expression of scaffold proteins (Fig. 8B) or in those expressing the HHV-8 ORF50 transactivator as a control (data not shown). These data suggest that the abnormal MCP localization induced by 35B2 can be overcome with high levels of scaffold proteins.
Fig 8.
Effect of overexpression of scaffold proteins on localization of the VZV MCP in infected cells. GPL cells were plated in media containing DMSO (A) or 20 μM 35B2 (B and C; same well) and transfected with pCMV-ORF33F expressing FLAG-tagged ORF33. One day after transfection, the cells were infected with V-Oka. Two days after infection, the cells were fixed with acetone. The FLAG-tagged ORF33 protein and ORF40 protein were visualized by IFA using anti-FLAG tag monoclonal antibody and rabbit anti-ORF40 polyclonal antibody as primary antibodies, Alexa Fluor 594-conjugated anti-mouse IgG Fab′ and FITC-conjugated anti-rabbit IgG Fab′ as secondary antibodies, and DAPI for counterstaining. Alexa Fluor 594, FITC, and DAPI signals are shown in red, green, and blue, respectively. There were cells with (C) and without (B) expression of FLAG-tagged ORF33 in the same well, since the transfection efficiency was 30 to 40%.
Effect of 35B2 on capsid formation.
To directly assess the effects of 35B2 treatment on capsid morphogenesis, ultrastructures in the infected cells were observed by transmission electron microscopy. Normal capsid formation was detected in 1 of 50 DMSO-treated cells (Fig. 9A and B), consistent with the multiplicity of infection (MOI) (∼0.02) used for this experiment. In contrast to what occurred in the DMSO-treated cells, no capsid-like structures were observed in >500 cells treated with 35B2, suggesting that 35B2 is an inhibitor of capsid formation (Fig. 9C and D).
Fig 9.
Nuclear ultrastructure in VZV-infected and 35B2-treated cells. HEL cells in T25 flasks were infected with cell-free virus (V-Oka) at an MOI of ∼0.02, cultured in the presence of DMSO (A and B) or 20 μM 35B2 (C and D), and harvested 46 h after infection. Panels B and D show magnified views of the regions indicated with boxes in panels A and C.
DISCUSSION
In this study, we screened 9,600 compounds and identified a derivative of pyrazolo [1,5-c]1,3,5-triazin-4-one (35B2) as an active anti-VZV compound. Analyses of virus clones selected for resistance against this compound indicate that it targets VZV MCP (ORF40) and thus represents a novel class of potential antiviral compounds. Treatment with 35B2 changed MCP from being widely distributed throughout the nucleus to being localized in a small number of discrete nuclear speckles. The abnormal distribution of MCP correlated with sensitivity to 35B2 treatment, since resistant clones did not show the phenotype. Based on electron microscopy (EM) observations, we consider it likely that 35B2 induces MCP aggregation and inhibits capsid assembly.
The capsid structures of an alphaherpesvirus (HSV-1), a betaherpesvirus (CMV), and a gammaherpesvirus (HHV-8) were observed by cryoelectron microscopy. Comparison of these structures identified many similarities and some minor differences (53, 54, 61). VP5 is the most abundant protein in the HSV-1 capsid, since the major components of the HSV-1 capsid are 150 hexons (each hexon consists of 6 VP5 molecules encoded by the UL19 gene, a VZV ORF40 homolog), 11 pentons that consist of 5 VP5 and 5 VP26 molecules encoded by the UL35 gene, a VZV ORF23 homolog, 320 triplexes (each triplex consists of 1 VP19C molecule encoded by the UL38 gene, a VZV ORF50 homolog, and 2 VP23 molecules encoded by the UL18 gene, a VZV ORF41 homolog, and 1 portal at the 12th pentonal position that consists of 12 molecules of the UL6 product, a VZV ORF54 homolog (19). Since the identities and similarities of MCPs between HSV-1 and VZV are 51% and 86%, respectively, and since the expression of VZV scaffold protein with HSV-1 MCP forms complete capsids (32), it is likely that the overall structure of VZV MCP is similar to that of HSV-1 VP5. Studies based on cryoelectron microscopy and bioinformatics identified 39 alpha helices of greater than 2.5 turns and 4 beta sheets in HSV-1 VP5 (61) and predicted three major structural domains, the upper, middle, and floor domains. The upper and middle domains interact with VP26 and the triplex, respectively (3, 4). The floor domain is thought to interact with the scaffold proteins, since mutations that complemented a mutation in the protease cleavage site of the scaffold proteins were identified mainly in the floor domain, mostly in the VP5 helixes, H1 (amino acids 22 to 42) and H2 (amino acids 58 to 72) (the mutations are highlighted in blue in Fig. 4) (10, 58). Furthermore, most HSV-1 mutants containing alterations in the I27 to V80 region of VP5 (highlighted in purple in Fig. 4) are lethal and exhibited abnormal formation of capsids (open capsid shells and/or small capsid-like particles), while HSV-1 mutants containing L71P and L75A (highlighted in light green) assembled no capsid-like structures, and those containing G59P, L65P, L67P, and F70A (highlighted in dark green) did not accumulate VP5 (20). Thus, it was proposed that H2 is important in capsid shell accretion and stabilization (20).
Since some mutations in the floor domain of HSV-1 VP5 decrease the stability of the protein, it was possible to consider that 35B2 unstabilized VZV MCP. However, 35B2 treatment did not result in an apparent decrease of MCP levels in ORF40-transfected cells or in VZV-infected cells. Two major observations in this study led us to hypothesize that 35B2 inhibits normal capsid formation through blockage of the proper interaction between MCP and the scaffold proteins.
First, the mutations in the 35B2-resistant clones are located in regions corresponding to the HSV-1 VP5 regions targeted by mutations complementing the scaffold protein mutation and those producing abnormal or no capsid structures (Fig. 4). Seven of the strong 35B2-resistant mutations are located within or near the region corresponding to VP5 H2. Since the time-lapse cryoelectron microscopic analysis suggested that the primary mechanism underlying capsid maturation is relative rotation of VP5 domains (18), it will be interesting to see whether 35B2 affects such rotations.
Second, 35B2 treatment led to aggregation of VZV MCP into discrete speckles in the vicinity of the nuclear membrane, and overexpression of scaffold protein restored normal MCP localization. Ultrastructural analysis revealed that 35B2 treatment resulted in some electron-dense areas in nuclei that might be aggregates of MCP. However, the aggregation of MCP is not unique to the defect in MCP. Accumulation of immature and/or abnormal capsids in the vicinity of the nuclear envelope was also observed in cells infected with HSV-1 mutants that have defects in the scaffold proteins (13) or in the UL17 and UL25 genes, which encode capsid-associated proteins (37, 41).
Our hypothesis seems to be inconsistent with other observations: there were no effects of 35B2 on the scaffold protein-mediated translocation of MCP from the cytoplasm to the nucleus or on the interaction between MCP and scaffold proteins in cells transiently transfected with the constructs for MCP and scaffold proteins. However, 35B2 treatment did not generate nuclear puncta in the transfected cells. It is likely that additional capsid components, such as ORF23, or host factors induced upon VZV infection are required to generate the MCP aggregates. Therefore, an alternative hypothesis would be that 35B2 induces conformational changes of MCP that affect capsid assembly. This study had a limitation in the detection of scaffold proteins in VZV-infected cells. It will be critical to see whether 35B2 causes changes in the steady-state levels of scaffold proteins and other capsid components as well as whether it inhibits the interaction of MCP with scaffold proteins or with other capsid components in infected cells.
Several antiviral compounds target herpesvirus capsid or nucleocapsid formation. For example, WAY-150138 {N-[3-chloro-4[(5-chloro-2,4-dimethoxyphenyl)carbamothioylamino]phenyl]-2-fluorobenzamide} specifically inhibits HSV encapsidation, and a related compound, compound I (2-fluoro-N-(4-{3-[1-(4-fluoro-phenyl)-ethyl]-thioureido}-phenyl)-benzamide), similarly affects VZV (29, 55). Some derivatives of [2,3-d]oxazinone inhibit the protease activity of the scaffold proteins of HSV, VZV, and CMV (11, 59). Dimerization of scaffold proteins is required for the protease activity; compounds that blocked their dimerization were recently reported for HHV-8 (42). Because 35B2 is the first antiviral that targets the conserved herpesvirus MCP, its novel antiviral mechanism can be exploited to target other herpesviruses.
In our next study, to further confirm the genetics of our sequencing analysis-based results and to systematically analyze which amino acid residues of VZV MCP are involved in the interaction with 35B2, it would be desirable to introduce various mutations into ORF40 using a VZV bacterial artificial chromosome (BAC) system. It will be important to do chemical structure-activity relation (SAR) analysis and to screen 35B2 derivatives to identify compounds with even stronger inhibitory activity against VZV and a target spectrum that includes other herpesviruses. Study of the binding kinetics of VZV MCP and its mutants with 35B2 and related compounds identified in the SAR analysis will help us to see whether 35B2 interacts with a tertiary structural element conserved among herpesviruses. Further studies are also required to see whether such anti-MCP compounds are effective in assays that represent various aspects of VZV infections in vivo, such as T lymphocyte cultures, skin raft/organ cultures, and SCID-hu mouse models (25, 28, 35, 46, 51).
In conclusion, we identified a novel anti-VZV compound that targets MCP and inhibits capsid formation. Given that MCP is structurally conserved among the Herpesviridae, this compound may provide a clue for the development of a pan-herpesvirus inhibitor.
ACKNOWLEDGMENTS
We thank Phil Pellett for his intellectual input, Tatsuo Suzutani for the V-Oka-YSR, CaQu, and Kanno strains, D. Scott Schmid for the MSP and Ellen strains, and Michiko Takayama for clinical isolates.
This work was supported by Research on Health Sciences Focusing on Drug Innovation grant SH54412 (to N.I.) from Japanese Human Science Foundation, Research Promotion of Emerging and Re-emerging Infectious Diseases grants H18-Shinko-013 and H21-Shinko-Ippan-009 (to N.I.) from the Ministry of Health, Labor and Welfare, and a Grant-in-Aid for Science (C) 22590426 (to N.I.) from the Ministry of Education, Culture, Science, Technology and Sports, Japan. None of the authors has competing interests.
Footnotes
Published ahead of print 29 August 2012
REFERENCES
- 1. Andrei G, et al. 2005. Susceptibilities of several clinical varicella-zoster virus (VZV) isolates and drug-resistant VZV strains to bicyclic furano pyrimidine nucleosides. Antimicrob. Agents Chemother. 49:1081–1086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Arvin AM. 2002. Antiviral therapy for varicella and herpes zoster. Semin. Pediatr. Infect. Dis. 13:12–21 [DOI] [PubMed] [Google Scholar]
- 3. Baker ML, et al. 2003. Architecture of the herpes simplex virus major capsid protein derived from structural bioinformatics. J. Mol. Biol. 331:447–456 [DOI] [PubMed] [Google Scholar]
- 4. Bowman BR, Baker ML, Rixon FJ, Chiu W, Quiocho FA. 2003. Structure of the herpesvirus major capsid protein. EMBO J. 22:757–765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Brunell PA, Casey HL. 1964. Crude tissue culture antigen for determination of varicella-zoster complement fixing antibody. Public Health Rep. 79:839–842 [PMC free article] [PubMed] [Google Scholar]
- 6. Chaudhuri V, Sommer M, Rajamani J, Zerboni L, Arvin AM. 2008. Functions of varicella-zoster virus ORF23 capsid protein in viral replication and the pathogenesis of skin infection. J. Virol. 82:10231–10246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chono K, et al. 2010. ASP2151, a novel helicase-primase inhibitor, possesses antiviral activity against varicella-zoster virus and herpes simplex virus types 1 and 2. J. Antimicrob. Chemother. 65:1733–1741 [DOI] [PubMed] [Google Scholar]
- 8. Cohrs RJ, Gilden DH. 2007. Prevalence and abundance of latently transcribed varicella-zoster virus genes in human ganglia. J. Virol. 81:2950–2956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dal Pozzo F, et al. 2008. Fluorescence-based antiviral assay for the evaluation of compounds against vaccinia virus, varicella zoster virus and human cytomegalovirus. J. Virol. Methods 151:66–73 [DOI] [PubMed] [Google Scholar]
- 10. Desai P, Person S. 1999. Second site mutations in the N-terminus of the major capsid protein (VP5) overcome a block at the maturation cleavage site of the capsid scaffold proteins of herpes simplex virus type 1. Virology 261:357–366 [DOI] [PubMed] [Google Scholar]
- 11. Flynn DL, Abood NA, Holwerda BC. 1997. Recent advances in antiviral research: identification of inhibitors of the herpesvirus proteases. Curr. Opin. Chem. Biol. 1:190–196 [DOI] [PubMed] [Google Scholar]
- 12. Fukui Y, et al. 2008. Establishment of a cell-based assay for screening of compounds inhibiting very early events in the cytomegalovirus replication cycle and characterization of a compound identified using the assay. Antimicrob. Agents Chemother. 52:2420–2427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gao M, et al. 1994. The protease of herpes simplex virus type 1 is essential for functional capsid formation and viral growth. J. Virol. 68:3702–3712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gates IV, et al. 2009. Quantitative measurement of varicella-zoster virus infection by semiautomated flow cytometry. Appl. Environ. Microbiol. 75:2027–2036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gilbert C, Bestman-Smith J, Boivin G. 2002. Resistance of herpesviruses to antiviral drugs: clinical impacts and molecular mechanisms. Drug Resist. Updat. 5:88–114 [DOI] [PubMed] [Google Scholar]
- 16. Grose C, Brunel PA. 1978. Varicella-zoster virus: isolation and propagation in human melanoma cells at 36 and 32 degrees C. Infect. Immun. 19:199–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hartline CB, et al. 2005. Inhibition of herpesvirus replication by a series of 4-oxo-dihydroquinolines with viral polymerase activity. Antiviral Res. 65:97–105 [DOI] [PubMed] [Google Scholar]
- 18. Heymann JB, et al. 2003. Dynamics of herpes simplex virus capsid maturation visualized by time-lapse cryo-electron microscopy. Nat. Struct. Biol. 10:334–341 [DOI] [PubMed] [Google Scholar]
- 19. Homa FL, Brown JC. 1997. Capsid assembly and DNA packaging in herpes simplex virus. Rev. Med. Virol. 7:107–122 [DOI] [PubMed] [Google Scholar]
- 20. Huang E, Perkins EM, Desai P. 2007. Structural features of the scaffold interaction domain at the N terminus of the major capsid protein (VP5) of herpes simplex virus type 1. J. Virol. 81:9396–9407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Inoue N, Dambaugh TR, Rapp JC, Pellett PE. 1994. Alphaherpesvirus origin-binding protein homolog encoded by human herpesvirus 6B, a betaherpesvirus, binds to nucleotide sequences that are similar to ori regions of alphaherpesviruses. J. Virol. 68:4126–4136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Inoue N, Winter J, Lal RB, Offermann MK, Koyano S. 2003. Characterization of entry mechanisms of human herpesvirus 8 by using an Rta-dependent reporter cell line. J. Virol. 77:8147–8152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Koshizuka T, Ota M, Yamanishi K, Mori Y. 2010. Characterization of varicella-zoster virus-encoded ORF0 gene–comparison of parental and vaccine strains. Virology 405:280–288 [DOI] [PubMed] [Google Scholar]
- 24. Koyano S, Mar EC, Stamey FR, Inoue N. 2003. Glycoproteins M and N of human herpesvirus 8 form a complex and inhibit cell fusion. J. Gen. Virol. 84:1485–1491 [DOI] [PubMed] [Google Scholar]
- 25. Ku CC, Padilla JA, Grose C, Butcher EC, Arvin AM. 2002. Tropism of varicella-zoster virus for human tonsillar CD4(+) T lymphocytes that express activation, memory, and skin homing markers. J. Virol. 76:11425–11433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Levin MJ, et al. 2003. Development of resistance to acyclovir during chronic infection with the Oka vaccine strain of varicella-zoster virus, in an immunosuppressed child. J. Infect. Dis. 188:954–959 [DOI] [PubMed] [Google Scholar]
- 27. Migliore M. 2010. FV-100: the most potent and selective anti-varicella zoster virus agent reported to date. Antivir. Chem. Chemother. 20:107–115 [DOI] [PubMed] [Google Scholar]
- 28. Moffat JF, Stein MD, Kaneshima H, Arvin AM. 1995. Tropism of varicella-zoster virus for human CD4+ and CD8+ T lymphocytes and epidermal cells in SCID-hu mice. J. Virol. 69:5236–5242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Newcomb WW, Brown JC. 2002. Inhibition of herpes simplex virus replication by WAY-150138: assembly of capsids depleted of the portal and terminase proteins involved in DNA encapsidation. J. Virol. 76:10084–10088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ogawa H, et al. 2007. Etiology of severe sensorineural hearing loss in children: independent impact of congenital cytomegalovirus infection and GJB2 mutations. J. Infect. Dis. 195:782–788 [DOI] [PubMed] [Google Scholar]
- 31. Okamoto S, et al. 2004. Rapid detection of varicella-zoster virus infection by a loop-mediated isothermal amplification method. J. Med. Virol. 74:677–682 [DOI] [PubMed] [Google Scholar]
- 32. Preston VG, et al. 1997. Efficient herpes simplex virus type 1 (HSV-1) capsid formation directed by the varicella-zoster virus scaffolding protein requires the carboxy-terminal sequences from the HSV-1 homologue. J. Gen. Virol. 78:1633–1646 [DOI] [PubMed] [Google Scholar]
- 33. Quinlivan ML, et al. 2011. Persistence of varicella-zoster virus viraemia in patients with herpes zoster. J. Clin. Virol. 50:130–135 [DOI] [PubMed] [Google Scholar]
- 34. Reichelt M, Brady J, Arvin AM. 2009. The replication cycle of varicella-zoster virus: analysis of the kinetics of viral protein expression, genome synthesis, and virion assembly at the single-cell level. J. Virol. 83:3904–3918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Rowe J, Greenblatt RJ, Liu D, Moffat JF. 2010. Compounds that target host cell proteins prevent varicella-zoster virus replication in culture, ex vivo, and in SCID-Hu mice. Antiviral Res. 86:276–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sahli R, Andrei G, Estrade C, Snoeck R, Meylan PR. 2000. A rapid phenotypic assay for detection of acyclovir-resistant varicella-zoster virus with mutations in the thymidine kinase open reading frame. Antimicrob. Agents Chemother. 44:873–878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Salmon B, Cunningham C, Davison AJ, Harris WJ, Baines JD. 1998. The herpes simplex virus type 1 U(L)17 gene encodes virion tegument proteins that are required for cleavage and packaging of viral DNA. J. Virol. 72:3779–3788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sauerbrei A, Taut J, Zell R, Wutzler P. 2011. Resistance testing of clinical varicella-zoster virus strains. Antiviral Res. 90:242–247 [DOI] [PubMed] [Google Scholar]
- 39. Schang LM. 2002. Cyclin-dependent kinases as cellular targets for antiviral drugs. J. Antimicrob. Chemother. 50:779–792 [DOI] [PubMed] [Google Scholar]
- 40. Schnute ME, et al. 2005. 4-Oxo-4,7-dihydrothieno[2,3-b]pyridines as non-nucleoside inhibitors of human cytomegalovirus and related herpesvirus polymerases. J. Med. Chem. 48:5794–5804 [DOI] [PubMed] [Google Scholar]
- 41. Scholtes L, Baines JD. 2009. Effects of major capsid proteins, capsid assembly, and DNA cleavage/packaging on the pUL17/pUL25 complex of herpes simplex virus 1. J. Virol. 83:12725–12737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Shahian T, et al. 2009. Inhibition of a viral enzyme by a small-molecule dimer disruptor. Nat. Chem. Biol. 5:640–646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Shigeta S, et al. 1983. Comparative efficacy of antiherpes drugs against various strains of varicella-zoster virus. J. Infect. Dis. 147:576–584 [DOI] [PubMed] [Google Scholar]
- 44. Shiraki K, et al. 1992. Comparison of antiviral assay methods using cell-free and cell-associated varicella-zoster virus. Antiviral Res. 18:209–214 [DOI] [PubMed] [Google Scholar]
- 45. Snoeck R, Andrei G, De Clercq E. 1999. Current pharmacological approaches to the therapy of varicella zoster virus infections: a guide to treatment. Drugs 57:187–206 [DOI] [PubMed] [Google Scholar]
- 46. Soong W, Schultz JC, Patera AC, Sommer MH, Cohen JI. 2000. Infection of human T lymphocytes with varicella-zoster virus: an analysis with viral mutants and clinical isolates. J. Virol. 74:1864–1870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Suzutani T, Ogasawara M, Shibaki T, Azuma M. 2000. Susceptibility of protein kinase (ORF47)-deficient varicella-zoster virus strains to anti-herpesvirus nucleosides. Antiviral Res. 45:79–82 [DOI] [PubMed] [Google Scholar]
- 48. Suzutani T, Saijo M, Nagamine M, Ogasawara M, Azuma M. 2000. Rapid phenotypic characterization method for herpes simplex virus and varicella-zoster virus thymidine kinases to screen for acyclovir-resistant viral infection. J. Clin. Microbiol. 38:1839–1844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Takayama M, Takayama N, Inoue N, Kameoka Y. 1996. Application of long PCR method of identification of variations in nucleotide sequences among varicella-zoster virus isolates. J. Clin. Microbiol. 34:2869–2874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Taylor SL, Kinchington PR, Brooks A, Moffat JF. 2004. Roscovitine, a cyclin-dependent kinase inhibitor, prevents replication of varicella-zoster virus. J. Virol. 78:2853–2862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Taylor SL, Moffat JF. 2005. Replication of varicella-zoster virus in human skin organ culture. J. Virol. 79:11501–11506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Tipples GA, et al. 2002. New variant of varicella-zoster virus. Emerg. Infect. Dis. 8:1504–1505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Trus BL, Gibson W, Cheng N, Steven AC. 1999. Capsid structure of simian cytomegalovirus from cryoelectron microscopy: evidence for tegument attachment sites. J. Virol. 73:2181–2192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Trus BL, et al. 2001. Capsid structure of Kaposi's sarcoma-associated herpesvirus, a gammaherpesvirus, compared to those of an alphaherpesvirus, herpes simplex virus type 1, and a betaherpesvirus, cytomegalovirus. J. Virol. 75:2879–2890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Visalli RJ, et al. 2003. Identification of small molecule compounds that selectively inhibit varicella-zoster virus replication. J. Virol. 77:2349–2358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Walters JN, Sexton GL, McCaffery JM, Desai P. 2003. Mutation of single hydrophobic residue I27, L35, F39, L58, L65, L67, or L71 in the N terminus of VP5 abolishes interaction with the scaffold protein and prevents closure of herpes simplex virus type 1 capsid shells. J. Virol. 77:4043–4059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Wang GQ, et al. 2006. Generation of a reporter cell line for detection of infectious varicella-zoster virus and its application to antiviral studies. Antimicrob. Agents Chemother. 50:3142–3145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Warner SC, Desai P, Person S. 2000. Second-site mutations encoding residues 34 and 78 of the major capsid protein (VP5) of herpes simplex virus type 1 are important for overcoming a blocked maturation cleavage site of the capsid scaffold proteins. Virology 278:217–226 [DOI] [PubMed] [Google Scholar]
- 59. Waxman L, Darke PL. 2000. The herpesvirus proteases as targets for antiviral chemotherapy. Antivir. Chem. Chemother. 11:1–22 [DOI] [PubMed] [Google Scholar]
- 60. Williams-Aziz SL, et al. 2005. Comparative activities of lipid esters of cidofovir and cyclic cidofovir against replication of herpesviruses in vitro. Antimicrob. Agents Chemother. 49:3724–3733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Zhou ZH, et al. 2000. Seeing the herpesvirus capsid at 8.5 A. Science 288:877–880 [DOI] [PubMed] [Google Scholar]