Abstract
The family Baculoviridae is a large group of insect viruses containing circular double-stranded DNA genomes of 80 to 180 kbp, which have broad biotechnological applications. A key feature to understand and manipulate them is the recognition of orthology. However, the differences in gene contents and evolutionary distances among the known members of this family make it difficult to assign sequence orthology. In this study, the genome sequences of 58 baculoviruses were analyzed, with the aim to detect previously undescribed core genes because of their remote homology. A routine based on Multi PSI-Blast/tBlastN and Multi HaMStR allowed us to detect 31 of 33 accepted core genes and 4 orthologous sequences in the Baculoviridae which were not described previously. Our results show that the ac53, ac78, ac101 (p40), and ac103 (p48) genes have orthologs in all genomes and should be considered core genes. Accordingly, there are 37 orthologous genes in the family Baculoviridae.
INTRODUCTION
The family Baculoviridae is composed of insect-specific DNA viruses containing covalently closed, double-stranded DNA (dsDNA) genomes that vary in size from approximately 80 to 180 kbp and encode 90 to 180 open reading frames (ORFs). This viral family is divided into four genera, Alphabaculovirus, Betabaculovirus, Gammabaculovirus, and Deltabaculovirus, which include lepidopteran-specific baculoviruses, lepidopteran-specific granuloviruses, hymenopteran-specific baculoviruses, and dipteran-specific baculoviruses, respectively (19, 20, 24, 25). The viral cycle presents a biphasic infection process generating progeny with two different phenotypes: budded viruses (BVs), which are produced at the initial stage of the multiplication cycle and are responsible for systemic infection inside the insect host, and occlusion-derived viruses (ODVs), which are produced in the last stage of the cycle within the infected cell and are required for the primary infection that takes place in the midgut epithelium cells of the insect host (5, 30, 40, 45). Mature ODVs are finally occluded in a protein matrix to form occlusion bodies (OBs), which protect them from the environment (23, 55).
The importance of the study of the Baculoviridae resides in the fact that they can be used as protein expression systems, models of genetic regulatory networks and genome evolution, putative nonhuman viral vectors for gene delivery, and biological control agents against insect pests (7, 12, 15, 21, 22, 28, 29, 37, 43, 47, 49, 59). Therefore, a genetic understanding of these viruses is a key point in their characterization. Thus, a set of common genes (known as core genes) which seem to be crucial factors for some of the main biological functions (transcription of viral late genes, production of the virion structure, infection of gut cells, abrogation of the host metabolism, and establishment of the infection) have been identified by comparisons among all baculovirus genomes sequenced (12, 15, 36, 50). To date, 33 genes are recognized as the set of encoding sequences shared by members of the family Baculoviridae (5, 6, 17, 19, 20, 24, 25, 35, 39, 44, 57). Core genes are commonly assumed to be orthologous genes as well. However, there are probably more orthologous sequences that may not be easily identified due to the accumulation of many mutations throughout divergent evolution. According to Fitch's definition, two genes of different biological entities can be considered orthologs if they diverged through a speciation event (12), whereas paralogy indicates the relationship between two sequences in a single organism after a duplication event. An accurate assessment of orthology is one of the most critical steps in studies of comparative genomics (14, 15, 36). A large variety of methods to predict orthology, focused principally on eukaryotic or prokaryotic organisms, have been developed in recent years (8, 13, 26, 32, 39). The vast majority of these methods use different algorithms to assign orthology by sequence comparisons of a query protein against predefined clusters of orthologous proteins available in online databases. Although these tools are useful and accurate, many are not applicable to organisms different from those for which they were developed; e.g., OrthoMCL and multiMSOAR 2.0 (32) identify only a few of the previously described orthologous genes in the family Baculoviridae (see references 8 and 14, respectively) or are limited to analyses of a few different genomes.
Traditionally, baculovirus orthology was assessed by sequence comparisons using BLASTP, BLASTN, or other standard NCBI tools. These tools allowed the assignment of the 33 published orthologous collections with various degrees of difficulty. It is important to note that the detection of orthologous proteins by using original GenBank records is a simple task when the level of sequence similarity is high and the gene has been annotated by the authors. However, a failure to detect orthologs should not be considered conclusive, because different situations may occur: (i) there may be proteins that are not annotated (NA), when one or more genomes have orthologs for the query protein but the corresponding sequences have not been annotated in GenBank; (ii) there may be remote orthology (RO), when one or more genomes have orthologs for the query protein but cannot be detected by using standard methods; and (iii) there may be a lack of orthology (LO), when one or more genomes truly lack orthologs for the query protein.
Taking the above-described situations into account, in the present work, we developed an algorithm to review or newly discover putative orthologous genes in addition to the 33 previously described orthologous collections for the Baculoviridae (5, 6, 17, 19, 20, 24, 25, 35, 39, 44, 57), using several additional bioinformatic approaches, such as PSI-Blast, BLASTP, tBlastN, and HaMStR (software that combines the Markov model with reciprocal best hits) (10); gene neighborhood conservation (38); and classical properties like protein size, the presence of transmembrane motifs, and additional protein analyses. This study provides new bioinformatic evidence to enrich the genetic knowledge of this important viral family.
MATERIALS AND METHODS
DNA and protein databases.
Currently, 58 complete genomes of baculoviruses have been deposited in the GenBank database (see Table S1 in the supplemental material). These genomes include 41 alphabaculoviruses, 13 betabaculoviruses, 3 gammabaculoviruses, and 1 deltabaculovirus. The proteomes of all viruses were extracted from the annotations in the corresponding GenBank files (www.ncbi.nlm.nih.gov). The nomenclature commonly found in the literature for baculoviral proteins and genes is sometimes ambiguous (e.g., double numbering). To avoid this issue, the proteins' names were standardized with an alphanumeric code of six characters: a three-letter code indicating the virus (see Table S1 in the supplemental material) and a three-number code corresponding to consecutive ORF numbering (e.g., ACN001 represents the first annotated protein in the Autographa californica multiple nucleopolyhedrovirus [MNPV] [AcMNPV] file). Thus, three different databases were created: the Global Proteome Database (GPD), containing all annotated proteins in the 58 baculovirus genomes; the Individual Genome Database (IGD), which stores individual nucleotide sequences of the same 58 baculoviruses; and the Individual Proteome Database (IPD), with 58 individual protein databases containing the annotated proteome of each virus. All these databases were indexed by using the formatdb routine from the NCBI BLAST standalone suite.
Multi PSI-Blast/tBlastN algorithm.
In order to obtain sets of orthologs of Baculoviridae, an ad hoc Perl routine, which combines the use of PSI-Blast, tBlastN, and BLASTP (2), was developed (see Fig. S1 in the supplemental material). The first step runs a PSI-Blast search using the annotated proteins of each baculovirus as a query against the GPD, being iterated until convergence. The PSI-Blast output file is then analyzed, and the hits accepted for each virus according to 15 different detection thresholds (an expect threshold of 1e−1 combined with an inclusion threshold of 1e−4, 1e−8, 1e−12, 1e−16, 1e−20, or 1e−40; an expect threshold of 1e−4 combined with an inclusion threshold of 1e−8, 1e−12, 1e−16, 1e−20, or 1e−40; and an expect threshold of 1e−8 combined with an inclusion threshold of 1e−12, 1e−16, 1e−20, or 1e−40) are added to a result list, designated the Multi PSI-Blast result (MPsiBr), which does not contain repeated proteins. The routine then verifies if all members of the four baculovirus genera have provided protein sequences for the MPsiBr. In this case, the process concludes, and the result is taken as a list of orthologous proteins. In situations where this does not occur, the routine performs a PSI-Blast search against the GPD by using all previously detected proteins of the corresponding genus as a query. Thus, the new results are added to the MPsiBr. The routine then applies three consecutive filters to analyze the MPsiBrs. The first filter selects all proteins that had a score of 54 to 58 orthologs (e.g., one protein of a particular virus has similarity with one protein of other virus species, and this result is repeated in 54 of the 58 possible comparisons). The second filter verifies and compares the lists of orthologs detected for each protein by using the 15 different thresholds and condenses them into a single list when the outcome files are identical (e.g., if three MPsiBrs for one protein of a particular virus obtained from the application of the routine using three different detection thresholds are coincident, the filter condenses the results into one MPsiBr). Finally, the third filter compares all the resulting MPsiBrs, and if it detects full coincidences among some of them, it merges the results into one file (e.g., if 50 MPsiBrs obtained from 50 proteins of 50 different viruses have exactly the same data sets, the filter condenses the result into only one postfiltered MPsiBr). If, at the end of this stage, some lists do not contain sequences for all viruses, an NA situation (sequences not annotated in GenBank genome files) may occur. As a consequence, the software performs a tBlastN search using all the proteins stored in the filtered MPsiBr list against the IGD of each absent virus. The nucleotide sequences detected (E value of <1e−5) are extended upstream and downstream until a start codon and a stop codon are found, respectively. The putative ORFs are then translated and used as a query to perform a BLASTP search against the previous MPsiBr to look for reciprocity. Finally, the NA protein is added to the corresponding IPD and GPD when the expected value is accepted. All tBlastN/BLASTP results were manually cured to avoid the addition of artifacts into databases.
Multi HaMStR.
In order to explore a remote orthology (RO) situation, the host orders (Lepidoptera, Hymenoptera, and Diptera) were considered. Thus, the potentially incomplete previous results for orthologs from 54 members (without sequences of Gammabaculovirus and Deltabaculovirus), 55 members (without sequences of Gammabaculovirus), or 57 members (without sequences of Deltabaculovirus) were selected. This set of proteins generated by the Multi PSI-Blast/Multi tBlastN algorithm was aligned by the use of PSI-Coffee software, using the t_coffee_msa and slow_pair parameters for multiple and pairwise steps, respectively (9, 27). Specific hidden Markov model-based profiles (HMM profiles) were then generated for each set of sequences by using HMMER3 local software (10). These inputs were used to find remote orthologs by the use of HaMStR v8.0 software (11), with the reciprocal best hit (rbh) and representative parameters (cutoff value of 1e−5 for hmmsearch v3.0). In order to avoid any bias due to phylogenetic proximity and to cover the complete universe of available baculoviral proteomes, the software was modified to incorporate the ability to use multiple reference species.
Sequence characterization for newly discovered core genes.
The previously undescribed baculovirus core genes obtained by the use of the Multi PSI-Blast/tBlastN and Multi HaMStR algorithms were analyzed and compared with their orthologs. Molecular weights were calculated by using Composition/Molecular Weight Calculation software (http://pir.georgetown.edu/pirwww/search/comp_mw.shtml) (31). The signal peptide and transmembrane helix were predicted by using SignalP 4.0 (http://www.cbs.dtu.dk/services/SignalP/) (42) and TMHMM-2.0 (http://www.cbs.dtu.dk/services/TMHMM-2.0/) (56), respectively. On the other hand, sequence identity and similarity studies were performed by an all-against-all strategy and by carrying out pairwise alignments with Clustal X software using default parameters and PAM350 (46). Identity was calculated as the percentage of identical residues in the pairwise alignment, and similarity was calculated as the sum of identities plus similarities (strong plus weak) and expressed as a percentage. Furthermore, the corresponding orthologs of newly discovered core genes detected in this work from all baculoviruses (24) were analyzed for the presence of protein motifs by using the technique of expectation maximization implemented by the MEME tool, with standard parameters (3). The putative motifs present in the 58 sequences studied (combined P value of <1e−5) were accepted. The distribution of motif dispositions and the corresponding sequence logos were provided by the MEME Web server (http://meme.sdsc.edu/meme/intro.html). In addition, to find putative conserved protein motifs for Ac53, the HHpred server (48) was used (the input was a Fasta multiple file of Ac53 orthologs, and the search options were select HMM databases, all; multiple sequence alignment (MSA) generation method, HHblits; maximum MSA generation iterations, 3; score secondary structure, yes; and alignment mode, local).
Additional studies to validate orthology postulations.
A Z-score approach for assessing the significance of similarity studies was carried out. First, sets of 58 amino acid sequences were generated for each Baculoviridae core protein with random composition and considering a length equal to the average length of the corresponding orthologous sequences (natural sequences [natS] and random sequences [rndS], respectively). All natS were aligned among them by using an all-against-all pairwise strategy. After this, all natS were aligned against the corresponding 58 rndS by using a pairwise strategy. Once the identity averages and the corresponding standard deviations from the alignments were obtained, the Z scores were calculated by using the following equation:
where InatS is the percent identity of each pairwise alignment of natS, ArndS/natS is the average percent identities obtained from pairwise alignments between each natS and each rndS, and σ is the standard deviation of ArndS/natS identities.
On the other hand, to determine the relationships among species, 37 individual core protein databases (ICPDs) were constructed. Relaxed BLASTP (E value = 100) was then performed by using individual proteins against the corresponding ICPD as a query. Finally, the minimum E values that showed reciprocity between pairs of viruses were selected and illustrated, involving all viral species in network graphs.
RESULTS
Multi PSI-Blast/HaMStR.
The development and execution of the algorithm based on Multi PSI-Blast/HaMStR allowed the detection of 35 orthologs in the family Baculoviridae. Of this total, 31 sequences were described previously (6, 35), 1 of these was recently postulated to be a core gene (odv-e25) (57), and 4 (ac53, ac78, ac101, and ac103) should be considered a new set of orthologous genes. In contrast, two of the accepted core genes (ac93 [p18] and ac100 [p6.9]) (6, 24, 35, 38) were not detected, being false negatives (Table 1).
Table 1.
The 37 genes shared by all baculovirusesa
| AcMNPV ORF | Gene designation | Description | NeabNPV ORF | CuniNPV ORF | Previous annotation | Annotation in this work | Method of detection |
|---|---|---|---|---|---|---|---|
| 6 | lef-2 | DNA replication/primase-associated factor | 58 | 25 | Core gene | Core gene | Multi PSI-Blast |
| 14 | lef-1 | DNA primase | 69 | 45 | Core gene | Core gene | Multi PSI-Blast |
| 22 | pif-2 | Required for per os infection (PIF-2) | 56 | 38 | Core gene | Core gene | Multi PSI-Blast |
| 40 | p47 | RNA polymerase subunit | 50 | 73 | Core gene | Core gene | Multi PSI-Blast |
| 50 | lef-8 | RNA polymerase subunit | 84 | 26 | Core gene | Core gene | Multi PSI-Blast |
| 53 | ac53 | Likely involved in nucleocapsid assembly/U-box/RING-like domain | 83 | 28 | α + β | Core gene | Multi PSI-Blast/HaMStR |
| 54 | vp1054 | Nucleocapsid protein | 89 | 8 | Core gene | Core gene | Multi PSI-Blast |
| 62 | lef-9 | RNA polymerase subunit | 39 | 59 | Core gene | Core gene | Multi PSI-Blast |
| 65 | dnapol | DNA replication | 12 | 91 | Core gene | Core gene | Multi PSI-Blast |
| 66 | Desmop | Present in nucleocapsid | 13 | 92 | Core gene | Core gene | Multi PSI-Blast |
| 68 | ac68 | Required for per os infection (PIF-6) | 40 | 58 | Core gene | Core gene | Multi PSI-Blast |
| 77 | Vlf1 | Involved in expression of the p10 and polh genes | 45 | 18 | Core gene | Core gene | Multi PSI-Blast |
| 78 | ac78 | Unknown function/transmembrane domain | 46 | 34 | α + β + γ | Core gene | Multi PSI-Blast/HaMStR |
| 80 | gp41 | Tegument protein | 48 | 33 | Core gene | Core gene | Multi PSI-Blast |
| 81 | ac81 | Unknown function | 49 | 106 | Core gene | Core gene | Multi PSI-Blast |
| 83 | p95 | Viral capsid-associated protein | 88 | 35 | Core gene | Core gene | Multi PSI-Blast |
| 89 | vp39 | Major capsid protein | 93 | 24 | Core gene | Core gene | Multi PSI-Blast |
| 90 | lef-4 | RNA polymerase subunit/capping enzyme | 63 | 96 | Core gene | Core gene | Multi PSI-Blast |
| 92 | p33 | Sulfhydryl oxidase | 8 | 14 | Core gene | Core gene | Multi PSI-Blast |
| 93† | p18 | Egress of nucleocapsids | 9 | 13 | Core gene | α + β + γ | Multi PSI-Blast/HaMStR |
| 94 | odv-e25 | ODV envelope protein | 10 | 15 | Core gene | Core gene | Multi PSI-Blast/HaMStR |
| 95 | helicase | Unwinding DNA | 62 | 89 | Core gene | Core gene | Multi PSI-Blast |
| 96 | ac96 | Required for per os infection (PIF-4) | 61 | 90 | Core gene | Core gene | Multi PSI-Blast |
| 98 | 38k | Required for nucleocapsid assembly | 60 | 87 | Core gene | Core gene | Multi PSI-Blast |
| 99 | lef-5 | Transcription initiation factor | 59 | 88 | Core gene | Core gene | Multi PSI-Blast |
| 100† | p6.9 | Nucleocapsid protein | 31 | 23 | Core gene | α + β + γ | Multi PSI-Blast/HaMStR |
| 101 | p40 | Subunit of protein complex | 32 | 22 | α + β | Core gene | Multi PSI-Blast/HaMStR |
| 103 | p48 | BV production and ODV envelopment | 34 | 55 | α + β + γ | Core gene | Multi PSI-Blast/HaMStR |
| 109 | odv-ec43 | Associated with ODV | 72 | 69 | Core gene | Core gene | Multi PSI-Blast |
| 115 | pif-3 | Required for per os infection (PIF-3) | 70 | 46 | Core gene | Core gene | Multi PSI-Blast |
| 119 | pif-1 | Mediates binding of ODV to midgut (PIF-1) | 80 | 29 | Core gene | Core gene | Multi PSI-Blast |
| 133 | alk exo | Involved in DNA recombination and replication | 36 | 54 | Core gene | Core gene | Multi PSI-Blast |
| 138 | p74 | Mediates binding of ODV to midgut (PIF-0) | 51 | 74 | Core gene | Core gene | Multi PSI-Blast |
| 142 | 49k | Required for BV production | 64 | 30 | Core gene | Core gene | Multi PSI-Blast |
| 143 | odv-e18 | ODV envelope protein | 66 | 31 | Core gene | Core gene | Multi PSI-Blast |
| 144 | odv-e27 | ODV envelope protein | 67 | 32 | Core gene | Core gene | Multi PSI-Blast |
| 148 | odv-e56 | ODV envelope protein (PIF-5) | 16 | 102 | Core gene | Core gene | Multi PSI-Blast |
Boldface type represents new core genes proposed in this work; † indicates accepted core genes not found by this strategy. The Greek letters α, β, and γ are used to indicate the genera Alpha-, Beta-, and Gammabaculovirus, respectively.
Other results from the algorithm application refer to the genes shared by sets of baculoviruses, except for some genera. Thus, nine genes were found in all members but Culex nigripalpus nucleopolyhedrovirus (NPV) (CuniNPV), including six sequences not previously described as orthologs in the genus Gammabaculovirus (ac61, ac75, ac106/107, ac108, ac131, and ac145) (Table 2). In contrast, the f-protein gene was found in all baculoviruses except gammabaculoviruses. Finally, in addition to 9 genes in lepidopteran-specific baculoviruses described previously, six new orthologs were found (ac64, ac67, ac110, ac129, ac139, and ac141) (Table 2).
Table 2.
All the genes shared by 57 (Alpha-, Beta-, and Gammabaculovirus), 55 (Alpha-, Beta-, and Deltabaculovirus), and 54 (Alpha- and Betabaculovirus) membersa
| AcMNPV ORF | Gene designation | Description | CpGV ORF | NeabNPV ORF | Previously reported genus(era) | Genera determined in this work | Method of Detection |
|---|---|---|---|---|---|---|---|
| Core genes for the genera Alpha-, Beta-, and Gammabaculovirus | |||||||
| 8 | polyhedrin granulin | OB protein | 1 | 1 | α + β + γ | α + β + γ | Multi PSI-Blast/HaMStR |
| 25 | dbp | DNA binding protein | 81 | 6 | α + β + γ | α + β + γ | Multi PSI-Blast/HaMStR |
| 37 | lef-11 | Required for expression of late genes | 58 | 7 | α + β + γ | α + β + γ | Multi PSI-Blast/HaMStR |
| 61 | fp/25k | Structural protein of BVs and ODVs | 118 | 54 | α + β | α + β + γ | Multi PSI-Blast |
| 75 | ac75 | Required for BV production | 108 | 43 | α + β | α + β + γ | Multi PSI-Blast/HaMStR |
| 106/107 | ac106/107 | Unknown function | 52 | 35 | α + β | α + β + γ | Multi PSI-Blast/HaMStR |
| 108 | ac108 | P11 protein, associated with the PIF complex | 56 | 72 | α + β | α + β + γ | Multi PSI-Blast/HaMStR |
| 131 | pe/pp34 | Involved in morphogenesis of polyhedral envelope | 23 | 53 | α | α + β + γ | Multi PSI-Blast |
| 145 | ac145 | OB protein, involved in oral infection | 9 | 67 | α + β | α + β + γ | Multi PSI-Blast/HaMStR |
| Core gene for the genera Alpha-, Beta-, and Deltabaculovirus | |||||||
| 23 | ac23 | Pathogenicity factor in alphabaculoviruses of group I; fusion protein in alphabaculoviruses of group II, betabaculoviruses, and gammabaculoviruses | 31 | 104b | α + β + δ | α + β + δ | Multi PSI-Blast/HaMStR |
| Core genes for the genera Alpha- and Betabaculovirus | |||||||
| 10 | pk-1 | Protein kinase | 3 | α + β | α + β | Multi PSI-Blast | |
| 13 | ac13 | Unknown function | 73 | α + β | α + β | Multi PSI-Blast | |
| 28 | lef-6 | Involved in late and very late gene expression | 80 | α + β | α + β | Multi PSI-Blast | |
| 35 | ubiquitin | Ubiquitin | 54 | α + β | α + β | Multi PSI-Blast | |
| 36 | pp31 | Late gene expression factor | 57 | α + β | α + β | Multi PSI-Blast | |
| 38 | ac 38 | ADP-ribose pyrophosphatase | 69 | α + β | α + β | Multi PSI-Blast | |
| 64 | gp37 | Chitin binding protein/spindle body protein | 13 | α | α + β | Multi PSI-Blast | |
| 67 | lef-3 | ssDNA binding protein | 113 | α + β | α + β | Multi PSI-Blast | |
| 82 | tlp | Unknown function | 102 | α + β | α + β | Multi PSI-Blast | |
| 102 | p12 | Involved in nuclear localization of G-actin | 84 | α + β | α + β | Multi PSI-Blast/HaMStR | |
| 110 | ac110 | Unknown function | 53 | α | α + β | Multi PSI-Blast | |
| 129 | p24 | ODV protein | 71 | α | α + β | Multi PSI-Blast | |
| 139 | me53 | DNA synthesis regulator, required for efficient BV production | 143 | α | α + β | Multi PSI-Blast | |
| 141 | Exon0 | Egress of nucleocapsids from the nucleus to the cytoplasm, interaction with β-tubulin and microtubules | 11 | α | α + β | Multi PSI-Blast | |
| 146 | ac146 | Unknown function | 8 | α + β | α + β | Multi PSI-Blast | |
| 147 | ie-1 | Immediate-early transactivator | 7 | α + β | α + β | Multi PSI-Blast |
Boldface type represents new core genes proposed by this work. The Greek letters α, β, γ, and δ are used to indicate the genera Alpha-, Beta-, Gamma-, and Deltabaculovirus, respectively. ssDNA, single-stranded DNA.
CuniNPV ORF.
In summary, the methodology carried out here was able to find 16 new orthologous genes, including 4 newly discovered core genes shared by all baculoviruses.
ac53 is a core gene.
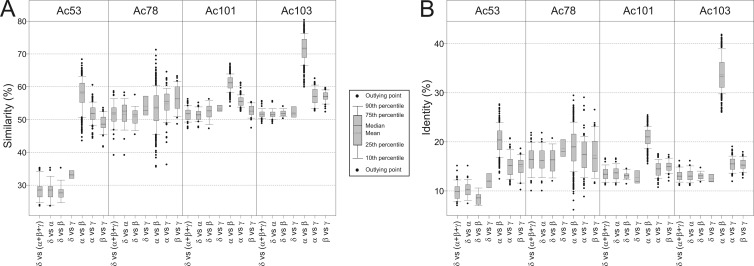
The ac53 gene was previously found in all baculoviruses except CuniNPV (6). The protein has an unknown function but seems to be involved in the correct assembly of the nucleocapsid (33). HHpred predicted that Ac53 and its orthologs have significant structural similarity to a U-box/RING-like domain typical of the E3 ubiquitin-protein ligase family (Pfam accession number PF05883) (probability = 99.94; E value = 1.6e−26; P value = 1.3e−31). Also, it was detected in the structure of BV (1, 53). The Multi PSI-Blast/HaMStR algorithm detected an orthologous sequence in CuniNPV (cuni28) that would encode a polypeptide of 31.2 kDa. Importantly, cuni28 is located upstream of lef-8, which is in accordance with the gene order in all baculoviruses (Fig. 1A). The encoded polypeptide showed maximum similarity with Helicoverpa armigera MNPV (HearMNPV), Mamestra configurata NPV-B (MacoNPV-B), and Adoxophyes orana granulovirus (AdorGV) (35.2%); similar values were found for previously accepted core genes (57). The amino acid identity was about 11% with respect to the corresponding orthologs from the same viruses (maximum of 15.1% with Trichoplusia ni single nucleopolyhedrovirus [TnSNPV]); a similar level sequence identity was found among some other interspecies-orthologous proteins (Fig. 2A). This low level of identity might be attributed to differences in the protein length because Cuni28 is approximately twice the size of the corresponding putative orthologs.
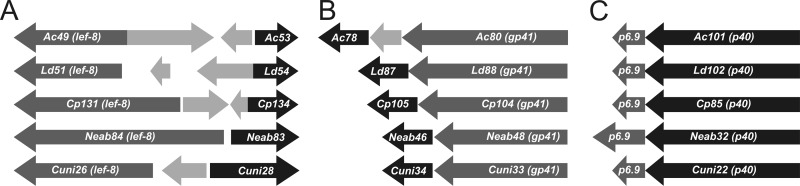
Fig 1.
Gene order. Shown are schematic physical maps for the ac53 (A), ac78 (B), and ac101 (C) individual gene clusters. AcMNPV, Lymantria dispar MNPV (LdMNPV), CpGV, NeabNPV, and CuniNPV are the prototypes representing alphabaculoviruses of group I, alphabaculoviruses of group II, betabaculoviruses, gammabaculoviruses, and deltabaculoviruses, respectively. The relative positions and orientations of these three gene clusters are conserved in all baculovirus genomes sequenced to date. The dark arrows represent the newly discovered core genes, the dark gray arrows indicate the previously accepted core genes, and the light gray arrows represent other ORFs described among the core genes illustrated.
Fig 2.
Sequence conservation. Box plots show the sequence conservation of newly discovered core genes. The percentages of similarity (A) and identity (B) for all protein pairs were calculated. To compare the CuniNPV-orthologous protein, all possible intergenus comparisons were considered (δ versus α + β + γ, δ versus α, δ versus β, δ versus γ, α versus β, α versus γ, and β versus γ, where α, β, γ, and δ indicate alphabaculoviruses, betabaculoviruses, gammabaculoviruses, and deltabaculoviruses, respectively). The boundary of the boxes closest to zero indicates the 25th percentile, the line within the box marks the median, the dashed line within the box marks the mean, and the boundary of the box furthest from zero indicates the 75th percentile. Error bars above and below the box indicate the 90th and 10th percentiles, respectively. The black circles indicate outlying points.
Z scores from gamma- and deltabaculovirus orthologs of Ac53 were in a narrow range (−1.86 to 5.73) (Fig. 3). Of the 219 total values, 182 were above zero (161 values between the 13th and 90th percentiles and 21 as superior outliers), and 34 were below zero (17 values as inferior outliers and 17 between 10th and 13th percentiles). Values below zero were observed among some standard orthologous proteins, e.g., Ac65 (DNA polymerase [DNApol]) and Ac66 (Desmoplakin) (Fig. 3).
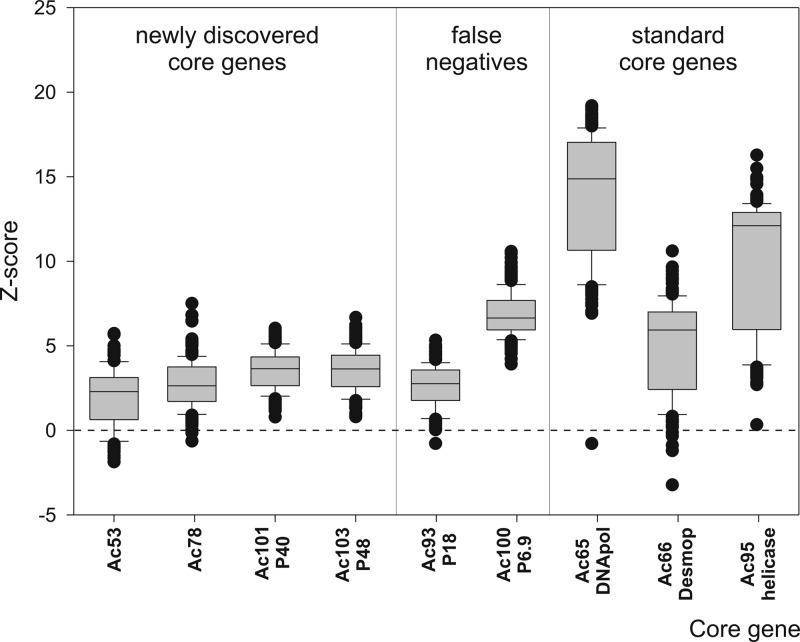
Fig 3.
Z scores. Box plots show the Z-score distribution for the four newly discovered (Ac53, Ac78, Ac101 [P40], and Ac103 [P48]), the two undetected (Ac93 [P18] and Ac100 [P6.9]), and the other three previously recognized (Ac65 [DNApol], Ac66 [Desmoplakin], and Ac95 [helicase]) core proteins. Z scores for all proteins were calculated, and only those for gammabaculoviruses and deltabaculoviruses are shown. The dashed line is only a reference to show that for Ac53, Ac78, Ac93, Ac65, and Ac66, several outliers have values below zero.
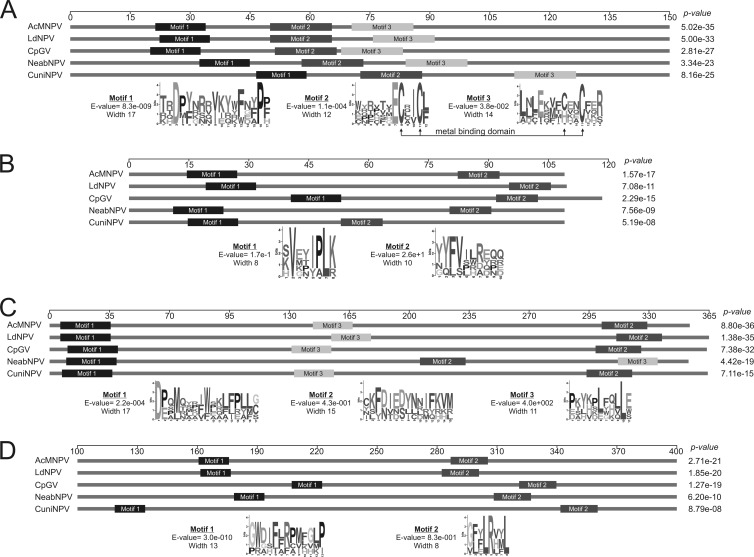
In order to find conserved protein motifs, we then analyzed the set of Ac53-orthologous proteins from baculoviruses. Thus, three motifs were detected (Fig. 4A). The first was located in the amino-terminal region (17 amino acids) and showed the highest level of conservation. In particular, this region has totally conserved aspartic acid and proline residues. The second motif was detected in the central region and contained two cysteines and one glutamine. Finally, the third motif was located in the carboxyl-terminal region, with a conserved cysteine. In spite of having different protein sizes, the 58 orthologs contained the three motifs in the same order. It is important to note that these motifs were included in the regions that showed the most significant values within the U-box/RING-like domain predicted by HHpred. Besides this, the RING-like domain contained a metal binding motif with a core pattern composed of [C]-x2-[C]-x23-26-[CH]-x2-[C]. In fact, the [C]-x2-[C] and [CH]-x2-[C] sequences were located in motifs 2 and 3, respectively.
Fig 4.
Protein motifs. The schematic map of Ac53 (A), Ac78 (B), Ac101 (C), and Ac103 (D) shows conserved motifs found by the MEME tool for the 58 orthologs of each newly discovered core protein. Only the prototypes representing alphabaculoviruses of group I (AcMNPV), alphabaculoviruses of group II (LdMNPV), betabaculoviruses (CpGV), gammabaculoviruses (NeabNPV), and deltabaculoviruses (CuniNPV) are shown. The relative positions of motif 1, motif 2, and motif 3 are indicated by black, dark gray, and light gray rectangles, respectively. The length and conservation for each motif are represented by sequence logos. In Ac53, a metal binding motif is indicated. An amino acid scale indicating the location of each motif is depicted above the groups of orthologs.
ac78 is a core gene.
Orthologs of ac78, a gene of unknown function, have been found in all lepidopteran- and hymenopteran-specific baculoviruses in previous works but have not been found in CuniNPV (6, 24, 35). The approach carried out here allowed the detection of an orthologous sequence in that virus: cuni34. It is important to note that Cuni34 (11.3 kDa) is an ODV structural protein in CuniNPV (41). The analyses of gene order revealed an arrangement similar to that for other baculoviruses, where gp41 is located upstream of this gene (Fig. 1B). With respect to similarity and identity studies, Cuni34 showed maximum sequence similarity with Neodiprion lecontei NPV (NeleNPV) and Adoxophyes honmai NPV (AdhoNPV) (58.6% and 58.2%, respectively) and maximum identity with Agrotis segetum NPV (AgseNPV) (21.6%); similar values were found between interspecies-orthologous proteins (Fig. 2). The cuni34 ORF encodes a protein of 108 amino acids, exactly the average length of all orthologs.
Z scores from gamma- and deltabaculovirus orthologs of Ac78 were in a narrow range (−0.51 to 7.63) (Fig. 3). Of the 219 total values, 176 were between the 10th and 90th percentiles, 22 were superior outliers, and 21 were inferior outliers (3 of them were below zero). Values below zero were observed among some standard orthologous proteins (e.g., Ac65 [DNApol] and Ac66 [Desmoplakin]) (Fig. 3).
The analyses of the conserved protein motifs revealed the presence of a transmembrane domain (TMM) in all the orthologs, which correlates with its association with the ODV envelope. In Cuni34, the TMM was located between amino acids 71 and 93 (data not shown). On the other hand, two motifs were found (Fig. 4B). One of them (10 amino acids) was the most conserved motif and was located in the amino-terminal region, except in Cydia pomonella GV (CpGV) (Cp105; central location). Taking into account that the first 30 amino acids of Cp105 did not have similarity with the other betabaculovirus sequences and considering a second methionine that appeared in the 31st position as a starting codon, the motif would be located in the same place as in the others. This motif presented two conserved amino acids (valine and leucine) in all sequences and a triad of isoleucine-proline-leucine for alphabaculovirus and deltabaculovirus proteins. The other motif was located in the carboxyl-terminal region of all baculoviruses except CuniNPV, in which it was detected in the central region.
ac101 is a core gene.
Orthologs of ac101 (p40 and odv-c42) were previously found in all lepidopteran-specific baculoviruses. In this work, we detected orthologous sequences in gammabaculoviruses (neab32, nele32, and nese35) and deltabaculoviruses (cuni22). Ac101 has an essential role in the viral cycle because it mediates the nuclear entry of P78/83 and is involved in nucleocapsid morphogenesis (51, 54). The cuni22 gene encodes a 42.2-kDa protein and has been found to be part of the ODV structure in CuniNPV (41). The cuni22 ORF has the same gene order as that of the other baculoviruses (upstream of p6.9) (Fig. 1C), which is in agreement with the conclusion that it is a core gene.
Cuni22 showed maximum sequence similarity with its ortholog from CpGV (56.2%; 51.4% with the other proteins). The identity values showed an average of 13.5% (maximum of 16.6% with AdhoNPV); similar data were found between interspecies-orthologous proteins (Fig. 2). The length of the protein encoded by cuni22 is 382 amino acids, only 12 residues longer than the average length for all the orthologous proteins.
Z scores from gamma- and deltabaculovirus orthologs of Ac101 were in a narrow range (0.79 to 6.04) (Fig. 3). Of the 219 total values, 178 were between the 10th and 90th percentiles, 21 were superior outliers, and 20 were inferior outliers.
The sequences detected here in gammabaculoviruses have not been described previously as orthologs of ac101. Only nese35 has been characterized as P40 (16). In fact, a simple search using the NCBI BLASTP tool with any of these proteins as a query gave five P40 proteins, belonging to the genera Alphabaculovirus (Ectropis obliqua NPV [EcobNPV], Spodoptera litura NPV [SpliNPV], and Orgyia leucostigma NPV [OrleNPV]) and Betabaculovirus (Pseudaletia unipuncta GV-Hawaiian [PsunGV] and Helicoverpa armigera GV [HearGV]), as results.
Three motifs were found in all the sequences analyzed (Fig. 4C). The first motif was located in the amino-terminal region (17 amino acids) in all sequences and presented a conserved aspartic acid. The second motif was located in the carboxyl-terminal region in all sequences except in the corresponding protein from Neodiprion abietis NPV (NeabNPV), in which it was located in the central region. In this case, the proteins analyzed did not show a high level of conservation of amino acid sequences. The third motif, which had one leucine conserved in all sequences, was located in the central region except in the ortholog of NeabNPV, in which it was located in the carboxyl-terminal region.
ac103 is a core gene.
The ac103 (p48) gene was previously found in all baculoviruses except CuniNPV (6, 24, 35). The protein encoded by this gene seems to be involved in the production of budded viruses and is required for nucleocapsid envelopment in ODVs (58). The algorithm application described here allows the detection of a sequence orthologous to ac103 in CuniNPV (cuni55), which encodes a protein that has been found in CuniNPV ODVs (41). Cuni55 (44.6 kDa) has only 1 amino acid less than the average length for all ortholog proteins. cuni55 is located close to lef-9 and ac68 but is not clustered as in the other baculoviruses. With respect to sequence studies, Cuni55 showed maximum similarity with OrleNPV and Apocheima cinerarium NPV (ApciNPV) (55.6% and 55.3%, respectively) and an average similarity of 51.7% with the other orthologous proteins. The average identity was 13.1% (maximum of 16.1% with EcobNPV); similar sequence identities were found for interspecies-orthologous protein comparisons (Fig. 2).
Z scores from gamma- and deltabaculovirus orthologs of Ac103 were in a narrow range (0.80 to 6.68) (Fig. 3). Of the 219 total values, 179 were between the 10th and 90th percentiles, 20 were superior outliers, and 20 were inferior outliers.
On the other hand, two motifs were found in all the proteins analyzed. The first motif was the most conserved motif and was located close to the amino-terminal region in all the sequences except in CpGV (Cp83), in which it was located in the central region. A situation similar to that of the ac78 and cp105 orthologs may occur. In this case, Cp83 presents a second methionine at the 47th position, which might be considered starting codon. If this is real, the protein would have the highest sequence similarity and a motif distribution identical to that of the other Betabaculovirus orthologs. The second motif was found in the carboxyl-terminal region for all orthologs and presented three conserved amino acids (two leucines and one arginine) (Fig. 4).
False negatives.
Two core proteins, Ac93 (P18) (recently described by Yuan et al. [57]) and Ac100 (P6.9) (a standard core protein) were not detected by the algorithm implemented in this work. However, Z-score analyses showed that both proteins exhibited narrow ranges of values (Ac93, −0.65 to 5.46; Ac100, 3.92 to 10.58), fitting into the broader ranges shown for other standard core proteins (Ac65, −0.78 to 19.20; Ac66, −3.22 to 10.61; Ac95, 0.34 to 16.28) (Fig. 3).
BLASTP protein relationships.
In addition to previous analyses, relaxed BLASTP searches for orthologous proteins of Ac53, Ac78, Ac101, and Ac103 against the corresponding ICPDs were performed. For comparison purposes, the same analysis was also carried out with orthologous proteins of Ac93. In order to relate all species in a network, the minimum E values obtained for pairs of viruses were selected (Fig. 5). It is important to note that the reciprocal E values were not always coincident because of the differences in protein size. In all studied proteins, the intragenus relationships showed values close to zero. In contrast, the intergenus relationships revealed different values according to the core protein considered.
Fig 5.
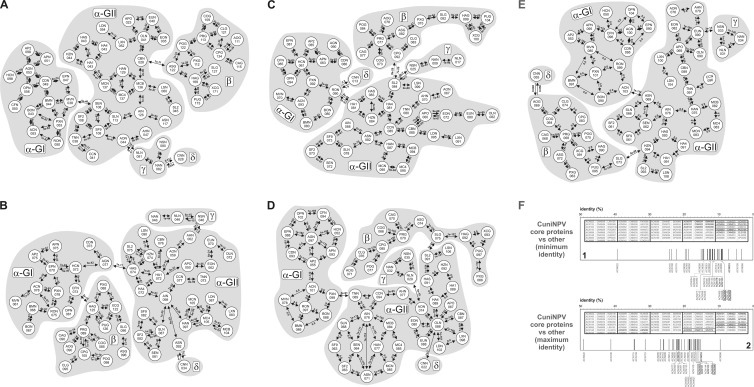
Protein relationship analyses. (A to E) Protein relationships among all baculoviruses derived from BLASTP analysis. The E value between pairs of species is indicated above each arrow. Gray-shaded areas correspond to the different subdivisions of the Baculoviridae: α-GI, alphabaculoviruses of group I; α-GII, alphabaculoviruses of group II; β, betabaculoviruses; γ, gammabaculoviruses; δ, deltabaculoviruses. (A) Ac53. (B) Ac78. (C) Ac93. (D) Ac101. (E) Ac103. (F) Minimum (1) and maximum (2) identity values between each deltabaculovirus core protein and the corresponding sequences of other baculovirus genera. The orthologous names are mentioned with the family prototype nomenclature. The 4 newly discovered core proteins, the 2 core proteins described previously by Yuan et al. (57), and the 31 standard core proteins are indicated with black characters, boxed black characters, and dark gray characters, respectively. The insets show the core proteins (AcMNPV nomenclature) (first column) in deltabaculoviruses (second column) and the corresponding species where orthologous sequences with minimum (1) or maximum (2) identity were found (third column). In all instances, six-character codes were used (see Table S1 in the supplemental material).
In view of this, Ac53 showed the following main links: Maruca vitrata MNPV (MaviMNPV)/Spodoptera exigua MNPV (SeMNPV) (alphabaculoviruses of group I with group II), Clanis bilineata NPV (ClbiNPV)/Agrotis segetum GV (AgseGV) (alphabaculoviruses of group II with betabaculoviruses), Adoxophyes orana NPV (AdorNPV)/NeleNPV (alphabaculoviruses of group II with gammabaculoviruses), and NeabNPV/CuniNPV (gammabaculoviruses with deltabaculovirus) (Fig. 5A). Meanwhile, Ac78 exhibited the following main nexus: Anticarsia gemmatalis MNPV (AgMNPV)/Helicoverpa armigera SNPV-NNg1 (HearSNPV-NNg1) (alphabaculoviruses of group I with group II), Choristoneura fumiferana MNPV (CfMNPV)/HearGV (alphabaculoviruses of group I with betabaculoviruses), AdhoNPV/Neodiprion sertifer NPV (NeseNPV) (alphabaculoviruses of group II with gammabaculoviruses), and AgseNPV/CuniNPV (alphabaculoviruses of group II with deltabaculovirus) (Fig. 5B). Ac101 showed the following main links: Plutella xylostella MNPV (PlxyMNPV)/TnSNPV (alphabaculoviruses of group I with group II), SpliNPV-G2/Spodoptera litura GV-K1 (SpliGV) (alphabaculoviruses of group II with betabaculoviruses), AdorNPV/NeleNPV (alphabaculoviruses of group II with gammabaculoviruses), and OrleNPV/CuniNPV (alphabaculoviruses of group II with deltabaculovirus) (Fig. 5C). On the other hand, Ac103 exhibited the following main nexus: AcMNPV/AgseNPV (alphabaculoviruses of group I with group II), Helicoverpa zea NPV (HezeNPV)/SpliGV (alphabaculoviruses of group II with betabaculoviruses), Euproctis pseudoconspersa NPV (EupsNPV)/NeseNPV (alphabaculoviruses of group II with gammabaculoviruses), and AdorGV/CuniNPV (betabaculoviruses with deltabaculovirus) (Fig. 5D). Finally, Ac93 showed the following main links: Rachiplusia ou MNPV (RoMNPV)/Helicoverpa armigera NPV-C1 (HearNPV-C1) (alphabaculoviruses of group I with group II), SpliNPV-G2/CpGV (alphabaculoviruses of group II with betabaculoviruses), SpliNPV-G2/NeseNPV (alphabaculoviruses of group II with gammabaculoviruses), and RoMNPV/CuniNPV (alphabaculoviruses of group I with deltabaculovirus) (Fig. 5E).
Otherwise, the minimum and maximum identities obtained from the global alignments among 37 CuniNPV-orthologous proteins against the corresponding sequences of the other baculoviruses were selected (Fig. 5F). This approach showed that the limit values for the newly discovered core proteins were similar to the results for other previously accepted core proteins.
DISCUSSION
Baculoviridae core genes are sequences shared by all known members of this viral family. This situation and the similarity among them suggest that these genome regions were present in the common ancestor. As is known, sequences differentially accumulate mutations according to the functional and structural domains of the protein encoded. Therefore, it is possible that certain regions of a gene diverge more than others, thus complicating overall similarity studies. For this reason, the main strategy for the detection of orthologous sequences should be based on local similarity approaches. In view of that, this was the focus of the work described here. Thus, the unbiased algorithm designed and implemented proved to be very efficient, allowing the detection of 94% of previously described core genes (31 of 33 genes) and the identification of 4 other encoding sequences (ac53, ac78, ac101, and ac103) never postulated in this category. In contrast, the ac93 and ac100 genes could not be detected.
An intriguing case is Ac53 and its orthologs. This sequence was the most remote of the newly discovered core proteins. In the search for potential biological functions, it was described previously that Ac53 plays a role in nucleocapsid assembly, an essential step for BV production (31). The finding of a putative U-box/RING-like domain (E3 ubiquitin ligase) supports such a role.
The ubiquitin-proteasome pathway is a well-known system in metazoans which is used for posttranslational targeting and degradation of proteins. It is remarkable that the E3 ubiquitin ligase is the third component of this system, containing a zinc binding motif (RING domain) essential for its function. This protein catalyzes the ligation of ubiquitin to the targets doomed for degradation by proteasomes. Programmed cell death is one of the numerous cellular processes in which ubiquitin posttranslational protein modification is involved. For instance, caspases and other proapoptotic proteins are substrates for this system (34). As a consequence, the U-box/RING-like domain of Ac53 and its orthologs might ensure efficient virus production through the inhibition of apoptosis. Similar functions were described previously for White spot syndrome virus and Spodoptera frugiperda Ascovirus 1a (4, 18).
ac93 and ac94 were the last two reported genes postulated to be shared by all baculoviruses (57). In this work, ac94 was detected as a core gene, adding support to such previous postulations. In contrast, ac93 was not detected and was considered a false negative. In view of this and reviewing the literature, it is possible that sequences similar to the sequence of this gene were previously found in alphabaculoviruses, betabaculoviruses, and gammabaculoviruses but were never identified in a deltabaculovirus until the postulation of cuni13 as the corresponding ortholog (57). The main basis of this assumption was the conservation of gene order, a biological feature not involved in the algorithm applied here but used to support the results.
The identification of ac93 orthologs in alpha- and betabaculoviruses is easy because BLASTP searches readily generated significant E values. In contrast, a more difficult task was the finding of orthologous sequences in gammabaculoviruses. For example, the BLASTP results (query, 54 alpha- and betabaculovirus orthologs of the Ac93 protein) were as follows: Neab9 was found 27 times (E values ranging from 8.2 to 1e−4; coverage of 59.4%), Nele17 was traced 28 times (E values ranging from 8.2 to 6e−6; coverage of 68.2%), and Nese25 was detected 40 times (E values ranging from 0.19 to 5e−7; coverage of 48.2%). Otherwise, all possible BLASTP searches produced a match for the deltabaculovirus only by using the Ro90 protein of RoMNPV as a query (E value of 2.2 and coverage of 22.9%). The above-described results were obtained by using a small database (9,273 letters) containing all putative orthologs derived from work reported previously by Yuan et al. (57). In contrast to this, the application of the algorithm described here did not detect Cuni13 using the Global Proteome Database, which contains all the annotated proteins in the 58 baculovirus genomes (2,316,379 letters), even when the less stringent threshold combination was used.
Despite the results described above, the conservation of gene order moderately supports the idea of orthology. Two additional studies, global pairwise alignments and Z-score analysis, were performed to support this assertion. Thus, the pairwise global alignments gave amino acid identity values from 8.8% (HearGV and PsunGV) to 16.7% (PlxyMNPV, RoMNPV, Spodoptera frugiperda MNPV-3AP2 [SfMNPV-3AP2], and SfMNVP-19) and similarities ranging from 41.4% (Choristoneura occidentalis GV [ChocGV] and CpGV) to 51.9% (AcMNPV, PlxyMNPV, RoMNPV, and SfMNPV-3AP2). In addition, the Z-score analysis exposed values showing a narrow distribution that was similar to the results obtained for the other newly described core proteins. All of these considerations suggest that ac93 and its orthologs accumulated mutations at a higher rate than the other core genes, consequently affecting the detection of similarity among them. Clearly, this is a notorious example of remote orthology, an extreme situation that requires manual inspection and the application of combined criteria for its recognition as a core gene.
ac100, known mostly as p6.9, was the other core gene not detected. P6.9 is a small basic protein with a great variation in size, from 48 to 112 residues depending on the species considered (52). Moreover, there are some baculoviral genomes deposited in the GenBank database that do not have a p6.9 annotation, as occurs for Clanis bilineata NPV, Spodoptera litura GV, and Neodiprion abietis NPV (35). However, a simple tBlastN search allowed the finding of the corresponding putative ORFs in ClbiNPV and SpliGV. Meanwhile, the tBlastN results for NeabNPV (query, P6.9 of Neodiprion lecontei NPV) required manual inspection to detect sequences with acceptable similarity values. In fact, two nearby sequences were identified by this approach but in different reading frames, making the automatic detection of orthology difficult.
Mention should also be made of the marked bias in amino acid composition. P6.9 sequence analysis revealed that 58 to 91% of all residues were represented by only three amino acids, arginine, serine, and glycine, in order of abundance. Besides, arginines and serines are distributed into clusters rich in the sequence RS. This particular composition also occurs in the middle region of most phosphoenolpyruvate (PEP)/PP34 proteins. As a consequence, the automated PSI Blast-based local search applied in this work generated a mixed result containing P6.9 sequences and fragments of PEP/PP34 or other proteins with the same peculiarity. This situation was dramatically enhanced in the subsequent iteration steps, producing a mixed result list containing more than 58 hits, which was rejected by the algorithm.
Concluding remarks.
The identification of orthologous sequences is a relevant topic in genetics. An understanding of genomic information can reveal biological roles of proteins. In this framework, baculoviruses are biological entities that are very useful in different aspects of life sciences, and their complete characterization should include the identification of ancestral, still functional genes. Viruses with large dsDNA genomes undergo evolution by structural mutations (horizontal gene transfers, deletions, and inversions) and by the accumulation of point mutations. The first process alters gene content, while the second leads to specialization in gene function. In view of this, the identification of genes shared by all the known members of the family Baculoviridae is a difficult task because some ORFs are not annotated in the corresponding gene files, because experimental viral proteomes are not completely known, and because a high level of sequence divergence and different gene contents may occur. In this work, a routine based on Multi PSI-Blast/tBlastN and Multi HaMStR was developed and carried out. Thus, ac53, ac78, ac101, and ac103 should be considered a new set of baculovirus orthologs because similar ORFs exist in all the genomes sequenced. Therefore, a total of 37 genes appear to be shared by all known members of this family. Furthermore, the strategy implemented here can be postulated to be a useful tool for the analysis of new genomic sequences for the Baculoviridae, for other viruses with large DNA genomes, or for other organisms.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by research funds from the Agencia Nacional de Promoción Científica y Técnica (ANPCyT) and the Universidad Nacional de Quilmes, Argentina. P.D.G. and M.N.B. are members of the research career of CONICET (Consejo Nacional de Investigaciones Científicas y Técnicas); M.J.G. and S.A.B.M. hold doctoral fellowships with CONICET, Argentina.
We acknowledge Andrés Romanowski for reading and discussing the manuscript.
Footnotes
Published ahead of print 29 August 2012
Supplemental material for this article may be found a http://jvi.asm.org/.
REFERENCES
- 1. Acharya A, Gopinathan K. 2002. Transcriptional analysis and preliminary characterization of ORF Bm42 from Bombyx mori nucleopolyhedrovirus. Virology 299:213–224 [DOI] [PubMed] [Google Scholar]
- 2. Altschul SF, et al. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bailey TL, Elkan C. 1994. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 2:28–36 [PubMed] [Google Scholar]
- 4. Bideshi DK, et al. 2006. Genomic sequence of Spodoptera frugiperda Ascovirus 1a, an enveloped, double-stranded DNA insect virus that manipulates apoptosis for viral reproduction. J. Virol. 80:11791–11805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Blissard GW, Rohrmann GF. 1990. Baculovirus diversity and molecular biology. Annu. Rev. Entomol. 35:127–155 [DOI] [PubMed] [Google Scholar]
- 6. Cohen DPA, Marek M, Davies BG, Vlak JM, van Oers MM. 2009. Encyclopedia of Autographa californica nucleopolyhedrovirus genes. Virol. Sin. 24:359–414 [Google Scholar]
- 7. Condreay JP, Kost TA. 2007. Baculovirus expression vectors for insect and mammalian cells. Curr. Drug Targets 8:1126–1131 [DOI] [PubMed] [Google Scholar]
- 8. Dessimoz C, et al. 2005. OMA, a comprehensive, automated project for the identification of orthologs from complete genome data: introduction and first achievements, p 61–72 In McLysath A, Huson DH. (ed), RECOMB 2005 Workshop on Comparative Genomics. LNBI 3678 of lecture notes in bioinformatics. Springer-Verlag, New York, NY [Google Scholar]
- 9. Di Tommaso P, et al. 2011. T-Coffee: a Web server for the multiple sequence alignment of protein and RNA sequences using structural information and homology extension. Nucleic Acids Res. 39:W13–W17 doi:10.1093/nar/gkr245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ebersberger I, Strauss S, von Haeseler A. 2009. HaMStR: profile hidden Markov model based search for orthologs in ESTs. BMC Evol. Biol. 9:157 doi:10.1186/1471-2148-9-157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Finn RD, Clements J, Eddy SR. 2011. HMMER Web server: interactive sequence similarity searching. Nucleic Acids Res. 39:W29–W37 doi:10.1093/nar/gkr367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Fitch WM. 1970. Distinguishing homologous from analogous proteins. Syst. Zool. 19:99–113 [PubMed] [Google Scholar]
- 13. Flicek P, et al. 2008. Ensembl 2008. Nucleic Acids Res. 36:D707–D714 doi:10.1093/nar/qKm988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gabaldón T. 2007. Evolution of proteins and proteomes, a phylogenetics approach. Evol. Bioinform. Online 24:51–56 [PMC free article] [PubMed] [Google Scholar]
- 15. Gabaldón T, Huynen MA. 2004. Prediction of protein function and pathways in the genome era. Cell. Mol. Life Sci. 61:930–944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Garcia-Maruniak A, et al. 2004. Sequence analysis of the genome of the Neodiprion sertifer nucleopolyhedrovirus. J. Virol. 78:7036–7051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hayakawa T, Rohrmann GF, Hashimoto Y. 2000. Patterns of genome organization and content in lepidopteran baculoviruses. Virology 278:1–12 [DOI] [PubMed] [Google Scholar]
- 18. He F, Fenner BJ, Godwin AK, Kwang J. 2006. White spot syndrome virus open reading frame 222 encodes a viral E3 ubiquitin ligase and mediates degradation of host tumor suppressor via ubiquitination. J. Virol. 80:3884–3892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Herniou EA, et al. 2001. Use of whole genome sequence data to infer baculovirus phylogeny. J. Virol. 75:8117–8126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Herniou EA, Olszewski JA, Cory JS, O'Reilly DR. 2003. The genome sequence and evolution of baculoviruses. Annu. Rev. Entomol. 48:211–234 [DOI] [PubMed] [Google Scholar]
- 21. Inceoglu AB, Kamita SG, Hammock BD. 2006. Genetically modified baculoviruses: a historical overview and future outlook. Adv. Virus Res. 68:323–360 [DOI] [PubMed] [Google Scholar]
- 22. Inceoglu AB, et al. 2001. Recombinant baculoviruses for insect control. Pest Manag. Sci. 57:981–987 [DOI] [PubMed] [Google Scholar]
- 23. Jackes RP. 1985. Stability of insect viruses in the environment, p 289–360 In Maromorosch E, Sherman K. (ed), Viral insecticides for biological control. Academic Press Inc, Orlando, FL [Google Scholar]
- 24. Jehle JA, et al. 2006. On the classification and nomenclature of baculoviruses: a proposal for revision. Arch. Virol. 151:1257–1266 [DOI] [PubMed] [Google Scholar]
- 25. Jehle JA, et al. 2006. Molecular identification and phylogenetic analysis of baculoviruses from Lepidoptera. Virology 346:180–193 [DOI] [PubMed] [Google Scholar]
- 26. Jensen LJ, et al. 2008. eggNOG: automated construction and annotation of orthologous groups of genes. Nucleic Acids Res. 36:D250–D254 doi:10.1371/Journal.pcbi.1000262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kemena C, Notredame C. 2009. Upcoming challenges for multiple sequence alignment methods in the high-throughput era. Bioinformatics 25:2455–2465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kost TA, Condreay JP. 1999. Recombinant baculoviruses as expression vectors for insect and mammalian cells. Curr. Opin. Biotechnol. 10:428–433 [DOI] [PubMed] [Google Scholar]
- 29. Kost TA, Condreay JP, Jarvis DL. 2005. Baculovirus as versatile vectors for protein expression in insect and mammalian cells. Nat. Biotechnol. 23:567–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kozlov EA, Levitina TL, Gusak NM. 1986. The primary structure of baculovirus inclusion body proteins. Evolution and structure-function aspects. Curr. Top. Microbiol. Immunol. 131:135–164 [DOI] [PubMed] [Google Scholar]
- 31. Krogh A, Larsson B, von Heijne G, Sonnhammer EL. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305:567–580 [DOI] [PubMed] [Google Scholar]
- 32. Li L, Stoeckert CJ, Jr, Roos DS. 2003. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 13:2178–2189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Liu C, et al. 2008. Autographa californica multiple nucleopolyhedrovirus ac53 plays a role in nucleocapsid assembly. Virology 382:59–68 [DOI] [PubMed] [Google Scholar]
- 34. Mace P, Shirley S, Day C. 2009. A sting in the tail: ubiquitin ligase function of inhibitor of apoptosis proteins. Aust. Biochemist 40:12–15 [Google Scholar]
- 35. Miele SAB, Garavaglia MJ, Belaich MN, Ghiringhelli PD. 2011. Baculovirus: molecular insights on their diversity and conservation. Int. J. Evol. Biol. 211:379424 doi:10.4061/2011/379424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Moreira D, Philippe H. 2000. Molecular phylogeny: pitfalls and progress. Int. Microbiol. 3:9–16 [PubMed] [Google Scholar]
- 37. Moscardi F. 1999. Assessment of the application of baculoviruses for control of lepidoptera. Annu. Rev. Entomol. 44:257–289 [DOI] [PubMed] [Google Scholar]
- 38. Notebaart RA, Huynen MA, Teusink B, Siezen RJ, Snel B. 2005. Correlation between sequence conservation and the genomic context after gene duplication. Nucleic Acids Res. 33:6164–6171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. O'Brien KP, Remm M, Sonnhammer EL. 2005. Inparanoid: a comprehensive database of eukaryotic orthologs. Nucleic Acids Res. 33:D476–D480 doi:10.1093/nar/gKi107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ohkawa T, Washburn JO, Sitapara R, Sid E, Volkman LE. 2005. Specific binding of Autographa californica M nucleopolyhedrovirus occlusion-derived virus to midgut cells of Heliothis virescens larvae is mediated by products of pif genes Ac119 and Ac022 but not by Ac115. J. Virol. 79:15258–15264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Perera O, Green TB, Stevens SM, Jr, White S, Becnel JJ. 2007. Proteins associated with Culex nigripalpus nucleopolyhedrovirus occluded virions. J. Virol. 81:4585–4590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Petersen TN, Brunak S, von Heijne G, Nielsen H. 2011. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods 8:785–786 [DOI] [PubMed] [Google Scholar]
- 43. Possee RD. 1997. Baculoviruses as expression vectors. Curr. Opin. Biotechnol. 8:569–572 [DOI] [PubMed] [Google Scholar]
- 44. Rohrmann GF. 2011. Baculovirus molecular biology, 2nd ed NCBI, Bethesda, MD [Google Scholar]
- 45. Rohrmann GF. 1992. Baculovirus structural proteins. J. Gen. Virol. 73:749–761 [DOI] [PubMed] [Google Scholar]
- 46. Schwartz RM, Dayhoff MO. 1979. Matrices for detecting distant relationships, p 353–358 In Dayhoff MO. (ed), Atlas of protein sequence and structure vol 5, suppl 3 National Biomedical Research Foundation, Washington, DC [Google Scholar]
- 47. Shi X, Jarvis DL. 2007. Protein N-glycosylation in the baculovirus-insect cell system. Curr. Drug Targets 8:1116–1125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Söding J. 2005. Protein homology detection by HMM-HMM comparison. Bioinformatics 21:951–960 [DOI] [PubMed] [Google Scholar]
- 49. Summers MD. 2006. Milestones leading to the genetic engineering of baculoviruses as expression vector systems and viral pesticides. Adv. Virus Res. 68:3–73 [DOI] [PubMed] [Google Scholar]
- 50. Sun XL, Peng HY. 2007. Recent advances in biological control of pest insect by using viruses in China. Virol. Sin. 22:158–162 [Google Scholar]
- 51. Vanarsdall AL, Pearson MN, Rohrmann GF. 2007. Characterization of baculovirus constructs lacking either the Ac101, Ac142, or the Ac144 open reading frame. Virology 367:187–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wang M, et al. 2010. Specificity of baculovirus P6.9 basic DNA-binding proteins and critical role of the C terminus in virion formation. J. Virol. 84:8821–8828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Wang R, et al. 2010. Proteomics of the Autographa californica nucleopolyhedrovirus budded virions. J. Virol. 84:7233–7242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Wang Y, et al. 2008. Autographa californica multiple nucleopolyhedrovirus nucleocapsid protein BV/ODV-C42 mediates the nuclear entry of P78/83. J. Virol. 82:4554–4561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Williams GV, Faulkner P. 1997. Cytological changes and viral morphogenesis during baculovirus infection, p 61–107 In Miller LK. (ed), The baculoviruses. Plenum Press Inc, New York, NY [Google Scholar]
- 56. Wu CH, et al. 2003. The Protein Information Resource. Nucleic Acids Res. 31:345–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Yuan M, et al. 2011. Identification of Autographa californica nucleopolyhedrovirus ac93 as a core gene and its requirement for intranuclear microvesicle formation and nuclear egress of nucleocapsids. J. Virol. 85:11664–11674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Yuan M, et al. 2008. A highly conserved baculovirus gene p48 (ac103) is essential for BV production and ODV envelopment. Virology 379:87–96 [DOI] [PubMed] [Google Scholar]
- 59. Zhang G. 1994. Research, development and application of Heliothis viral pesticide in China. Resourc. Environ. Yangtze Valley 3:1–6 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.