Abstract
The mechanisms of astrovirus pathogenesis are largely unknown, in part due to a lack of a small-animal model of disease. Using shotgun sequencing and a custom analysis pipeline, we identified two novel astroviruses capable of infecting research mice, murine astrovirus (MuAstV) STL1 and STL2. Subsequent analysis revealed the presence of at least two additional viruses (MuAstV STL3 and STL4), suggestive of a diverse population of murine astroviruses in research mice. Complete genomic characterization and subsequent phylogenetic analysis showed that MuAstV STL1 to STL4 are members of the mamastrovirus genus and are likely members of a new mamastrovirus genogroup. Using Rag1−/− mice deficient in B and T cells, we demonstrate that adaptive immunity is required to control MuAstV infection. Furthermore, using Stat1−/− mice deficient in innate signaling, we demonstrate a role for the innate immune response in the control of MuAstV replication. Our results demonstrate that MuAstV STL permits the study of the mechanisms of astrovirus infection and host-pathogen interactions in a genetically manipulable small-animal model. Finally, we detected MuAstV in commercially available mice, suggesting that these viruses may be present in academic and commercial research mouse facilities, with possible implications for interpretation of data generated in current mouse models of disease.
INTRODUCTION
Recent studies in mouse models have highlighted complex interactions between viruses, genetic susceptibility, and disease (reviewed in reference 59). To characterize the enteric virome of specific-pathogen-free (SPF)-housed research mice in an unbiased manner, we used shotgun sequencing, which revealed the presence of multiple novel astroviruses. Astroviruses are nonenveloped, positive-sense, polyadenylated RNA viruses often associated with gastrointestinal (GI) disease (11, 44). Human astrovirus 1 (HAstV1) to HAstV8 are an important cause of gastroenteritis in pediatric inpatients and outpatients, HIV-infected and immunocompromised populations, and the elderly (9, 10, 12, 16, 21, 22, 24, 28, 39, 44, 48, 53, 63). In addition, human astroviruses have been associated with sporadic outbreaks of gastroenteritis in immunocompetent adults (4, 44, 47). As a viral agent of pediatric gastroenteritis, HAstV is reportedly second only to rotavirus (12, 44), with a seroprevalence of neutralizing antibodies to HAstV1 nearing 90% by the age of 9 (35). To date, astroviruses have been isolated from a number of hosts, including wild and domestic animals, marine mammals, and birds, and new astrovirus-susceptible hosts continue to be identified (11, 34, 42, 49, 52).
Astrovirus-related disease is not limited only to the GI tract. Astroviruses are implicated as the cause of hepatitis in ducks and encephalomyelitis in minks (5, 18, 20). In humans, an astrovirus was identified in the central nervous system of an immunocompromised child with X-linked agammaglobulinemia who died of encephalitis (51). Furthermore, we previously reported the presence of human astrovirus MLB2 in the plasma of a febrile child (27). Despite the prevalence of human astrovirus infection and the potential for extraintestinal disease, there are no specific treatment protocols for astrovirus infection, and no vaccine exists. In addition, little is known about the molecular mechanisms of astrovirus infection, replication, disease pathogenesis, or immunity.
The family Astroviridae is divided into the mamastrovirus and avastrovirus genera, which are characterized by the ability to infect mammals and avian species, respectively (44). The mamastrovirus genus is further subdivided into two genogroups based on the complete amino acid sequence of open reading frame 2 (ORF2) and distinguishes genogroups I and II based on a mean amino acid genetic distance (p-dist) of 0.671 ± 0.016 between the groups, with intragenogroup distances ranging from 0.338 to 0.783 (31).
Across both mamastrovirus and avastrovirus genera, the astrovirus genome ranges from 6.1 to 7.7 kb in length, not including the 3′ polyadenylated tail, and contains three ORFs as well as 5′ and 3′ untranslated regions (UTRs) (44). ORF1a encodes a polypeptide of 920 to 935 amino acids (aa) containing several conserved motifs, including a serine protease (11, 44). A highly conserved heptanucleotide motif and downstream hairpin structure at the ORF1a-ORF1b junction generates a −1 frameshift, leading to the translation of an ORF1a-ORF1b polypeptide which is later cleaved into polypeptides corresponding to ORF1a and ORF1b (11, 29, 41, 44). ORF1b encodes a polypeptide of approximately 515 to 528 aa, containing the RNA-dependent RNA polymerase (RdRP) (40, 44). ORF2 encodes a polypeptide consisting of 672 to 816 aa which contains the viral capsid (11, 40, 44).
Human case studies have highlighted the importance of the adaptive immune system in the control of mamastrovirus disease (51, 63). The turkey model of avastrovirus pathogenesis has been used to characterize astrovirus disease and immune response (32, 33) but lacks the ability to easily dissect host determinants of astrovirus infection and immunology. Furthermore, while mamastrovirus tissue culture systems do exist (23, 38), the ability to elucidate molecular mechanisms that control mamastrovirus infection and pathogenesis has been hampered by a lack of a small-animal model.
In this paper, we report the first complete genome sequences of astroviruses in research mice, murine astrovirus (MuAstV) STL1 and STL2. Subsequent analysis revealed the presence of at least two additional MuAstV STL viruses, suggestive of a diverse population of related astroviruses. Phylogenetic analysis of ORF2 indicated that the MuAstV STL viruses group with several other newly described astroviruses in a genetic cluster distinct from mamastrovirus genogroups I and II. MuAstV STL is capable of infecting laboratory mice, which represent a genetically manipulable small-animal model in which to study astrovirus infection. Initial experiments using this model of mamastrovirus infection revealed a role for both innate and adaptive immunity in the control of astrovirus replication. Finally, we show that MuAstV is present in commercially available mice, with potential implications for existing disease models.
MATERIALS AND METHODS
Nucleic acid preparation and 454 sequencing.
Shotgun sequencing was performed as follows: stool samples were collected from C57BL/6J mice and resuspended in 6 volumes of phosphate-buffered saline (PBS) (14). The sample was centrifuged to pellet particulate matter, and the supernatant fluids were then passed through a 0.45-μm-pore-size filter. Total nucleic acid was isolated from 200 μl primary stool filtrate using an Ampliprep DNA extraction machine (Roche) according to the manufacturer's instructions. To enable subsequent detection of both RNA and DNA viruses, total nucleic acid from each sample was reverse transcribed and amplified as previously described (61). Briefly, RNA templates were reverse transcribed using primerA containing a 16-nucleotide (nt) specific sequence followed by 9 random nucleotides for random priming. The 16-nt specific sequence is unique for each sample and served as a barcode in assigning sequencing reads to a sample. Sequenase (United States Biochemical) was used for second-strand cDNA synthesis and for random-primed amplification of DNA templates using PrimerA. Each sample was then subjected to 40 cycles of PCR amplification using PrimerB containing the same 16-nt specific sequence as in the corresponding PrimerA. Amplification products were pooled, adaptor ligated, and sequenced on the 454 GS-FLX Titanium platform (454 Life Sciences).
Detection and analysis of viral sequences using VirusHunter software.
Sequences were analyzed using VirusHunter, a customized pipeline, as described previously (64). Briefly, sequence reads were assigned to samples based on the unique barcode sequences (i.e., PrimerB sequences). For further analysis, primer sequences were trimmed off and the sequence reads were clustered using CD-HIT (43) to identify redundant reads. Sequences were clustered on the basis of 95% identity over 95% sequence length, and the longest sequence from each cluster was picked as the representative sequence. Then, sequences were masked by RepeatMasker (http://www.repeatmasker.org). If a sequence did not contain a stretch of at least 50 consecutive non-“N” nucleotides or if greater than 40% of the total length of the sequence was masked, it was removed from further analysis (i.e., “filtered”). Good-quality sequences after filtering were sequentially compared against (i) the human genome using BLASTn; (ii) the GenBank nt database using BLASTn; or (iii) the GenBank nr database using BLASTx (2). Minimal e-value cutoffs of 1e−10 and 1e−5 were applied for BLASTn and BLASTx, respectively. Sequences were phylotyped as human, mouse, fungal, bacterial, phage, viral, or other based on the identity of the top BLAST hit. Sequences without any significant hit to any of the databases were placed in the “unassigned” category. If a sequence aligned to both a virus and a member of another kingdom (e.g., a bacterium or fungus) with the same e value, it was classified as “ambiguous.” All eukaryotic viral sequences were further classified into viral families based on the taxonomy identification of the best hit.
All viral sequences and unassigned sequences from each sample were assembled into contigs using Newbler (454 Life Sciences) with default parameters. If a sample was sequenced multiple times, all available sequencing data were used to try to obtain the best assembly.
Genome amplification and confirmatory sequencing.
Total RNA was extracted from the murine stool samples previously used for shotgun sequencing with an RNeasy minikit (Qiagen, Valencia, CA) to generate complete genomic sequences. To generate partial genome sequences corresponding to ORF2, total RNA was extracted from murine stool samples harvested from immunocompetent mice housed in different SPF facilities at Washington University. One microgram of RNA was used as the template for rapid amplification of cDNA ends (RACE) reactions to generate the 5′ and 3′ genome ends with 5′ RACE and 3′ RACE kits (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. To generate full genomic sequences, one microgram of RNA was used as the template for cDNA synthesis using a SuperScript III first-strand synthesis kit and an oligo(dT)12–20 primer (Invitrogen) according to the manufacturer's instructions. Sequences were then amplified using Elongase enzyme mix (Invitrogen) and primers 5′-CCAAGAAAGAGGCACTAGTGGCACTC-3′ and 5′-GTTTTTTTTTTTTTTTTTTTTTGCCAATTTTTATGCCAATTATATCACCC-3′. Partial genome sequences corresponding to ORF2 were amplified using a 3′RACE kit and gene-specific primer 5′-CTTTGGAGGGGHGGACCAA-3′. The resulting PCR products were gel purified and ligated into a pCR4-TOPO TA sequencing vector (Invitrogen). Universal M13 forward and reverse primers were used for sequencing, and primer walking was applied as needed. Four clones were used to construct consensus sequence MuAstV STL1 with 2-to-4-fold redundancy using Geneious Pro v5.0.4. Three clones were used to construct the MuAstV STL2 consensus sequence with 2-to-4-fold redundancy. MuAstV STL3 and STL4 consensus sequences were each generated from three clones with 2-to-4-fold redundancy. Predicted ORFs were identified using Geneious. All protein motifs were predicted using Pfam (17).
Phylogenetic analysis.
Sequences from astrovirus genomic segments encoding ORF1a, -1b, and -2, available in GenBank as of 26 July 2012, were translated. These sequences were aligned using ClustalX 2.0.12 (37). Phylogenetic inference was performed with maximum parsimony using PAUP 4b10 (57) and maximum likelihood using RAxML (55) and a BLOSUM62 transition matrix with 1,000 bootstrap replicates. The resulting phylogenetic trees were visualized using FigTree 1.3.1 (http://tree.bio.ed.ac.uk/software/figtree). MEGA 5.05 was used for distance estimation (uncorrected p-dist) (58).
Mice.
C57BL/6J, B6.Rag1−/− (45), and Stat1−/− (13) mice were bred and housed in an enhanced-barrier SPF facility at Washington University in St. Louis in compliance with federal and institutional guidelines (8). B6.Rag1−/− mice were also purchased from The Jackson Laboratory (Bar Harbor, ME). All studies were performed using age-matched female mice between 8 and 12 weeks of age.
For MuAstV STL detection in commercially available mice, B6.Rag1−/− mice (B6.129S7-Rag1tm1Mom/J; catalog no. 002216) (45) were ordered from The Jackson Laboratory facility AX30, Rag2−/− mice (129S6/SvEvTac-Rag2tm1Fwa; catalog no. RAG2-F) (54) were ordered from Taconic facility IBU25, and Fox Chase SCID mice (CB17/Icr-Prkdcscid/IcrCrl; catalog no. 236) (6) were ordered from Charles River facility W09. Upon arrival, mice were maintained in their commercial transport container until they were sacrificed for sample collection on the same day.
Measurement of viral RNA by TaqMan qPCR.
Total RNA was extracted from individual stool pellets using an RNeasy minikit (Qiagen) or from tissue samples using TRIzol reagent (Invitrogen). One-tenth of the total stool RNA or 1 μg of tissue RNA was reverse transcribed using ImpromII reverse transcriptase (RT) (Promega, Madison, WI) and random primers (Invitrogen) to yield cDNA. Triplicate quantitative PCRs (qPCR) were performed using 1/10 of the cDNA, primers specific to an 80-nt region of ORF1b (sense, 5′-TACATCGAGCGGGTGGTCGC-3′; antisense, 5′-GTGTCACTAACGCGCACCTTTTCA-3′), and a TaqMan probe (Applied Biosystems, Foster City, CA) with the sequence 5′-TTTGGCATGTGGGTTAA-3′ containing a 5′ 6-carboxyfluorescein (FAM) dye label, 3′ nonfluorescent quencher (NFQ), and minor groove binder (MGB). The number of genome copies per sample was determined by comparison to a standard curve (generated by a 10-fold dilution of target-containing plasmid in yeast tRNA [Invitrogen]). For stool samples, the number of genome copies per sample was multiplied by 100 to account for dilution from total RNA originally extracted from the stool pellet and is reported as the number of genome copies per stool pellet.
Nucleotide sequence accession numbers.
The sequences for MuAstV STL1, STL2, STL3, and STL4 have been deposited in GenBank and have been assigned accession numbers JX544743 to JX544746.
RESULTS
Detection of novel astroviruses using shotgun sequencing.
To examine the mouse virome in an unbiased manner, we generated fecal RNA and DNA libraries from three immunocompetent wild-type mice, shotgun sequenced them, and used VirusHunter (14–16, 25, 26, 50, 64) to analyze the resulting reads. We identified 132 sequences (76, 21, and 35 from mice A, B, and C, respectively) with significant similarity to known astrovirus sequences. No other viral reads were identified. The viral reads, as well as reads with no detectable homology to sequences in public databases, detected in the feces of mouse A were used to assemble a 6,748-nucleotide (nt) contig with 9-fold coverage. A BLASTx (2) search of the NCBI nr database identified this contig to be a highly divergent astrovirus with at most 60% identity to a 205-aa region of the most closely related virus, HAstV-6 isolate Katano (Fig. 1A). Assembly of reads from the feces of mouse B generated three contigs ranging from 1,024 to 2,463 nt with 89.1% to 95.2% identity to mouse A contig 1 (Fig. 1B). Assembly of reads detected in the feces of mouse C generated four contigs ranging from 288 to 3,095 nt with 99.0% to 99.7% nt identity to the 6,748-nt contig from mouse A (data not shown). These data suggested the presence of at least two novel astroviruses in our research mouse facility.
Fig 1.
Identification of novel astrovirus sequences. (A) Comparison between the longest contig assembled from mouse A and HAstV6 isolate Katano (HM237363), the most closely related virus. *, percent amino acid identity and length of the best aligned homologous region compared to the reference genome sequence. (B) Comparison between the longest contig assembled from mouse A and contigs assembled from mouse B. **, percent nucleotide identity across the indicated contig.
Generation and sequencing of complete and partial murine astrovirus genomes.
Subsequent analysis of the fecal specimens from mouse A and mouse B utilizing RACE reactions and traditional Sanger sequencing generated the complete consensus genomes of the two mouse astroviruses MuAstV STL1 and STL2 (Fig. 2). Excluding the poly(A) tail, MuAstV STL1 and STL2 were 6,817 and 6,806 nt long, respectively, and shared 93.4% nt identity across their genomes.
Fig 2.
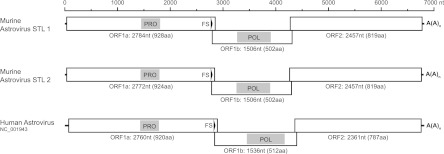
Schematic of the MuAstV STL genome. Predicted open reading frames (ORFs) are represented by boxes and the 5′ and 3′ UTRs are represented by a black line. The genome coordinates are noted at the top. Gray boxes depict predicted domains based on the corresponding amino acid sequence. The black arrowheads indicate the conserved heptanucleotide frameshift motif (AAAAAAC). Human astrovirus 1 is depicted for comparison. PRO, trypsin-like peptidase domain; FS, frameshift signal; POL, RdRP; A(A)n, 3′ poly(A) tail; nt, nucleotides.
To begin to assess MuAstV diversity, we collected fecal samples from two nonobese diabetic mice, housed in two different research mouse facilities at Washington University. A primer corresponding to the highly conserved region upstream of ORF2 was used in a 3′ RACE reaction to generate ORF2 consensus sequences from these samples, here called MuAstV STL3 and STL4. MuAstV STL3 and STL4 showed 95.3% nt identity to each other and 89.6% to 91.4% nt identity to MuAstV STL1 and STL2. These data demonstrate the presence of at least four novel astroviruses in research mouse facilities at Washington University.
The genome organization of MuAstV STL is consistent with those of other mamastroviruses.
MuAstV STL1 and STL2 were predicted to contain a 5′ UTR, three ORFs (ORF1a, -1b, and -2), a 3′ UTR, and a poly(A) tail. The 5′ UTR was determined to be 46 nt in length. The 3′ UTRs of MuAstV STL1 and STL2 were determined to be 90 and 91 nt in length, respectively. ORF1a was predicted to encode a 928 (MuAstV STL1)- or 924 (MuAstV STL2)-aa protein, with each containing a trypsin-like peptidase domain as revealed by Pfam analysis (17) and showing significant similarity (minimal e-value cutoff of 1e−5) to those of known astroviruses by BLASTp (2). The differences in four amino acids occurred downstream of the putative protease motif. The 58-nt ORF1a-ORF1b junction of MuAstV STL1 and STL2 contained the heptanucleotide frameshift signal (AAAAAAC) conserved in all astroviruses (11, 29, 41, 44). Furthermore, FSFinder analysis (46) confirmed that the downstream sequence may form a stem-loop structure required for a −1 ribosomal frameshift to lead to ORF1a-ORF1b translation (7, 19, 44). The first amino acid in frame with the frameshift signal was predicted to be the start position for ORF1b of MuAstV STL1 and STL2. ORF1b of MuAstV STL1 and STL2 is predicted to encode a 502-aa protein containing an RdRP domain, consistent with other members of the Astroviridae family (11, 44).
MuAstV STL contained a sequence that is highly conserved among mammalian astroviruses upstream of ORF2, C UUU GGA GGG GAG GAC CAA AAG CUC UUC AUG GGC, which encompasses the ORF2 start codon (in bold) and is suggested to be a promoter for subgenomic RNA synthesis (44, 60). As in most other mamastroviruses (11), MuAstV STL had an 8-nt region of overlap at the end of ORF1b and beginning of ORF2, with ORF2 maintained in the same frame as ORF1a. ORF2 of MuAstV STL1 and STL2 was predicted to encode an 819-aa protein containing the structural capsid protein. Collectively, these data demonstrate that the genome organization of MuAstV STL1 and STL2 is consistent with those of other members of the Astroviridae family and of the mamastrovirus genus in particular.
MuAstV STL viruses are members of a putative new mamastrovirus genogroup.
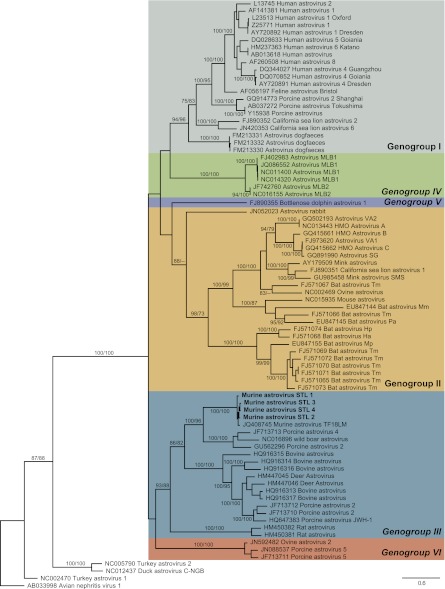
To evaluate the relationship between MuAstV STL1 to STL4 and other known astroviruses, we performed phylogenetic analysis using translated complete amino acid sequences from ORF2 of 72 mamastroviruses (Fig. 3). Phylogenetic analyses of MuAstV STL1 and STL2 performed for ORF1a and -1b showed similar relationships between these novel astroviruses and known astroviruses (data not shown). In all analyses, the MuAstV STL viruses clustered with the mamastrovirus genus and not the avastrovirus genus, demonstrating that the MuAstV STL viruses are mamastroviruses.
Fig 3.
MuAstV STL is a member of a putative third genogroup of mamastroviruses. Genomic sequences corresponding to ORF2 were translated and aligned. Phylogenetic analyses were performed using maximum-parsimony (MP) and maximum-likelihood (ML) methods with 1,000 bootstrap replicates. The bootstrap support (ML/MP) is indicated adjacent to each branch of the best-scoring ML tree where significant. Corresponding sequences from representative avastroviruses were included for comparison. GenBank accession numbers immediately precede each sequence name.
The MuAstV STL viruses were most closely related to several recently characterized porcine and wild boar astroviruses but shared only 33% to 36% amino acid identity in the capsid region and 63% to 67% amino acid identity in the RdRP region. This clade also included an astrovirus sequence found in laboratory mice in Cincinnati, OH, and recently uploaded to GenBank (murine astrovirus strain TF18LM) but was highly divergent from an astrovirus sequence previously detected in a wild house mouse (Mus musculus; mouse astrovirus M-52/USA/2008) (49), indicating that the potential population of murine astroviruses is quite diverse.
In analyses of ORF2, the MuAstV STL viruses, along with porcine and wild boar astroviruses, formed a distinct clade with p-dist values of 0.762 ± 0.010 and 0.789 ± 0.010 with respect to mamastrovirus genogroups I and II, respectively. Intragroup p-dist values were 0.548 ± 0.010, 0.629 ± 0.011, and 0.641 ± 0.009 for mamastrovirus genogroups I and II and the MuAstV STL-containing clade, respectively. These p-dist values suggested that the MuAstV STL viruses may be members of a new genogroup within the mamastrovirus genus.
A sensitive and robust qRT-PCR assay for MuAstV detection.
As MuAstV STL1 and STL2 were originally identified in the feces of asymptomatic wild-type mice, we examined whether MuAstV STL was present in the feces of other mice from the same SPF research mouse colony. To quantify the number of MuAstV STL1 and STL2 genome copies in tissues and feces, we designed a TaqMan-based quantitative reverse transcriptase PCR (qRT-PCR) assay (Fig. 4A), which targeted a 72-bp region of the RdRP conserved between MuAstV STL1 and STL2. Across multiple experiments, the assay consistently detected from 106 to 101 genome copies, and 1 genome copy was detected in 2 of 3 technical replicates consistent with Poisson distribution statistics, demonstrating that this assay was both sensitive and robust.
Fig 4.
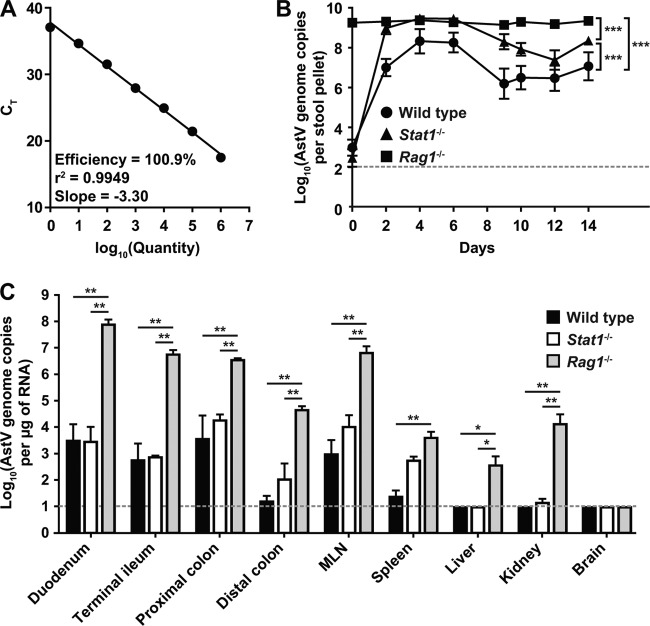
Adaptive and innate immune responses contribute to the control of MuAstV infection. (A) Standard curve of MuAstV STL qRT-PCR targeting a conserved region within the RdRP. Cycle threshold (CT) is plotted as a function of the quantity of input plasmid containing the target region. Data are means ± standard errors of the means determined for triplicate samples and are representative of the results of at least three experiments. (B and C) Wild-type, Stat1−/−, and Rag1−/− mice were cohoused for 14 days. (B) Stool samples were collected every 2 days and analyzed for MuAstV STL genome copy numbers by qRT-PCR. (C) At day 14, mice were sacrificed and the quantity of viral RNA in the indicated tissues was determined by qRT-PCR. Viral RNA levels shown represent means ± standard errors of the means determined for six mice pooled from two independent experiments. The dotted line indicates the limit of detection for the assay. Statistical significance was analyzed by two-way analysis of variance (ANOVA) with a Bonferroni posttest correction. *, P < 0.05; **, P < 0.001; ***, P < 0.0001.
Innate and adaptive immunity contribute to the control of MuAstV replication.
Given that previous human studies have implicated the adaptive immune system as essential in the control of astrovirus pathogenesis (63), we measured the number of astrovirus genome copies present in the feces of mice deficient in B and T cells due to a mutation in Recombination Activating Gene 1 (Rag1) (45). While these mice exhibited no overt signs of illness, we detected up to 109 astrovirus genome copies per fecal pellet. Notably, 21/21 Rag1−/− mice that we screened were positive for astrovirus (data not shown). These data suggest that adaptive immunity is important for restricting MuAstV replication.
To assess the role of innate and adaptive immunity in restricting MuAstV replication, we examined the time course of natural astrovirus infection in wild-type mice and mice deficient in signal transducer and activator of transcription 1 (Stat1) (13). As previous studies have shown that astrovirus infections may spread beyond the GI tract (5, 51), we chose to examine infection of extraintestinal tissues as well as intestinal infection and fecal shedding. In order to ensure infection by the same viruses, we cohoused wild-type and Stat1−/− mice with infected Rag1−/− mice for 14 days.
Consistent with our screening data from Rag1−/− mice, we observed high levels of MuAstV shedding in fecal samples from Rag1−/− mice at day 0 and at all time points tested (Fig. 4B). In contrast, low levels of MuAstV were observed in the feces of both wild-type and Stat1−/− mice at day 0, prior to cohousing. After 2 days of cohousing, elevated levels of MuAstV genome copies were detected in feces from wild-type and Stat1−/− mice, with Stat1−/− mice shedding significantly more MuAstV than wild-type mice at day 2 (P < 0.05). Overall, levels of MuAstV shedding over the course of the experiment differed significantly by genotype (P < 0.0001 for two-way comparisons). These data suggest that both the innate and adaptive immune systems contribute to the control of MuAstV replication.
We next analyzed the tissue distribution of MuAstV STL after 14 days of cohousing. We detected high levels of genome copies in the GI tract of Rag1−/− mice (Fig. 4C), consistent with our observation that Rag1−/− mice shed up to 109 genome copies per stool pellet. The quantity of viral genome copies detected in the GI tract of wild-type and Stat1−/− mice was significantly lower than in the Rag1−/− mice (P < 0.001 for all GI tract tissues tested). MuAstV RNA was detected in the liver and kidney of Rag1−/− mice but not in the liver or kidney of wild-type or Stat1−/− mice. While we occasionally detected low levels of MuAstV RNA in the spleen of Stat1−/− mice, we did not detect any genome copies in the spleen of wild-type mice. We were unable to detect MuAstV STL in the brain of any mouse tested. In aggregate, these data suggest a role for both the innate and adaptive immune systems in the control of astrovirus infection and replication.
MuAstV is present in mice purchased from commercial mouse vendors.
Since we had detected MuAstV STL in our research mouse facility, we assessed whether MuAstV was present in mice purchased from commercial vendors. As we had observed extremely high levels of MuAstV STL in the feces of Rag1−/− mice, we decided to assess the presence of MuAstV in commercially available mice with adaptive immune deficiencies. Of the available options from three commercial vendors, we purchased Rag1−/− mice, Rag2−/− mice (54), and SCID mice (6).
Consistent with our previous findings in Rag1−/− mice from our academic research mouse facility, we observed extremely high levels of MuAstV in fecal and tissue samples from Rag1−/− and Rag2−/− mice from commercial mouse facilities (Fig. 5). While we did detect a limited number of MuAstV genome copies in the feces of two out of three SCID mice, none were detected in tissue samples. These mice may have been truly negative for MuAstV or may have been infected with a strain that we were unable to detect using our MuAstV STL-specific primers and probe. Overall, the presence of MuAstV in both academic and commercial mouse facilities strongly suggests that MuAstV is a common agent in laboratory mice, likely present in many research mouse facilities.
Fig 5.
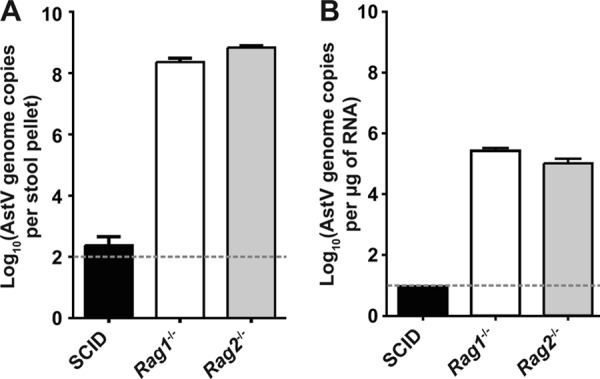
MuAstV detection in commercially available mice. Mice lacking B and T cells were ordered from three vendors in the United States and immediately sacrificed upon delivery. Viral genomes in (A) fecal samples or (B) mesenteric lymph nodes were quantitated using a qRT-PCR. Viral RNA levels shown represent means ± standard errors of the means of the results determined for three mice. The dotted lines denote the limit of detection for each assay.
DISCUSSION
We present here the complete genome sequence of two novel astroviruses, MuAstV STL1 and STL2, initially discovered in immunocompetent mice housed in a specific-pathogen-free academic mouse facility. MuAstV STL is a member of the mamastrovirus genus and highly divergent from the previously described astrovirus found in a wild house mouse (49). Analysis of MuAstV natural infection revealed a requirement for adaptive immunity in the control of MuAstV replication and spread. The discovery and complete genomic characterization of MuAstV STL, an astrovirus capable of infecting research mice, provide an opportunity to investigate biology and immunology of a mammalian astrovirus, with implications for our understanding of human astrovirus disease mechanisms.
A new model of astrovirus infection.
Turkey astrovirus 2 (TAstV2) has been used to study astrovirus pathogenesis in young turkeys and has been the only small-animal model of astrovirus infection and pathogenesis to date (33, 44). TAstV2, as a member of the avastrovirus genus, is genetically distinct from the mamastroviruses, including human astroviruses. Here, we have shown that the mamastrovirus MuAstV STL is capable of infecting research mice, an ideal system in which to examine astrovirus infection and host-virus interactions. Indeed, our initial studies during the course of natural MuAstV infection in mice demonstrated that the adaptive immune system is essential in restricting murine astrovirus replication, as has been reported for human astrovirus replication and persistence (51, 63). In addition, we also used this new model to identify a role for Stat1 in controlling astrovirus replication. Thus, this animal model is the first genetically manipulable small-animal model with which the molecular mechanisms of astrovirus infection may be elucidated.
MuAstV STL belongs to a potentially novel mamastrovirus genogroup.
Our phylogenetic analysis of the mamastroviruses, including several recently described astroviruses, provides support for the addition of a third potential genogroup which comprises MuAstV STL as well as astroviruses that infect rats, pigs, deer, and cattle but not (to date) humans. The significance of the viruses within this putative new genogroup remains to be seen. At least one strain within this new putative genogroup has been associated with diarrheal disease in its host (36), suggesting that members of this proposed genogroup may have the potential to exhibit signs of classical human astrovirus disease.
In addition to the putative genogroup III to which MuAstV STL belongs, our analysis provides support for the creation of up to three additional mamastrovirus genogroups. AstV MLB1 and STL2, bottlenose dolphin astrovirus, and ovine astrovirus 2 and porcine astrovirus 5 all have deep branches and p-dist values greater than 0.671 with respect to the other genogroups. The creation of three new genogroups would serve to reduce the intragenogroup p-dist values, which previously overlapped with the intergenogroup values, likely due to the more limited number of astrovirus sequences available for analysis at the time the current taxonomy was established. The recent characterization of a number of novel astroviruses added resolution to our analysis of phylogenetic relationships and allowed us to detect a total of six potential genogroups within the mamastrovirus genus.
Immune modulation of astrovirus infection.
The mechanisms by which the immune system responds to astrovirus infection are not well defined. A role for T- and B-cell-mediated immunity in controlling AstV infection is supported by case studies describing chronic astrovirus infection in two T-cell-deficient children and astrovirus-associated encephalitis in a child with X-linked agammaglobulinemia (51, 63). Experiments using TAstV2 have demonstrated that antibodies are not essential to controlling viral infection (32), but the importance of the humoral response itself has not been examined for mamastroviruses. Using MuAstV, we demonstrated that B and T cells are required to control astrovirus replication in multiple tissues as well as fecal shedding. Consistent with observations in T-cell-deficient children, we detected persistent astrovirus infection and shedding in the absence of B and T cells. Further studies are needed to assess the importance of each cell type in the modulation of astrovirus infection, but it is clear that the adaptive immune system is required for control of astrovirus replication.
While adaptive immunity plays a dominant role in the control of MuAstV infection, our results also provide evidence for a role for innate immunity. We observed increased MuAstV replication and shedding in Stat1−/− mice compared to wild-type mice. Stat1 is required for type I and II interferon signaling, and Stat1−/− mice are more susceptible to viral infection (13). Stat1 has also been shown to be essential for the expression of inducible nitric oxide synthase (iNOS) in intestinal epithelial cells (56), and nitric oxide activity is induced in the intestines of TAstV2-infected turkey embryos, thus suppressing astrovirus replication (32). It will be interesting to assess the role of iNOS during the course of mamastrovirus replication as well.
Research implications of prevalent astrovirus infection.
The ability to interpret data generated in small-animal models of disease pathogenesis requires an accurate representation of the immune response to perturbation. Subclinical infection may alter immune phenotypes and baseline homeostasis (1, 3, 8). For example, in the case of mouse hepatitis virus (MHV) infection, in which clinical signs of disease are not apparent in immunocompetent adults, the inadvertent exposure of nonobese diabetic mice to MHV led to a decrease in diabetes incidence (62). Due to the potential for confounding experimental outcomes, homeostasis-altering agents such as MHV are generally excluded from SPF facilities.
MuAstV was identified in mice purchased from several commercial vendors in the United States as well as in mice in several mouse research facilities at Washington University, suggesting that it may be present in many academic and commercial research facilities. Indeed, the sequence of murine astrovirus strain TF18LM isolated from a research facility in Cincinnati, OH, was similar to that of MuAstV STL by phylogenetic analysis.
While MuAstV was found in asymptomatic mice, its effects on host immunity and physiology are not known. Importantly, differences in astrovirus presence between facilities, or even between groups of experimental and control animals, may contribute to altered outcomes. Our previous studies have demonstrated the importance of the combination of genetic susceptibility and environmental factors in disease phenotypes. In particular, we showed that murine norovirus, which was originally discovered as a common contaminant in research mouse facilities, triggers Crohn's disease-like pathology in mice hypomorphic for the autophagy protein Atg16L1 (8, 30). Standard methods for animal health monitoring currently account only for known pathogens; however, our identification of multiple viruses in research mice suggests that this approach does not detect all possible contributors to disease phenotypes. An unbiased approach to animal health monitoring might reveal additional members of the mouse virome and contribute to the reliability and interpretation of data from the animal models on which our understanding of disease mechanisms so heavily relies.
ACKNOWLEDGMENTS
This research was supported by National Institutes of Health grants RO1 AI054483 and AI084887 (H.W.V.), Crohns and Colitis Foundation grant 3132 (H.W.V.), and training grant T32AI007163 (C.C.Y.).
We thank members of the Virgin laboratory for their comments during the preparation of the manuscript and Darren Kreamalmeyer for his assistance with mouse husbandry.
A patent disclosure for MuAstV STL1, STL2, STL3, and STL4 has been filed by H.W.V., D.W., J.L., G.Z., T.S.S., L.B.T., and C.C.Y.
Footnotes
Published ahead of print 5 September 2012
REFERENCES
- 1. Abt MC, et al. 2012. Commensal bacteria calibrate the activation threshold of innate antiviral immunity. Immunity 37:158–170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410 [DOI] [PubMed] [Google Scholar]
- 3. Barton ES, et al. 2007. Herpesvirus latency confers symbiotic protection from bacterial infection. Nature 447:326–329 [DOI] [PubMed] [Google Scholar]
- 4. Belliot G, Laveran H, Monroe SS. 1997. Outbreak of gastroenteritis in military recruits associated with serotype 3 astrovirus infection. J. Med. Virol. 51:101–106 [DOI] [PubMed] [Google Scholar]
- 5. Blomström A-L, Widén F, Hammer A-S, Belák S, Berg M. 2010. Detection of a novel astrovirus in brain tissue of mink suffering from shaking mink syndrome by use of viral metagenomics. J. Clin. Microbiol. 48:4392–4396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bosma MJ, Carroll AM. 1991. The SCID mouse mutant: definition, characterization, and potential uses. Annu. Rev. Immunol. 9:323–350 [DOI] [PubMed] [Google Scholar]
- 7. Brierley I, Gilbert RJC, Pennell S. 2008. RNA pseudoknots and the regulation of protein synthesis. Biochem. Soc. Trans. 36:684–689 [DOI] [PubMed] [Google Scholar]
- 8. Cadwell K, et al. 2010. Virus-plus-susceptibility gene interaction determines Crohn's disease gene Atg16L1 phenotypes in intestine. Cell 141:1135–1145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Coppo P, Scieux C, Ferchal F, Clauvel J, Lassoued K. 2000. Astrovirus enteritis in a chronic lymphocytic leukemia patient treated with fludarabine monophosphate. Ann. Hematol. 79:43–45 [DOI] [PubMed] [Google Scholar]
- 10. Cunliffe NA, et al. 2002. Detection and characterisation of human astroviruses in children with acute gastroenteritis in Blantyre, Malawi. J. Med. Virol. 67:563–566 [DOI] [PubMed] [Google Scholar]
- 11. De Benedictis P, Schultz-Cherry S, Burnham A, Cattoli G. 2011. Astrovirus infections in humans and animals—molecular biology, genetic diversity, and interspecies transmissions. Infect. Genet. Evol. 11:1529–1544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dennehy PH, et al. 2001. A prospective case-control study of the role of astrovirus in acute diarrhea among hospitalized young children. J. Infect. Dis. 184:10–15 [DOI] [PubMed] [Google Scholar]
- 13. Durbin JE, Hackenmiller R, Simon MC, Levy DE. 1996. Targeted disruption of the mouse Stat1 gene results in compromised innate immunity to viral disease. Cell 84:443–450 [DOI] [PubMed] [Google Scholar]
- 14. Finkbeiner SR, et al. 2008. Metagenomic analysis of human diarrhea: viral detection and discovery. PLoS Pathog. 4:e1000011 doi:10.1371/journal.ppat.1000011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Finkbeiner SR, Le BM, Holtz LR, Storch GA, Wang D. 2009. Detection of newly described astrovirus MLB1 in stool samples from children. Emerg. Infect. Dis. 15:441–444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Finkbeiner SR, et al. 2009. Identification of a novel astrovirus (Astrovirus VA1) associated with an outbreak of acute gastroenteritis. J. Virol. 83:10836–10839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Finn RD, et al. 2010. The Pfam protein families database. Nucleic Acids Res. 38:D211–D222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gavier-Widén D, et al. 2004. Investigations into shaking mink syndrome: an encephalomyelitis of unknown cause in farmed mink (Mustela vison) kits in Scandinavia. J. Vet. Diagn. Invest. 16:305–312 [DOI] [PubMed] [Google Scholar]
- 19. Giedroc DP, Cornish PV. 2009. Frameshifting RNA pseudoknots: structure and mechanism. Virus Res. 139:193–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gough RE, Borland ED, Keymer IF, Stuart JC. 1985. An outbreak of duck hepatitis type II in commercial ducks. Avian Pathol. 14:227–236 [DOI] [PubMed] [Google Scholar]
- 21. Gray JJ, Wreghitt TG, Cubitt WD, Elliot PR. 1987. An outbreak of gastroenteritis in a home for the elderly associated with astrovirus type 1 and human calicivirus. J. Med. Virol. 23:377–381 [DOI] [PubMed] [Google Scholar]
- 22. Grohmann GS, et al. 1993. Enteric viruses and diarrhea in HIV-infected patients. Enteric Opportunistic Infections Working Group. N. Engl. J. Med. 329:14–20 [DOI] [PubMed] [Google Scholar]
- 23. Herrmann JE, Hudson RW, Perron-Henry DM, Kurtz JB, Blacklow NR. 1988. Antigenic characterization of cell-cultivated astrovirus serotypes and development of astrovirus-specific monoclonal antibodies. J. Infect. Dis. 158:182–185 [DOI] [PubMed] [Google Scholar]
- 24. Herrmann JE, Taylor DN, Echeverria P, Blacklow NR. 1991. Astroviruses as a cause of gastroenteritis in children. N. Engl. J. Med. 324:1757–1760 [DOI] [PubMed] [Google Scholar]
- 25. Holtz LR, Finkbeiner SR, Kirkwood CD, Wang D. 2008. Identification of a novel picornavirus related to cosaviruses in a child with acute diarrhea. Virol. J. 5:159–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Holtz LR, et al. 2009. Klassevirus 1, a previously undescribed member of the family Picornaviridae, is globally widespread. Virol. J. 6:86–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Holtz LR, et al. 2011. Astrovirus MLB2 viremia in febrile child. Emerg. Infect. Dis. 17:2050–2052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Jeong HS, Jeong A, Cheon D-S. 2012. Epidemiology of astrovirus infection in children. Korean J. Pediatr. 55:77–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Jiang B, Monroe SS, Koonin EV, Stine SE, Glass RI. 1993. RNA sequence of astrovirus: distinctive genomic organization and a putative retrovirus-like ribosomal frameshifting signal that directs the viral replicase synthesis. Proc. Natl. Acad. Sci. U. S. A. 90:10539–10543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Karst SM, Wobus CE, Lay M, Davidson J, Virgin HW. 2003. STAT1-dependent innate immunity to a Norwalk-like virus. Science 299:1575–1578 [DOI] [PubMed] [Google Scholar]
- 31. King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ. (ed). 2011. Virus taxonomy: classification and nomenclature of viruses. Ninth report of the International Committee on Taxonomy of Viruses Elsevier Academic Press, London, United Kingdom [Google Scholar]
- 32. Koci MD, Kelley LA, Larsen D, Schultz-Cherry S. 2004. Astrovirus-induced synthesis of nitric oxide contributes to virus control during infection. J. Virol. 78:1564–1574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Koci MD, et al. 2003. Astrovirus induces diarrhea in the absence of inflammation and cell death. J. Virol. 77:11798–11808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Koci MD, Seal BS, Schultz-Cherry S. 2000. Molecular characterization of an avian astrovirus. J. Virol. 74:6173–6177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Koopmans MP, Bijen MH, Monroe SS, Vinje J. 1998. Age-stratified seroprevalence of neutralizing antibodies to astrovirus types 1 to 7 in humans in The Netherlands. Clin. Diagn. Lab. Immunol. 5:33–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lan D, et al. 2011. Molecular characterization of a porcine astrovirus strain in China. Arch. Virol. 156:1869–1875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Larkin MA, et al. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948 [DOI] [PubMed] [Google Scholar]
- 38. Lee TW, Kurtz JB. 1981. Serial propagation of astrovirus in tissue culture with the aid of trypsin. J. Gen. Virol. 57:421–424 [DOI] [PubMed] [Google Scholar]
- 39. Lewis DC, Lightfoot NF, Cubitt WD, Wilson SA. 1989. Outbreaks of astrovirus type 1 and rotavirus gastroenteritis in a geriatric in-patient population. J. Hosp. Infect. 14:9–14 [DOI] [PubMed] [Google Scholar]
- 40. Lewis TL, Greenberg HB, Herrmann JE, Smith LS, Matsui SM. 1994. Analysis of astrovirus serotype 1 RNA, identification of the viral RNA-dependent RNA polymerase motif, and expression of a viral structural protein. J. Virol. 68:77–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lewis TL, Matsui SM. 1995. An astrovirus frameshift signal induces ribosomal frameshifting in vitro. Arch. Virol. 140:1127–1135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Li L, et al. 2011. The fecal viral flora of California sea lions. J. Virol. 85:9909–9917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Li W, Godzik A. 2006. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22:1658–1659 [DOI] [PubMed] [Google Scholar]
- 44. Mendez E, Arias CF. 2007. Astroviruses, p 981–999 In Knipe D, Howley P. (ed), Fields virology, 5th ed Lippincott, Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 45. Mombaerts P, et al. 1992. RAG-1-deficient mice have no mature B and T lymphocytes. Cell 68:869–877 [DOI] [PubMed] [Google Scholar]
- 46. Moon S, Byun Y, Kim HJ, Jeong S, Han K. 2004. Predicting genes expressed via −1 and +1 frameshifts. Nucleic Acids Res. 32:4884–4892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Oishi I, et al. 1994. A large outbreak of acute gastroenteritis associated with astrovirus among students and teachers in Osaka, Japan. J. Infect. Dis. 170:439–443 [DOI] [PubMed] [Google Scholar]
- 48. Palombo EA, Bishop RF. 1996. Annual incidence, serotype distribution, and genetic diversity of human astrovirus isolates from hospitalized children in Melbourne, Australia. J. Clin. Microbiol. 34:1750–1753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Phan TG, et al. 2011. The fecal viral flora of wild rodents. PLoS Pathog. 7:e1002218 doi:10.1371/journal.ppat.1002218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Presti RM, et al. 2009. Quaranfil, Johnston Atoll, and Lake Chad viruses are novel members of the family Orthomyxoviridae. J. Virol. 83:11599–11606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Quan PL, et al. 2010. Astrovirus encephalitis in boy with X-linked agammaglobulinemia. Emerg. Infect. Dis. 16:918–925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Reuter G, Pankovics P, Boros A. 2011. Identification of a novel astrovirus in a domestic pig in Hungary. Arch. Virol. 156:125–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Shastri S, Doane AM, Gonzales J, Upadhyayula U, Bass DM. 1998. Prevalence of astroviruses in a children's hospital. J. Clin. Microbiol. 36:2571–2574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Shinkai Y, et al. 1992. RAG-2-deficient mice lack mature lymphocytes owing to inability to initiate V(D)J rearrangement. Cell 68:855–867 [DOI] [PubMed] [Google Scholar]
- 55. Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22:2688–2690 [DOI] [PubMed] [Google Scholar]
- 56. Stempelj M, Kedinger M, Augenlicht L, Klampfer L. 2007. Essential role of the JAK/STAT1 signaling pathway in the expression of inducible nitric-oxide synthase in intestinal epithelial cells and its regulation by butyrate. J. Biol. Chem. 282:9797–9804 [DOI] [PubMed] [Google Scholar]
- 57. Swofford DL. 2002. PAUP*: Phylogenetic analysis using parsimony (*and other methods), v. 4.0. Sinauer Associates, Sunderland, MA [Google Scholar]
- 58. Tamura K, et al. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28:2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Virgin HW, Todd JA. 2011. Metagenomics and personalized medicine. Cell 147:44–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Walter JE, et al. 2001. Molecular characterization of a novel recombinant strain of human astrovirus associated with gastroenteritis in children. Arch. Virol. 146:2357–2367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Wang D, et al. 2003. Viral discovery and sequence recovery using DNA microarrays. PLoS Biol. 1:E2 doi:10.1371/journal.pbio.0000002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Wilberz S, Partke HJ, Dagnaes-Hansen F, Herberg L. 1991. Persistent MHV (mouse hepatitis virus) infection reduces the incidence of diabetes mellitus in non-obese diabetic mice. Diabetologia 34:2–5 [DOI] [PubMed] [Google Scholar]
- 63. Wood DJ, David TJ, Chrystie IL, Totterdell B. 1988. Chronic enteric virus infection in two T-cell immunodeficient children. J. Med. Virol. 24:435–444 [DOI] [PubMed] [Google Scholar]
- 64. Zhao G, et al. 2011. The genome of Yoka poxvirus. J. Virol. 85:10230–10238 [DOI] [PMC free article] [PubMed] [Google Scholar]