Abstract
Cysteines were introduced into the membrane-proximal external region (MPER) of the paramyxovirus F protein. A disulfide bond formed, and the mutant protein was expressed at the cell surface but was fusion inactive. Reduction of the disulfide bond restored fusion activity. The data indicate that in addition to dissociation of the three-helix bundle stalk domain of prefusion F, the MPER region also needs to separate for F to be able to refold and cause fusion.
TEXT
The Paramyxoviridae family includes some of the great and ubiquitous disease-causing viruses of humans and animals and consists of enveloped negative-stranded RNA viruses that enter cells by fusing their envelope at neutral pH with the plasma membrane of the target cell (with the exception of a few isolates of human metapneumovirus that require low pH for fusion [4, 13]). Virus fusion is mediated by the F protein that folds initially to form a trimeric metastable prefusion form that is triggered to undergo large-scale irreversible refolding events to form the trimeric postfusion conformation. It is thought that F refolding couples the energy released with membrane fusion (reviewed in reference 7).
The atomic structures of the prefusion form of the parainfluenza virus 5 (PIV5) F protein and the postfusion form of the human parainfluenza virus 3 F protein have been determined (14, 15). The structure of prefusion F and data from biochemical experiments have indicated that F refolds via at least two intermediate structures (9–11, 15). These intermediates include a temperature-arrested intermediate that involves changes to a region of F that forms a 3-helix bundle (3-HB) stalk domain and in the monomer is known as HRB, and a prehairpin intermediate in which the fusion peptide (FP) and heptad repeat A (HRA) are released and the FP is inserted into the target membrane with HRA, forming a 3-HB such that F bridges the viral and cellular membranes. The last step of fusion involves the zippering up of HRB onto HRA to form a stable 6-HB and formation of the postfusion form. Although considerable evidence exists for the prehairpin intermediate (6, 9), the only data on the formation of the temperature-arrested intermediate comes from peptide inhibition studies in which the N-1 peptide (derived from HRA) binds to F and inhibits fusion on the binding of the receptor-binding protein hemagglutinin-neuraminidase (HN) to sialic acid on cells. There is currently little information on the seven residues of F known as the membrane-proximal external region (MPER) (17) that link the transmembrane domain to HRB. It is known that the MPER region cannot be switched among paramyxovirus F proteins with retention of fusion activity (17) and that although some deletions in the F MPER region can be made that retain biological activity, insertion of residues cannot be tolerated for retention for fusion activity (1, 16).
The introduction of disulfide bonds to inhibit movement of domains with concomitant inhibition of fusion has been studied previously for the measles virus F protein and influenza virus hemagglutinin (3, 5, 8). In the case of measles virus F protein, Lee and colleagues (8), using a model of the measles prefusion F protein based on the atomic coordinates of the PIV5 F structure (15), introduced two pairs of mutations in measles virus F in the expectation that an intersubunit disulfide bond between residues 452 and 460 would link the F head to its stalk and that a disulfide bond between residues 307 and 448 would link adjacent loops in the head domains. A disulfide bond formed between residues 452 and 460 that reversibly blocked fusion activity, whereas cysteine at residues 307 and 448 did not form a disulfide bond and the F protein was malfolded. Even with an atomic structure, introduction of cysteine residues may lead to slightly incorrect bond angles.
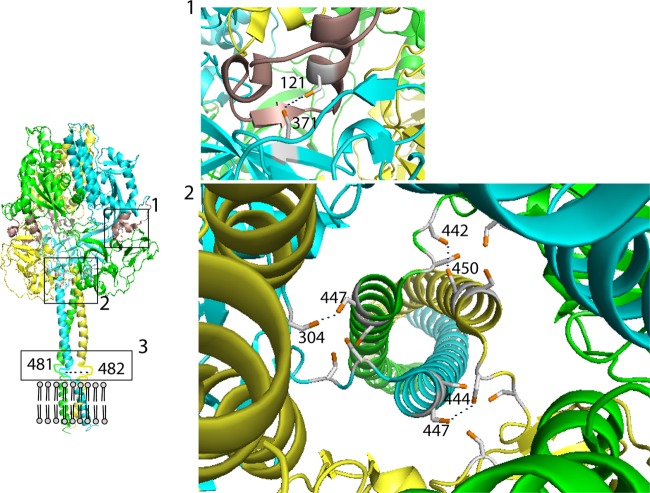
To probe the movement of PIV5 F domains during refolding, we introduced cysteine residues in locations predicted to form intermonomer disulfide bonds in the PIV5 F protein. Due to the fact that F is a trimer, cysteine double mutants were made that were predicted to form disulfide bonds. These were in the MPER (residues T481C and T482C), the interface of the prefusion head and stalk (residues L442C and S450C, I444C and L447C, and Q304C and L447C), and between the fusion peptide and the head of an adjacent monomer (V121C and L371C) (Fig. 1).
Fig 1.
Positions of the predicted disulfide bonds in the PIV5 F trimer. The soluble ectodomain (residues 1 to 477) of the prefusion F trimer is shown (15). The structure of the MPER (residues 478 to 484) is not known and is indicated as flexible lines. The structure of the transmembrane (TM) domain has been predicted to be helical by biochemical studies (2) and is indicated as helices. The gray circles and lines indicate the lipid bilayer. The positions for residues 481 and 482 in the MPER are indicated on the trimer. The three monomers are colored green, blue, and yellow. The FP is colored brown. Panel 1 shows the positions for cysteine 121 in the fusion peptide on one monomer and for cysteine 371 in domain II on another monomer of the trimer. Panel 2 shows the top view of a trimer, indicating the positions for the three pairs of cysteines at positions 304 and 447, 442 and 450, and 444 and 447, which are hypothesized to form intersubunit disulfide bonds, indicated by dotted black lines.
F cleavage, surface expression, and fusion of the cysteine mutants.
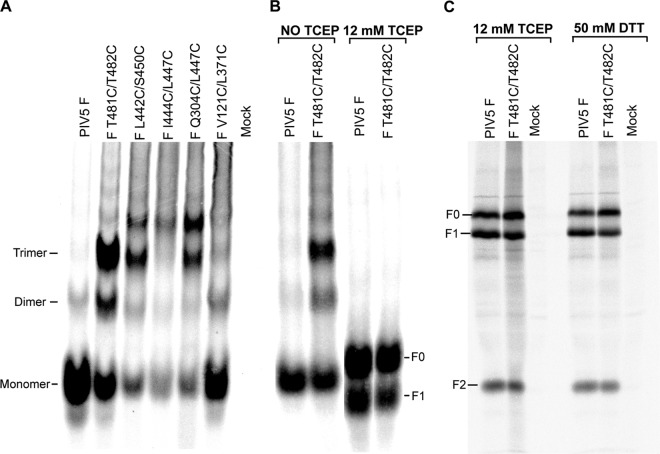
F protein is synthesized as a precursor, F0, that is cleaved in the trans-Golgi network to F1 and F2. The rate of cleavage correlates with the level of expression at the cell surface, and poor cleavage is indicative of malfolding of the F protein. The cysteine double mutants were constructed in the pCAGGS mammalian expression vector, transiently expressed in HeLa CD4 LTR βgal cells, metabolically labeled with 35S-Promix, immunoprecipitated, and analyzed by SDS-PAGE as described previously (17). The mutants F T481C/T482C and F V121C/L371C were cleaved into F1 and F2 at levels comparable to that of wild-type (wt) F protein, whereas F L442C/S450C, F I444C/L447C, and F Q304C/L447C showed reduced amounts of F cleavage (Fig. 2). Cell surface expression levels of the F cysteine mutants were determined by flow cytometry as described previously (17) (Table 1). The amounts of F expressed at the cell surface mirrored the amounts of F cleavage, indicating that only two of the mutants reached the cell surface to significant levels (F T481C/T482C and F V121C/L371C). To determine the extent of cell-cell fusion caused by the mutants, a quantitative luciferase reporter assay was performed as described previously (17). F V121C/L371C caused fusion at levels comparable to that of wt F whereas the other mutants, including F T481C/T482C, despite its good surface expression level, did not cause detectable fusion (Table 1).
Fig 2.
Cleavage of the cysteine mutants into F1 and F2. HeLa CD4 LTR βgal cells were transfected with pCAGGS F, metabolically labeled with 35S-Promix, incubated for 120 min (chase), and immunoprecipitated with an anti-F polyclonal antibody, and polypeptides were analyzed by SDS-PAGE on 15% gels. Radioactivity was detected by a Fuji phosphorimager.
Table 1.
Normalized surface expression and fusion activity of cysteine double mutants
| Construct | Surface expression (MFI [% wt])a | Fusion (RLU [% wt])b |
|---|---|---|
| PIV5 F | 100.0 | 100.0 |
| F T481C/T482C | 71.4 ± 2.7 | 4.3 ± 1.0 |
| F L442V/S450C | 14.3 ± 1.6 | 2.9 ± 0.6 |
| F I444C/L447C | 5.3 ± 0.4 | 1.2 ± 0.1 |
| F Q304C/L447C | 13.9 ± 2.1 | 3.1 ± 0.1 |
| F V121C/L371C | 49.9 ± 2.8 | 124 ± 4.6 |
| None (mock) | 0.6 ± 0.7 | 1.9 ± 0.6 |
Surface expression levels of the cysteine mutants were determined by flow cytometry of transfected HeLa CD4 LTR βgal cells using monoclonal antibody (MAb) F1a. The mean fluorescence intensity (MFI) is indicated as a percentage of the wt F level. Values are shown as means ± standard errors of the means except for the value for PIV5 F.
Fusion activity of the cysteine mutants was determined by a luciferase assay. Vero cells were transfected with plasmids expressing PIV5 F and HN and luciferase under the control of the T7 promoter. At 20 h posttransfection, the transfected cells were overlaid with BSR cells that stably express T7 polymerase. After 8 h, the cells were lysed, and luciferase activity was measured in a luminometer. Relative light units (RLU) for mutants are expressed as a percentage of wt F RLU. Values are shown as means ± standard errors of the means except for the value for PIV5 F.
Formation of the disulfide bond for the mutant F T481C/T482C.
To investigate whether the cysteine mutants formed disulfide bonds, F was analyzed by SDS-PAGE on 3.5% gels under nonreducing conditions (12). Wild-type F migrated as monomers, whereas F T481C/T482C showed extensive amounts of trimer species in addition to some monomers and dimers, suggesting the formation of a disulfide bond (Fig. 3A). This electrophoretic pattern is very similar to that of chemically cross-linked F (12). However, F V121C/L371C migrated predominantly as a monomer, similar to wt F, suggesting that this mutant does not form a disulfide bond. F L442C/S450C, F I444C/L447C, and F Q304/L447C showed overall smaller amounts of protein on the gel, with small amounts of trimer species and other slower-mobility species that may represent disulfide-linked aggregates (Fig. 3A). Taking together the reduced cleavage of F, its reduced surface expression, and the results of the gel analysis suggest that the last three mutants express malfolded protein.
Fig 3.
Formation of disulfide bonds. HeLa CD4 LTR βgal cells were transfected with wt F and the F cysteine mutants, radiolabeled, and immunoprecipitated as described in the text. (A) wt F and the F cysteine mutants were analyzed by SDS-PAGE on 3.5% gels under nonreducing conditions. (B) wt F and cysteine mutant F T481C/T482C were left untreated or treated with TCEP (12 mM) and analyzed on 3.5% gels. Samples were electrophoresed several lanes apart on the gel to prevent leakage of TCEP across to untreated lanes, and the image is derived from two separate sections of the same slab gel. (C) wt F and cysteine mutant F T481C/T482C were treated with TCEP (12 mM) or dithiothreitol (DTT) (50 mM) and analyzed on 15% gels.
To demonstrate that formation of a disulfide bond has a specific effect on the F fusion activity, it is necessary to reduce the disulfide bond. At the biochemical level, it was found that addition of the reducing agent tris(2-carboxyethyl) phosphine (TCEP) converted F T481C/T482C to monomers consisting of a mixture of F0 plus F1 and F2 (Fig. 3B and C). For comparison, SDS-PAGE of F reduced using 50 mM dithiothreitol is shown (Fig. 3C). In Fig. 3B, unreduced F has a different mobility from F0 and F1 presumably because the unreduced F has disulfide bonds that confer rigidity and prevent the molecule from having the same Stoke's radius on SDS denaturation as reduced F0 and F1 species.
Restoration of fusion on reduction of the disulfide bond.
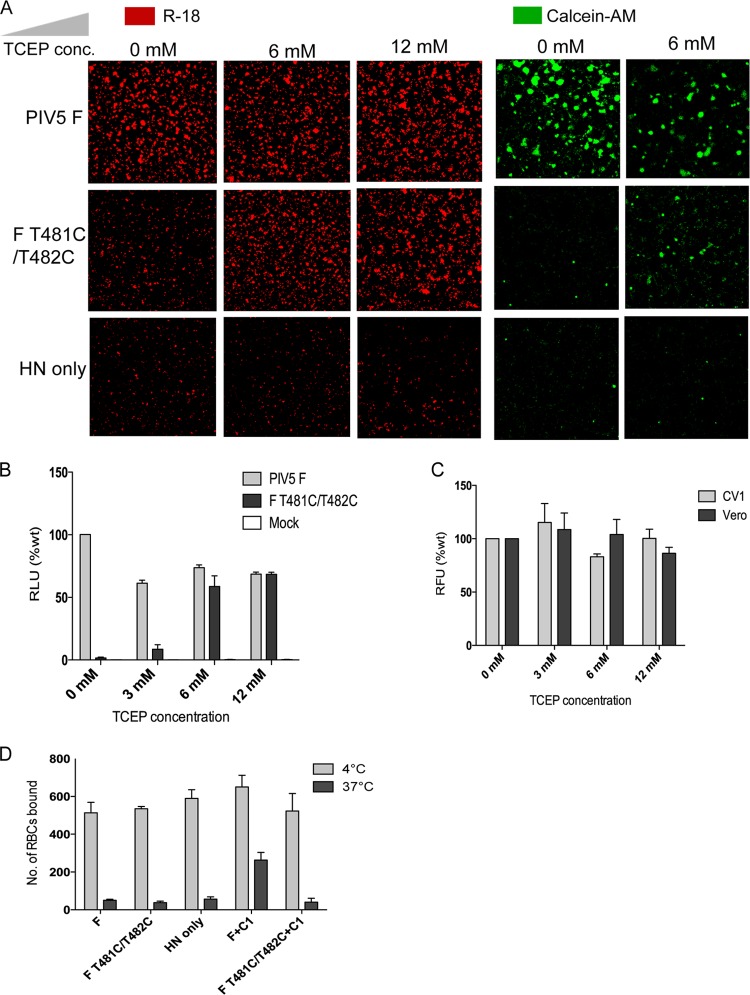
As F T481C/T482C forms a disulfide bond and is unable to cause fusion, we tested whether reduction of the disulfide bond would restore fusion activity. CV1 cells expressing F and HN were analyzed for fusion by a dye transfer assay using chicken red blood cells (RBCs) labeled with either the membrane dye octadecyl rhodamine (R-18) or the aqueous dye Calcein-AM (9). As shown in Fig. 4A, fusion mediated by F T481C/T482C could be restored by treatment of the CV1 cells with 6 mM TCEP. A somewhat deleterious effect of TCEP on fusion caused by wt PIV5 F was observed (Fig. 4A, Calcein-AM), which explains the low level of Calcein-AM labeling of cells expressing F T481C/T482C on treatment with 6 mM TCEP. A luciferase assay to quantify fusion (17) was also performed in which TCEP was added during overlay of target cells. Fusion caused by F T481C/T482C increased to wt levels in a dose-dependent manner on treatment with TCEP (Fig. 4B). TCEP did not have an effect on the viability of CV1 cells or Vero cells (Fig. 4C) as determined by an alamarBlue-based cell viability assay.
Fig 4.
Reduction of disulfide bond in F T481C/T482C by TCEP restores fusion. (A) Chicken RBCs labeled with the lipidic dye R-18 (red) or the aqueous dye Calcein-AM (green) were bound to CV1 cells transfected with plasmids expressing PIV5 F and HN at 4°C. Unbound RBCs were washed with cold phosphate-buffered saline (PBS). The samples were then transferred to 37°C for 15 min in the presence or absence of the indicated concentrations of TCEP to allow fusion of bound RBCs. Samples were returned to 4°C, washed with cold PBS, and visualized by confocal microscopy. (B) Luciferase assay to measure fusion in the absence or presence of the indicated concentrations of TCEP. Vero cells expressing PIV5 F and HN and luciferase under the control of the T7 promoter at 20 h posttransfection were overlaid with BSR cells that stably express T7 polymerase. The overlay medium contained the indicated concentrations of TCEP. After 8 h, the cells were lysed, and luciferase activity (in relative light units [RLU]) was measured. The luciferase activity for mutants is expressed as a percentage of the luciferase activity for wt F. (C) Viability of CV1 cells and Vero cells in the presence of TCEP. Cells treated with indicated concentrations of TCEP were treated with alamarBlue reagent, and relative fluorescence units (RFU) were measured. (D) RBC retention assay in the presence and absence of the C1 peptide, as previously described (10). Briefly, CV1 cells grown on glass coverslips were transfected as described previously (17) with 1 μg each of pGEM expression plasmids containing HN and with either F or F T481C/T482C. At 18 h posttransfection, the cells were incubated with 0.1% hematocrit of octadecyl rhodamine-labeled chicken RBCs at 4°C for 1 h. Unbound RBCs were removed by washing five times with cold PBS. The samples were shifted to 37°C for 15 min. Where indicated, 50 μg of C-1 peptide was added during RBC binding or during the 37°C incubation. Samples were transferred back to 4°C and washed with cold PBS. Retained RBCs were counted using a Carl Zeiss Pascal scanning confocal microscope. Values are means plus standard errors (error bars) of three trials.
A disulfide bond in the membrane-proximal region blocks formation of the F prehairpin intermediate.
It has been shown previously that addition of the C-1 peptide, which is derived from the HRB sequence, blocks fusion by binding to the prehairpin intermediate, and this can be measured by the retention of RBCs in which F trimers have inserted their FP but cannot form the 6-HB (9). As shown in Fig. 4D, whereas wt F caused retention of RBCs at 37°C in the presence of C-1 peptide, F T481C/T482C could not form the prehairpin intermediate at 37°C and did not retain RBCs. Attempts to use the N-1 peptide to inhibit the temperature-arrested intermediate of fusion were thwarted by the insolubility of N-1 peptide upon spontaneously forming trimers.
In summary, we demonstrate that PIV5 F can be retained in its prefusion conformation by the formation of a disulfide bond in the membrane-proximal region. Fusion can be restored on reducing the disulfide bonds. It is thought that there is rotational as well as translational motion of the paramyxovirus F protein stalk in the course of conversion of the prefusion to postfusion forms (reviewed in reference 11). Although rotational motion of the stalk as a whole is not restricted in F T481C/T482C, the translational motion of the individual C termini with respect to other C termini of a trimer is blocked. Previous studies have hypothesized the need for paramyxovirus F protein HRB 3-HB to dissociate (15). This study shows that the MPER that is immediately C terminal to HRB also needs to dissociate for F to be able to refold.
ACKNOWLEDGMENTS
This research was supported in part by National Institutes of Health research grants AI-23173 (to R.A.L.) and GM-61050 (to T.S.J.). S.A.C was an Associate and R.A.L. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
Published ahead of print 5 September 2012
REFERENCES
- 1. Baker KA, Dutch RE, Lamb RA, Jardetzky TS. 1999. Structural basis for paramyxovirus-mediated membrane fusion. Mol. Cell 3:309–319 [DOI] [PubMed] [Google Scholar]
- 2. Bissonnette ML, Donald JE, DeGrado WF, Jardetzky TS, Lamb RA. 2009. Functional analysis of the transmembrane domain in paramyxovirus F protein-mediated membrane fusion. J. Mol. Biol. 386:14–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Godley L, et al. 1992. Introduction of intersubunit disulfide bonds in the membrane-distal region of the influenza hemagglutinin abolishes membrane fusion activity. Cell 68:635–645 [DOI] [PubMed] [Google Scholar]
- 4. Herfst S, et al. 2008. Low-pH-induced membrane fusion mediated by human metapneumovirus F protein is a rare, strain-dependent phenomenon. J. Virol. 82:8891–8895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kemble GW, Bodian DL, Rose J, Wilson IA, White JM. 1992. Intermonomer disulfide bonds impair the fusion activity of influenza virus hemagglutinin. J. Virol. 66:4940–4950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kim YH, et al. 2011. Capture and imaging of a prehairpin fusion intermediate of the paramyxovirus PIV5. Proc. Natl. Acad. Sci. U. S. A. 108:20992–20997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lamb RA, Jardetzky TS. 2007. Structural basis of viral invasion: lessons from paramyxovirus F. Curr. Opin. Struct. Biol. 17:427–436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lee JK, Prussia A, Snyder JP, Plemper RK. 2007. Reversible inhibition of the fusion activity of measles virus F protein by an engineered intersubunit disulfide bridge. J. Virol. 81:8821–8826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Russell CJ, Jardetzky TS, Lamb RA. 2001. Membrane fusion machines of paramyxoviruses: capture of intermediates of fusion. EMBO J. 20:4024–4034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Russell CJ, Kantor KL, Jardetzky TS, Lamb RA. 2003. A dual-functional paramyxovirus F protein regulatory switch segment: activation and membrane fusion. J. Cell Biol. 163:363–374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Russell CJ, Luque LE. 2006. The structural basis of paramyxovirus invasion. Trends Microbiol. 14:243–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Russell R, Paterson RG, Lamb RA. 1994. Studies with cross-linking reagents on the oligomeric form of the paramyxovirus fusion protein. Virology 199:160–168 [DOI] [PubMed] [Google Scholar]
- 13. Schowalter RM, Smith SE, Dutch RE. 2006. Characterization of human metapneumovirus F protein-promoted membrane fusion: critical roles for proteolytic processing and low pH. J. Virol. 80:10931–10941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yin H-S, Paterson RG, Wen X, Lamb RA, Jardetzky TS. 2005. Structure of the uncleaved ectodomain of the paramyxovirus (hPIV3) fusion protein. Proc. Natl. Acad. Sci. U. S. A. 102:9288–9293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yin HS, Wen X, Paterson RG, Lamb RA, Jardetzky TS. 2006. Structure of the parainfluenza virus 5 F protein in its metastable, prefusion conformation. Nature 439:38–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhou J, Dutch RE, Lamb RA. 1997. Proper spacing between heptad repeat B and the transmembrane domain boundary of the paramyxovirus SV5 F protein is critical for biological activity. Virology 239:327–339 [DOI] [PubMed] [Google Scholar]
- 17. Zokarkar A, Lamb RA. 2012. The paramyxovirus fusion protein C-terminal region: mutagenesis indicates an indivisible protein unit. J. Virol. 86:2600–2609 [DOI] [PMC free article] [PubMed] [Google Scholar]