Abstract
The double-stranded RNA bacteriophage ϕ6 is an extensively studied prokaryotic model system for virus assembly. There are established in vitro assembly protocols available for the ϕ6 system for obtaining infectious particles from purified protein and RNA constituents. The polymerase complex is a multifunctional nanomachine that replicates, transcribes, and translocates viral RNA molecules in a highly specific manner. The complex is composed of (i) the major structural protein (P1), forming a T=1 icosahedral lattice with two protein subunits in the icosahedral asymmetric unit; (ii) the RNA-dependent RNA polymerase (P2); (iii) the hexameric packaging nucleoside triphosphatase (NTPase) (P4); and (iv) the assembly cofactor (P7). In this study, we analyzed several ϕ6 virions and recombinant polymerase complexes to investigate the relative copy numbers of P2, P4, and P7, and we applied saturated concentrations of these proteins in the self-assembly system to probe their maximal numbers of binding sites in the P1 shell. Biochemical quantitation confirmed that the composition of the recombinant particles was similar to that of the virion cores. By including a high concentration of P2 or P7 in the self-assembly reaction mix, we observed that the numbers of these proteins in the resulting particles could be increased beyond those observed in the virion. Our results also suggest a previously unidentified P2-P7 dependency in the assembly reaction. Furthermore, it appeared that P4 must initially be incorporated at each, or a majority, of the 5-fold symmetry positions of the P1 shell for particle assembly. Although required for nucleation, excess P4 resulted in slower assembly kinetics.
INTRODUCTION
All double-stranded RNA (dsRNA) viruses with polyhedral capsids replicate their genomes within a protein shell known as a polymerase complex. This complex is essential for the recognition and encapsidation of viral genomic precursor molecules, genome replication and transcript synthesis, and the formation of the core of the mature virion (41, 44).
Bacteriophage ϕ6, which belongs to the family Cystoviridae (2), is a well-studied prokaryotic model system for dsRNA virus assembly, with an established in vitro self-assembly system to produce infectious viral nucleocapsids from purified protein and nucleic acid constituents (43). The ϕ6 virion core is composed of a dodecahedral polymerase complex enclosing the three genomic dsRNA segments (S, M, and L segments) (6, 45). This core is covered by a layer of nucleocapsid surface shell protein (P8) arranged on a T=13 icosahedral lattice and a protein-lipid envelope layer anchoring the receptor-binding spikes (3, 6, 21).
During ϕ6 virion formation, the empty polymerase complex (procapsid [PC]) is first assembled from proteins P1 (85 kDa), P2 (75 kDa), P4 (35 kDa), and P7 (17 kDa) (30). The dodecahedral PC framework is composed of 120 copies of protein P1 arranged as 60 asymmetric dimers on a T=1 icosahedral lattice (6, 20). This type of capsid organization is common for dsRNA viruses but has not been observed in any other type of virus (41). Purified ϕ6 P1 is a monomer, but the equivalent protein from a related cystovirus, ϕ8, forms tetramers in solution (25, 43). Protein P2 is an RNA-dependent RNA polymerase catalyzing genome replication and transcription (19, 26, 27). In dsRNA viruses of the family Reoviridae, such as orthoreovirus (52), rotavirus (29), rice dwarf virus (33), aquareovirus (11), and gram carp reovirus (10), the polymerase subunit localizes to the interior of the T=1 shell, close to 5-fold axes of icosahedral symmetry, resulting in a maximal number of 12 polymerase subunits per particle. However, a recent cryo-electron tomography study of recombinant ϕ6 PCs suggests that the polymerase (P2) resides near the 3-fold axes in the PC interior, with 20 potential binding sites per particle (46). Nemecek et al. (34) observed that P2 is assembled randomly at the proposed binding locations, with a mean copy number of eight subunits per recombinant PC. Protein P4 is arranged as a ring-shaped hexamer and functions as a packaging nucleoside triphosphatase (NTPase) (14, 18, 24, 28, 39, 40). P4 hexamers localize to the 5-fold symmetry positions on the outer surface of the PC, with a copy number of 12 (hence, there are 72 copies of P4 per PC) (14, 20). The interaction between a P4 hexamer and the P1 shell is relatively unstable; the amount of P4 in ϕ6 PCs can be reduced significantly by detergent treatment (40). Furthermore, it has been determined (34) that a single recombinant PC preparation can be composed of particles containing 0 to 12 P4 hexamers, with the majority of particles having either one or five 5-fold symmetry positions occupied. Protein P7 is an assembly cofactor that accelerates PC assembly and is essential for the formation of infectious virions (42, 43). P7 forms elongated dimers in solution (23); however, recent structural studies indicate that P7 subunits are arranged as monomers around the 3-fold axes of empty PCs (35). Thus, the multimeric state of P7 has not been established.
The self-assembly of complete PCs has been achieved in vitro by the condensation of purified protein constituents (Fig. 1). The resulting particles are competent for all enzymatic functions of the PC, including NTP hydrolysis-driven single-stranded RNA (ssRNA) packaging, replication of the packaged RNA molecules to the dsRNA form, and subsequent RNA transcription from the dsRNA templates (43). Incomplete PCs containing the main structural protein, P1, with one or two additional protein components, can be obtained using the recombinant expression system (14, 17, 19, 38) or by omitting minor protein subunits from the self-assembly reaction mix (43).
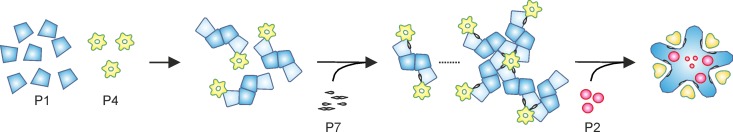
Fig 1.
Model for the self-assembly pathway of ϕ6 PC. Monomers of protein P1 (blue) interact with hexamers of P4 (yellow) to form (P1)4(P4)6 intermediates. Protein P7 (gray), which predominantly forms dimers in solution, stabilizes P1-P4 interactions and the formation of (P1)4(P4)6(P7)n complexes (43). Protein P2 (purple) does not have a measurable influence on the kinetics of PC assembly, suggesting late incorporation into the particle. The PC shell is composed of 120 copies of P1. The different conformers of P1 are presented in light and dark blue. P4 hexamers are located at the icosahedral 5-fold symmetry positions on the surface of the P1 shell. P2 and P7 are located in the interior of the capsid, near the icosahedral 3-fold symmetry axes (34, 35, 46). The theoretical maximal copy numbers for P2, P4, and P7 are 20, 72, and 60, respectively. The binding sites for P2 and P7 may overlap, reducing the actual number of polypeptides that can be incorporated into the PC.
The ϕ6 PC assembly pathway has been established based on kinetic studies of in vitro assembly reactions (43). The nucleation of PC assembly is dependent on the P4 hexamer (Fig. 1), and P7 accelerates the initial rate of assembly, whereas protein P2 does not have a detectable effect on the kinetics (42, 43). Two early assembly intermediates relating to nucleation are formed: the (P1)4(P4)6 intermediate, composed of two P1 dimers and one P4 hexamer (observed only in the absence of P7), and the (P1)4(P4)12(P7)n intermediate, where P7 stabilizes a complex containing a P1 tetramer connecting two P4 hexamers (Fig. 1). The formation of the latter intermediate is the initial rate-limiting step of PC assembly (43).
The structures and functions of proteins P2 and P4 are well characterized (7, 28, 50), but high-resolution structural information on the ϕ6 virion core is not available. The existing information regarding the stoichiometry, location, and occupancy of PC proteins was obtained primarily from electron microscopy studies (20, 21, 34, 46). The recent observation of the P2 location near the 3-fold symmetry axes of the PC, as well as the apparently low occupancy of P2 at the 20 potential binding locations of the recombinant PC particle (34, 46), encouraged us to systematically analyze the average copy numbers of proteins P2, P4, and P7 in several recombinant PC and ϕ6 virion preparations. We used the in vitro self-assembly system for ϕ6 PCs to probe the number of maximal binding sites for P2 and P7 in the ϕ6 PC structure. Through applying saturated concentrations of P2 and P7, we observed that the numbers of these proteins in PCs could be increased beyond those observed in virions and recombinant PCs. Our results also suggest a dependence between P2 and P7 incorporation, although both proteins can be assembled into particles independently (43). Furthermore, an excess of P4 over the other protein components in the PC self-assembly reaction mix significantly affected the initial rate of assembly but did not substantially change the copy number of P4 in the resulting particles. This implies that a productive assembly reaction requires P4 to initially be incorporated at each, or most, of the 5-fold symmetry positions.
MATERIALS AND METHODS
Bacterial strains and plasmids.
Escherichia coli strain BL21(DE3) was used to produce proteins P2 and P7 (48), E. coli strain HMS174 (DE3) was used for P4 recombinant protein expression (48), and the P1P4 particle and complete PCs were produced in E. coli strain JM109 (51). Pseudomonas syringae pv. phaseolicola HB10Y was used as a host for pseudomonas phage ϕ6 (49).
Plasmids pEM2, pJTJ7, and pEM7 encode proteins P2, P4, and P7, respectively (27, 36, 43). P1P4 particles were produced from plasmid pLM358 (19) for further P1 protein purification. Plasmids pLM687 (32) and pLM450 (19), containing the ϕ6 L segment cDNA, were used to produce recombinant PCs.
ϕ6 PC component purification and particle isolation.
Proteins P1, P2, P4, and P7 were purified as previously described (23, 24, 27, 43). Proteins P1, P4, and P7 were stored at −80°C, whereas protein P2 was stored at 4°C or −80°C. Protein concentrations were determined by the Bradford assay (5), using bovine serum albumin as a standard, or directly from the protein sample by using the absorbance at 280 nm. Recombinant PCs from the in vivo assembly system were isolated as previously described and stored at −80°C (16, 19). Bacteriophage ϕ6 was propagated and isolated according to the methods of Bamford et al. (1).
In vitro assembly of ϕ6 PCs.
The purified proteins P1, P2, P4, and P7 were combined in the desired relative proportions, 1 mM ATP was applied to stabilize ring-shaped P4 hexamers, and the assembly reaction was initiated by adding 50 mM Tris-HCl, pH 8.0, containing 6% (wt/vol) polyethylene glycol (PEG) 4000 (43). The reaction was carried out at room temperature for 90 min, with a P1 concentration of 0.15 or 0.2 mg/ml. Subsequently, the self-assembled PCs were separated from unassembled subunits by rate-zonal centrifugation in a linear gradient of 10 to 30% sucrose in 20 mM Tris-HCl, pH 8.0, 150 mM NaCl (Sorvall SW50.1 rotor at 114,000 × g, 90 min, 10°C). A BioComp gradient fractionator was used for fraction collection. Kinetic analysis of in vitro assembly reactions was carried out at room temperature with a P1 concentration of 0.075 mg/ml. Changes in light scattering during the reaction were recorded at 350 nm (bandwidth, 10.0 nm) in 5-s increments, using a V560 UV-visible (UV-vis) spectrophotometer (Jasco).
Quantitation of relative protein amounts.
The protein components of the recombinant PC preparations, ϕ6 virions, and sedimentation fractions in in vitro assembly reaction mixtures were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (37). The gels were subsequently stained with Coomassie brilliant blue (Serva Blue R), followed by removal of excess stain with 10% acetic acid. An Epson Perfection 4490 photo scanner was used to record the images of SDS-polyacrylamide gels. The quantitative estimation of proteins was performed on images of SDS-polyacrylamide gels via densitometric analysis of band signal intensities by Aida Image Analyzer software (Raytest; Isotopenmessgeräte GmbH). Standard curves for each PC protein were made to establish the relationships between signal intensity and protein quantity. The linear range of the standard covers the quantitative intensities observed in this study. Subsequently, the relative copy number of each PC protein in the sample (virions, recombinant PCs, or self-assembled PCs) was estimated based on the band signal intensities and the molecular weights of the proteins. The copy numbers of minor proteins P2, P4, and P7 were normalized to the copy number of the main structural protein P1, which is constant at 120 copies per PC.
RESULTS
Biochemical quantitation of relative protein amounts in ϕ6 PCs and virions.
Eighteen independently produced and purified recombinant PC samples were analyzed by SDS-PAGE and subsequent staining with Coomassie brilliant blue. The intensities of the bands corresponding to proteins P1, P2, P4, and P7 were quantitated, and the measured signal intensities were converted to relative copy numbers. A reliable relationship between the protein band intensity in a Coomassie brilliant blue-stained gel and relative protein quantity was achieved through preparation of a standard curve for each PC protein (see Fig. S1 in the supplemental material). Although each protein bound to Coomassie brilliant blue slightly differently (as indicated by the differences in the slopes of the lines in Fig. S1), the intensity of the staining correlated linearly with the amount of protein in a band of a Coomassie brilliant blue-stained SDS-polyacrylamide gel. The amount of protein per band for P1 was 0.5 to 10 μg (see Fig. S1A), and the amounts for P2, P4, and P7 were approximately 0.2 to 3.5 μg per band (see Fig. S1B). Consequently, it was possible to estimate the relative amounts of different protein components in a given PC preparation. These were converted to copy numbers by use of the known molecular weights of the proteins and the fact that the P1 copy number is 120. Similar analyses were also carried out for 13 independently produced and purified ϕ6 virion samples to identify possible differences in recombinant PCs and native PCs formed during virus assembly. It appeared that both PCs and virions contained approximately 10 copies of P2, with little variation between the different preparations (Table 1). On average, there were 10 or 11 P4 hexamers and 36 or 40 copies of P7 in recombinant PCs or virions, respectively. Slightly more variation was observed in the estimated P4 and P7 copy numbers than in the P2 copy number.
Table 1.
Relative copy numbers of proteins P2, P4, and P7 in virions and recombinant ϕ6 PCsa
| Particle type | Mean copy no. ± SD |
||
|---|---|---|---|
| P2 | P4 | P7 | |
| ϕ6 virions | 10 ± 1.3 | 66 ± 12 | 40 ± 6 |
| Recombinant PCs | 10 ± 1.6 | 62 ± 13 | 36 ± 12 |
The numbers of virions and PCs analyzed were 13 and 18, respectively.
Experimental setup for in vitro assembly analysis.
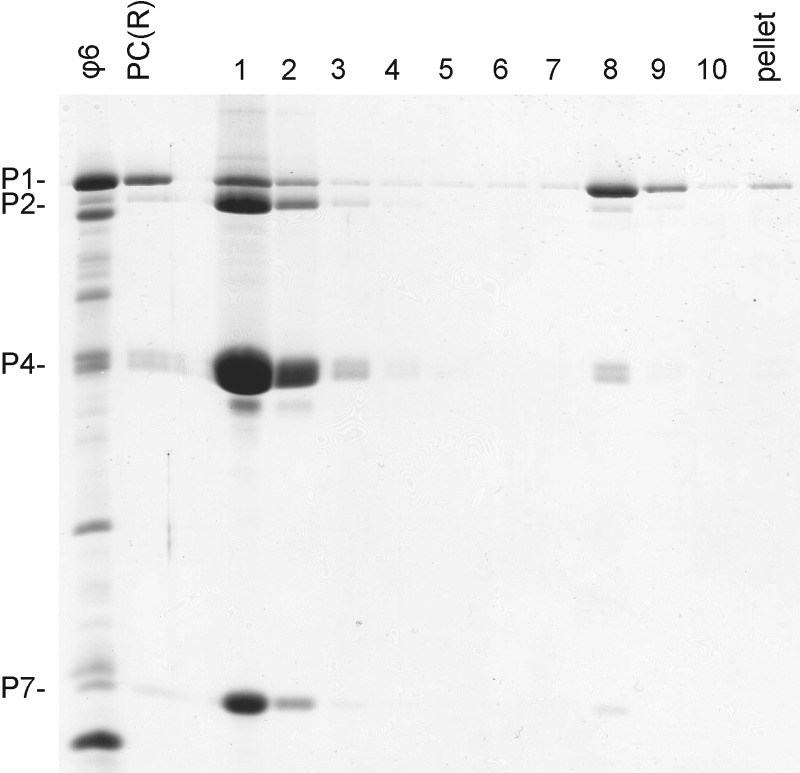
The number of potential binding sites for minor protein components in the ϕ6 PC shell was probed by titrating the amount of each protein in the in vitro assembly system followed by quantitation of the relative protein amounts in the self-assembly products. Original assembly reactions were performed using proteins P1, P2, P4, and P7 at a molar ratio of 120:12:72:45 (60 P1 dimers:12 P2 monomers:12 P4 hexamers:45 P7 monomers), corresponding to a mass ratio of 70:6:17:5. Minor protein component amounts were individually adjusted from 1/10 (for P2) or 1/4 (for P4 and P7) to up to 10-fold above the original concentration, while the concentrations of the other components were kept constant. After a 90-min self-assembly reaction, the PCs formed were separated from the unassembled material by rate-zonal centrifugation. To confirm the analytical capacity of the sedimentation assay, we prepared an assembly reaction mixture with a 10-fold molar excess of proteins P2, P4, and P7 (120:120:720:450 [P1:P2:P4:P7]). After sedimentation, the assembled PCs were detected in fractions 8 and 9, clearly separated from the unassembled protein subunits in the top fractions (Fig. 2). Little protein aggregation was observed in the pellet. Thus, rate-zonal sedimentation could be used to analyze titration reactions without significant cross-contamination between the fractions containing assembled and unassembled material.
Fig 2.
Separation capacity of 10 to 30% sucrose gradient. Proteins present in ϕ6 PCs were combined in a molar ratio of 120:120:720:450 (P1:P2:P4:P7) for an in vitro assembly reaction. In vitro-assembled PCs were separated from the unassembled subunits by use of a linear 10 to 30% sucrose gradient. Fractions 1 to 10 and the pellet were collected using a BioComp gradient fractionator and then analyzed by SDS-PAGE. Recombinant PCs [PC(R)] from E. coli and purified ϕ6 virions were added as protein size markers. The mobilities of the PC proteins are indicated on the left.
The number of potential binding sites for P2 exceeds the number of P2 molecules in ϕ6 virions.
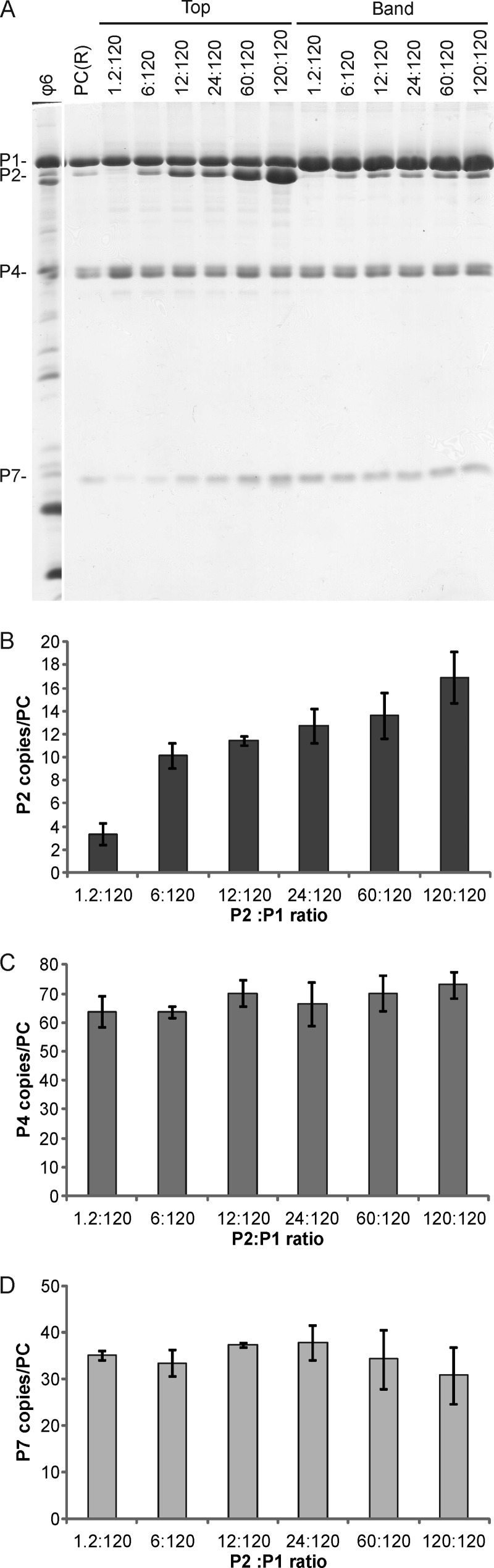
Protein P2 has been suggested to reside close to the 3-fold symmetry axes of the ϕ6 PC, with a proposed 20 binding sites per particle, although no PC with a full occupancy of P2 has been detected (34, 46). We probed the number of potential P2 binding sites by titrating the amount of P2 in the in vitro assembly reaction mix for ϕ6 PCs. The range of the P2:P1 molar ratio in the reactions was 1.2:120 to 120:120. Changes in the initial P2 amount were clearly reflected in the amount of P2 in the self-assembled PCs (Fig. 3A). The calculated copy numbers for protein P2 in the self-assembly products of different reactions suggested that approximately 12 copies of P2 were incorporated per PC in the 12:120 reaction and that up to 19 copies per PC were incorporated in the 120:120 reaction (Fig. 3B). This indicates that P2 binding sites in ϕ6 PCs exceed the number of P2 molecules in virions or in in vivo-assembled PCs (Table 1). However, changes in the amount of P2 did not significantly alter the overall PC production or the relative amounts of P4 (Fig. 3C) and P7 (Fig. 3D) proteins. The average copy numbers of P4 and P7 obtained under these conditions, i.e., 65 and 35, respectively, are similar to those detected in recombinant ϕ6 PCs (Table 1).
Fig 3.
Incorporation of P2 into in vitro-assembled PCs. In vitro assembly reactions for ϕ6 PCs were carried out with increasing amounts of P2. (A) The assembled PCs were separated from the unassembled protein subunits by rate-zonal centrifugation, and the light-scattering band and top fractions of the gradient were collected and analyzed by SDS-PAGE. Recombinant PCs [PC(R)] from E. coli and purified ϕ6 virions were added as protein size markers. The PC proteins are indicated on the left, and the P2:P1 molar ratio in each reaction mix is shown at the top. The relative copy numbers of P2 (B), P4 (C), and P7 (D) in the self-assembly products are shown with bar graphs. The error bars represent standard deviations of the means for three replicates.
High occupancy of P4 is required for formation of ϕ6 PCs.
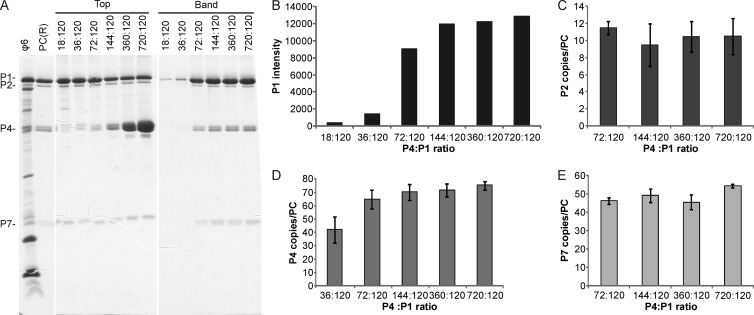
Protein P4 is necessary for the nucleation of ϕ6 PC assembly in vitro (43), but it is not established whether a single P4 hexamer is sufficient to induce the formation of a complete P1 shell. To probe this issue, we carried out a ϕ6 PC self-assembly reaction using various amounts of P4, with P4:P1 molar ratios of 18:120 to 720:120. The yield of self-assembled PCs was low if the molar ratio of P4 and P1 was below that observed in virions, as more protein subunits were detected in the top fraction (Fig. 4A). When the relative P4 amount was increased, PC production significantly increased until the P1 concentration became the limiting factor (Fig. 4B). Approximately 70 P4 subunits were incorporated per PC when a 10-fold P4 excess was applied in the reaction mix (720:120 molar ratio) (Fig. 4D), which approximately corresponds to the theoretical number of 12 P4 hexamers. Interestingly, the relative P4 copy number in PCs remained reasonably stable regardless of the initial amount of P4 in the assembly reaction mix. This suggests that for PC formation, P4 should initially associate with the P1 shell at each, or at least most, 5-fold position. Furthermore, changes in the initial P4 concentration did not significantly change the number of P2 or P7 protein molecules in the formed PCs (Fig. 4C and E).
Fig 4.
Incorporation of P4 into in vitro-assembled PCs. In vitro assembly reactions for ϕ6 PCs were carried out with increasing amounts of P4. (A) The assembled PCs were separated from the unassembled subunits by rate-zonal centrifugation, and the collected top and band fractions of the gradient were analyzed by SDS-PAGE. Recombinant PCs [PC(R)] from E. coli and purified ϕ6 virions were added as protein size markers. The PC proteins are indicated on the left, and the P4:P1 molar ratio in each reaction mix is shown at the top. The band intensities for assembled protein P1 (B) and the relative copy numbers of proteins P2 (C), P4 (D), and P7 (E) in the self-assembly products are shown with bar graphs. Quantitation for P2, P4, and P7 is shown only if the intensity of the corresponding band was sufficient. The error bars represent standard deviations of the means for three replicates.
The number of P7 binding sites in ϕ6 PCs can exceed the copy number in native ϕ6 virions.
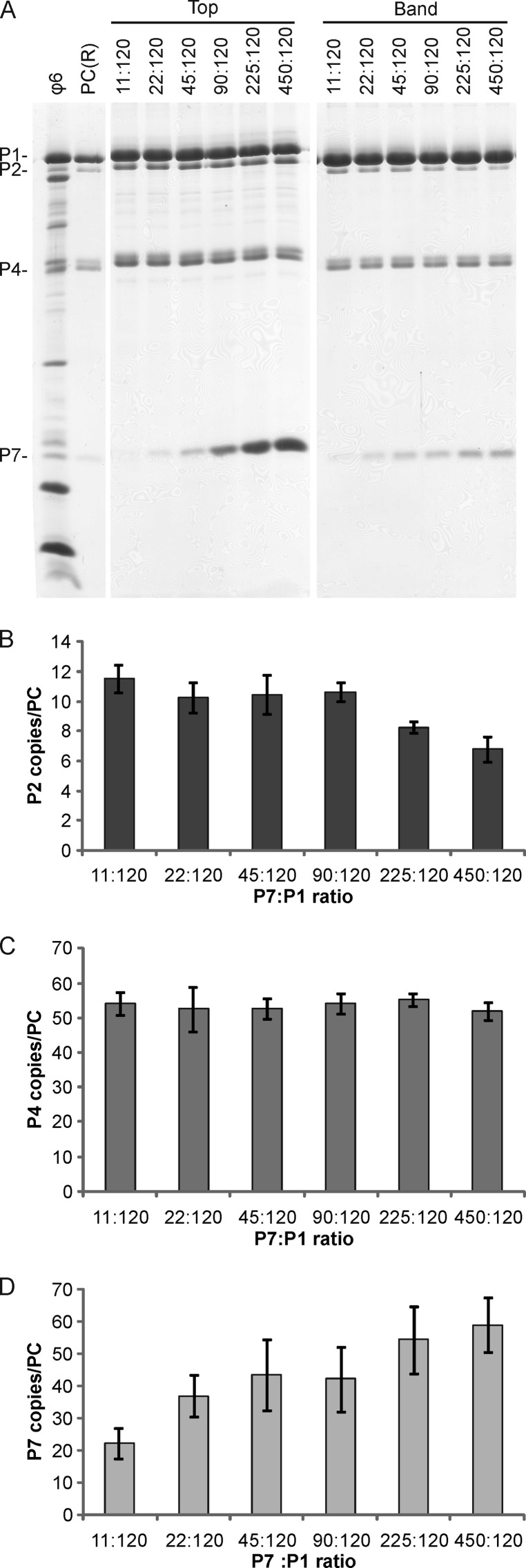
Titration of the P7 protein was carried out with P7:P1 molar ratios of 11:120 to 450:120. An increase in the relative copy number of P7 was detected with increasing P7 concentrations in the reaction mixtures. The normalized P7 copy number in the particles recovered from reaction mixtures with the largest amount of P7 (450:120 molar ratio) (Fig. 5) was approximately 1 1/2 times that observed in ϕ6 virions (Table 1), resulting in approximately 59 copies of P7 per PC (Fig. 5D). This suggested that additional P7 binding sites may exist in the ϕ6 virion. One interesting phenomenon was that the number of P2 molecules in the self-assembled PCs declined with gradually increasing P7 concentrations in the reaction mixtures (Fig. 5A and B). This suggested that previously unidentified dependencies may exist between proteins P2 and P7. Changes in P7 concentration did not have any significant effect on particle production or P4 incorporation (Fig. 5C). Cross-linking of the purified P7 protein revealed dimers, but a weak signal corresponding to the size of a P7 trimer could also be detected (see Fig. S2 in the supplemental material).
Fig 5.
Incorporation of protein P7 into in vitro-assembled PCs. In vitro assembly reactions for ϕ6 PCs were carried out with increasing amounts of protein P7. (A) The assembled PCs were separated from the unassembled protein subunits by rate-zonal centrifugation, and the collected top and band fractions of the gradient were analyzed by SDS-PAGE. Recombinant PCs [PC(R)] from E. coli and purified ϕ6 virions were added as protein size markers. The PC proteins are indicated on the left, and the P7:P1 molar ratio in each reaction mix is shown at the top. Relative copy numbers of proteins P2 (B), P4 (C), and P7 (D) in the self-assembly products are shown with bar graphs. The error bars represent standard deviations of the means for three replicates.
An imbalance in the P4:P1 molar ratio has prominent effects on PC assembly kinetics.
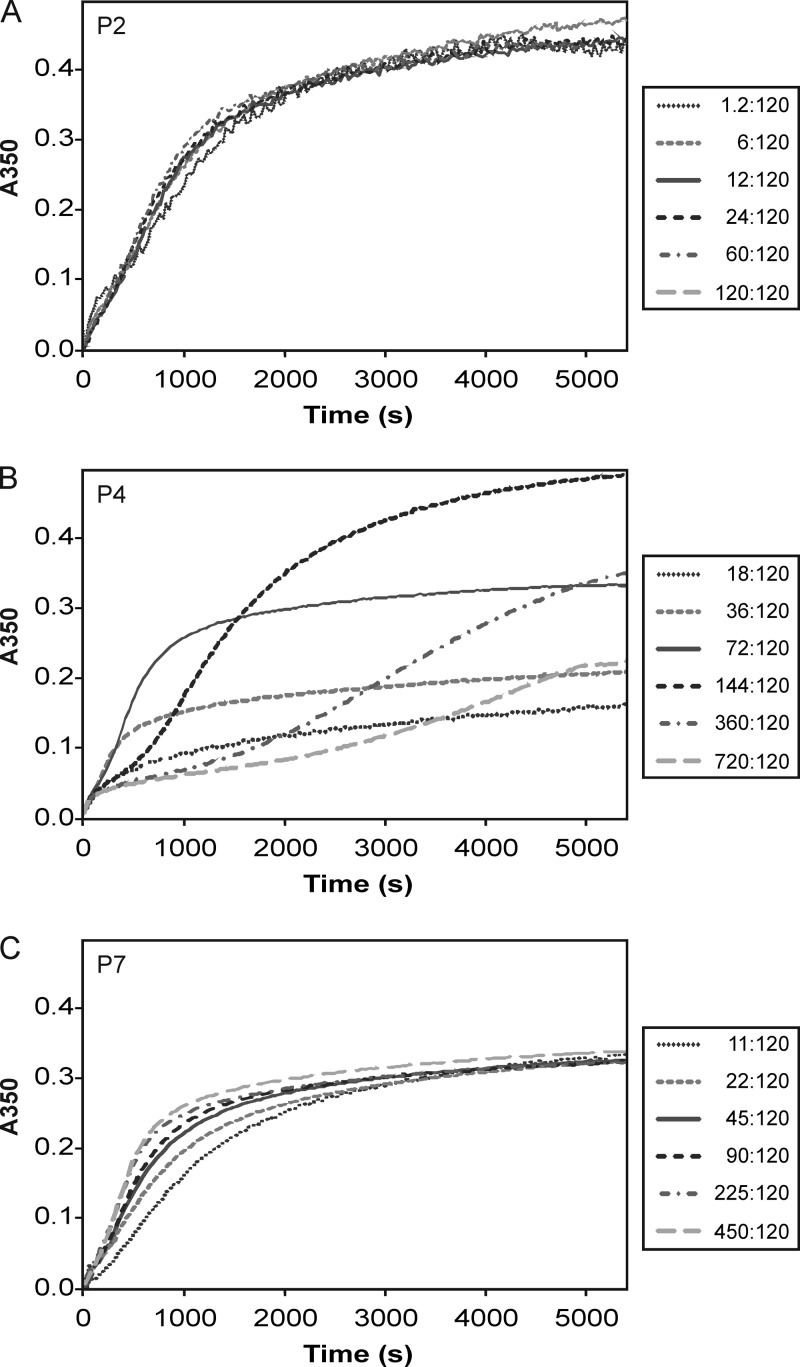
Due to the observed changes in particle composition and yields, we carried out measurements of the assembly reaction kinetics by following changes in light scattering (A350) during the self-assembly reaction and applying reaction conditions (protein ratios) similar to those described above. No significant differences could be detected in the shapes of the kinetic curves for reaction mixtures with different amounts of P2 (Fig. 6A), and an increase in P7 concentration resulted in faster initial reaction kinetics (Fig. 6C). Despite the different initial rates in kinetic curves for the P7 titration reaction, the final levels of light scattering were approximately the same for all reactions (Fig. 6C). These results are consistent with previous observations (42, 43) and confirm the role of P7 as an assembly cofactor.
Fig 6.
Increase of light scattering in solutions of proteins P1, P2, P4, and P7 after initiation of the assembly reaction. The molar ratios of P2:P1 (A), P4:P1 (B), and P7:P1 (C) for each reaction are indicated.
Increasing the P4:P1 ratio beyond the theoretical stoichiometry (72:120) resulted in a significant and previously undescribed change in the initial reaction kinetics (Fig. 6B). Reactions with a 72:120 P4:P1 molar ratio resulted in an S-shaped curve with a short initial phase, related to the nucleation of the assembly, followed by an exponential growth phase and a stationary phase (Fig. 6B). With higher P4 concentrations, the initial and exponential phases of the kinetic curves became progressively longer (Fig. 6B).
Effect of order of protein addition to the assembly mixture on the composition of the end product.
The order in which the assembly reaction components are combined can potentially affect the composition of the end product. Protein components of the assembly reaction were mixed in combinations of two or three proteins, and the reactions were initiated by applying 6% PEG. After a 60-min incubation, the missing protein component(s) was added and the incubation was continued for an additional 60 min. A P7 signal could not be detected in PC assemblies when P7 was added to preformed P1P2P4 or P1P4 particles (Fig. 7, lanes 5 and 7) (42), confirming its internal location and involvement in the formation of early assembly intermediates (Fig. 1). However, a small amount of P2 (an average of one copy per PC) was detected in reaction products when P2 was added to preformed assembly reaction mixtures missing P2 (Fig. 7, lanes 6 and 7). This may be due to continuation of the assembly reaction at the stage of P2 addition, allowing P2, which according to the kinetic data interacts with late assembly intermediates (Fig. 1), to be incorporated into the particle with a low efficiency. Interestingly, the formation of soluble particles was severely compromised if P1, P2, and P7 were preincubated without P4 in the presence of PEG (Fig. 7, lane 8), whereas postponed addition of P1 to a mixture of the other assembly reaction components did not have any effect on particle formation (Fig. 7, compare lanes 3 and 4). Analysis of the sedimentation profile of the former reaction revealed P1-specific material in the pellet fraction (data not shown), suggesting a role for P4 in directing P1 to the correct assembly pathway, preventing aggregation.
Fig 7.
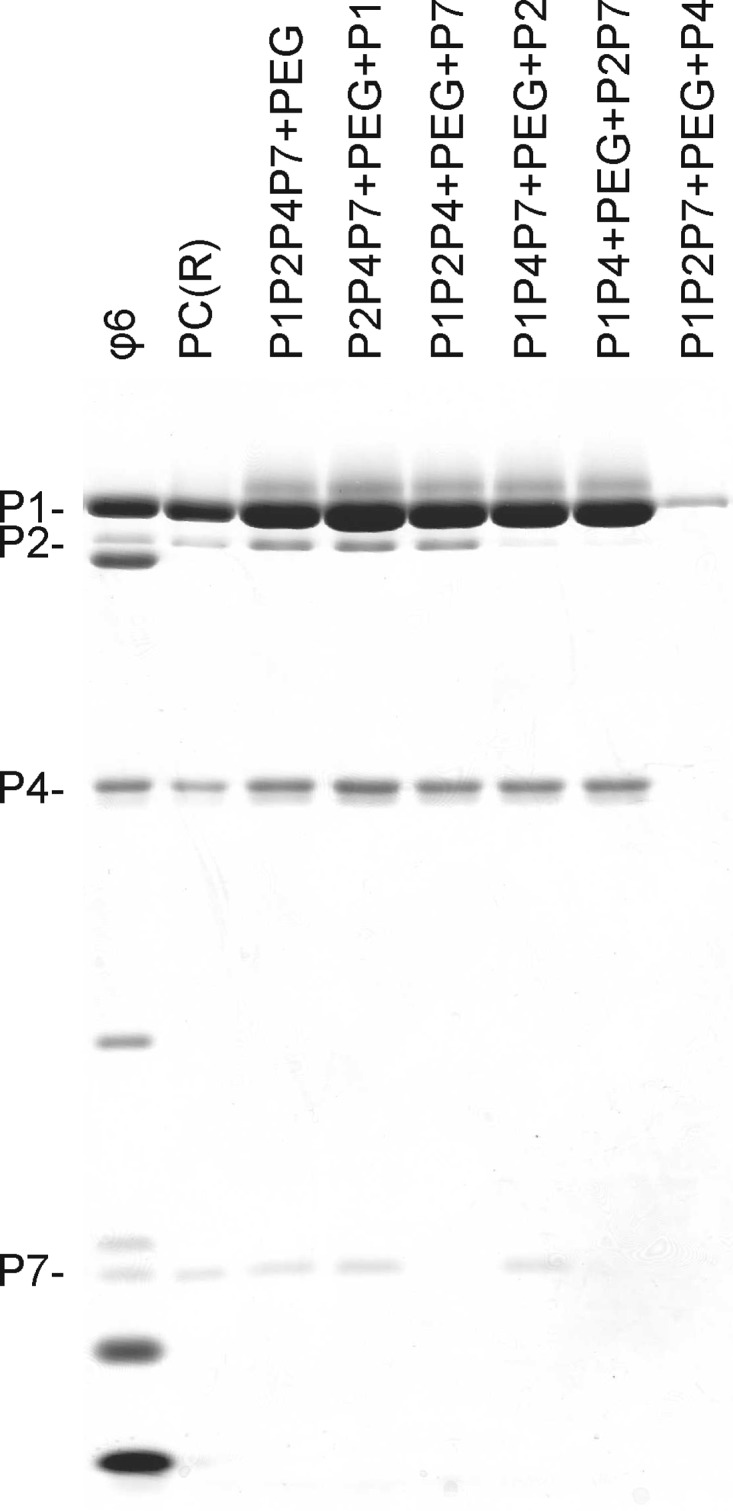
Dependency of in vitro PC production on the temporal order of reaction component addition. Standard assembly reactions were carried out in the absence of P1 (lane 4), P2 (lane 6), P4 (lane 8), P7 (lane 5), or P2 and P7 (lane 7) and in the presence of 6% PEG for 60 min. Subsequently, the missing protein component(s) was added and the incubation was continued for 60 min. The final molar ratio of proteins P1, P2, P4, and P7 was 120:12:72:45 in all reaction mixtures. The assembled PCs were separated from the unassembled protein subunits by rate-zonal centrifugation, and the light-scattering fractions of the gradients were analyzed by SDS-PAGE. Recombinant PCs [PC(R)] from E. coli and purified ϕ6 virions were added as protein size markers.
DISCUSSION
The bacteriophage ϕ6 PC, with an in vitro assembly, ssRNA packaging, replication, and transcription system, is a well-established model for dsRNA viruses. However, the stoichiometry of the protein components within the ϕ6 PC has not been established. Early biochemical studies based on quantification of individual proteins by use of radioactively labeled virus proposed that there are 100, 14, 111, and 92 copies of P1, P2, P4, and P7, respectively, in the virions of ϕ6 (13). Cryo-electron microscopy data have located minor protein components at the 5-fold (P4) or 3-fold (P2 and P7) axis of icosahedral symmetry (14, 35, 46). Here we describe a Coomassie brilliant blue staining-based biochemical quantification method for measurement of ϕ6 PC protein components (see Fig. S1 in the supplemental material). Unlike methods dependent on radioactive labeling of the proteins, the method applied in this study allows for the analysis of viral particles produced under established optimal production and purification conditions.
The assembly of ϕ6 recombinant PCs can be achieved by coexpression of the particle components in a heterologous organism and is not dependent upon additional cellular or viral factors (17). In this study, we confirmed that the composition of recombinant PCs is similar to that of virions (17) (Table 1), although the relative quantities of PC proteins, particularly proteins P4 and P7, varied slightly from preparation to preparation.
In reoviruses, the main enzymatic components (polymerase and helicase) relating to RNA replication and transcription reside in the proximity of the 5-fold vertices underneath the icosahedral capsid shell; the maximum copy number of the polymerase is proposed to be 12, which also restricts the number of genome segments to 12 (10, 11, 29, 33, 52). We observed 8 to 12 (mean of 10) P2 polymerases per particle in both ϕ6 virion and recombinant PC particles (Table 1). However, changes in the initial amount of P2 in the self-assembly system were clearly reflected in the number of P2 subunits incorporated into the final assembly product (Fig. 3B). This suggests that the P2 copy number is dependent upon the concentration of P2 in the environment during PC formation. When we used high initial P2 concentrations in the assembly reaction mix, up to 19 P2 subunits were incorporated into the PC, which supports the observation of Sen et al. (46) that P2 has 20 potential binding sites within the PC. One possible explanation for the lower P2 copy number in wild-type PCs could be the relatively low expression of P2 due to the translational coupling of gene2 to gene7 (31). Apparently, full occupancy of the P2 binding sites is not necessary for viral reproduction, and the three viral genome segments can be replicated and transcribed efficiently by PCs possessing only approximately half of the polymerase binding sites occupied. It may be more important for the efficient reproduction of the virus that the polymerase subunit, which can potentially convert any cellular ssRNA into dsRNA (26), be sequestered efficiently from the cytoplasm in particles than that there be a full occupancy of polymerase subunits in the PC.
Interestingly, the number of P7 binding sites in the ϕ6 PCs also exceeded the number in native virions or recombinant PCs. Approximately 59 copies of protein P7 were observed per particle when the initial P7:P1 ratio in the assembly reaction was high (Fig. 5D), whereas approximately 36 or 40 copies were detected in recombinant PCs or ϕ6 virions, respectively (Table 1). The observed 59 P7 subunits could occupy almost all of the theoretical 60 binding sites in the PC (three subunits at each of the 20 3-fold symmetry axes of the PC) that were also indicated in a recent cryo-electron microscopy study (35). Apparently, the association of P7 with other PC components could stabilize the trimeric state detected for cross-linked, purified P7 (see Fig. S2 in the supplemental material).
High P7 concentrations resulted in reduced P2 incorporation into the PCs (Fig. 5A and B). Since P2 does not participate in the formation of the nucleation complex, it is likely incorporated into the PC during a later stage of assembly (43) (Fig. 1). The P7-induced increase in the rate of assembly (42, 43) (Fig. 6C) could potentially decrease P2 incorporation by making a shorter time window for P2 binding to a transiently exposed binding site in the growing PC shell. This would explain why a similar dependency between P2 and P7 could not be detected for reaction mixtures with a high concentration of P2 (Fig. 3D). Alternatively, P2 and P7 could compete for the same binding site in the PC interior, as suggested by Nemecek et al. (35). This could also be an explanation for the relatively low copy number of P7 in recombinant PCs and virions (Table 1).
The detected additional binding sites for P2 and P7 are likely specific. Even at the highest applied concentrations, there were only 0.1 P7 and 0.02 P2 molecule per 17.4 × 106 Å3, the internal volume of ϕ6 PCs (20). Thus, it is unlikely that these proteins would be captured within the PC shell by chance under the conditions applied.
Protein P4 is a ring-shaped hexamer essential for the nucleation of the ϕ6 PC self-assembly reaction (43). It also mediates the viral genome packaging (40). Our analyses of the ϕ6 PC assembly also suggest that a high occupancy of P4 (Fig. 4D) could be crucial for initial particle formation. This implies that the P4 hexamer nucleating the PC assembly is not sufficient for formation of the complete shell, supporting a model in which the PC assembly proceeds from the (P1)4(P4)12(P7)n nucleation complex to complete capsids via P4-assisted P1 incorporation (see Fig. S3A in the supplemental material). This deviates from the requirements of the tailed dsDNA bacteriophages, where only a single portal complex is required to build the PC (4, 9, 12, 15, 22, 47). However, the number of P4 molecules in the recombinant ϕ6 PCs is typically significantly below full occupancy (Table 1) (14, 34). This is apparently due to the dissociation of P4 from the PC during purification. We could recover particles with practically full occupancy of P4 hexamers (12 hexamers per particle) only when a substantial excess of P4 was present during the isolation of the particle (Fig. 4D).
Previous kinetic analyses have revealed that P4 is critical in the rate-limiting step of PC assembly (43). Interestingly, an excess of P4 over the other protein components in the assembly reaction mix decreased rather than accelerated the rate of assembly (Fig. 6B). One possible explanation for such a phenomenon is that in the presence of an excess of P4, the (P1)4(P4)12(P7)n nucleation complexes accumulate, but such intermediates cannot interact with each other to build a capsid (see Fig. S3B in the supplemental material). Dissociation of one of the P4 hexamers from the complex was required to produce a (P1)4(P4)6(P7)n subassembly which could productively interact with the nucleation complex and subsequent assembly intermediates. Furthermore, it appeared that in the absence of P4, P1 may adopt an unproductive conformation that cannot be rescued by later addition of P4 (Fig. 7). Consequently, a correct ratio of P4 to P1 during ϕ6 infection is important for efficient PC production.
The effect of P7 on assembly kinetics confirmed its role as an assembly cofactor: addition of P7 increased the initial rate of assembly (Fig. 6C) (42, 43) in a concentration-dependent manner. In contrast to P4 (Fig. 6B), excess P7 in the assembly reaction mix did not cause any adverse effects on the reaction kinetics (Fig. 6C) or PC formation (Fig. 5). The amounts of P2 and P7 in the PCs were significantly diminished if these proteins were added to preassembled particles (Fig. 7), which is in accordance with their internal location (34, 35).
A generally accepted model for virus capsid shell assembly involves conformational switching, i.e., protein subunits change their conformation upon binding to a growing structure, consequently creating a new binding site for another subunit (8). This mechanism, also known as protein-determined cooperative assembly, ensures fidelity and efficiency of virus assembly, producing highly symmetrical complete virion shells with well-defined protein stoichiometries. In this study, we specifically addressed the question of how minor capsid-associated proteins are incorporated into virions. Our results indicate that incorporation of the minor capsid subunits into the PC of bacteriophage ϕ6 is not controlled by conformational switching; the growing of the P1 shell is not dependent upon the accurate incorporation of P2 and P7 subunits. Rather, their inclusion appears to depend merely on their binding affinities for specific sites in the growing capsid shell and their relative concentrations during the assembly of the virion.
Production of the minor protein components during ϕ6 infection appears insufficient to lead to a full occupancy of the potential binding sites (Table 1), and consequently, the particles produced are not symmetrical, even though the protein P1 shell utilizes icosahedral symmetry. This has significantly hampered the acquisition of high-quality structural information on the ϕ6 PC, and the localization of the minor protein constituents has been particularly difficult. Apparently, the self-assembled particles with a high occupancy of P2 and P7 reported here may offer an opportunity to define the locations and interactions of these proteins more precisely by use of electron microscopy or crystallography.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by Academy Research Fellow grants 250113 and 256069 (to M.M.P.) and by Academy professor research funding (grants 256197 and 256518 to D.H.B.) from the Academy of Finland. X.S. is a fellow in the Viikki Doctoral Program in Molecular Biosciences. We thank the University of Helsinki for support to the EU ESFRI Instruct Centre for Virus Production and Purification.
We thank R. Tarkiainen for technical assistance. We are grateful to D. Nemecek, J. Qiao, L. Mindich, A. C. Steven, and J. B. Heymanna for sharing their data prior to publication.
Footnotes
Published ahead of print 29 August 2012
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1. Bamford DH, Ojala PM, Frilander M, Walin L, Bamford JKH. 1995. Isolation, purification, and function of assembly intermediates and subviral particles of bacteriophages PRD1 and ϕ6. Methods Mol. Genet. 6:455–474 [Google Scholar]
- 2. Bamford DH, Poranen MM. 2011. Family Cystoviridae, p 515–518 In King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ. (ed), Virus taxonomy: ninth report of the International Committee on Taxonomy of Viruses. Elsevier Academic Press, Oxford, United Kingdom [Google Scholar]
- 3. Bamford DH, Romantschuk M, Somerharju PJ. 1987. Membrane fusion in prokaryotes: bacteriophage ϕ6 membrane fuses with the Pseudomonas syringae outer membrane. EMBO J. 6:1467–1473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bazinet C, King J. 1985. The DNA translocating vertex of dsDNA bacteriophage. Annu. Rev. Microbiol. 39:109–129 [DOI] [PubMed] [Google Scholar]
- 5. Bradford MM. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248–254 [DOI] [PubMed] [Google Scholar]
- 6. Butcher SJ, Dokland T, Ojala PM, Bamford DH, Fuller SD. 1997. Intermediates in the assembly pathway of the double-stranded RNA virus ϕ6. EMBO J. 16:4477–4487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Butcher SJ, Grimes JM, Makeyev EV, Bamford DH, Stuart DI. 2001. A mechanism for initiating RNA-dependent RNA polymerization. Nature 410:235–240 [DOI] [PubMed] [Google Scholar]
- 8. Caspar DL. 1980. Movement and self-control in protein assemblies. Quasi-equivalence revisited. Biophys. J. 32:103–138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chen DH, et al. 2011. Structural basis for scaffolding-mediated assembly and maturation of a dsDNA virus. Proc. Natl. Acad. Sci. U. S. A. 108:1355–1360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cheng L, Fang Q, Shah S, Atanasov IC, Zhou ZH. 2008. Subnanometer-resolution structures of the grass carp reovirus core and virion. J. Mol. Biol. 382:213–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cheng L, et al. 2010. Backbone model of an aquareovirus virion by cryo-electron microscopy and bioinformatics. J. Mol. Biol. 397:852–863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Choi KH, et al. 2008. Insight into DNA and protein transport in double-stranded DNA viruses: the structure of bacteriophage N4. J. Mol. Biol. 378:726–736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Day LA, Mindich L. 1980. The molecular weight of bacteriophage ϕ6 and its nucleocapsid. Virology 103:376–385 [DOI] [PubMed] [Google Scholar]
- 14. de Haas F, Paatero AO, Mindich L, Bamford DH, Fuller SD. 1999. A symmetry mismatch at the site of RNA packaging in the polymerase complex of dsRNA bacteriophage ϕ6. J. Mol. Biol. 294:357–372 [DOI] [PubMed] [Google Scholar]
- 15. Fokine A, et al. 2004. Molecular architecture of the prolate head of bacteriophage T4. Proc. Natl. Acad. Sci. U. S. A. 101:6003–6008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Frilander M, Bamford DH. 1995. In vitro packaging of the single-stranded RNA genomic precursors of the segmented double-stranded RNA bacteriophage ϕ6: the three segments modulate each other's packaging efficiency. J. Mol. Biol. 246:418–428 [DOI] [PubMed] [Google Scholar]
- 17. Gottlieb P, Strassman J, Bamford DH, Mindich L. 1988. Production of a polyhedral particle in Escherichia coli from a cDNA copy of the large genomic segment of bacteriophage ϕ6. J. Virol. 62:181–187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gottlieb P, Strassman J, Frucht A, Qiao XY, Mindich L. 1991. In vitro packaging of the bacteriophage ϕ6 ssRNA genomic precursors. Virology 181:589–594 [DOI] [PubMed] [Google Scholar]
- 19. Gottlieb P, Strassman J, Qiao XY, Frucht A, Mindich L. 1990. In vitro replication, packaging, and transcription of the segmented double-stranded RNA genome of bacteriophage ϕ6: studies with procapsids assembled from plasmid-encoded proteins. J. Bacteriol. 172:5774–5782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Huiskonen JT, et al. 2006. Structure of the bacteriophage ϕ6 nucleocapsid suggests a mechanism for sequential RNA packaging. Structure 14:1039–1048 [DOI] [PubMed] [Google Scholar]
- 21. Jäälinoja HT, Huiskonen JT, Butcher SJ. 2007. Electron cryomicroscopy comparison of the architectures of the enveloped bacteriophages ϕ6 and ϕ8. Structure 15:157–167 [DOI] [PubMed] [Google Scholar]
- 22. Jiang W, et al. 2006. Structure of epsilon15 bacteriophage reveals genome organization and DNA packaging/injection apparatus. Nature 439:612–616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Juuti JT, Bamford DH. 1997. Protein P7 of phage ϕ6 RNA polymerase complex, acquiring of RNA packaging activity by in vitro assembly of the purified protein onto deficient particles. J. Mol. Biol. 266:891–900 [DOI] [PubMed] [Google Scholar]
- 24. Juuti JT, Bamford DH, Tuma R, Thomas GJ., Jr 1998. Structure and NTPase activity of the RNA-translocating protein (P4) of bacteriophage ϕ6. J. Mol. Biol. 279:347–359 [DOI] [PubMed] [Google Scholar]
- 25. Kainov DE, Butcher SJ, Bamford DH, Tuma R. 2003. Conserved intermediates on the assembly pathway of double-stranded RNA bacteriophages. J. Mol. Biol. 328:791–804 [DOI] [PubMed] [Google Scholar]
- 26. Makeyev EV, Bamford DH. 2000. The polymerase subunit of a dsRNA virus plays a central role in the regulation of viral RNA metabolism. EMBO J. 19:6275–6284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Makeyev EV, Bamford DH. 2000. Replicase activity of purified recombinant protein P2 of double-stranded RNA bacteriophage ϕ6. EMBO J. 19:124–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mancini EJ, et al. 2004. Atomic snapshots of an RNA packaging motor reveal conformational changes linking ATP hydrolysis to RNA translocation. Cell 118:743–755 [DOI] [PubMed] [Google Scholar]
- 29. McClain B, Settembre E, Temple BR, Bellamy AR, Harrison SC. 2010. X-ray crystal structure of the rotavirus inner capsid particle at 3.8 Å resolution. J. Mol. Biol. 397:587–599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mindich L, Davidoff-Abelson R. 1980. The characterization of a 120 S particle formed during ϕ6 infection. Virology 103:386–391 [DOI] [PubMed] [Google Scholar]
- 31. Mindich L, et al. 1988. Nucleotide sequence of the large double-stranded RNA segment of bacteriophage ϕ6: genes specifying the viral replicase and transcriptase. J. Virol. 62:1180–1185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mindich L, Qiao X, Onodera S, Gottlieb P, Frilander M. 1994. RNA structural requirements for stability and minus-strand synthesis in the dsRNA bacteriophage ϕ6. Virology 202:258–263 [DOI] [PubMed] [Google Scholar]
- 33. Miyazaki N, et al. 2010. The functional organization of the internal components of Rice dwarf virus. J. Biochem. 147:843–850 [DOI] [PubMed] [Google Scholar]
- 34. Nemecek D, Heymann JB, Qiao J, Mindich L, Steven AC. 2010. Cryo-electron tomography of bacteriophage ϕ6 procapsids shows random occupancy of the binding sites for RNA polymerase and packaging NTPase. J. Struct. Biol. 171:389–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Nemecek D, Qiao J, Mindich L, Steven AC, Heymann JB. Packaging accessory protein P7 and polymerase P2 have mutually occluding binding sites inside the bacteriophage ϕ6 procapsid. J. Virol., in press. doi:10.1128/JVI.01347-01312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ojala PM, Juuti JT, Bamford DH. 1993. Protein P4 of double-stranded RNA bacteriophage ϕ6 is accessible on the nucleocapsid surface: epitope mapping and orientation of the protein. J. Virol. 67:2879–2886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Olkkonen VM, Bamford DH. 1989. Quantitation of the adsorption and penetration stages of bacteriophage ϕ6 infection. Virology 171:229–238 [DOI] [PubMed] [Google Scholar]
- 38. Paatero AO, Mindich L, Bamford DH. 1998. Mutational analysis of the role of nucleoside triphosphatase P4 in the assembly of the RNA polymerase complex of bacteriophage ϕ6. J. Virol. 72:10058–10065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Paatero AO, Syvaoja JE, Bamford DH. 1995. Double-stranded RNA bacteriophage ϕ6 protein P4 is an unspecific nucleoside triphosphatase activated by calcium ions. J. Virol. 69:6729–6734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Pirttimaa MJ, Paatero AO, Frilander MJ, Bamford DH. 2002. Nonspecific nucleoside triphosphatase P4 of double-stranded RNA bacteriophage ϕ6 is required for single-stranded RNA packaging and transcription. J. Virol. 76:10122–10127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Poranen MM, Bamford DH. 2012. Assembly of large icosahedral double-stranded RNA viruses. Adv. Exp. Med. Biol. 726:379–402 [DOI] [PubMed] [Google Scholar]
- 42. Poranen MM, Butcher SJ, Simonov VM, Laurinmaki P, Bamford DH. 2008. Roles of the minor capsid protein P7 in the assembly and replication of double-stranded RNA bacteriophage ϕ6. J. Mol. Biol. 383:529–538 [DOI] [PubMed] [Google Scholar]
- 43. Poranen MM, Paatero AO, Tuma R, Bamford DH. 2001. Self-assembly of a viral molecular machine from purified protein and RNA constituents. Mol. Cell 7:845–854 [DOI] [PubMed] [Google Scholar]
- 44. Poranen MM, Tuma R. 2004. Self-assembly of double-stranded RNA bacteriophages. Virus Res. 101:93–100 [DOI] [PubMed] [Google Scholar]
- 45. Semancik JS, Vidaver AK, Van Etten JL. 1973. Characterization of segmented double-helical RNA from bacteriophage ϕ6. J. Mol. Biol. 78:617–625 [DOI] [PubMed] [Google Scholar]
- 46. Sen A, et al. 2008. Initial location of the RNA-dependent RNA polymerase in the bacteriophage ϕ6 procapsid determined by cryo-electron microscopy. J. Biol. Chem. 283:12227–12231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Simpson AA, et al. 2000. Structure of the bacteriophage ϕ29 DNA packaging motor. Nature 408:745–750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Studier FW, Moffatt BA. 1986. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J. Mol. Biol. 189:113–130 [DOI] [PubMed] [Google Scholar]
- 49. Vidaver AK, Koski RK, Van Etten JL. 1973. Bacteriophage ϕ6: a lipid-containing virus of Pseudomonas phaseolicola. J. Virol. 11:799–805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wright S, Poranen MM, Bamford DH, Stuart DI, Grimes JM. 2012. Noncatalytic ions direct the RNA-dependent RNA polymerase of bacterial double-stranded RNA virus ϕ6 from de novo initiation to elongation. J. Virol. 86:2837–2849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Yanisch-Perron C, Vieira J, Messing J. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103–119 [DOI] [PubMed] [Google Scholar]
- 52. Zhang X, Walker SB, Chipman PR, Nibert ML, Baker TS. 2003. Reovirus polymerase λ3 localized by cryo-electron microscopy of virions at a resolution of 7.6 Å. Nat. Struct. Biol. 10:1011–1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.