Abstract
HLA-B*81:01 and HLA-B*39:10 alleles have been associated with viremic control in HIV-1 subtype C infection. Both alleles restrict the TL9 epitope in p24 Gag, and cytotoxic-T-lymphocyte (CTL)-mediated escape mutations in this epitope have been associated with an in vitro fitness cost to the virus. We investigated the timing and impact of mutations in the TL9 epitope on disease progression in five B*81:01- and two B*39:10-positive subtype C-infected individuals. Whereas both B*39:10 participants sampled at 2 months postinfection had viruses with mutations in the TL9 epitope, in three of the five (3/5) B*81:01 participants, TL9 escape mutations were only detected 10 months after infection, taking an additional 10 to 15 months to reach fixation. In the two remaining B*81:01 individuals, one carried a TL9 escape variant at 2 weeks postinfection, whereas no escape mutations were detected in the virus from the other participant for up to 33 months postinfection, despite CTL targeting of the epitope. In all participants, escape mutations in TL9 were linked to coevolving residues in the region of Gag known to be associated with host tropism. Late escape in TL9, together with coevolution of putative compensatory mutations, coincided with a spontaneous increase in viral loads in two individuals who were otherwise controlling the infection. These results provide in vivo evidence of the detrimental impact of B*81:01-mediated viral evolution, in a single Gag p24 epitope, on the control of viremia.
INTRODUCTION
Human leukocyte antigen (HLA) class 1 is the strongest predictor of HIV-1 disease progression (15, 44). Furthermore, cytotoxic-T-lymphocyte (CTL) responses have been associated with control of viremia, in both acute and chronic HIV-1 infections (3, 21, 23, 26, 31, 32). However, individuals carrying beneficial HLAs do not always have delayed disease progression (13). Understanding the relationship between the virus and the host, and how HLA differentially influences disease progression, remains a central question in HIV research and optimal vaccine immunogen design (40).
CTL responses targeting p24 Gag have been associated with long-term control of HIV-1 replication (1, 2, 19, 21, 31). Indeed, HLA alleles associated with control of viremia preferentially present p24 Gag epitopes and in acute infection (2, 55). An analysis of HLA-B*57/58:01 individuals from the same cohort as characterized for the present study found that individuals who targeted the TW10 epitope had better virological control, which was not lost following immune escape (7). Aside from HLA-B*57/58:01, there is limited information on the impact of CTL immune escape in individuals controlling infection in African cohorts, which may differ from Caucasian cohorts due to differences in HLA allelic frequency. HLA B7 supertype is very common in Black Africans compared to Caucasians (20, 22). The HLA-B7 supertype includes HLA-B*07:02, HLA-B*39:10, HLA-B*42:01, HLA-B*42:02, and HLA-B*81:01 (34) and, within this supertype, only HLA-B*81:01 and HLA-B*39:10 alleles are consistently associated with the control of HIV-1 in subtype C chronic infection (34). The TL9 epitope in p24 (Gag residues 180 to 188) is immunodominant, and although all individuals positive for B7 alleles can restrict this epitope, the magnitude of TL9-specific responses and the frequency of the corresponding polymorphisms in this epitope are higher in HLA-B*81:01- and HLA-B*39:10-positive individuals compared to HLA-B*42:01-positive individuals. This difference in recognition and selective pressure could be due to minor variations in these HLAs (18) or differences in HLA specificity (34), which may contribute to their differential role in influencing disease progression.
Mutations in residues E177, Q182, and T186, located in or proximal to the TL9 epitope, have been associated with immune escape in the B*81:01-, B*42:01-, and B*39:10-positive individuals (34). In vitro studies have shown that the E177D/A and T186S mutations incur a fitness cost to the virus (47, 54, 56), whereas the Q182S mutation reverts after transmission to HLA-mismatched individuals, suggesting that it is not beneficial to the virus (39). We provide here a detailed analysis of individuals with alleles belonging to the HLA-B7 supertype recruited during acute/early infection and monitored for 3 years. We report on the timing and frequency of escape in the TL9 epitope and elucidate the impact of escape on disease progression in HLA-B*81:01- and HLA-B*39:10-positive HIV-1 subtype C-infected participants.
MATERIALS AND METHODS
Study subjects.
The CAPRISA 002 Acute Infection Cohort enrolled subjects within 3 months of infection and monitored them until the initiation of antiretroviral therapy according to South African National Guidelines (defined at that time as CD4+ < 200 cells/μl or AIDS defining illness) (53). Women were enrolled from both the HIV-negative cohort and other seroincidence cohorts in Durban, South Africa. HIV-1 infection was identified using rapid antibodies assay and PCR and was confirmed using an enzyme immunoassay test as previously described (53). CD4+ T cells counts were assessed using a FACSCalibur flow cytometer, while viral loads were measured using a Cobas Amplicor HIV-1 monitor test (v1.5; Roche Diagnostics, Branchburg, NJ). Samples were collected at enrollment, weekly for 3 weeks, fortnightly until 3 months, monthly until a year, and quarterly thereafter. Written informed consent was obtained from all participants. This study received ethical approval from the University of KwaZulu-Natal, the University of Witwatersrand, and the University of Cape Town.
HIV-1 plasma RNA isolation and sequencing.
RNA was isolated from plasma samples using either a Magna-Pure compact nucleic extractor (Roche) or manually using a QIAamp viral RNA extraction minikit (Qiagen) for samples with plasma viral loads of <2,000 copies per ml. A SuperScript III reverse transcription kit (Invitrogen) and two gene-specific primers—Gag D reverse (′5-AAT TCC TCC TAT CAT TTT TGG-3′; HXB positions 2382 to 2402) and Nef OR (′5-AGG CAA GCT TTA TTG AGG-3′; HXB positions 9608 to 9625) for gag and nef, respectively—were used to transcribe cDNA from RNA. First- and second-round PCR and sequencing primers for Gag were as previously described by Chopera et al. (7). Using only the Gag B forward and Gag B reverse primers, only p24 gag sequences were generated. Sequences were assembled using Chromaspro (Technelysium, Pty., Ltd.) and aligned using the CLUSTAL W in Bioedit (default settings) (52).
IFN-γ ELISpot assay.
HIV-1-specific T-cell responses across the entire HIV-1 clade C proteome were quantified by interferon gamma (IFN-γ) enzyme-linked immunospot (ELISpot) assay using freshly isolated and prepared peripheral blood mononuclear cells as previously described (38). TL9 responses that were detected at the single-peptide level within peptide pools were confirmed in a second ELISpot assay and monitored longitudinally for these participants as previously described (27). Responses were considered positive if there were >100 spot-forming units (SFU)/106 cells (after subtraction of the background).
HLA typing.
HLA class I genotyping was performed as previously described (8). Exons 2, 3, and 4 were sequenced using Atria Allele SEQR kits (Abbott Diagnostics) and Assign SBT 3.5 software (Conexio Genomics).
Statistical analyses.
Statistical analysis and graphical presentations were implemented in Prism 5.0 (GraphPad Software, Inc.). To control for fluctuations in viral loads and CD4 counts, the 12- and 36-month set points of these parameters were calculated by taking the median of measurements at three time points closest to 12 and 36 months, respectively. To identify networks of interacting sites in the alignment, approaches coupling phylogenetic and Bayesian network models in Spidermonkey (45) were used (www.datamonkey.org). Repeated inferences with ancestral states sampled from the posterior distribution were used to evaluate robustness (11) in identifying networks of interacting sites in an alignment using Bayesian network techniques, which assumes that coevolving sites will tend to acquire mutations along the same set of branches (45). Statistical analysis of significance was based on Mann-Whitney two-tailed t test.
Nucleotide sequence accession numbers.
Sequences for the present study were submitted to GenBank under accession numbers JX434413 to JX34485 and JX434486 to JX434595.
RESULTS
Of the 62 women recruited into the CAPRISA 002 acute infection study, five were HLA-B*81:01 positive and two were B*39:10 positive (Table 1). Women were recruited at a median time point of 6 weeks postinfection (interquartile range [IQR] = 4 to 8 weeks). To evaluate gag evolution and the timing of escape in the B*81:01- and B*39:10-restricted TL9 epitope, sequencing was performed on samples collected at the earliest time point and every 3 months postinfection until 3 years or initiation of antiretroviral (ARV) therapy according to national guidelines (CD4+ T-cell counts < 200 cells/μl or AIDS-defining illness). Samples from additional time points were sequenced to further refine the timing of escape.
Table 1.
HLA type 1for the five B*81:01- and two B*39:10-positive individuals in this study
| PIDa | HLA type(s) |
||
|---|---|---|---|
| Allele pair |
HLA-C | ||
| HLA-A | HLA-B | ||
| CAP0129 | 26:01, 80:01 | 18:01, 81:01 | 02:02, 04:01 |
| CAP0222 | 30:01, 33:03 | 53:01, 81:01 | 04:01, 04:01 |
| CAP0225 | 01:01, 30:01 | 42:02, 81:01 | 17:01, 18:01 |
| CAP0262 | 01:01, 66:02 | 42:01, 81:01 | 17:01, 18:01 |
| CAP0277 | 30:09, 43:01 | 58:02, 81:01 | 04:01, 04:01 |
| CAP0278 | 30:01, 43:01 | 39:10, 42:02 | 12:03, 17:01 |
| CAP0289 | 30:02, 68:01 | 39:10, 58:02 | 06:02, 12:03 |
PID, participant identification number.
Influence of HLA-B*81:01 and HLA-B*39:10 on early and chronic disease progression.
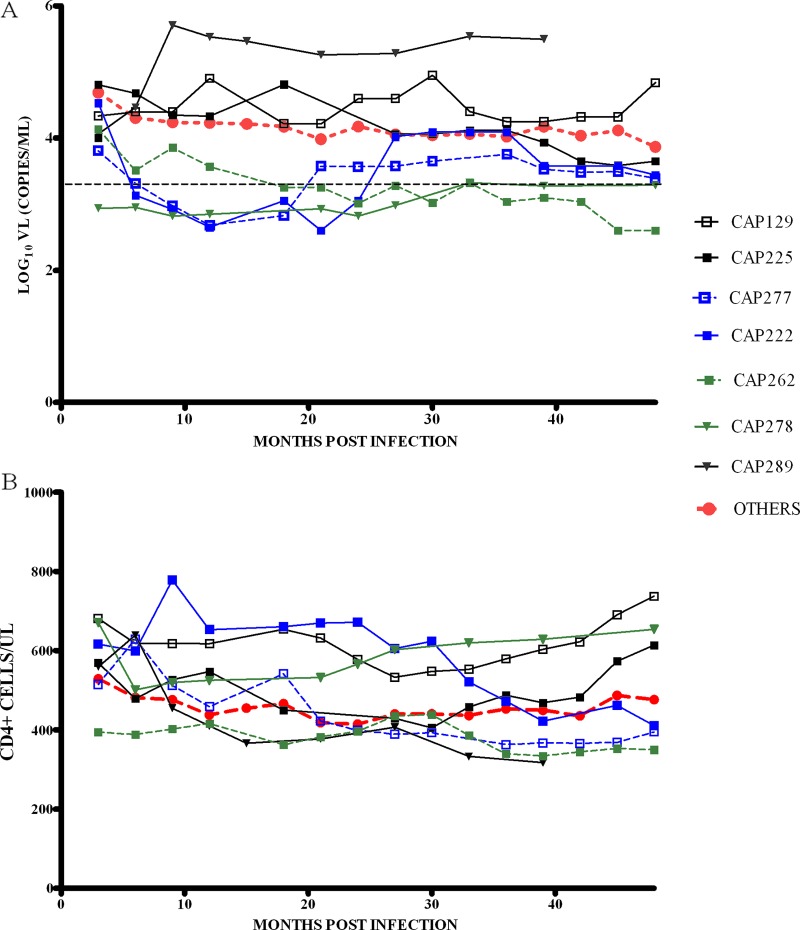
To understand disease progression in HLA-B*81:01/B*39:10-positive participants, we analyzed the trajectory of viral loads and CD4+ T-cell counts over a 4-year postinfection period (Fig. 1).
Fig 1.
(A) Viral load and (B) CD4+ T-cell count trajectories for the first 48 months of infection for five HLA-B*81:01-positive participants (CAP129, CAP222, CAP225, CAP262 and CAP277; represented by squares) and two B*39:10-positive participants (CAP278 and CAP289; represented by triangles). CAP289 had not reached 48 months postinfection by the time of this study. The trajectory of median viral loads and median CD4+ T-cell counts for all other CAPRISA participants (OTHERS), excluding B*39:10/81:01 and B*57/58:01, are shown as dotted red lines. The broken black line represents the viral load level of 2,000 copies/ml, defining controllers. Viral loads and CD4+ T-cell counts were smoothed at each time point (medians of measurements taken at three time points closest to intervals 3, 6, 9, 12, 15, etc., up to 48 months postinfection).
Of the two B*39:10-positive participants, one was a progressor (CAP289) and the other (CAP278) a controller. The B*39:10 progressor maintained high viral loads from the first year of infection (>500,000 copies/ml), with lower median CD4+ T cells compared to the rest of the cohort. The B*39:10 controller maintained viral loads below 2,000 copies/ml and had high CD4+ T cells (>600 cells/μl) after 4 years of infection. Of the five B*81:01-positive participants, one controlled her viral load after 18 months of infection (CAP262); two participants who initially controlled viral replication below 2,000 copies/ml for the first 18 to 24 months of infection had viral loads that increased by 1 log over a 6-month period and thereafter (CAP222 and CAP277). By 4 years postinfection, these individuals had lower CD4 T-cell counts compared to individuals with nonprotective alleles (i.e., B*81:01/57/58:01/39:10-negative participants). Of the two remaining B*81:01 individuals, CAP129 had a relatively high viral load, though with increase in CD4+ T-cell counts between years 3 and 4 postinfection, and CAP225 had typical viral loads which declined between years 3 and 4 postinfection with a concomitant increase in CD4+ T-cell count.
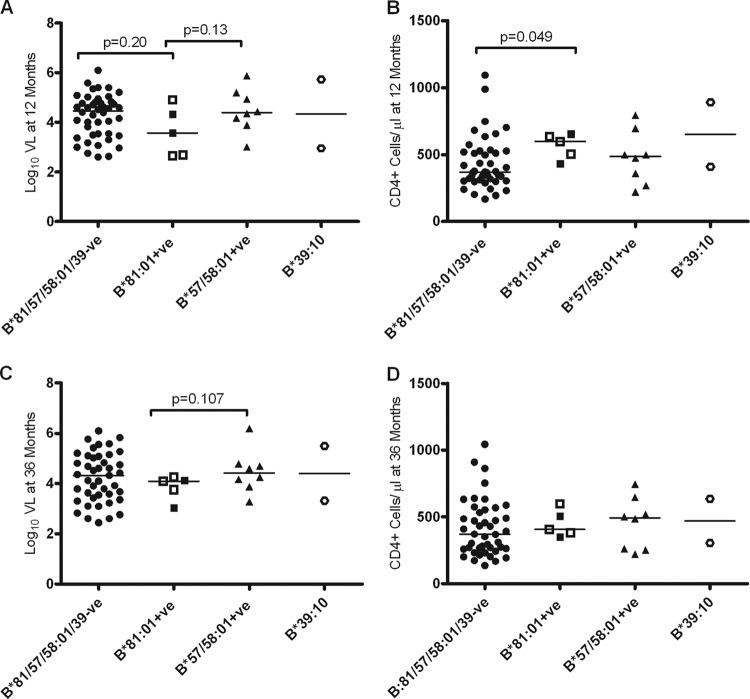
We then investigated the influence of HLA-B*81:01 and HLA-B*39:10 on viral load set point and CD4+ T-cell counts at 1 and 3 years postinfection. B*57/58:01 participants were analyzed separately since these “protective” alleles are often associated with better control of HIV-1 replication (41, 42). Although the median viral load at set point (1 year postinfection) was lower in B*81:01-positive participants compared to those with nonprotective alleles (i.e., B*81:01/57/58:01/39:10-negative participants), this was not significant (P = 0.2) (Fig. 2A; median, 3.57 log10 copies/ml [IQR = 2.7 to 4.6] versus 4.46 log10 copies/ml [IQR = 3.49 to 4.8]). There were, however, significantly higher CD4+ T-cell counts for the B*81:01-positive participants compared to individuals with nonprotective alleles at 1 year postinfection (P = 0.049) (Fig. 2B; median, 597 cells/μl [IQR = 467 to 644] versus 369 cells/μl [IQR = 305 to 525]). To determine whether these HLA alleles affected disease progression in chronic infection, we analyzed both viral loads and CD4+ T-cell counts at 3 years postinfection. By 3 years postinfection, 12 participants had initiated ARV therapy and, in order to not bias the analysis toward the controllers, we used the last available viral load and CD4 count data for these subjects prior to their commencing ARV therapy. We found no significant differences in either viral load or CD4+ T-cell counts at 3 years postinfection (Fig. 2) or 4 years (data not shown) in B*81:01-positive participants compared to those with nonprotective alleles, suggesting that this potential benefit detected at 12 months was not sustained in chronic infection.
Fig 2.
Association between viral load (VL) and CD4+ T-cell counts prognostic markers and HLA-B*81:01 and HLA-B*39:10 alleles at 12 and 36 months postinfection. (A and C) Log10 VL values compared between individuals without and with HLA-B*81:01, HLA-B*39:10, or HLA-B*57:03/58:01 alleles at 12 and 36 months postinfection; (B and D) CD4+ T-cell counts compared between individuals without and with HLA-B*81:01, HLA-B*39:10, or HLA-B*57/58:01 alleles at 12 and 36 months postinfection, respectively. B*81:01-positive participants shown as empty symbols are those positive for the Cw*04:01 allele. Both B*39:10-positive participants are also positive for the Cw*12:03 allele. Viral load and CD4+ T-cell counts were calculated as medians of measurements taken at three time points closest to these intervals. To not enrich for slow progressors, we included the last attained median values for viral load and CD4+ T-cell counts before the initiation of ARV therapy for individuals who had received treatment prior to 12 months (n = 7) or 36 months (n = 12) postinfection.
We further investigated whether B*81:01-positive participants controlled infection better than B*57/58:01-positive participants. Although at 1 year, the CD4+ T-cell counts tended to be higher and viral loads lower in B*81:01-positive participants compared to B*57/58:01-positive participants, this difference was not significant, and this trend was not sustained at 3 years (Fig. 2C and D).
Of the two B*39:10-positive participants, one was a controller, whereas the other was a typical disease progressor (Fig. 1). The controller had a viral load at 1 year of 2.95 log10 copies/ml compared to 5.72 log10 copies/ml for the progressor, while their respective CD4+ T-cell counts were 568 and 410 cells/μl (Fig. 2).
Linkage to HLA-Cw alleles did not impact disease progression.
In subtype C infection, a good clinical outcome in individuals positive for B*81:01 and B*39:10 alleles has been linked to an additive impact of the Cw*04:01 and Cw*12:03 alleles, respectively (33). Of the five B*81:01-positive participants, three participants were positive for the Cw*04:01 allele, of which two were controllers (CAP222 and CAP277) and the third was a typical progressor (CAP129). However, of the two B*81:01-positive participants who did not carry the Cw*04:01 allele, one was a typical progressor (CAP225), while the other was a controller (CAP262). Although both of the B*39:10-positive participants were also positive for the Cw*12:03 allele (Table 1), one was a progressor and the other was a controller (Fig. 1). These results suggest that the differential disease progression in B*81:01- or B*39:10-positive participants was not simply due to the HLA-Cw allelic combinations.
Kinetics of p24-Gag-TL9 escape in B*39:10- and B*81:01-positive participants.
Substitutions in positions E177, Q182, and T186 Gag residues that are located within or proximal to the TL9 epitope (Gag180-188) result in the loss of immune recognition in B*81:01/B*39:10-positive individuals, with the predominant polymorphisms associated with HLA-mediated TL9 restriction in vivo being E177D, Q182S, and T186S (16, 34, 39, 47).
In B*81:01-positive participants, mutations in TL9 generally emerged later in infection (Table 2). The first mutation to emerge was at position 182. In three participants who selected this mutation postinfection, the Q182X (where X = S/T/A/H) was detected between at 70 and 83 weeks postinfection in CAP222, at 50 and 61 weeks in CAP225, and at 48 and 50 weeks in CAP277. In two of the three participants (CAP222 and CAP277), additional escape mutations were detected within 6 to 12 months after the emergence of the escape mutation at position 182 and reached fixation only 10 to 15 months later (Table 2). The T186S mutation emerged either alone as in CAP277 or concurrently with E177D, as in CAP222.
Table 2.
HLA-B*81:01, HLA-B*39:10, and other B7 allelic restricted sequence variations in and proximal to the TL9 epitope and putative compensatory mutationsa
| PIDb | wpic | TL9 mutations |
Other mutations |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| El77 | T180 | Q182 | T186 | L188 | T190 | H219 | I223 | A248 | M250 | I256 | ||
| CAP129 | 2 | – | – | T | – | – | – | – | H | T | – | – |
| B*81:01 | 23 | – | – | T | – | – | – | – | H | T | – | – |
| 32 | – | – | S | – | – | – | – | H | T | – | – | |
| 37 | – | S | – | – | – | – | H | T | – | – | ||
| 42 | – | – | S | – | – | – | – | H | T | – | – | |
| 54 | – | – | S | – | – | – | – | H | T | – | – | |
| 106 | – | – | S | – | – | – | – | H | T | – | – | |
| CAP222 | 56 | – | – | – | – | – | – | – | A | – | – | – |
| B*81:01 | 70 | – | – | – | – | – | – | – | A | – | – | – |
| 83 | – | – | S (6/9) | – | – | – | – | A | – | – | V | |
| 95 | – | – | S (1/6) | – | – | – | – | A | – | – | V | |
| 108 | – | – | S (3/11) | – | – | – | – | A | – | – | V (3/11) | |
| 108 | D | A | S | S (1/11) | – | – | – | A | – | – | – | |
| 108 | D | – | S | S (7/11) | – | – | – | V (2/7) | – | – | – | |
| 122 | D | – | S | S (13/13) | – | – | – | V 13/13 | – | – | – | |
| 133 | D | – | S | S | – | – | – | V (13/13) | – | – | – | |
| 148 | D | – | S | S | – | – | – | V | – | – | – | |
| 161 | D | – | S | S | – | – | – | V | – | – | – | |
| 174 | D | – | S | S | – | – | – | V (13/13) | – | – | – | |
| 190 | D | – | S | S | – | – | – | V | – | – | – | |
| CAP225 | 50 | – | – | – | – | – | – | – | – | – | – | – |
| B*81:01 | 61 | – | – | S (10/15) | – | – | – | – | – | – | – | V (10/15) |
| 74 | – | – | S | – | – | – | – | – | – | – | V | |
| 85 | – | – | S (3/15) | – | – | – | – | – | – | – | V (3/15) | |
| 98 | – | – | S | – | – | – | – | – | – | – | V | |
| 117 | – | – | S | – | – | – | – | – | – | – | V | |
| 130 | – | – | S | – | – | – | – | – | – | – | V | |
| 145 | – | – | S | – | – | – | – | – | – | – | V | |
| 158 | – | – | S | – | – | – | – | – | – | – | V | |
| 171 | – | – | S | – | – | – | – | – | – | – | V | |
| 183 | – | – | S | – | – | – | – | – | – | – | V | |
| CAP277 | 16 | – | – | – | – | – | – | – | – | – | I | – |
| B*81:01 | 41 | – | – | – | – | – | – | – | – | – | I | – |
| 46 | – | – | – | – | – | – | – | – | – | I | – | |
| 50 | – | – | H | – | – | – | – | – | – | I | – | |
| 54 | – | – | T | – | – | – | – | – | – | I | – | |
| 58 | – | – | T | – | – | – | – | – | – | I | – | |
| 71 | – | – | A | – | – | – | – | – | – | I | – | |
| 84 | – | – | S | – | – | – | – | – | – | I | – | |
| 97 | – | – | S (3/13) | – | – | – | – | – | – | I (3/13) | – | |
| 97 | – | – | S | S (6/13) | – | – | – | – | – | – (6/13) | – | |
| 97 | – | – | – | – | – | – | – | – | – | – (4/13) | – | |
| 110 | – | – | S | S | – | – | – | – | T | – | – | |
| 123 | – | – | S | S | – | – | – | – | T | – | – | |
| 137 | – | – | S | S | – | – | – | – | T | – | – | |
| CAP262 | 70 | – | – | – | – | – | – | – | – | – | – | – |
| B*81:01 | 84 | – | – | – | – | – | – | – | – | – | – | – |
| B*42:01 | 95 | – | – | – | – | – | – | – | – | – | – | – |
| 109 | – | – | – | – | – | – | – | – | – | – | – | |
| 122 | – | – | – | – | – | – | – | – | – | – | – | |
| 136 | – | – | – | – | – | – | – | – | – | – | – | |
| CAP278 | 9 | D | – | S | S | – | – | – | V | – | – | – |
| B*39:10 | 54 | D | – | S | S | – | – | – | V | – | – | – |
| 85 | D | – | S | S | – | – | Q | V | – | – | – | |
| CAP289 | 8 | – | – | A | – | – | – | – | V | – | – | V |
| B*39:10 | 24 | – | – | A | – | – | – | – | V | – | – | V |
| 28 | – | – | G | – | – | – | – | V | – | – | V | |
| 41 | – | – | G | – | – | – | – | V | – | – | V | |
| CAP061 | 14 | – | – | – | – | – | – | Q | V | – | – | – |
| B*42:01 | 33 | – | – | – | – | – | – | Q | V | – | – | – |
| C*08:02 | 51 | – | – | – | – | – | – | Q | V | – | – | – |
| 76 | – | – | – | – | F | – | Q | V | – | I | – | |
| 102 | – | – | – | – | – | – | – | V | – | – | – | |
| CAP282 | 11 | – | – | – | – | – | – | – | – | – | – | V |
| B*42:01 | 35 | – | – | – | – | – | – | – | – | – | – | V |
| 53 | – | – | T | – | – | – | – | – | – | – | V | |
The commonly evolving amino acid residues are indicated in lowercase italics in the peptide 176-SeGATPqDLNtML-188. –, No residue substitution or else the incoming residue is indicated; ND, not done or not available. Where applicable, the ratio of the number of mutations detected/total is indicated in parentheses.
PID, participant identification number.
wpi, weeks postinfection.
In the fourth participant, CAP129, sampled prior to a detectable HIV antibody response, the Q182T mutation was already present. The mutation remained stable for up to 28 weeks; however, by 32 weeks the Q182S mutation emerged. Since the Q182X mutation in the TL9 epitope was detected in the earliest sample sequenced from three of the 37 CAPRISA participants who were negative for HLA-B7 supertype alleles, it is possible that this mutation was present in the transmitted virus. This is supported by the fact that there was no evidence of an immune response to this epitope in acute infection (Table 3). There was no sequence evolution in the TL9 epitope in the fifth B*81:01 participant (CAP262).
Table 3.
Temporal pattern of TL9-specific IFN-γ ELISpot assay responses inB7-positive participants
| PIDa | HLAb | wpic | SFU/106 cellsd | Last WT and first TL9 mutatione |
|---|---|---|---|---|
| CAP129 | A*26:01, A*80:01; B*18:01, B*81:01; Cw*02:02, Cw*04:01 | 2 | Neg | Q182T |
| 4 | Neg | |||
| 6 | Neg | |||
| 13 | Neg | |||
| 23 | Neg | Q182T | ||
| 32 | ND | Q182S | ||
| CAP222 | A*30:01, A*33:03; B*53:01, B*81:01; Cw*04:01, Cw*04:01 | 6 | 245 | |
| 8 | 103 | |||
| 9 | 1,568 | |||
| 11 | 368* | |||
| 15 | 168 | |||
| 17 | 243 | |||
| 70 | ND | WT | ||
| 83 | ND | WT/Q182S (mixed) | ||
| 108 | 1,233 | Q182S/and E177D with T186S (mixed) | ||
| 122 | ND | E177D, Q182S, T186S (fixed) | ||
| CAP225 | A*01:01, A*30:01; B*81:01, B*42:02; Cw*17:01, Cw*18:01 | 3 | 9,078 | |
| 4 | 888 | |||
| 5 | 475 | |||
| 7 | 1,288 | |||
| 9 | 1,058 | |||
| 14 | 203 | |||
| 18 | 3,328 | |||
| 21 | 1,513 | |||
| 24 | 1,463 | |||
| 29 | 2,080 | |||
| 33 | 818 | |||
| 37 | 468 | |||
| 50 | 1,710 | WT | ||
| 61 | ND | WT/Q182S (mixed) | ||
| 85 | Neg | WT/Q182S (mixed) | ||
| 145 | 1,221 | Q182S (fixed) | ||
| CAP262 | A*01:01, A*66:02; B*42:01, B*81:01; Cw*17:01, Cw*18:01 | 7 | 365 | |
| 14 | Neg | |||
| 16 | 215 | |||
| 18 | 250 | |||
| 26 | 105 | |||
| 136 | ND | WT | ||
| CAP277 | A*30:09, A*43:01; B*58:02, B*81:01; Cw*04:01, Cw*04:01 | 37 | 6,147 | |
| 46 | ND | WT | ||
| 50 | ND | Q182H | ||
| 97 | 193 | WT/Q182S/T186S (mixed) | ||
| CAP278 | A*30:01, A*43:01; B*39:10, B*42:02; Cw*12:03, Cw*17:01 | 8 | 117 | ND |
| 9 | ND | E177D, Q182S, and T186S | ||
| 10 | Neg | |||
| CAP289 | A*30:02, 68:01; B*39:10, B*58:02; Cw*06:02, Cw*12:03 | 8 | ND | Q182A |
| 9 | Neg | |||
| 28 | ND | Q182G | ||
| CAP037 | A*23:01, A*24:02; B*07:02, B*53:01; Cw*17:01, Cw*17:01 | 122 | 273 | WT |
| CAP040 | A*30:01, A*30:02; B*15:10, B*42:01; Cw*03:04, Cw*17:01 | 106 | 293 | WT |
| CAP220 | A*30:04, A*74:01; B*42:01, B*42:01; Cw*17:01, Cw*17:01 | 68 | 373 | WT |
| CAP239 | A*01:23, A*29:02; B*42:01, B*58:01; Cw*06:02, Cw*17:01 | 67 | 258 | WT |
| CAP269 | A*02:05, A*68:02; B*07:02, B*58:02; Cw*06, Cw*07 | 120 | Neg | WT |
| CAP282 | A*30:01, A*66:01; B*42:01, B*58:02; Cw*:ND | 23 | 129 | |
| 35 | ND | WT | ||
| 53 | ND | Q182T first observed at 12 months postinfection | ||
| 60 | Neg |
PID, participant identification number.
HLAs known to restrict TL9 are underlined.
wpi, weeks postinfection.
ND, not done; Neg, negative; SFU, spot-forming units.
WT, wild type. Mixed, a population in which more than one viral variant was identified; fixed, a population in which only one viral variant was identified.
The viruses from the two B*39:10-positive participants had a different pattern of escape. In one B*39:10 participant (CAP278), all three mutations (E177D, Q182S, and T186S) were already present in the earliest sequences available at 9 weeks postinfection, with no further evolution within the epitope during 22 months of follow-up. In the second B*39:10 participant (CAP289), the Q182A mutation was present at 8 weeks postinfection and evolved to Q182G at 28 weeks postinfection.
To evaluate the likelihood of the HLA-B*39:10-associated mutations in residues E177, Q182, and T186 being present on the transmitted virus in CAP278, we estimated the frequency of TL9 mutations in participants negative for HLA-B7 supertype and Cw*08:02 alleles that recognize this epitope (35) in the CAPRISA 002 cohort. We found no sequences from either acute or chronic infection with either the two (Q182S with T186S) or the three (E177D, Q182S, and T186S) concurrent mutations. Furthermore, the triple mutation was not present in any of the sequences analyzed from 507 HIV-1 subtype C-infected adults in Durban, KwaZulu-Natal, South Africa (30, 48).
Together, these results show that the TL9 escape in B*81:01 generally occurs later and can take approximately 1 year from the first mutations for escape in this epitope to reach fixation. Furthermore, we show that the three concurrent E177D, Q182S, and T186S site mutations are rare and might have arisen due to early escape in one of the two B*39:10-positive participants, although it cannot be excluded that some of these mutations were transmitted variants from a B7 supertype donor.
HLA-B*81:01 and HLA-B*39:10 exert more selection pressure on TL9 than HLA-B*42 and HLA-B*07:02.
Previous studies have shown that the strength of immune responses targeting the TL9 epitope differ according to the HLAs belonging to HLA-B7 supertype with HLA-B*81:01 being most dominant, followed by HLA-B*39:10 and then HLA-B*42:01 (34). Since three participants in the present study were also positive for the HLA-B42 allele, we wanted to know the frequency of HLA-mediated sequence evolution in the TL9 epitope in participants carrying HLA-B7 supertype (n = 17), excluding the HLA-B*81:01/39:10-positive participants. Analysis of sequences at 18 months postinfection revealed that only two of the 13 B*42:01-positive participants developed polymorphisms in the epitope (Table 2), whereas no mutations were detected in the four B*07:02-positive participants (Table 3). In one of the two B*42:01-positive participants who selected mutations in the TL9 epitope, an escape mutation L188F emerged between 51 and 76 weeks; however, this mutation was not fixed since it was not detected at 102 weeks postinfection (Table 2). In the second participant, the Q182T mutation was selected between 48 and 53 weeks postinfection, while four of the five B*81:01-positive individuals selected for 182S. A recent study analyzing 2,126 chronically HIV-1 subtype C-infected, antiretroviral naive adults showed that unlike the B*81:01 allele that select against the Q182T mutation, the B*42:01 allele selected for Q182T (6). Thus, the differences in escape at residue 182 provide further support that B*81:01 restriction is responsible for the changes in the TL9 epitopes. These results support the findings of Leslie et al. (34), which demonstrated that B*81:01 and B*39:10 alleles select for mutations in the TL9 epitope over the HLA-B*42:01 allele. Overall, these findings emphasize the differential role of closely related HLA in the timing of epitope escape and sequence evolution.
Late sequence evolution in TL9 is not due to the lack of CTL responses in acute infection.
A lack of CTL pressure in acute infection has been suggested to partly explain why some epitopes remain invariant through acute infection and only evolve late in infection (24). IFN-γ ELISpot data are available for 13 participants with HLA-B7 supertype alleles (Table 3). Responses to the TL9 epitope were identified in nine participants, with the magnitude of responses in B*81:01-positive participants being generally higher, ranging from >100 up to 9,078 SFU/106 cells, compared to other B7 participants, which ranged from about >100 to 373 SFU/106 cells.
Of the five B*81:01-positive participants, four targeted the TL9 epitope with responses detected in acute infection for three participants and at 9 months for one participant (CAP277) (earliest sample tested). Three B*81:01-positive participants had strong TL9 responses (1,569 to 9,078 SFU/106 cells) and, despite these strong responses, escape only occurred 10 to 12 months after the first detectable responses. We did not detect escape during the 33 months of follow-up in the fourth B*81:01-positive (CAP262) participant with detectable but low TL9 responses (250 SFU/106 cells). The one B*81:01-positive participant (CAP129) who harbored an escape mutant at 2 weeks postinfection did not have detectable responses to TL9 in the first 6 months of infection. Only one of the B*39:10-positive participants had detectable responses to TL9 during acute infection (CAP278). The peptide used to detect responses corresponded to the wild-type sequence, and it is possible that responses in individuals harboring TL9 escape viruses in very early infection could be missed due to differences in the viral mutant and the peptide used.
Other than the B*81:01/39:10-positive participants, immune responses at later time points (>12 months postinfection) were generated for six of the 19 participants positive for other HLA-B7 supertype and Cw*08:02 alleles known to target the TL9 epitope. Low-magnitude responses (129 to 273 SFU/106 cells) were detected in five of the six participants tested; however, only one of these participants (CAP282) developed an escape mutant (Q182T).
Similar to a previous study in this cohort (43), fluctuations in immune response were not necessarily related to immune escape. Due to the limited availability of specimens, it was not possible to accurately determine the effect of sequence changes on immune responses. However, loss of responses with concomitant sequence change was detected in three individuals (CAP225, CAP278, and CAP282).
Sequence evolution in the TL9 epitope is associated with covarying sites outside of the epitope.
Studies have found significant associations between mutations in the TL9 epitope and highly variable residues (Gag residues 138, 228, 248, 252, and 256) located in or flanking the three tropism-determining loops of p24 involved in the recognition of host factors necessary for virus replication or inhibition (loops 1 to 3 located at Gag residues 137 to 147, 214 to 225, and 248 to 254, respectively) (5, 10, 28, 47). We investigated whether we could detect mutations in these tropism-determining loops that were coselected with TL9-associated mutations. We identified variation in five residues, at positions 219, 223, 248, 250, and 256, with the residues 219 and 223 located in the second loop, which contains the cyclophilin A binding sites, and residues 248, 250, and 256 located in or proximal to the third loop (Table 2).
We investigated coselection using approaches that couple phylogenetic and Bayesian network models in Spidermonkey (45) using 463 CAPRISA longitudinal population sequences, including more than 120 clones from four B*81:01-positive participants at selected time points (Table 2) and found significant correlation between various sites: 186S with 177D, 182S with 256V, and 186S with 250M (posterior probability > 0.5) (Table 4). In the longitudinal analysis in CAP277, the emergence of T186S mutation was accompanied with evolution of sites, 248 and 250 (Table 2). Interestingly, every sequence carrying the TL9 double mutations (Q182S and T186S) was associated with reversion of the M250I mutation, suggesting a selective reversion to accommodate the double TL9 mutations. Similarly, the fixation of the triple mutations E177D, Q182S, and T186S at around 122 weeks in CAP222 was preceded by significant toggling in the epitope, as well as a reversion in a putative compensatory site V223A (Table 2) (P = 0.0035), suggesting that the evolution of mutation V223A was under selective reversion (12). Furthermore, in CAP222, as well as in CAP225, the evolution of the single Q182S mutation in the TL9 epitope was accompanied by the emergence of a Gag I256V mutation (Table 2). In three other participants (CAP129, CAP278, and CAP289) combinations of mutations (Q182T/A248T, E177D/Q182S/T186S/I223V, and Q182A/I223V/I256V, respectively) were present in the first sample analyzed. Sequence evolution at site 190 has been shown to compensate for escape in TL9 epitope through the T186S mutation (56, 57). However, we found no sequence evolution at this residue (T190) in any of the B7-positive individuals or in any of the other participants investigated here (Table 2).
Table 4.
Sites covarying with each other in three of the four individuals who evolved sequences in the B*81:01-restricted TL9 epitope
| PIDa | HXB2 position site |
Posterior probability (P) | |
|---|---|---|---|
| 1 | 2 | ||
| CAP222 | 186S | 177D | 0.73 |
| CAP222 | 182S | 256V | 0.73 |
| CAP225 | 182S | 256V | 0.73 |
| CAP277 | 186S | 250M | 0.79 |
PID, participant identification number.
The timing of the sequence evolution influenced the differential disease progression among the B*81:01- and B*39:10-positive participants.
In vitro studies have demonstrated that substitution in the TL9 epitope residue T186 alone (56, 57) or in both T186 and E177 (47) incurs a fitness cost to the virus, whereas mutations at residue Q182 revert to consensus after transmission to HLA-mismatched recipients (39). Moreover, a recent study demonstrated that in a subtype C Gag backbone Q182S incurs a fitness cost to the virus in vitro (57).
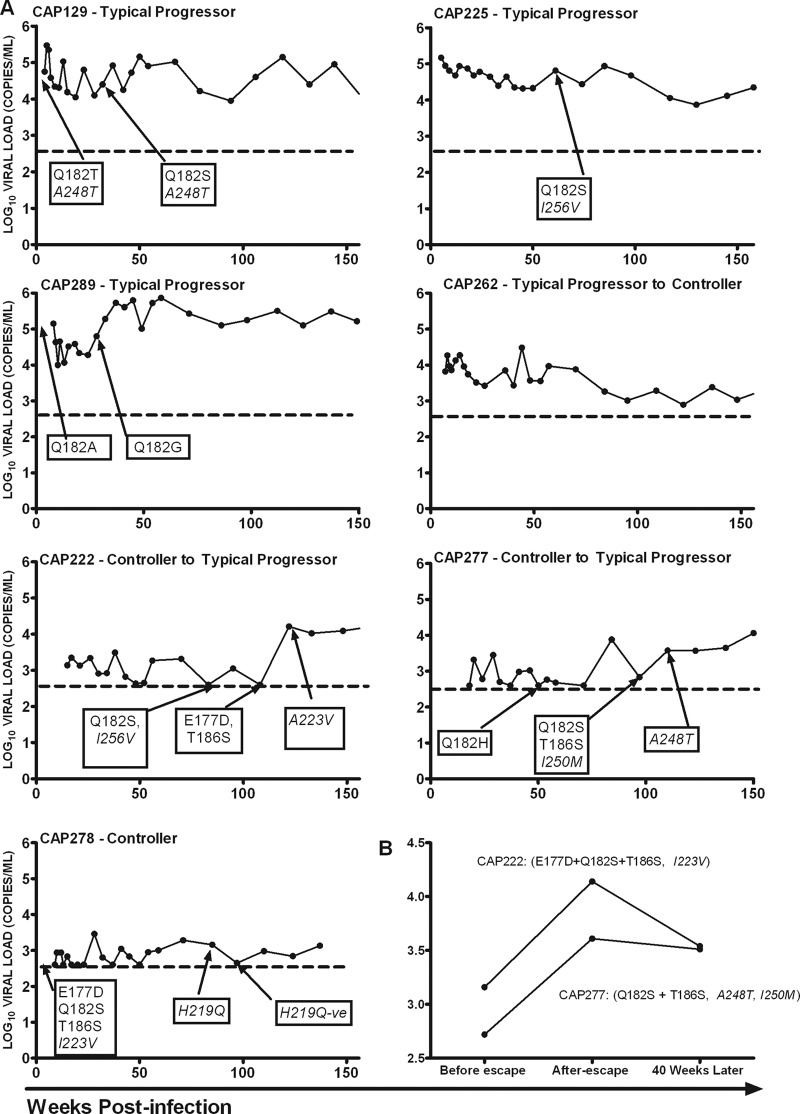
In order to understand CTL-mediated escape and its impact on disease progression, we investigated the temporal relationship between sequence evolution in the TL9 epitope and viral loads and CD4+ T-cell counts in B*81:01- and B*39:10-positive participants (Fig. 3A). No escape was detected in virus from CAP262, who elicited immune responses to TL9 in early infection and gradually controlled viral load to levels below 2,000 copies/ml after 24 months postinfection. We found that evolution of a single site, Q182 in the TL9 epitope in tandem with putative compensatory mutations A248T or I256V, did not affect viral loads or CD4+ T-cell counts in the three typical progressors—CAP129, CAP225, and CAP289—for at least the first 24 months after escape. These participants maintained consistently high viral loads for at least the first 3 years postinfection. Interestingly, characteristic of escape from a dominant response (9, 14, 23), the spontaneous increase in viral load and reduction in CD4+ T-cell counts in two individuals controlling viremia, CAP222 and CAP277, coincided with late sequence evolution in three (E177D, Q182S, and T186S) and two (Q182S with T186S) sites, respectively, together with reversion of putative compensatory residues I/V223A and M250I, respectively. Moreover, when the cost of escape in these two participants was estimated by comparing the median viral load measurements for 9 months before escape and 9 months later, we found that multiple residue escape was associated with a 1-log10 increase in viral load (Fig. 3B). This higher viral load was sustained for approximately 9 months after escape, after which it subsided, although to a level higher than before the escape (Fig. 3B). When viral loads in these participants at 4 years postinfection were compared to those who evolved single site escape mutation and to the median value for the rest of the cohort (i.e., individuals with nonprotective alleles, including B*81:01/57/58:01/39:10-negative participants) (Fig. 1), we found that both CAP277 and CAP222 still had lower viral loads. Interestingly, the B*39:10-positive participant (CAP278), who had the same mutations in acute infection, maintained low viral loads below 2,000 copies/ml over the 3-year period.
Fig 3.
(A) Viral load trajectories in the first 156 weeks (36 months) postinfection in B*81:01-positive (CAP129, CAP225, CAP222, CAP262, and CAP277) and B*39:10-positive (CAP278 and CAP289) participants, illustrating the timing of TL9-associated escape. The broken black line illustrates the limit of detection of the assay (400 copies/ml or 2.6 log10). Putative compensatory mutations are shown in italics. (B) Cost of multiple sequence evolution in or flanking the TL9 epitope in individuals who were controlling the infection, as measured by the change in viral load and longer follow-up showing the reduction in the maximum viral load but to levels higher than before escape. Four values of log10 viral loads over 9 months were used to calculate the median viral load before multiple residue escapes, after, and 40 weeks later. Putative compensatory mutations are shown in italics.
DISCUSSION
The B-7 HLA restriction alleles, including HLA-B*07:02, HLA-B*39:10, HLA-B*42:01, HLA-B*42:02, and HLA-B*81:01, are common in Black Africans and were found at a frequency of 40% (25/62) in the CAPRISA cohort. The TL9 epitope, located in p24 Gag, is an immunodominant epitope restricted by this HLA supertype. These HLAs are less common in Caucasians and the differential CTL targeting of Gag in Caucasians compared to Black South Africans is largely due to differences in frequency of TL9 recognition (22). The significance of evaluating this epitope is further emphasized by accumulating evidence showing that HLA-B*39:10 and HLA-B*81:01 alleles are associated with better infection outcomes in subtype C infection in southern Africa (30, 33). Aside from well-characterized epitopes associated with viral control, such as the B*57/58:01-restricted TW10, there is little known about the influence of epitope escape on HIV infection of Africans. We report here on a longitudinal study of subtype C-infected women from Kwazulu-Natal, South Africa, with acute to chronic infections. This study allowed us to determine the timing of escape in TL9 and identify coevolving sites, and it provided insights into the impact of these mutations on disease progression.
Previous studies have shown that individuals carrying any of the closely related HLA-B7 supertype alleles can mount a CD8+ T-cell response that targets the TL9 epitope (25, 34), with the most dominant being HLA-B*81:01, followed by HLA-B*39:10 and then HLA-B*42:01 (34). Although B42 alleles may have contributed to the selection of mutations in TL9 in each of the two participants positive for B*81:01 in combination with B42 allele (Table 1), there were far fewer polymorphisms in viruses in individuals positive for only B*42:01/42:02 alleles. Our results confirm previous observations that B*81:01 exert the greatest selective pressure on the virus (34) (Table 5), at least for the first 18 months postinfection. Different patterns of escape and generally higher TL9 specific immune responses seen in B*81:01-positive individuals compared to other B7-positive individuals confirm previously reported differences in recognition and selection pressure by the different HLAs belonging to the HLA-B7 supertype (18, 34).
Table 5.
Differences in the rate of escape in participants positive for the HLA- B7 supertype and Cw*08:02 alleles that target TL9 epitope
| Timea (mo postinfection) | No. of participants with indicated alleleb |
||||
|---|---|---|---|---|---|
| B*39:10 (n = 2) | B42 (n = 14) | B*07:02 (n = 4) | B*81:01 (n = 5) | Cw*08:02 (n = 1) | |
| Enrollment | 1 and 1* | 0 | 0 | 1* | 0 |
| <3 | 0 | 0 | 0 | 0 | 0 |
| 3–6 | 0 | 0 | 0 | 0 | 0 |
| 6–12 | 1* | 1† | 0 | 1 and 1* | 0 |
| 12–18 | 0 | 1‡ | 0 | 2 | 0 |
The time corresponds to when the first mutation was observed in TL9 epitope in the CAPRISA (AI02) cohort.
*, Participants with mutation at enrollment but with later further evolution of the mutation; †, mutation Q182T; ‡, mutation L188F was transient.
We documented the emergence of known B*81:01 escape mutations (E177D, Q182S, and T186S) (34) and identified a common pattern of evolution with the Q182S mutation generally emerging prior to T186S. Unlike the B*57/58:01-restricted TW10 escape, which usually occurs rapidly and during acute and early infection (7, 9, 37), escape in women with a B*81:01 background was usually late and took nearly a year to reach fixation. In late infection, slow escape has been postulated to be due to a number of factors, including low selection pressure, an increased diversity of specific responses, and raised viral fitness cost (17, 36, 49). Indeed, in vitro studies have demonstrated that T186S mutation alone (56) or in combination with E177D (56, 57) incurs a fitness cost to the virus. Interestingly, we found that late evolution of T186S either alone or in combination with E177D coincided with an increase in viral load and although viral replication was subsequently brought under control, it returned to a level higher than before escape. This reflects the complex balance between the host and the virus, where immune escape results in loss of control and is thus detrimental to the host; however, the impact of the mutation on viral replication capacity provides some benefit. Of interest, Wright et al. (56, 57) demonstrated that individuals carrying both 182S and/or 190X and 186S had higher replication capacities than individuals with 186S alone, although in our study we did not observe a single 186S and/or 190X mutation.
Although escape mutations that reduce viral fitness often select for variants that affect structural and/or reduced function (10, 46, 49, 50), the development of mutations that compensate for the fitness cost in the TL9 epitope are not well described. However, although in vitro studies for the Q182S mutation alone yielded discordant replication results, experiments in a subtype C Gag backbone found that Q182S incurred a fitness cost to the virus and same level of replication for either T186S alone or both T186S and Q182S (57). A recent cross-sectional study on sequences from 662 individuals infected with HIV-1, subtype C, found that polymorphism at I256 was significantly associated with the B*81:01-restricted Q182 and T186 sites (10). Here, we found concomitant evolution of I256V in each sequence carrying the Q182X mutation, providing further support that this mutation compensates for the fitness cost due to sequence evolution at site 182. However, the replication cost for this site mutation with the coevolving site remains to be determined in vitro.
In vitro studies have demonstrated that mutations in residues H219, I223, and M228 compensate for the fitness cost due to the T242N escape mutation in B*57/58:01 restricted TW10, whereas A248T and M250I have been associated with TW10 escape (4, 10, 37). We found an overlap in sites associated with B*81:01/39:10-restricted TL9. The development of both Q182S and T186S mutations in TL9 was associated with reversion to M250. Similarly, we found that fixation of the three escape residues at 2 years postinfection in CAP222 was associated with selective reversion of the transmitted mutation I/V223A in the cyclophilin binding loop, whereas the H219Q mutation in Gag subsequently developed 17 months postinfection in one of the controllers (CAP278). These common evolutionary pathways associated with TW10/TL9 restriction may be due to functional and structural interactions between the region harboring these epitopes (37, 51). These results support the model proposed by Crawford et al. (10), in which exposed capsid variable-residues generically compensate for fitness cost due to selection in structurally and functionally conserved capsid residues. The findings described here also support common compensatory pathways of viral evolution in variable domains associated with host tropism.
In conclusion, these findings expand our understanding of why some individuals with so-called beneficial alleles do not control viral replication. We provide here in vivo evidence that late escape at multiple sites both in and proximal to the TL9 epitope is associated with increased viremia in subtype C HIV-1-infected individuals, and we expand our knowledge regarding characteristics of Gag epitopes that are important to take into account when designing vaccine strategies.
ACKNOWLEDGMENTS
We thank the participants and the clinical and laboratory staff at CAPRISA for the specimens.
This study was funded by National Institute of Allergy and Infectious Diseases (NIAID), National Institutes of Health, U.S. Department of Health and Human Services grant U19 A151794, by NIAID International Research in Infectious Diseases grant 1R01AI078936, and by the Technology Innovation Agency.
Footnotes
Published ahead of print 29 August 2012
REFERENCES
- 1. Berger CT, et al. 2011. High-functional-avidity cytotoxic T lymphocyte responses to HLA-B-restricted Gag-derived epitopes associated with relative HIV control. J. Virol. 85:9334–9345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Borghans JA, Molgaard A, de Boer RJ, Kesmir C. 2007. HLA alleles associated with slow progression to AIDS truly prefer to present HIV-1 p24. PLoS One 2:e920 doi:10.1371/journal.pone.0000920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Borrow P, et al. 1997. Antiviral pressure exerted by HIV-1-specific cytotoxic T lymphocytes (CTLs) during primary infection demonstrated by rapid selection of CTL escape virus. Nat. Med. 3:205–211 [DOI] [PubMed] [Google Scholar]
- 4. Brockman MA, et al. 2007. Escape and compensation from early HLA-B57-mediated cytotoxic T-lymphocyte pressure on human immunodeficiency virus type 1 Gag alter capsid interactions with cyclophilin A. J. Virol. 81:12608–12618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Carlson JM, et al. 2008. Phylogenetic dependency networks: inferring patterns of CTL escape and codon covariation in HIV-1 Gag. PLoS Comput. Biol. 4:e1000225 doi:10.1371/journal.pcbi.1000225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Carlson JM, et al. 2012. Widespread impact of HLA restriction on immune control and escape pathways of HIV-1. J. Virol. 86:5230–5243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chopera DR, et al. 2011. Virological and immunological factors associated with HIV-1 differential disease progression in HLA-B*58:01-positive individuals. J. Virol. 85:7070–7080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chopera DR, et al. 2008. Transmission of HIV-1 CTL escape variants provides HLA-mismatched recipients with a survival advantage. PLoS Pathog. 4:E10000033 doi:10.1371/journal.ppat.1000033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Crawford H, et al. 2009. Evolution of HLA-B*5703 HIV-1 escape mutations in HLA-B*5703-positive individuals and their transmission recipients. J. Exp. Med. 206:909–921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Crawford H, et al. 2011. The hypervariable HIV-1 capsid protein residues comprise HLA-driven CD8+ T-cell escape mutations and covarying HLA-independent polymorphisms. J. Virol. 85:1384–1390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Delport W, Poon AF, Frost SD, Kosakovsky Pond SL. 2010. Datamonkey 2010: a suite of phylogenetic analysis tools for evolutionary biology. Bioinformatics 26:2455–2457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Delport W, Scheffler K, Seoighe C. 2008. Frequent toggling between alternative amino acids is driven by selection in HIV-1. PLoS Pathog. 4:e1000242 doi:10.1371/journal.ppat.1000242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Emu B, et al. 2008. HLA class I-restricted T-cell responses may contribute to the control of human immunodeficiency virus infection, but such responses are not always necessary for long-term virus control. J. Virol. 82:5398–5407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Feeney ME, et al. 2004. Immune escape precedes breakthrough human immunodeficiency virus type 1 viremia and broadening of the cytotoxic T-lymphocyte response in an HLA-B27-positive long-term-nonprogressing child. J. Virol. 78:8927–8930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Fellay J, et al. 2007. A whole-genome association study of major determinants for host control of HIV-1. Science 317:944–947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Frater AJ, et al. 2007. Effective T-cell responses select human immunodeficiency virus mutants and slow disease progression. J. Virol. 81:6742–6751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ganusov VV, et al. 2011. Fitness costs and diversity of the cytotoxic T lymphocyte (CTL) response determine the rate of CTL escape during acute and chronic phases of HIV infection. J. Virol. 85:10518–10528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Geldmacher C, et al. 2009. Minor viral and host genetic polymorphisms can dramatically impact the biologic outcome of an epitope-specific CD8 T-cell response. Blood 114:1553–1562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Goepfert PA, et al. 2008. Transmission of HIV-1 Gag immune escape mutations is associated with reduced viral load in linked recipients. J. Exp. Med. 205:1009–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gonzalez-Galarza FF, Christmas S, Middleton D, Jones AR. 2011. Allele frequency net: a database and online repository for immune gene frequencies in worldwide populations. Nucleic Acids Res. 39:D913–D919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Goonetilleke N, et al. 2009. The first T cell response to transmitted/founder virus contributes to the control of acute viremia in HIV-1 infection. J. Exp. Med. 206:1253–1272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Goulder PJ, et al. 2000. Differential narrow focusing of immunodominant human immunodeficiency virus Gag-specific cytotoxic T-lymphocyte responses in infected African and Caucasoid adults and children. J. Virol. 74:5679–5690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Goulder PJ, et al. 1997. Late escape from an immunodominant cytotoxic T-lymphocyte response associated with progression to AIDS. Nat. Med. 3:212–217 [DOI] [PubMed] [Google Scholar]
- 24. Goulder PJ, Watkins DI. 2004. HIV and SIV CTL escape: implications for vaccine design. Nat. Rev. Immunol. 4:630–640 [DOI] [PubMed] [Google Scholar]
- 25. Goulder PJ, Watkins DI. 2008. I. Watkins. Impact of MHC class I diversity on immune control of immunodeficiency virus replication. Nat. Rev. Immunol. 8:619–630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Goulder PJR, et al. 2001. Evolution and transmission of stable CTL escape mutations in HIV infection. Nature 412:334–338 [DOI] [PubMed] [Google Scholar]
- 27. Gray CM, et al. 2009. Human immunodeficiency virus-specific gamma interferon enzyme-linked immunospot assay responses Targeting specific regions of the proteome during primary subtype C infection are poor predictors of the course of viremia and set point. J. Virol. 83:470–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hatziioannou T, Cowan S, von Schwedler UK, Sundquist WI, Bieniasz PD. 2004. Species-specific tropism determinants in the human immunodeficiency virus type 1 capsid. J. Virol. 78:6005–6012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Honeyborne I, et al. 2007. Control of human immunodeficiency virus type 1 is associated with HLA-B*13 and targeting of multiple gag-specific CD8+ T-cell epitopes. J. Virol. 81:3667–3672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kiepiela P, et al. 2004. Dominant influence of HLA-B in mediating the potential co-evolution of HIV and HLA. Nature 432:769–774 [DOI] [PubMed] [Google Scholar]
- 31. Kiepiela P, et al. 2007. CD8+ T-cell responses to different HIV proteins have discordant associations with viral load. Nat. Med. 13:46–53 [DOI] [PubMed] [Google Scholar]
- 32. Koup RA, et al. 1994. Temporal association of cellular immune responses with the initial control of viremia in primary human immunodeficiency virus type 1 syndrome. J. Virol. 68:4650–4655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Leslie A, et al. 2010. Additive contribution of HLA class I alleles in the immune control of HIV-1 infection. J. Virol. 84:9879–9888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Leslie A, et al. 2006. Differential selection pressure exerted on HIV by CTL targeting identical epitopes but restricted by distinct HLA alleles from the same HLA supertype. J. Immunol. 177:4699–4708 [DOI] [PubMed] [Google Scholar]
- 35. Llano A, Frahm N, Brander C. 2009. How to optimally define optimal cytotoxic T lymphocyte epitopes in HIV infection? Los Alamos National Laboratory, Theoretical Biology and Biophysics Group, Los Alamos, NM [Google Scholar]
- 36. Loh L, Petravic J, Batten CJ, Davenport MP, Kent SJ. 2008. Vaccination and timing influence SIV immune escape viral dynamics in vivo. PLoS Pathog. 4:e12 c17 doi:10.1371/journal.ppat.0040012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Martinez-Picado J, et al. 2006. Fitness cost of escape mutations in p24 Gag in association with control of human immunodeficiency virus type 1. J. Virol. 80:3617–3623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Masemola A, et al. 2004. Hierarchical targeting of subtype C human immunodeficiency virus type 1 proteins by CD8+ T cells: correlation with viral load. J. J. Virol. 78:3233–3243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Matthews PC, et al. 2008. Central role of reverting mutations in HLA associations with human immunodeficiency virus set point. J. Virol. 82:8548–8559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. McMichael A, McCutchan FE. 2010. Host genetics and viral diversity: report from a global HIV vaccine enterprise working group. Nat. Proc. 2010:4797.2 [Google Scholar]
- 41. Migueles SA, et al. 2000. HLA B*5701 is highly associated with restriction of virus replication in a subgroup of HIV-infected long-term nonprogressors. Proc. Natl. Acad. Sci. U. S. A. 97:2709–2714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Miura T, et al. 2009. HLA-B57/B*5801 human immunodeficiency virus type 1 elite controllers select for rare Gag variants associated with reduced viral replication capacity and strong cytotoxic T-lymphocyte recognition. J. Virol. 83:2743–2755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Mlotshwa M, et al. 2010. Fluidity of HIV-1-specific T-cell responses during acute and early subtype C HIV-1 infection and associations with early disease progression. J. Virol. 84:12018–12029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Pereyra F, et al. 2010. The major genetic determinants of HIV-1 control affect HLA class I peptide presentation. Science 330:1551–1557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Poon AF, Lewis FI, Frost SD, Kosakovsky Pond SL. 2008. Spidermonkey: rapid detection of co-evolving sites using Bayesian graphical models. Bioinformatics 24:1949–1950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Prado JG, et al. 2009. Functional consequences of human immunodeficiency virus escape from an HLA-B*13-restricted CD8+ T-cell epitope in p1 Gag protein. J. Virol. 83:1018–1025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Rolland M, et al. 2010. Amino acid covariation in HIV-1 Gag subtype C: HLA-mediated selection pressure and compensatory dynamics. PLoS One 5:12463 doi:10.1371/journal.pone.0012463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Rousseau CM, et al. 2008. HLA class I-driven evolution of human immunodeficiency virus type 1 subtype c proteome: immune escape and viral load. J. Virol. 82:6434–6446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Schneidewind A, et al. 2008. Structural and functional constraints limit options for cytotoxic-T-lymphocyte escape in the immunodominant HLA-B27-restricted epitope in human immunodeficiency virus type 1 capsid. J. Virol. 82:5594–5605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Schneidewind A, et al. 2007. Escape from the dominant HLA-B27-restricted cytotoxic T-lymphocyte response in Gag is associated with a dramatic reduction in human immunodeficiency virus type 1 replication. J. Virol. 81:12382–12393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Tang C, Ndassa Y, Summers MF. 2002. Structure of the N-terminal 283-residue fragment of the immature HIV-1 Gag polyprotein. Nat. Struct. Biol. 9:537–543 [DOI] [PubMed] [Google Scholar]
- 52. Thompson JD, Higgins DG, Gibson TJ. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. van Loggerenberg F, et al. 2008. Establishing a cohort at high risk of HIV infection in South Africa: challenges and experiences of the CAPRISA 002 acute infection study. PLoS One 3:e1954 doi:10.1371/journal.pone.0001954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. von Schwedler UK, Stray KM, Garrus JE, Sundquist WI. 2003. Functional surfaces of the human immunodeficiency virus type 1 capsid protein. J. Virol. 77:5439–5450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Wang YE, et al. 2009. Protective HLA class I alleles that restrict acute-phase CD8+ T-cell responses are associated with viral escape mutations located in highly conserved regions of human immunodeficiency virus type 1. J. Virol. 83:1845–1855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Wright JK, et al. 2010. Gag-protease-mediated replication capacity in HIV-1 subtype C chronic infection: associations with HLA type and clinical parameters. J. Virol. 84:10820–10831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Wright JK, et al. 2012. Impact of HLA-B*81-associated mutations in HIV-1 Gag on viral replication capacity. J. Virol. 86:3193–3199 [DOI] [PMC free article] [PubMed] [Google Scholar]