Abstract
Epstein-Barr virus infection has been epidemiologically associated with the development of multiple autoimmune diseases, particularly systemic lupus erythematosus and multiple sclerosis. Currently, there is no known mechanism that can account for these associations. The germinal-center (GC) model of EBV infection and persistence proposes that EBV gains access to the memory B cell compartment via GC reactions by driving infected cells to differentiate using the virus-encoded LMP1 and LMP2a proteins, which act as functional homologues of CD40 and the B cell receptor, respectively. The ability of LMP2a, when expressed in mice, to allow escape of autoreactive B cells suggests that it could perform a similar role in infected GC B cells, permitting the survival of potentially pathogenic autoreactive B cells. To test this hypothesis, we cloned and expressed antibodies from EBV+ and EBV− memory B cells present during acute infection and profiled their self- and polyreactivity. We find that EBV does persist within self- and polyreactive B cells but find no evidence that it favors the survival of pathogenic autoreactive B cells. On the contrary, EBV+ memory B cells express lower levels of self-reactive and especially polyreactive antibodies than their uninfected counterparts do. Our work suggests that EBV has only a modest effect on the GC process, which allows it to access and persist within a subtly unique niche of the memory compartment characterized by relatively low levels of self- and polyreactivity. We suggest that this might reflect an active process where EBV and its human host have coevolved so as to minimize the virus's potential to contribute to autoimmune disease.
INTRODUCTION
Epstein-Barr virus (EBV) is a B cell transforming virus that nevertheless establishes a benign, lifelong, latent infection in the resting memory B cells of ≥90% of the human population worldwide (23). It has been suggested to play a role in both neoplastic and autoimmune diseases. There is good evidence linking EBV with cancer. In addition to its transforming capacity, the virus is carried latently by several lymphomas and carcinomas, although, paradoxically, the transforming latent proteins are often not expressed. Evidence linking EBV with autoimmune disease is less strong, being limited primarily to epidemiological associations between EBV seropositivity and disease. The most convincing case is for a link with multiple sclerosis (MS) (2) and systemic lupus erythematosus (SLE) (9). The experience of acute EBV infection (AIM), in particular, appears to increase the risk of developing MS by ∼20-fold (3), while EBV carriage has also been shown to be an independent risk factor for SLE (14, 24). EBV appears to be particularly strongly linked to juvenile forms of these diseases (1, 13). In contrast to the extensive range of evidence for a causal link between EBV and cancer, there is no known mechanism that explains how EBV may contribute to the pathogenesis of autoimmune diseases. Generally, the explanations take the form of suggesting that EBV's capacity to persist in and/or transform B cells could lead to a break in tolerance (10, 20, 29).
One widely accepted model of the mechanism by which EBV persists is known as the germinal-center (GC) model (31). This model proposes that EBV employs the sequential expression of four virus-encoded latency transcription programs to establish persistent infection. It is thought that these programs, known as growth (latency 3), default (latency 2), EBNA1-only (latency 1), and latency (latency 0), respectively, drive infected resting naïve B cells to become proliferating blasts, participate in GC reactions, and finally enter the resting memory B cell compartment, where the cells occasionally divide as part of memory B cell homeostasis. In this way, EBV infects a pool of long-lived quiescent cells in which it can persist latently for the life of the host. An alternative model has been proposed by Kuppers and Rajewsky (the direct-infection model), which suggests that EBV directly infects memory B cells (18). Although it was proposed over 10 years ago, no evidence has subsequently been provided to explain the mechanism behind this model. Specifically, it does not account for the four well-defined transcription programs/states of latent EBV infection, intermediate states between newly infected and persistently infected memory B cells have not been identified in vivo, and the model does not account for the origin of EBV-infected tumors. Furthermore, predictions of the direct-infection model were incorrect (27) or supported the GC model (6). In comparison, the GC model is supported by the observation that the expression of the four transcription programs is closely restricted to B cells of particular developmental stages (4) and anatomic locations in Waldeyer's ring (25), as predicted. This in itself also explains the origins and viral phenotype of EBV-associated lymphoma (for a detailed discussion, see reference 32). Importantly, subsequent work has shown that EBV-infected cells bearing the functional and phenotypic characteristics of GC B cells physically reside in GCs and express the default transcription program (25) (unlike the direct-infection model, which predicts that they should express the growth program [27]). This was a direct prediction of the GC model because the default program involves the expression of two latent membrane proteins, LMP1 and LMP2a. Extensive work in vitro and with transgenic mice had shown that these proteins have the capacity to mimic the signals required to rescue a GC B cell into memory (5, 35).
This discussion generates two important questions. First, how do we resolve the contradiction between the potent signaling capacities of LMP1 and LMP2a and the apparently normal appearance of the infected GC B cells and the resulting memory cells? Second, does this model provide a potential mechanism to explain how EBV could be a predisposing factor in autoimmune disease? Of the viral proteins expressed in the GC, LMP1 is a membrane-associated molecule that functions as a ligand-independent, constitutively active mimic of CD40 and is both transforming in vitro and tumorigenic in vivo when constitutively expressed alone (17, 37). LMP2a is a functional homologue of the B cell receptor (BCR) and conveys an antiapoptotic signal via Bcl-2 family members (21). When constitutively expressed alone, this signal is sufficient to permit even BCR-negative B cells to survive, enter GCs, undergo somatic hypermutation (SHM), and persist in the periphery (5, 6). This demonstrates that LMP2a is able to replace tonic BCR survival signals. By providing an LMP2a-mediated prosurvival signal, EBV could rescue forbidden clones from the proapoptotic environment of the GC, allowing them to differentiate into the memory B cell compartment. Indeed, Swanson-Mungerson et al. (30) and Wang et al. (38) have shown that LMP2a, when expressed alone and constitutively in transgenic mice, allowed self-reactive B cells to bypass anergy induction and preplasma cell checkpoints and, under some conditions, produce high levels of self-reactive antibodies. Curiously, there is no evidence for these behaviors in persistently infected individuals, where we have shown that LMP1 and LMP2a expression is tightly regulated and they are always coexpressed. This again raises the possibility that the net effect of EBV in GC B cells is modest and has led us to suggest that the transgenic mouse results are artifactual consequences of expressing these molecules constitutively in the absence of the intact virus (26). They may better reflect rare pathogenic conditions rather than normal persistence.
It is has been reported that as many as 63% of normal memory B cells are weakly self-reactive, with ∼23% being polyreactive, i.e., reacting with a broad range of antigens. We foresee three possible scenarios for EBV-infected memory B cells in vivo. EBV could, through LMP2a signaling, subvert the selection process that eliminates self-reactive B cells in the GC, allowing a high percentage of EBV-infected memory B cells to be self-reactive, including cells expressing potentially pathogenic antibodies. Alternatively, if LMP2a signaling bypasses the antigen selection process entirely, then the accumulation of deleterious mutations in the BCR could abrogate its binding capacity, in which case we would see the loss of self-reactivity in the infected compartment. Lastly, the presence of EBV may have a minimal impact on the normal processes of antigen-driven selection, in which case EBV should be present in approximately the same fraction of self-reactive cells as normal memory B cells.
In the present work, we have attempted to distinguish these alternatives. Specifically, we wanted to address whether EBV persists within self-reactive memory B cells and, if so, whether it persists preferentially in them and allows the survival of potentially pathogenic self-reactive cells. To achieve this, we profiled the self- and polyreactive properties of BCRs expressed by EBV+ memory B cells present during the acute phase of infection to determine if the virus affects their development and behavior.
MATERIALS AND METHODS
B cells from donor patients with AIM.
This study was performed in accordance with the institutional review board protocols of the Tufts Medical Center and the University of Massachusetts. Peripheral blood mononuclear cells (PBMCs) were obtained from donors at the time that they presented with symptoms of acute infectious mononucleosis (AIM) as described previously (12). PBMCs (2 × 107/ml) were stained with allophycocyanin-conjugated anti-human CD19 (Dako) and phycoerythrin-conjugated anti-human IgD (BD Pharmingen). Single memory B (CD19+ IgD−) cells were sorted with a Cytomation MoFlo fluorescence-activated cell sorter (FACS) into 20 μl of iScript reverse transcriptase buffer (Bio-Rad, Inc.) in 96-well plates, immediately frozen on dry ice, and stored at −80°C.
cDNA synthesis and limiting-dilution analysis.
Total RNA was reverse transcribed in the original sorting plate by using the iScript cDNA synthesis kit (Bio-Rad). Five microliters of cDNA was used as the template for EBER1 PCR in a limiting-dilution format to determine the frequency of EBV-infected cells as previously described (12). For single-cell analysis, PBMCs from selected donors were stained as described above, and single memory B cells were sorted into 96-well plates containing 20 μl/well cDNA iScript master mix (Bio-Rad). cDNA synthesis was performed according to the protocols included by the supplier. The EBV status of the single sorted cells was first assessed by EBER1 PCR on a 5-μl aliquot of the cDNA. The remainder was used for amplification of light and heavy Ig V region genes.
Single-cell PCR amplification of Ig V region genes.
Protocols for Ig gene PCR were adapted from references 33 and 39. Both protocols utilize a two-round nested PCR to amplify paired IgH and IgL V regions from single cells by using multiplex primers capable of amplifying V(D)J genes from all families. The first round of PCR used primers originally described by Wang and Stollar (39). A master mixture was prepared with 6 μl (10×) PfuTurbo PCR buffer, 1.6 μl (10 mM) deoxynucleoside triphosphates, 0.5 μl each of the VH primer mix and the VL primer mix, and 0.5 μl each of the CHI and CLI primer mixes (20 pmol/μl each primer), along with 1 μl PfuTurbo (Stratagene) and enough nuclease-free H2O to bring the final volume to 52 μl per well. Eight microliters of cDNA was added to each well, and the plates were sealed with a foil sealer (Denville Scientific, Inc.), centrifuged briefly, and placed in a thermal cycler. The thermal cycling conditions for the first round of amplification were as follows: 95°C for 2 min, followed by three cycles of preamplification (94°C for 45 s, 45°C for 45 s, 72°C for 1 min 45 s), 30 cycles of amplification (94°C for 45 s, 72°C for 1 min 45 s), a final extension step of 72°C for 10 min, and an indefinite hold at 4°C. First-round PCR products were then used immediately as templates for second-round Ig PCRs or frozen at −80°C until further use.
A second round of amplification was performed in order to acquire sufficient PCR product for sequencing and identification of the individual V(D)J genes composing the VH and VL regions of a given cell. This round used separate reaction mixtures to amplify VH and VL PCR products. For the VH amplifications, a master mixture was composed of 8 μl HotStar HiFidelity Taq polymerase 5× buffer; 0.5 μl of HotStar HiFidelity DNA polymerase (Qiagen); 1 μl each of primers CμII, CγII, and CαII; 1 μl each of primers CκII and CλII; and 1 μl each of several VH or VL primers (each primer at 20 pmol/μl). Second-round PCR products were electrophoresed on 2% agarose gels. V gene PCR products were extracted using the QIAquick gel extraction kit (Qiagen) and sequenced. Sequences were aligned with the closest germ line sequences using the IMGT/V-QUEST database (http://www.imgt.org/IMGT_vquest/share/textes/) to identify individual V(D)J genes.
In parallel, a second round of PCR was also performed according to the protocol of Tiller et al. (33), using the first-round PCR products as a template. Amplification of Ig variable regions relied on primers with restriction sites, allowing subsequent cloning into expression vectors. For VH amplifications, a master mixture was composed of 8 μl HotStar HiFidelity Taq polymerase 5× buffer, 0.5 μl of HotStar HiFidelity DNA polymerase (Qiagen), 0.5 μl of a gene-specific 3′ SalI JH primer, 0.5 μl of a gene-specific 5′ AgeI VH primer (each primer at 20 pmol/μl), and enough nuclease-free H2O to bring the final volume to 36 μl per well. A separate master mixture was used for the amplification of VL regions using a gene-specific 3′ BsiWI Jκ primer or a 3′ XholI Cλ primer and a 5′ AgeI Vκ or a 5′ AgeI Vλ primer. Thirty-six microliters of the appropriate master mix was added to each well of 96-well PCR plates, along with 4 μl of the first-round PCR product. The thermal cycling conditions were 50 cycles of 94°C for 30 s, 58°C for 30 s, and 72°C for 55 s and then 4°C indefinitely. All cycling was performed using a Perkin-Elmer GeneAmp PCR System 9600.
Expression vector cloning.
Purified IgH and IgL variable gene PCR products were cloned into expression vectors that contained a multiple cloning site upstream of fixed Igγ1, Igκ, or Igλ constant regions, allowing the expression of full-length recombinant antibodies. Igγ1, Igκ, and Igλ expression vectors were acquired from Hedda Wardemann's lab and have been published previously (33, 34). Digestions were performed by using the appropriate restriction enzymes (New England BioLabs) for each PCR product, and PCR products were purified subsequent to digestion using a QIAquick 96 PCR purification kit (Qiagen) and QIAVac 96. Digested expression vectors were treated with Antarctic Phosphatase (NEB) to reduce background colony formation, and linearized vectors were purified using QIAquick kits (Qiagen). Ten microliters of competent Escherichia coli TOP10 bacteria was transformed at 42°C with 1 μl of the ligation product, and clones were selected on the basis of resistance to ampicillin. Selected clones were grown overnight in 10-ml cultures of ampicillin-containing LB, and constructs were recovered using QIAprep miniprep kits (Qiagen).
Sequence analysis.
Constructs were sequenced following ligation to confirm the identity of the PCR insert, as well as to compile sequence statistics. Using the IMGT/V-QUEST database, sequences were aligned with germ line V(D)J genes with the highest identity, allowing the identification of hypermutations, insertions, and deletions. Primer binding regions were disregarded. The length and number of positively charged amino acids within the IgH complementarity-determining regions (CDRs) were determined using the IMGT/V-QUEST database. A binomial distribution model described by M. Uduman et al. (available at http://clip.med.yale.edu/sel/index.php) was used to analyze the probability that an excess or scarcity of replacement (R) mutations had occurred in IgH variable regions.
Antibody production.
Human embryonic kidney (HEK) 293T cells were cultured under standard conditions in Dulbecco's modified Eagle's medium (DMEM; Gibco) supplemented with 10% ultralow IgG (Invitrogen), 1 mM sodium pyruvate (Gibco), 100 μg/ml streptomycin, 100 U/ml penicillin G, and 0.25 μg/ml amphotericin (Gibco). For transfections, 5 × 106 cells were plated in 10 ml of DMEM in 150-mm TC plates (Falcon; BD) and grown to 75% confluence. On the day of transfections, the medium was aspirated, cells were gently washed 1× with phosphate-buffered saline (PBS; Gibco), and then the medium was replaced with 10 ml of prewarmed DMEM/Nut-SP, which is standard DMEM supplemented with 2 mM l-glutamine, 1× antibiotic-antimycotic mixture (ABAM; Gibco), and 1× Nutridoma-SP (Roche). Individual transfection solutions were prepared by mixing 5 to 20 μg of each respective IgH and IgL plasmid along with polyethyleneimine (PEI; Sigma) at a 3:1 ratio of micrograms of PEI to micrograms of DNA, all in 2 ml of DMEM/Nut-SP. These were vortexed for 10 s and incubated for 10 min at room temperature. Transfection solutions were added, dropwise, to plates of 293T cells. Supernatants were collected on days 3 and 6. On day 3, the medium was replaced with 10 ml of DMEM/Nut-SP. Supernatants were cleared by centrifugation at 2,000 rpm for 5 min. Sodium azide was added to a final concentration of 0.1 g/liter. Supernatants were stored at 4°C.
Recombinant antibodies were purified with protein G-Sepharose beads (Amersham; GE). Up to 25 ml of supernatant was incubated with 25 μl of protein G-Sepharose beads overnight at 4°C under rotation. Supernatants and beads were then centrifuged at minimal speed for 5 min, and supernatants were discarded. Beads were transferred to chromatography spin columns (Bio-Rad) and washed several times with PBS. Antibodies were then eluted in two fractions with 200 μl each of 0.1 M glycine at a pH of 2.5 to 3.0. Eluates were collected in microcentrifuge tubes that contained 20 μl 1 M Tris at pH 8.0 with 0.5% sodium azide and stored at 4°C. Antibody concentrations were determined by ELISA according to the protocols of Tiller et al. (33).
HEp-2 ELISAs and IFAs.
HEp-2 ELISAs (QUANTALite; INOVA Diagnostics, Inc.) and IFAs (Bion Enterprises, Ltd.) were performed as previously described (33, 34). Briefly, for ELISAs, antibodies were tested at a concentration of 10 μg/ml and 4:1 serial dilutions in PBS. High-positive-, low-positive-, and negative-control sera provided by the manufacturer were included in all assays. For IFAs, 20 μl of antibody at a concentration of 50 μg/ml was incubated for 30 min before being washed in PBS and incubated for a further 30 min with fluorescein isothiocyanate-labeled goat anti-human Ig. Control staining included antinuclear staining serum and negative-control serum provided by the manufacturer. Slides were examined using the ImageXpress Micro System (Molecular Devices).
Polyreactivity ELISAs.
Polyreactivity ELISAs were performed as previously described (33, 34). Antibodies were evaluated at 1 μg/ml and at 4:1 serial dilutions in PBS. High-binding ELISA plates (Costar) were coated at 50 μl/well with an individual antigen, i.e., 5 μg/ml insulin (Sigma) or 10 μg/ml of lipopolysaccharide (LPS; E. coli serotype O55:B5; Sigma), salmon sperm double-stranded DNA (dsDNA; Invitrogen), or single-stranded DNA (ssDNA). ssDNA was prepared by boiling dsDNA for 30 min and freezing it at −20°C. Controls for polyreactivity included high-polyreactivity antibody ED38, low-polyreactivity antibody eiJB40, and negative-control antibody mG053 (control plasmids; a kind gift of Hedda Wardemann). Antibodies with absorbances comparable to or higher than that of low-positive-control antibody eiJB40 against more than one antigen were scored as polyreactive.
Reactivity to MOG antigen.
Antibodies were tested for reactivity against native myelin oligodendrocyte glycoprotein (MOG) with a flow cytometry-based assay. MO3.13 human oligodendroglial cells were stabily transduced with human MOG lentivirus or MOG-negative control virus. The cells were sorted for MOG expression and incubated with the monoclonal antibodies at a 10-μg/ml antibody concentration for 1 h at room temperature. Goat anti-human-Alexa647 (Invitrogen) antibody was used for detection. Secondary antibody alone and an antibody against an irrelevant protein were used as negative controls. Anti-human MOG antibody 818C5 was used as a positive control. Reactivity was expressed as the ratio of the MFI of staining on MOG+ versus MOG− cells.
Reactivity to EBV antigens.
Antibodies were screened by immunoblot analysis of extracts from HH514 cells that had been induced into the lytic cycle as described previously (19).
Statistics.
P values for Ig gene repertoire analysis, positive charges in IgH CDR3 regions, and antibody reactivities were calculated by Fisher's Exact test or chi-square test. P values for IgH CDR3 length and frequency of somatic hypermutations were calculated by two-tailed unpaired Student's t test.
RESULTS
EBV is present in self-reactive memory B cells during AIM.
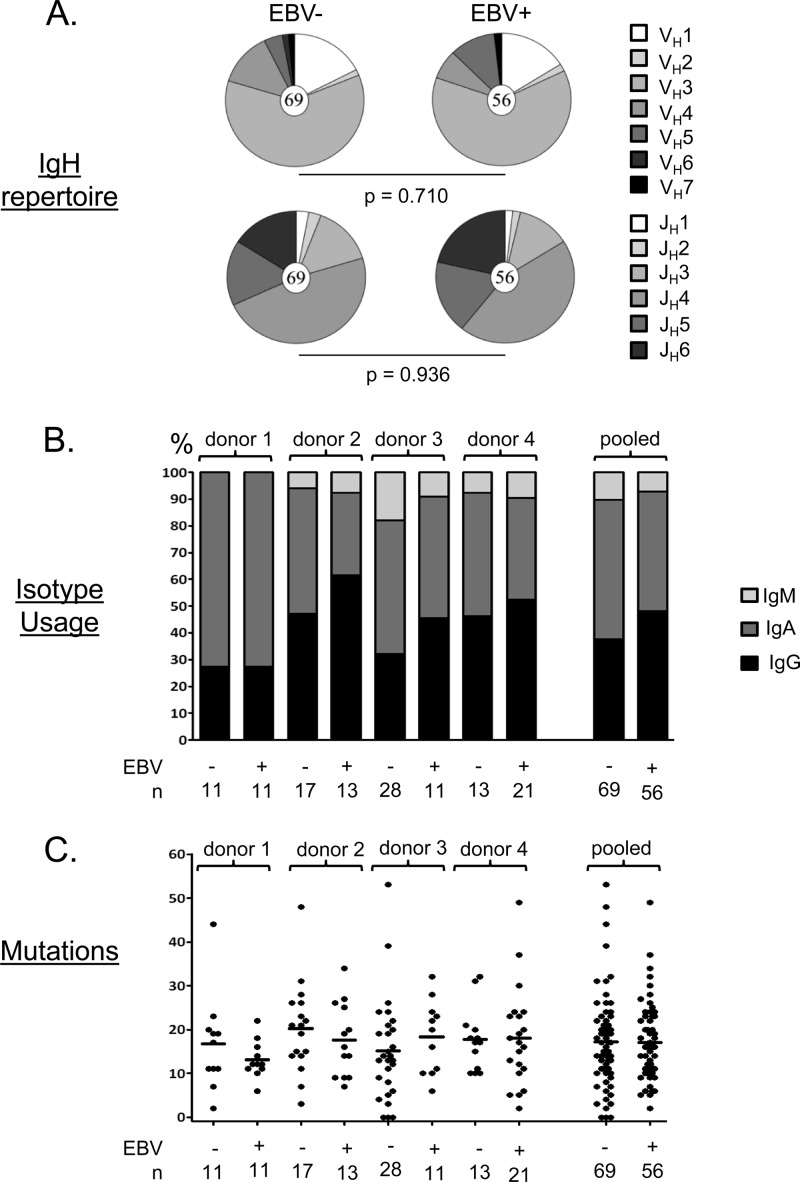
To investigate the role of EBV in the development of autoimmune disease, we have characterized the autoreactive profile of EBV-infected memory B cells from patients with AIM, a known risk factor for developing diseases such as MS. Single memory B cells (IgD− CD19+) from four patients were sorted by FACS, and cDNA was prepared (Fig. 1A). EBV-positive memory cells were distinguished on the basis of detection by PCR of the abundant EBV-encoded EBER1 transcripts. The remaining cDNA was used as a template for multiplex PCRs to amplify and clone the Ig heavy (IgG, IgA, or IgM)- and light (Cκ or Cλ)-chain variable regions (Fig. 1B). The amplicons were then cloned, and the paired IgH and IgL antibodies were expressed in 293T cells. In all, we obtained 56 antibodies from EBV+ memory B cells (by donor, n = 11, 13, 11, and 21). These antibodies had a distribution of isotypes, VH and JH usage (although JH6 was consistently higher in the EBV+ group), and SHMs similar to those of the matched EBV− controls (Fig. 2).
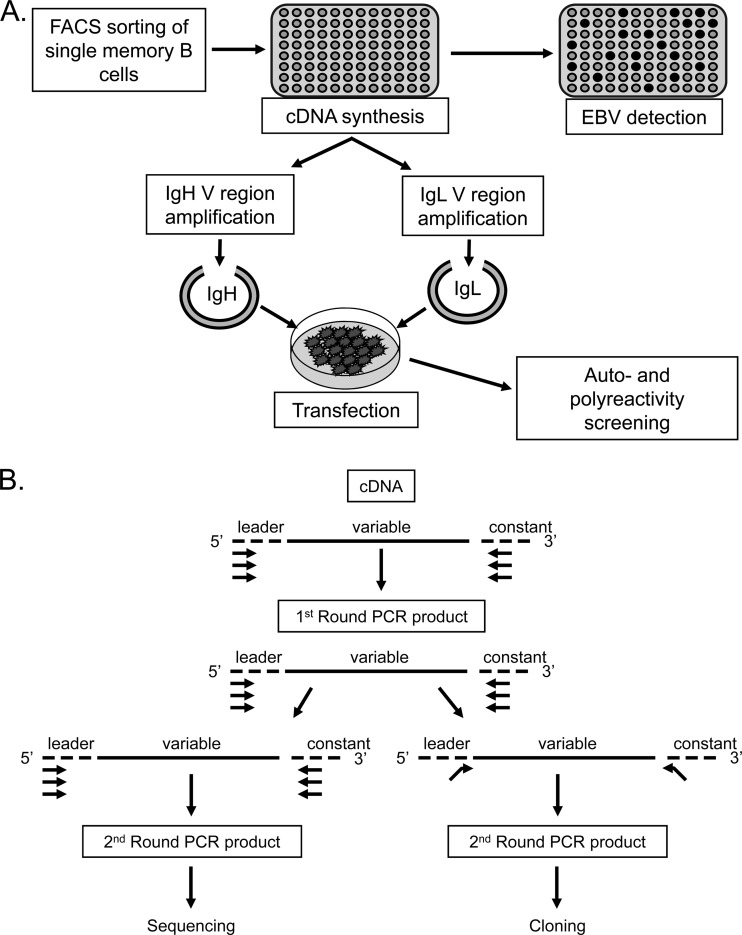
Fig 1.
Schematic representation of the cloning and expression methods used in this study. (A) Single CD19+ IgD− memory cells were sorted into 96-well plates, and cDNA was prepared. A fraction of this cDNA was then used to distinguish EBV+ from EBV− cells on the basis of the detection of the abundant EBV small RNA EBER1. The remaining cDNA was used for PCR amplification of Ig heavy- and light-chain variable regions (see below), which were cloned into expression vectors and transfected into 293T cells essentially as described by Tiller et al. (33). The resulting secreted antibodies were then purified and used for self- and polyreactivity assays. (B) PCR amplification of heavy- and light-chain variable regions (adapted from references 33 and 39). The first round of PCR used multiplex primers. A second round of amplification was performed in order to acquire sufficient PCR product for sequencing and identification of the individual V(D)J genes composing the VH and VL regions of a given cell. This round used separate reactions to amplify VH and VL PCR products. Sequences were aligned with the closest germ line sequences using the IMGT/V-QUEST database (http://www.imgt.org/IMGT_vquest/share/textes/). In parallel, PCR was also performed using the first-round products as a template and primers with restriction sites to allow subsequent cloning into expression vectors according to the protocol of Tiller et al. (33).
Fig 2.
Summary of the properties of the cloned antibodies. (A) Each pie chart shows the distribution of VH and JH gene usage for all the cloned antibodies tested. The total number of sequences analyzed is shown in the center circle. No significant differences were seen, although the frequency of JH6 was consistently higher in the EBV+ population. (B) Isotype usage for all of the cloned antibodies tested by donor. Note that the population studied is CD19+ IgD− memory B cells and therefore excludes IgM+ IgD+ memory cells, which we have shown previously to lack EBV (15). (C) Absolute numbers of somatic mutations in individual IgH genes from all four donors. Each dot represents an independent antibody. Horizontal lines indicate averages. Note that unlike our previous cohort (28), we did not see a significant difference in the number of mutations between antibodies derived from EBV+ and EBV− memory cells in this smaller cohort.
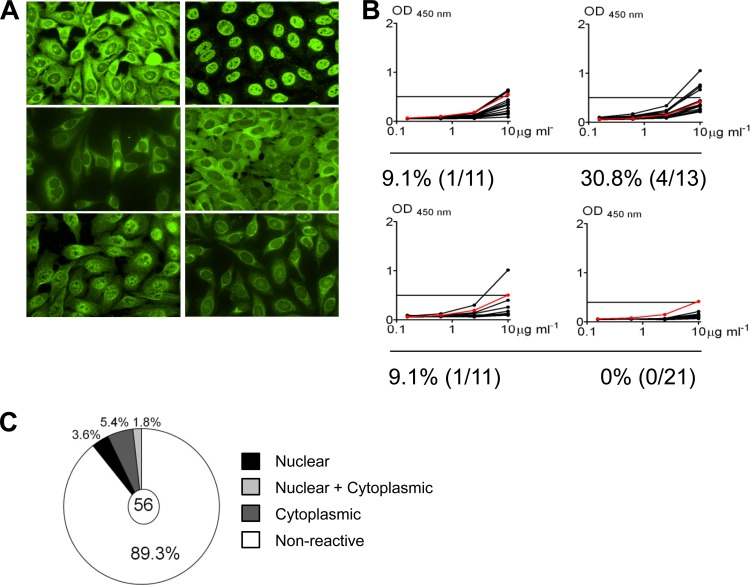
To examine the reactivity of these antibodies, we employed two assays—binding to fixed whole HEp-2 cells by enzyme-linked immunosorbent assay (ELISA) or by immunofluorescence assay (IFA). These assays have routinely been used to detect self-reactive antibodies in the serum of patients with autoimmune diseases (7) and more recently in the B cells of healthy individuals (34, 40). An antibody is considered to be self-reactive if it scores positive in both tests. The results are summarized in Fig. 3. In all, 26.8% (15/56) were positive by IFA (Fig. 3A) and approximately one-half of these (10.7%) were also positive in the ELISA (6/57) (Fig. 3B and C), with individual variation for the double positives ranging from 0% in donor 4 to 30.8% in donor 2. The antibodies scored as self-reactive demonstrated a variety of cytoplasmic, nuclear, or mixed staining patterns by IFA (Fig. 3C), suggesting that infected memory B cells are not reactive to one particular self antigen. Two (3.6%) of 56 demonstrated true antinuclear staining patterns more typically associated with autoimmune disease. However, the binding of these antibodies, like that of all of the positive antibodies, was relatively weak in the ELISA compared with that of self-reactive antibodies, such as those seen in SLE or the high-polyreactivity control antibody ED38. Thus, although EBV can persist in autoreactive memory B cells, we saw no evidence that it could rescue cells that express pathogenic antibodies.
Fig 3.
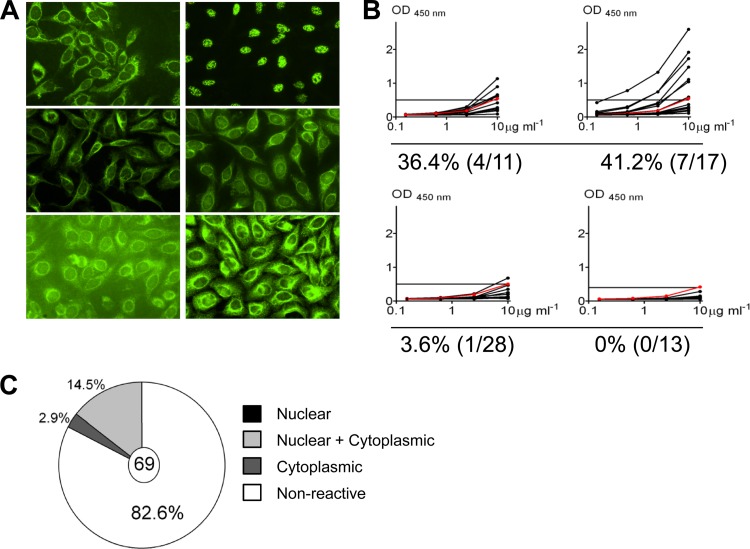
Self-reactivity of antibodies derived from EBV+ memory B cells. (A) Representative IFA staining patterns on HEp-2 cells. (B) ELISA for binding to HEp-2 cell lysates. Each black line represents an independent antibody. Horizontal lines indicate cutoff (optical density [OD] at 450 nm) for positive reactivity determined by comparison with low-polyreactivity positive-control antibody eiJB40 (red line). (C) Pie chart showing the fraction of antibodies positive by both IFA and ELISA and the breakdown of their staining patterns.
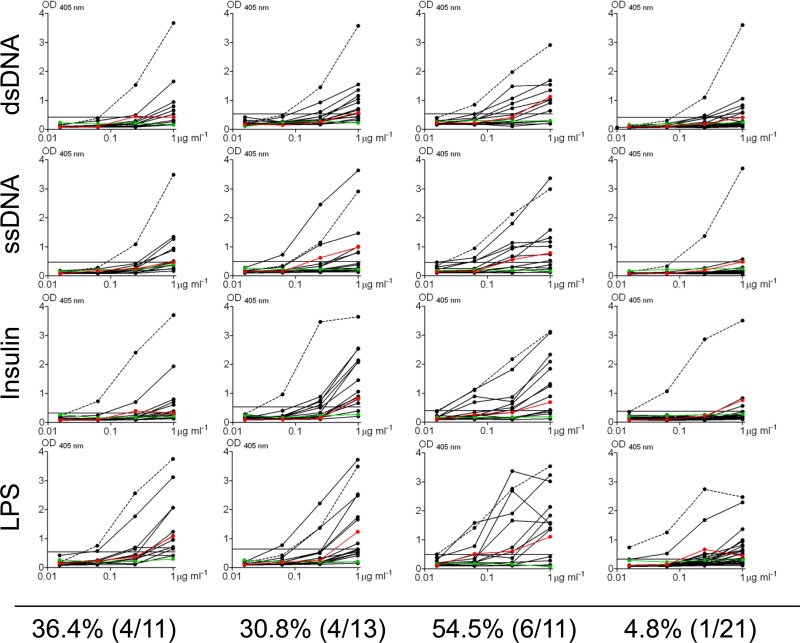
To further evaluate the self-reactivity of EBV+ memory B cells, the antibodies were also profiled for their reactivity to a defined panel of four antigens, including dsDNA, ssDNA, insulin, and LPS. Antibodies that reacted significantly with more than one antigen were scored as polyreactive (Fig. 4). The frequency of polyreactive antibodies varied between donors (ranging from 4.8% in donor 4 to 54.5% in donor 3), and they all showed only weak reactivity. Overall, ∼27% of EBV+ memory B cells were found to express polyreactive antibodies. We conclude, therefore, that EBV is able to persist within self-reactive and polyreactive memory B cells.
Fig 4.
Polyreactivity profiles of antibodies derived from EBV+ memory B cells. Antibodies were tested for polyreactivity by ELISA with dsDNA, ssDNA, insulin, and LPS. Each black line represents an independent antibody. Controls for polyreactivity included the high-polyreactivity antibody ED38 (dotted line), the low-polyreactivity antibody eiJB40 (red line), and negative-control antibody mG053 (green line). Antibodies with absorbances comparable to or higher than that of low-polyreactivity positive-control antibody eiJB40 against more than one antigen were scored as polyreactive. OD, optical density.
EBV does not preferentially persist in self-reactive B cells.
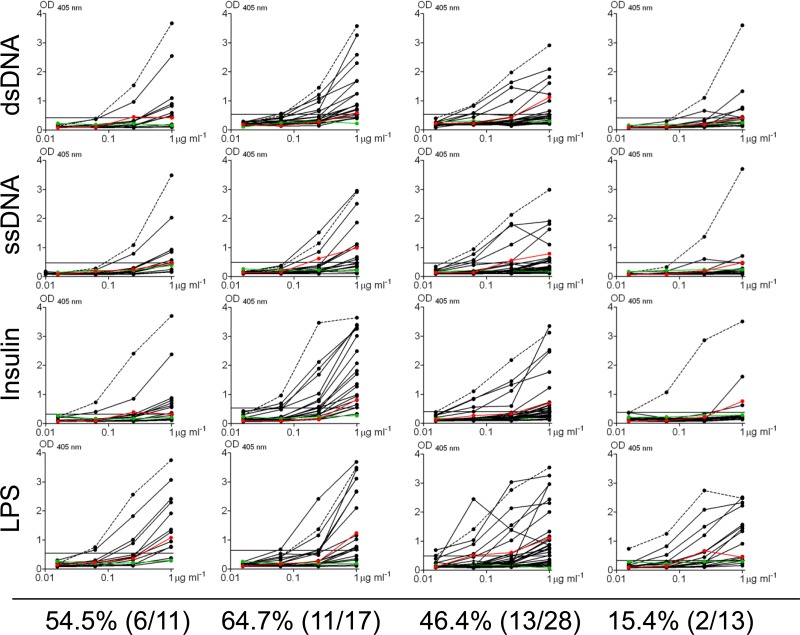
Weakly self-reactive B cells have been described previously in peripheral memory B cells at levels ranging to as high as 63% of the cells and with strengths similar to that which we have observed for the EBV+ component of the compartment (34). This raises the possibility that EBV does not influence the repertoire of the memory B cells it persists in. This is consistent with the idea that EBV simply “inhabits” this compartment without impacting its biology. To test this, we raised a second panel of antibodies, this time from the EBV− memory B cells from the same four donors. In all, we cloned and expressed 69 such antibodies (by donor, n = 11, 17, 28, and 13) and tested them in the HEp-2 ELISA and IFA (Fig. 5) and in a polyreactivity assay (Fig. 6). We found 17.4% to be self-reactive, i.e., positive by both ELISA and IFA (range, 0 to 41.2%) and 46.4% (range, 15.4 to 64.7%) to be polyreactive. This is quite different from what has been reported for healthy individuals, where the frequency of self-reactivity was 46.8% (range, 31.8 to 62.8%) and that of polyreactivity was 22.7% (range, 22.2 to 23.3%) (34). Specifically, it suggests a significant decrease in the self-reactivity and an increase in the polyreactivity of memory B cells during AIM.
Fig 5.
Self-reactivity of antibodies derived from EBV− memory B cells. (A) Representative IFA staining patterns on HEp-2 cells. (B) ELISA for binding to HEp-2 cell lysates. Each black line represents an independent antibody. Horizontal lines indicate the cutoff (optical density [OD] at 450 nm) for positive reactivity determined by comparison with low-polyreactivity positive-control antibody eiJB40 (red line). (C) Pie chart showing the fraction of antibodies positive by both IFA and ELISA and the breakdown of their staining patterns.
Fig 6.
Polyreactivity profiles of antibodies derived from EBV− memory B cells. Antibodies were tested for polyreactivity by ELISA with dsDNA, ssDNA, insulin, and LPS. Each black line represents an independent antibody. Controls for polyreactivity included high-polyreactivity positive-control antibody ED38 (dotted line), low-polyreactivity positive-control antibody eiJB40 (red line), and negative-control antibody mG053 (green line). Antibodies with absorbances comparable to or higher than that of low-polyreactivity positive-control antibody eiJB40 against more than one antigen were scored as polyreactive.
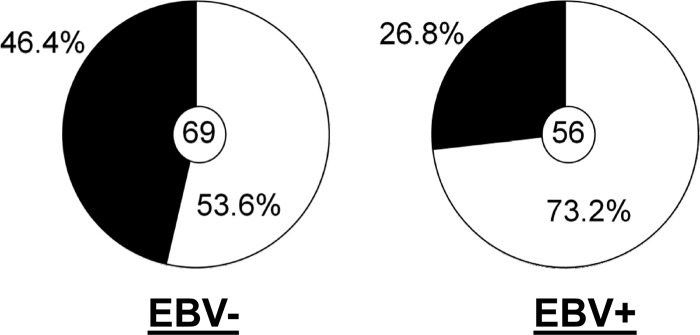
A summary comparing the results obtained with all of the EBV+ and EBV− memory B cells is shown in Table 1. Although we observed only a modest difference in autoreactivity between the two on the basis of the HEp2 assays, there was a markedly and significantly lower number of polyreactive cells in the EBV+ compartment (Fig. 7; 26.8% versus 46.4%; P = 0.027), such that the EBV+ memory compartment during AIM looks remarkably similar to the entire memory compartment in healthy individuals (26.8% versus 22.7% [from reference 34]).
Table 1.
Summary of reactivities of MAbs derived from EBV+ and EBV− memory B cells
| Donor | No. of Abs | % (no./total) |
|
|---|---|---|---|
| Self-reactive (HEp-2) | Polyreactive | ||
| IM1 EBV+ | 11 | 9.1 (1/11) | 36.4 (4/11) |
| IM1 EBV− | 11 | 36.4 (4/11) | 54.5 (6/11) |
| IM2 EBV+ | 13 | 30.8 (4/13) | 30.8 (4/13) |
| IM2 EBV− | 17 | 41.2 (7/17) | 64.7 (11/17) |
| IM3 EBV+ | 11 | 9.1 (1/11) | 54.5 (6/11) |
| IM3 EBV− | 28 | 3.6 (1/28) | 46.4 (13/28) |
| IM4 EBV+ | 21 | 0 (0/21) | 4.8 (1/21) |
| IM4 EBV− | 13 | 0 (0/13) | 15.4 (2/13) |
| EBV+ | 56 | 10.7 (6/56)a | 26.8 (15/56)b |
| EBV− | 69 | 17.4 (12/69)a | 46.4 (32/69)b |
P = 0.32.
P = 0.027.
Fig 7.
Summary of the frequencies of polyreactive antibodies for all EBV+ and EBV− memory B cells. The pie charts summarize the frequencies of polyreactivity for all of the antibodies tested from Fig. 4 and 6 (P = 0.027). The value in each center circle is the total number of independent antibodies analyzed.
We conclude that AIM is associated with disruption of the self- and polyreactive profile of memory B cells. Furthermore, EBV is able to persist in self-reactive B cells; however, none of those that we examined showed the characteristics of pathogenic autoreactivity; rather, they are skewed away from polyreactivity.
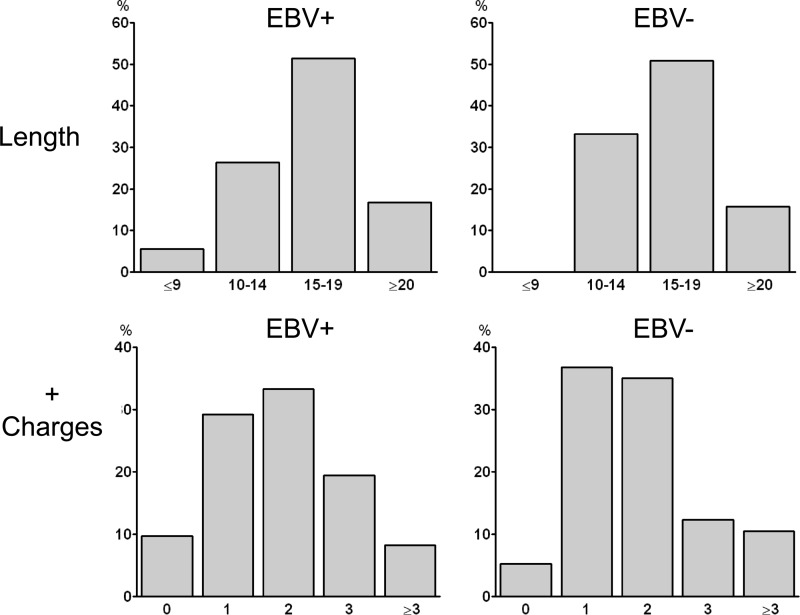
It is believed that self- and polyreactive antibodies tend to have longer CDR3 regions, with an increased frequency of positively charged amino acids (22). We sequenced our panel of antibodies and evaluated them for both traits. We found no significant difference in either of these traits between the EBV+ and EBV− compartments (Fig. 8). This again suggests that EBV does not persist in pathologically self-reactive memory B cells.
Fig 8.
Analysis of CDR3 regions for evidence of self-reactivity. (Top) Comparison of CDR3 lengths. The averages were 16.0 for the EBV+ population and 16.4 for the EBV− population (P = 0.54). (Bottom) Measurement of the number of positively charged amino acids in the CDR3s. The distributions were not significantly different (P = 0.63).
We conclude that the presence of EBV has a modest effect on the repertoire of the memory B cells in which it persists, skewing them away from polyreactivity.
EBV+ memory B cells do not recognize antigens associated with MS or with the virus itself.
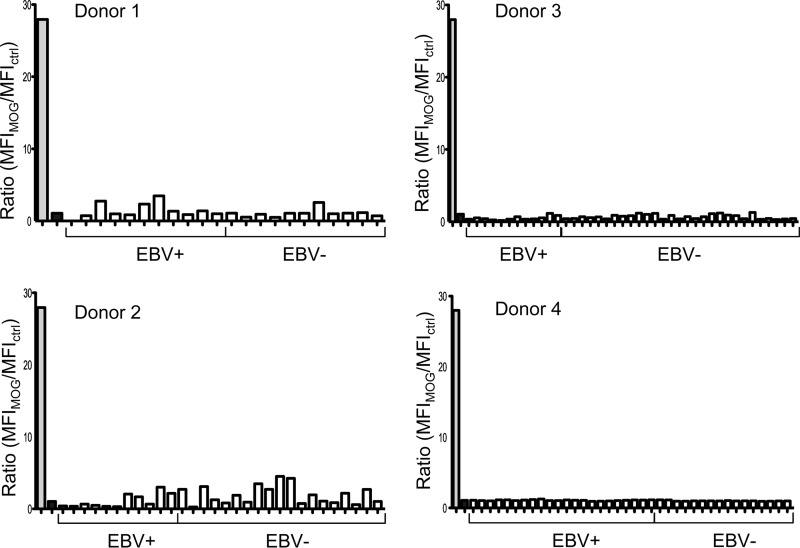
AIM has been shown to be a risk factor for the subsequent development of MS. One possible mechanism proposed is that EBV allows the survival of self-reactive memory B cells, producing the antibodies characteristic of MS. We therefore tested our panel of antibodies for reactivity to the MS-related antigen MOG. The results are shown in Fig. 9. None of the 125 antibodies derived from EBV+ and EBV− memory B cells demonstrated reactivity to MOG above the background levels. We conclude that there is no indication of the production of MOG reactivity during AIM, and this is true of both the EBV+ and EBV− components of the compartment.
Fig 9.
Reactivity of antibodies derived from donor-matched EBV+ and EBV− memory B cells to MOG. Reactivity was assessed with a flow cytometry-based assay using an oligodendrocyte cell line expressing MOG. Positive (anti-MOG antibody)- and negative (irrelevant antibody)-control (ctrl) antibody binding is shown by the gray and black bars, respectively. Reactivity is expressed as the ratio of the MFI of staining on MOG+ cells to the MFI of staining on MOG− cells.
When EBV infects B cells, it does so by binding CD21 on resting B cells. It has been argued that in vivo infection might be favored if the target B cell had a BCR with specificity for EBV, since CD21 and the BCR synergistically activate B cells, which is a requisite component of EBV-driven transformation. To test this hypothesis, we screened our panel of antibodies against structural components of the virus and other viral antigens and found no evidence that the antibodies produced by the EBV+ memory B cells are directed against viral antigens (data not shown).
Analysis of SHM patterns for evidence of antigen selection of EBV+ memory B cells.
One possible explanation for the reduction of polyreactivity that we observed among EBV+ memory B cells is that the viral latent proteins expressed in the GC, especially LMP2a, might alleviate the cells from antigen-driven selection. This would allow them to accumulate deleterious mutations in the CDRs, which could impair binding. We have previously reported, however, that EBV+ memory B cells show patterns of SHM consistent with having undergone selection in the GC, based on a multinomial analysis for antigen selection. However, subsequent advances in modeling affinity maturation have called into question the validity of this technique, as it is confounded by high levels of “cross talk.” This phenomenon refers to erroneously high estimates of the relative frequency of R mutations in CDRs due to a relative paucity of R mutations in the sequence as a whole, an occurrence that results in a high frequency of false positives (11). A more recent model utilizes a focused binomial-test- and Z-score-based method to detect the presence of negative or positive selection with improved sensitivity and specificity. To reexamine whether EBV+ memory B cells have circumvented antigen selection pressures, we analyzed their SHMs using these updated models.
The focused binomial test was used first to analyze mutation patterns of IgH sequences from EBV+ memory B cells to detect antigen selection on a single-cell basis. Four (7.1%) of the 56 sequences were scored as having an excess of RCDR mutations compared to those expected under conditions of no selection. This is comparable to the expected type I error rate at an α value of 0.05, suggesting that these results may reflect antigen selection or simply the intrinsic effects of SHM. In order to increase sensitivity while maintaining specificity, mutations from all 56 sequences were pooled. This demonstrated an overall excess of RCDR mutations (231 observed versus 169 expected; P = 0.008), suggesting that EBV+ memory B cells, in aggregate, have indeed experienced antigen selection, confirming our previous conclusion. We obtained a similar result with the EBV− memory B cells; the focused binomial test detected an excess of RCDR mutations in 7.6% (5/66) of single uninfected cells and also demonstrated an excess of RCDR mutations (285 observed versus 195 expected; P = 0.0016) among the pooled mutations from all of the sequences. We conclude that EBV+ memory B cells appear to have undergone antigen selection to an extent similar to that of normal memory B cells.
DISCUSSION
In this study, we have shown that EBV is able to persist in B cells producing low-affinity self-reactive and polyreactive antibodies. However, during AIM, the EBV+ memory cells show a marked decrease in polyreactivity compared to that of their EBV− counterparts. Furthermore, we found no evidence to suggest that during acute infection the virus was associated with rescuing self-reactive B cells with the potentially pathogenic character of those found in SLE or MS. Counterintuitively, our results suggest that AIM is associated with deregulation of self-reactive B cells, but this is occurring in the EBV− compartment, where the level of polyreactivity is almost double that found in healthy controls, which is actually similar to what we find in the EBV+ compartment. This suggests that EBV may play an active role in limiting the production of polyreactive memory B cells. We know that EBV has evolved strategies to minimize the risk of cancer, presumably because it is to the advantage of the virus to persist in a healthy host. For example, the virus persists in a quiescent state in memory B cells, where the growth-promoting latent genes are turned off and the viral epitopes on transformed B cells recognized by CTLs are conserved (16), guaranteeing that an aberrantly proliferating blast will be killed. Given EBV's potential to cause autoimmune disease, it is conceivable that the virus has evolved strategies to minimize this risk by limiting its own persistence in polyreactive memory B cells during acute infection. For example, polyreactive B cells may be more likely to receive BCR stimulation, triggering the lytic cycle of the virus and eliminating them. If this is correct, then the threat of developing autoimmune disease from AIM comes not from virus-infected B cells but from the disruption of the immune response caused by the acute infection.
We have shown here that the infected memory compartment is skewed against polyreactivity and previously that the repertoire of these cells is skewed against IgD+ IgM+ memory cells, specifically, that EBV preferentially persists in GC rather than marginal-zone-derived memory B cells and that the cells are enriched for the expression of JH6. How this skewing arises is unclear; however, it suggests that EBV accesses a subtly unique niche within the memory B cell compartment. This niche consists of GC-derived, isotype-switched memory cells that preferentially lack polyreactivity but nevertheless include a small proportion of cells expressing antibodies capable of binding dsDNA and other nuclear antigens and with IFA staining patterns traditionally associated with autoimmune disease. By finding that EBV persists within weakly self-reactive memory B cells, our work demonstrates that EBV can gain access to a sensitive niche of the human immune system that could represent an initial step toward the subsequent development of autoimmunity. Thus, our studies do not exclude the possibility that one of these self-reactive B cells could eventually become pathogenic at a later time as a consequence of the presence of EBV.
Our accumulated experience of analyzing the immunoglobulins expressed by EBV-infected memory B cells leads to the conclusion that EBV persists in memory B cells with a frequency and level of self-reactivity similar to, albeit lower than, those of donor-matched uninfected cells and with similar V(D)J repertoires, CDR3 properties, SHM patterns, and evidence of antigen selection. These results suggest a model in which EBV+ B cells remain susceptible to “normal” selection pressures while transiting GCs. While in the GC, EBV-infected cells express the latent proteins LMP1 and LMP2a (4, 25). Nevertheless, the GC B cells retain all of the characteristic phenotypic and functional characteristics of GC B cells (25), consistent with the conclusion from this work that the expression of these proteins does not drastically disrupt normal GC behavior. Nevertheless, in a number of experimental studies both in vitro and in transgenic mice (5, 29, 30, 35), these molecules have been shown to possess the signaling properties of CD40 and the BCR and may be sufficient to completely override antigen selection in the GC. This would be expected to produce an observable impact on the repertoire of EBV+ memory B cells compared to that of their uninfected counterparts, which we have not seen. Furthermore, transgenic-mouse studies have suggested that LMP1 prevents B cells from accessing the GC and instead causes lymphoma (17, 35), while LMP2a has been shown to break tolerance mechanisms. We have criticized these studies (26) in part because the transgenes are usually expressed from the Eμ enhancer, causing the proteins to be constitutively expressed in B cells, unlike the intact virus, where their expression is tightly regulated. Furthermore, we have suggested that it is inappropriate to study these genes in isolation since they are almost always expressed together in vivo. Recently, a transgenic-mouse study in which both LMP1 and LMP2a were expressed was published (36). Those authors reported none of the dramatic effects reported for the single transgenes. It was especially striking that despite the expression of both LMP1 and LMP2a, the B cells were able to transit the GC normally and undergo normal antigen selection and affinity maturation, as measured by the ability of such mice to produce high-affinity antibodies to 2,4,6-trinitrophenyl–keyhole limpet hemocyanin. These studies exactly mirror the results here, suggesting that the expression of LMP1 and LMP2a in the GC has, at most, modest effects on the behavior of the B cells and strongly suggesting that in order to become pathogenic, signaling from LMP1 and LMP2a needs to be uncoupled. In this light, it is worth noting that we have detected isolated LMP2a expression in the circulating memory B cells of up to 18% of SLE patients (8), which almost never occurs in healthy donors, suggesting that aberrant expression is correlated with autoimmunity.
One caveat to our work is that we have only studied memory-B cells derived from donors experiencing acute infection. It is possible that the immunological niche that the virus persists in changes as the infection moves into the chronic stage. Similarly, we cannot exclude the possibility that EBV preferentially favors the survival of highly autoreactive B cells during the chronic stage of infection and perhaps only in individuals already predisposed to develop SLE or MS.
Recently, work has been published suggesting that EBV can drive the maturation of newly infected naïve B cells into memory in vitro, i.e., in the absence of antigen and a GC reaction (10a). If correct, this would offer an alternate pathway for EBV to access the memory B cell compartment. Curiously, the authors did not show the SHM patterns of the putative in vitro EBV-driven memory-B cells nor did they provide an analysis of the R/S ratios in the CDRs. This was a surprising omission, since it would seem to be central to their claims, i.e., that these cells have acquired a memory cell phenotype in the absence of antigen, to demonstrate that these cells had unselected R/S ratios.
Our work supports an emerging counterintuitive consensus that EBV+ B cells remain subject to antigen selection, despite the well-characterized capabilities of LMP1 and LMP2a to circumvent it. This prompts the intriguing question that if the EBV default program does not function to autonomously drive infected cells into the memory compartment, what advantage does it provide to the virus? Several possibilities present themselves. The first is that their main role may be simply to act as insurance to ensure the survival of EBV-infected cells in the competitive environment of the GC. Alternatively, their role could be to provide modulating signals to ensure that infected cells become memory rather than plasma cells. An intriguing idea is that LMP1 and LMP2a actually provide signals that result in the observed reduced polyreactivity compared to that of the uninfected population, thus reducing the possible risk of developing autoimmune disease associated with AIM. In conclusion, our work suggests that EBV persists in a subtly unique niche of the memory compartment characterized by relatively low levels of self- and polyreactivity. Our work argues against a central role for EBV-infected memory B cells in deregulating autoimmune responses during acute infection, as EBV appears to act as a benign passenger during the GC and memory phases of its life cycle.
ACKNOWLEDGMENTS
We especially thank Hedda Wardemann for all her help in setting up the experimental system used here, including providing the necessary reagents, vectors and advice. We thank David Stollar for advice on the modification of the PCR protocol, and we thank Steve Kwok and Allen Parmelee for help with flow cytometry.
This work was supported by Public Health Service grants R01 CA65883 and R01 AI18757 to D.T.L.
Footnotes
Published ahead of print 5 September 2012
REFERENCES
- 1. Alotaibi S, Kennedy J, Tellier R, Stephens D, Banwell B. 2004. Epstein-Barr virus in pediatric multiple sclerosis. JAMA 291:1875–1879 [DOI] [PubMed] [Google Scholar]
- 2. Ascherio A, Munger KL. 2010. 99th Dahlem conference on infection, inflammation and chronic inflammatory disorders: Epstein-Barr virus and multiple sclerosis: epidemiological evidence. Clin. Exp. Immunol. 160:120–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ascherio A, Munger KL. 2007. Environmental risk factors for multiple sclerosis. Part I: the role of infection. Ann. Neurol. 61:288–299 [DOI] [PubMed] [Google Scholar]
- 4. Babcock GJ, Hochberg D, Thorley-Lawson AD. 2000. The expression pattern of Epstein-Barr virus latent genes in vivo is dependent upon the differentiation stage of the infected B cell. Immunity 13:497–506 [DOI] [PubMed] [Google Scholar]
- 5. Caldwell RG, Wilson JB, Anderson SJ, Longnecker R. 1998. Epstein-Barr virus LMP2A drives B cell development and survival in the absence of normal B cell receptor signals. Immunity 9:405–411 [DOI] [PubMed] [Google Scholar]
- 6. Casola S, et al. 2004. B cell receptor signal strength determines B cell fate. Nat. Immunol. 5:317–327 [DOI] [PubMed] [Google Scholar]
- 7. Egner W. 2000. The use of laboratory tests in the diagnosis of SLE. J. Clin. Pathol. 53:424–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gross AJ, Hochberg D, Rand WM, Thorley-Lawson DA. 2005. EBV and systemic lupus erythematosus: a new perspective. J. Immunol. 174:6599–6607 [DOI] [PubMed] [Google Scholar]
- 9. Harley JB, Harley IT, Guthridge JM, James JA. 2006. The curiously suspicious: a role for Epstein-Barr virus in lupus. Lupus 15:768–777 [DOI] [PubMed] [Google Scholar]
- 10. Harley JB, James JA. 2006. Epstein-Barr virus infection induces lupus autoimmunity. Bull. NYU Hosp. Jt. Dis. 64:45–50 [PubMed] [Google Scholar]
- 10a. Heath E, et al. 2012. Epstein-Barr virus infection of naïve B cells in vitro frequently selects clones with mutated immunoglobulin genotypes: implications for virus biology. PLoS Pathog. 8:e1002697 doi:10.1371/journal.ppat.1002697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hershberg U, Uduman M, Shlomchik MJ, Kleinstein SH. 2008. Improved methods for detecting selection by mutation analysis of Ig V region sequences. Int. Immunol. 20:683–694 [DOI] [PubMed] [Google Scholar]
- 12. Hochberg D, et al. 2004. Acute infection with Epstein-Barr virus targets and overwhelms the peripheral memory B-cell compartment with resting, latently infected cells. J. Virol. 78:5194–5204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. James JA, et al. 1997. An increased prevalence of Epstein-Barr virus infection in young patients suggests a possible etiology for systemic lupus erythematosus. J. Clin. Invest. 100:3019–3026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. James JA, et al. 2001. Systemic lupus erythematosus in adults is associated with previous Epstein-Barr virus exposure. Arthritis Rheum. 44:1122–1126 [DOI] [PubMed] [Google Scholar]
- 15. Joseph AM, Babcock GJ, Thorley-Lawson DA. 2000. EBV persistence involves strict selection of latently infected B cells. J. Immunol. 165:2975–2981 [DOI] [PubMed] [Google Scholar]
- 16. Khanna R, et al. 1997. Evolutionary dynamics of genetic variation in Epstein-Barr virus isolates of diverse geographical origins: evidence for immune pressure-independent genetic drift. J. Virol. 71:8340–8346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kulwichit W, et al. 1998. Expression of the Epstein-Barr virus latent membrane protein 1 induces B cell lymphoma in transgenic mice. Proc. Natl. Acad. Sci. U. S. A. 95:11963–11968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kurth J, et al. 2000. EBV-infected B cells in infectious mononucleosis: viral strategies for spreading in the B cell compartment and establishing latency. Immunity 13:485–495 [DOI] [PubMed] [Google Scholar]
- 19. Meij P, et al. 1999. Restricted low-level human antibody responses against Epstein-Barr virus (EBV)-encoded latent membrane protein 1 in a subgroup of patients with EBV-associated diseases. J. Infect. Dis. 179:1108–1115 [DOI] [PubMed] [Google Scholar]
- 20. Pender MP. 2003. Infection of autoreactive B lymphocytes with EBV, causing chronic autoimmune diseases. Trends Immunol. 24:584–588 [DOI] [PubMed] [Google Scholar]
- 21. Portis T, Longnecker R. 2004. Epstein-Barr virus (EBV) LMP2A mediates B-lymphocyte survival through constitutive activation of the Ras/PI3K/Akt pathway. Oncogene 23:8619–8628 [DOI] [PubMed] [Google Scholar]
- 22. Radic MZ, Weigert M. 1994. Genetic and structural evidence for antigen selection of anti-DNA antibodies. Annu. Rev. Immunol. 12:487–520 [DOI] [PubMed] [Google Scholar]
- 23. Rickinson AB, Kieff E. 2001. Epstein-Barr virus, p 2575–2628 In Knipe DM, et al. (ed), Fields virology, 4th ed, vol 2 Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 24. Rothfield NF, Evans AS, Niederman JC. 1973. Clinical and laboratory aspects of raised virus antibody titres in systemic lupus erythematosus. Ann. Rheum. Dis. 32:238–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Roughan JE, Thorley-Lawson DA. 2009. The intersection of Epstein-Barr virus with the germinal center. J. Virol. 83:3968–3976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Roughan JE, Torgbor C, Thorley-Lawson DA. 2010. Germinal center B cells latently infected with Epstein-Barr virus proliferate extensively but do not increase in number. J. Virol. 84:1158–1168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Siemer D, et al. 2008. EBV transformation overrides gene expression patterns of B cell differentiation stages. Mol. Immunol. 45:3133–3141 [DOI] [PubMed] [Google Scholar]
- 28. Souza TA, Stollar BD, Sullivan JL, Luzuriaga K, Thorley-Lawson DA. 2007. Influence of EBV on the peripheral blood memory B cell compartment. J. Immunol. 179:3153–3160 [DOI] [PubMed] [Google Scholar]
- 29. Swanson-Mungerson M, Longnecker R. 2007. Epstein-Barr virus latent membrane protein 2A and autoimmunity. Trends Immunol. 28:213–218 [DOI] [PubMed] [Google Scholar]
- 30. Swanson-Mungerson MA, Caldwell RG, Bultema R, Longnecker R. 2005. Epstein-Barr virus LMP2A alters in vivo and in vitro models of B-cell anergy, but not deletion, in response to autoantigen. J. Virol. 79:7355–7362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Thorley-Lawson DA. 2001. Epstein-Barr virus: exploiting the immune system. Nat. Rev. Immunol. 1:75–82 [DOI] [PubMed] [Google Scholar]
- 32. Thorley-Lawson DA, Gross A. 2004. Persistence of the Epstein-Barr virus and the origins of associated lymphomas. N. Engl. J. Med. 350:1328–1337 [DOI] [PubMed] [Google Scholar]
- 33. Tiller T, et al. 2008. Efficient generation of monoclonal antibodies from single human B cells by single cell RT-PCR and expression vector cloning. J. Immunol. Methods 329:112–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Tiller T, et al. 2007. Autoreactivity in human IgG+ memory B cells. Immunity 26:205–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Uchida J, et al. 1999. Mimicry of CD40 signals by Epstein-Barr virus LMP1 in B lymphocyte responses. Science 286:300–303 [DOI] [PubMed] [Google Scholar]
- 36. Vrazo AC, Chauchard M, Raab-Traub N, Longnecker R. 2012. Epstein-Barr virus LMP2A reduces hyperactivation induced by LMP1 to restore normal B cell phenotype in transgenic mice. PLoS Pathog. 8:e1002662 doi:10.1371/journal.ppat.1002662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wang D, Liebowitz D, Kieff E. 1985. An EBV membrane protein expressed in immortalized lymphocytes transforms established rodent cells. Cell 43:831–840 [DOI] [PubMed] [Google Scholar]
- 38. Wang H, et al. 2006. EBV latent membrane protein 2A induces autoreactive B cell activation and TLR hypersensitivity. J. Immunol. 177:2793–2802 [DOI] [PubMed] [Google Scholar]
- 39. Wang X, Stollar BD. 2000. Human immunoglobulin variable region gene analysis by single cell RT-PCR. J. Immunol. Methods 244:217–225 [DOI] [PubMed] [Google Scholar]
- 40. Wardemann H, et al. 2003. Predominant autoantibody production by early human B cell precursors. Science 301:1374–1377 [DOI] [PubMed] [Google Scholar]