Abstract
PA-X is a fusion protein of influenza A virus encoded in part from a +1 frameshifted X open reading frame (X-ORF) in segment 3. We show that the X-ORFs of diverse influenza A viruses can be divided into two groups that differ in selection pressure and likely function, reflected in the presence of an internal stop codon and a change in synonymous diversity. Notably, truncated forms of PA-X evolved convergently in swine and dogs, suggesting a strong species-specific effect.
TEXT
It was recently determined that segment 3 of influenza A virus (IAV) encodes a second protein, made in part from a +1 open reading frame (ORF) embedded within the PA gene. This ORF is accessed via ribosomal frameshifting to produce a fusion protein, denoted PA-X, whose N-terminal 191 amino acids are derived from the PA ORF, while the C-terminal sequence, most commonly 61 amino acids long, is derived from the +1 frameshifted “X-ORF” (Fig. 1). PA-X represses cellular RNA polymerase II-mediated gene expression in transfection reporter systems and reduces pathogenicity in a mouse model of influenza virus infection (6). A key evolutionary signature of the +1 X-ORF is a marked reduction in synonymous diversity in frame 0 of the PA gene overlapping the +1 X-ORF (3); this is indicative of selective constraint against nonsynonymous changes in PA-X and hence of functional importance. However, some strains of influenza A virus possess stop codons in the X-ORF, leading to a truncated PA-X protein. The most notable examples are the human 2009 pandemic H1N1 viruses (H1N1pdm), which possess a TGG (Trp)-to-TAG (stop) nonsense mutation at codon 42 in the X-ORF, resulting from a synonymous mutation in the PA gene (6). To determine how PA-X has evolved across IAV as a whole and particularly to infer whether changes in protein function are associated with instances of cross-species transmission, we performed an evolutionary analysis of PA-X in 10,164 influenza A viruses that represent 12 different avian and mammalian viral lineages and many antigenic subtypes. For comparison we analyzed 190 isolates of influenza B virus.
Fig 1.
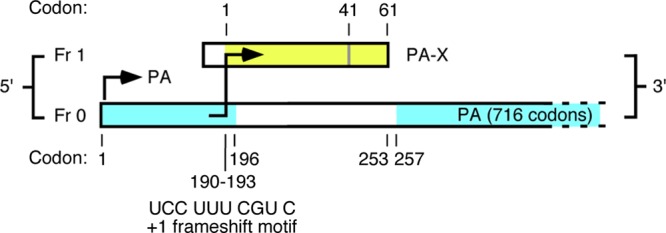
Influenza A virus segment 3 showing open reading frames for PA (in frame 0) and for the PA-X ORF (in frame 1), with the frameshift motif shown. Yellow shading indicates the region of the X-ORF that is translated; blue shading indicates structural domains of PA (1, 4, 10). The PA-X open reading frame encodes either 61 or 41 amino acids as indicated. Note that the X-ORF product lies largely within a linker region between the PA N- and C-terminal domains. (Reprinted from reference 6 with permission of the publisher).
We assembled all available nonidentical PA protein-coding sequences from GenBank and translated them in frame 1 to reveal the frameshifted portion of the PA-X protein. These data comprised avian influenza (all subtypes), human H3N2, human seasonal H1N1, human H1N1pdm, swine “classical” (CS) H1N1, swine “Eurasian” (EA) H1N1, swine “triple-reassortant” (TR) H1N1, equine H3N8, equine H7N7, canine H3N8 (equine H3N8-derived), canine H3N2 (avian H3N2-derived), bat influenza, and influenza B viruses (data set sizes are shown in Table 1). Sequences were aligned manually, after which a phylogenetic tree was inferred using the neighbor-joining method available in the MEGA5 program (8) and employing the Kimura 2-parameter substitution model. The highly divergent influenza B virus sequences were excluded from this analysis. Occurrences of stop codon mutations at X-ORF codon 42 were then mapped onto the phylogeny. To determine the selection pressures acting on PA-X, we estimated the numbers of synonymous (dS) and nonsynonymous (dN) substitutions per site. As the strongest signal for functional constraint on the +1 X-ORF is a reduction in the synonymous diversity in frame 0 of PA (6), we focused on the evolutionary pressures in this frame, comparing the X-ORF to the rest of the PA gene. Within these gene regions we estimated dS, dN, and the ratio dN/dS using the CODEML method available in the PAML package (9).
Table 1.
Frequency of stop codon mutations and the numbers of synonymous and nonsynonymous nucleotide substitutions per site for different influenza virusesa
| Virusb | nd | No. truncatede |
dN for: |
dS for: |
ds X-ORF/ ds rest | dN/dS X-ORF frame 0 | dN/dS +1 X-ORFi | ||
|---|---|---|---|---|---|---|---|---|---|
| X-ORF frame 0c | Rest frame 0 | X-ORF frame 0 | Rest frame 0 | ||||||
| Avian influenza | 4,361 (80) | 19 (2) | 0.50 | 0.52 | 4.08 | 16.44 | 0.25 | 0.12 | 3.20 |
| Human H3N2 | 2,296 (49) | 8 (5) | 0.09 | 0.09 | 0.53 | 1.23 | 0.43 | 0.16 | 4.92 |
| Human H1N1s | 853 (50) | 3 (1) | 0.10 | 0.11 | 0.93 | 1.62 | 0.57 | 0.10 | 3.88 |
| Human H1N1pdm | 1,916 (56) | 1,914 (1,914) | 0.09 | 0.10 | 0.68 | 0.57 | 1.19 | 0.13 | 4.36 |
| Swine CS | 121 (52) | 6 (3) | 0.17 | 0.17 | 2.18 | 2.18 | 1.00 | 0.08 | 6.94 |
| Swine CS–stopf | 99 (53) | 98 (98) | 0.15 | 0.16 | 1.95 | 1.67 | 1.17 | 0.08 | 21.29 |
| Swine EA | 152 (60) | 0 (0) | 0.33 | 0.28 | 3.18 | 3.60 | 0.88 | 0.10 | 3.95 |
| Swine TR | 248 (73) | 225 (225) | 0.46 | 0.30 | 3.18 | 2.76 | 1.15 | 0.14 | 5.58 |
| Equine H3N8 | 80 (35) | 0 (0) | 0.12 | 0.09 | 0.25 | 0.73 | 0.34 | 0.49 | 1.15 |
| Equine H7N7g | 2 (2) | 2 (0) | 0.01 | 0.005 | 0.23 | 0.19 | 1.21 | 0.06 | ∞ |
| Bat influenzag | 2 (2) | 2 (0) | 0.01 | 0.005 | 0.14 | 0.13 | 1.08 | 0.05 | 0.59 |
| Canine H3N8 | 26 (6) | 25 (24) | 0.03 | 0.01 | 0.07 | 0.05 | 1.40 | 0.41 | 1.26 |
| Canine H3N2 | 8 (6) | 8 (7) | 0.03 | 0.01 | 0.16 | 0.05 | 3.20 | 0.18 | 4.13 |
| Influenza B | 190 (34) | 190 (AUh) | 0.05 | 0.06 | 1.07 | 1.11 | 0.96 | 0.05 | NAj |
Virus groups with truncated PA-X proteins and associated data are in italics.
Human H1N1s, seasonal H1N1; human H1N1pdm, 2009 pandemic H1N1; swine CS, “classical” swine H1N1; swine EA, Eurasian avian H1N1-like swine H1N1; swine TR, triple-reassortant swine H1N1.
Reading frame encoding the PA protein.
First number is the total data set. The representative sample size used for the analysis of dN and dS is shown in parentheses.
First number is the number of viruses from the total data set that are truncated because of premature stop codons. The number of viruses with stop codon mutations at codon 42 is shown in parentheses.
The classical swine viruses can be divided into two groups representing those with and without the stop codon mutation at codon 42. Note that there has been a single reversion to the non-stop codon state in the stop codon group.
Because the very small sample size these sequences were analyzed with MEGA5 rather than PAML.
AU, sequence alignment is uncertain in this region.
For the full-length X-ORF the analysis of dN/dS was performed on all 61 amino acids present in the protein, while for sequences encoding the truncated protein this analysis was restricted to the 41 amino acids prior to the position corresponding to the stop codon.
NA, contains too many stop codons for meaningful analysis.
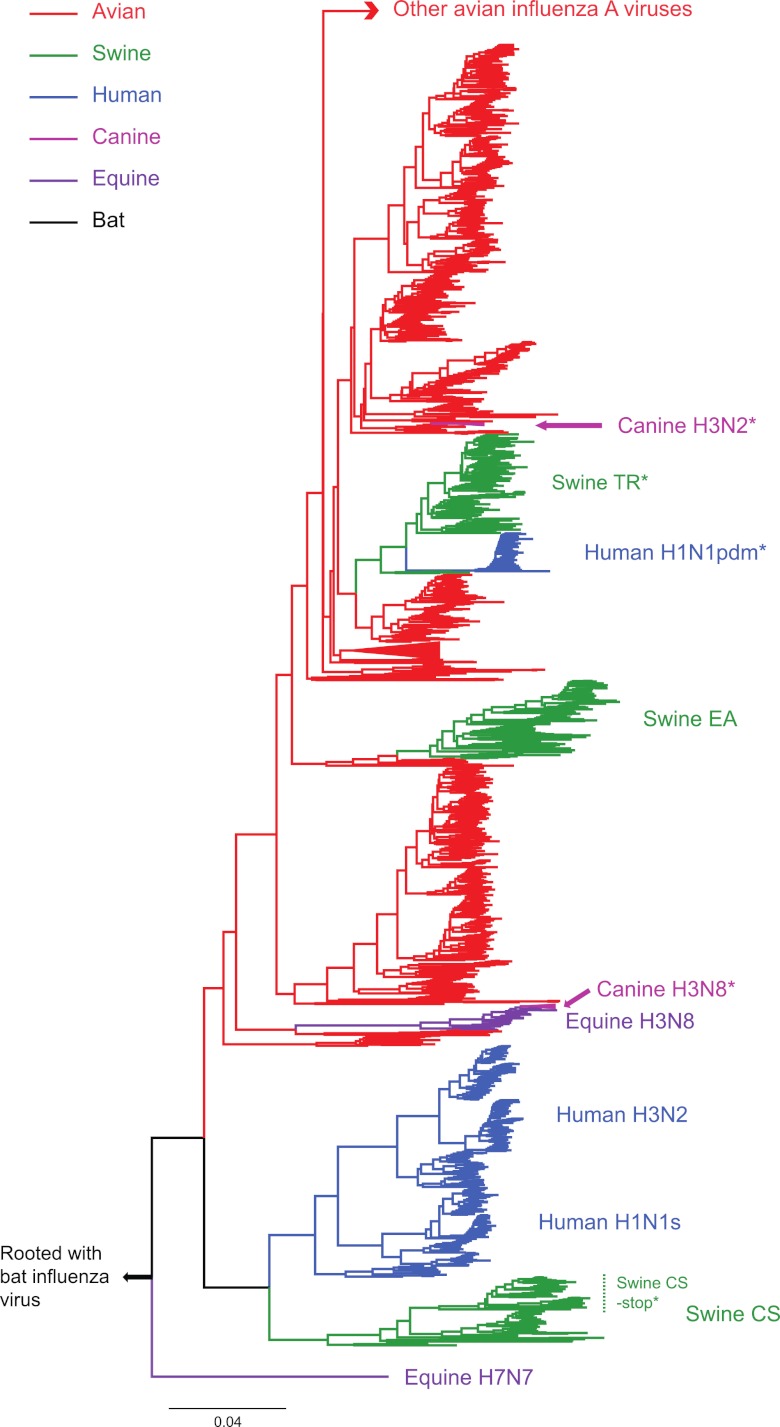
Of the 10,164 IAV sequences analyzed, 2,310 were truncated due to nonsense mutations, of which 2,279 (99%) occurred at X-ORF codon 42 (Fig. 2). These truncated PA-X proteins were associated with seven IAV groups: human H1N1pdm, swine TR H1N1, one cluster of swine CS H1N1 viruses, equine H7N7, canine H3N8, canine H3N2, and bat influenza virus (italics in Table 1), although the truncated proteins in the very small equine H7N7 and bat influenza virus data sets are due to stop codon mutations other than those at codon 42. The CS H1N1 viruses are particularly noteworthy, as the truncated form of PA-X occurs in a cluster of viruses sampled between 1985 and 2009, which fall within a group of generally older (1930 to 2006) swine CS viruses that possess a full-length PA-X (Fig. 2). This phylogenetic pattern suggests that swine CS viruses with the truncated form of PA-X were directly derived from those with full-length PA-X sequences. In addition, as the origins of the PA segment in human H1N1pdm lie with the TR swine influenza virus (2), the stop codon mutation at X-ORF codon 42 in this virus was clearly inherited from swine.
Fig 2.
Phylogenetic tree of the full-length PA gene (frame 0) of influenza A virus showing the occurrence of the stop codon mutation at codon 42 in the X-ORF. Because of the very large size of the data set, this tree was inferred using a subsample of 3,281 PA sequences that differed by at least 0.005 nucleotide substitution per site. Clusters are colored according to host species, with those containing the stop codon mutation at codon 42 marked by asterisks. All horizontal branch lengths are drawn to a scale of nucleotide substitutions per site.
Overall, our phylogenetic analysis suggests that the nonsense mutation at X-ORF codon 42 was fixed at least four times independently—twice in swine and twice in dogs. Uniquely, however, the stop codon change in the canine H3N2 viruses is due to a TGG-to-TGA mutation caused by a nonsynonymous mutation in frame 0 of the PA gene. In comparison, the “PA-X” sequence in influenza B virus is characterized by multiple stop codon mutations, which is also the case for the two available bat influenza viruses. Hence, a functional PA-X protein most likely evolved after the divergence of influenza A viruses from influenza B/bat influenza viruses.
The presence of a full-length PA-X protein in avian influenza virus, human H3N2, human 1918 pandemic and seasonal H1N1, swine EA, and equine H3N8 was associated with a reduction in dS in the region of the PA gene (frame 0) overlapping the +1 X-ORF compared to the rest of the PA gene (reflected in the “dS X-ORF/dS rest” column in Table 1). For example, in the case of the largest data set, representing avian influenza sequences, dS in the region of PA gene covering the X-ORF is 0.25 times that of dS in the remainder of the PA gene. In contrast, there was no such reduction in dS in those groups that possess truncated PA-X proteins, as was also the case for influenza B virus. For example, in human H1N1pdm, dS in the X-ORF was 1.19 times that of dS in rest of the PA gene (Table 1). The CS H1N1 viruses were again noteworthy; although most of the earlier-sampled CS viruses possess a full-length PA-X, there was little reduction in dS in the region of the PA gene covering the X-ORF. This, coupled with the fact that most recent CS sequences harbor a truncated PA-X, suggests that this protein may be of less functional importance in classical swine influenza viruses. Finally, it was also striking that the overall dN/dS ratio for the +1 X-ORF was substantially higher than that for the PA gene encoded in frame 0 (Table 1). Although accurately estimating selection pressures in sequences with dual reading frames is notoriously difficult (7), this observation indicates that PA-X is able to tolerate a high number of amino acid changes. As synonymous mutations in frame 0 will either be nonsynonymous or nonsense mutations in frame 1, this is likely to be the case for many viral proteins encoded by +1 ORFs (5).
The conservation of the decanucleotide frameshift motif (Fig. 1) and the maintenance of the full-length +1 X-ORF in the majority of IAV genomes infecting diverse host species (6) suggest that PA-X is important for influenza A virus biology. Interestingly, there are other possible synonymous mutations in the PA gene that would result in a PA-X truncated more severely than the commonly observed 41-amino-acid product of X-ORF. That these are not often seen suggests that stop codon mutations at X-ORF codon 42 renders the protein functionally distinct from products of 61-codon X-ORF isolates. As there is no evidence for decreased synonymous variability in the overlapping 0 frame of these truncated isolates, there are likely to be few selective constraints on X-ORF in these particular lineages. It is therefore probable that the protein domains encoded by the truncated X-ORFs have lost or altered functionality compared to the PA-Xs encoded by full-length X-ORFs. This hypothesis will need to be evaluated experimentally, especially in the context of particular host species. Indeed, as X-ORF protein truncation appears to be associated with IAV lineages circulating in particular hosts, i.e., pigs and dogs, there may be some species specificity to the evolution of a truncated PA-X protein. That a 41-amino-acid X-ORF protein evolved convergently in both IAV subtypes that infect dogs (H3N2 and H3N8) is particularly noteworthy and suggests that the truncation of this protein may be associated with the adaptation and emergence of influenza virus in this host species.
Footnotes
Published ahead of print 5 September 2012
REFERENCES
- 1. Dias A, et al. 2009. The cap-snatching endonuclease of influenza virus polymerase resides in the PA subunit. Nature 458:914–918 [DOI] [PubMed] [Google Scholar]
- 2. Garten RJ, et al. 2009. Antigenic and genetic characteristics of swine-origin 2009 A(H1N1) influenza viruses circulating in humans. Science 325:197–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Gog JR, et al. 2007. Codon conservation in the influenza A virus genome defines RNA packaging signals. Nucleic Acids Research. 35:1897–1907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. He X, et al. 2008. Crystal structure of the polymerase PA(C)-PB1(N) complex from an avian influenza H5N1 virus. Nature 454:1123–1126 [DOI] [PubMed] [Google Scholar]
- 5. Holmes EC, Lipman DJ, Zamarin D, Yewdell JW. 2006. Comment on “Large-scale sequence analysis of avian influenza isolates.” Science 313:1573 (Reply, 313:1573.) [DOI] [PubMed] [Google Scholar]
- 6. Jagger BW, et al. 2012. An overlapping protein-coding region in influenza A virus segment 3 modulates the host response. Science 337:199–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sabath N, Landan G, Graur D. 2008. A method for the simultaneous estimation of selection intensities in overlapping genes. PLoS One 3:e3996 doi:10.1371/journal.pone.0003996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Tamura K, et al. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28:2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yang Z. 2007. PAML 4: phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 24:1586–1591 [DOI] [PubMed] [Google Scholar]
- 10. Yuan P, et al. 2009. Crystal structure of an avian influenza polymerase PA(N) reveals an endonuclease active site. Nature 458:909–913 [DOI] [PubMed] [Google Scholar]