Abstract
A tyrosine-sulfated CCR5-mimetic peptide, CCR5mim1, inhibits HIV-1 infection more efficiently than sulfopeptides based on the CCR5 amino terminus. Here we characterized sulfopeptide chimeras of CCR5mim1 and the heavy-chain CDR3 of the antibody PG16. Two chimeras bound a range of envelope glycoproteins and neutralized HIV-1 more efficiently than CCR5mim1. An immunoadhesin form of one of these, CCR5mim2-Ig, synergized with CD4-Ig to neutralize HIV-1. These sulfopeptides are among the broadest and most potent CCR5-mimetic peptides described to date.
TEXT
Human immunodeficiency virus type 1 (HIV-1) entry requires cellular expression of CD4 and a coreceptor, principally CCR5 or CXCR4 (1, 5, 11). Virion association with CD4 triggers conformational changes in the HIV-1 envelope glycoprotein gp120 that promote high-affinity association with the coreceptor (21, 23). An acidic, tyrosine-sulfated region of the CCR5 amino terminus is critical for gp120 association, and most or all functional HIV-1 and simian immunodeficiency virus (SIV) coreceptors have amino termini bearing multiple sulfotyrosines (7–9). These sulfotyrosines interact with conserved pockets in the C4 region and at the base of the V3 loop of HIV-1 gp120 (3, 4, 10, 12). Coreceptor-binding site antibodies with tyrosine-sulfated antigen-combining regions also bind these conserved pockets, including E51, the most potent of the CD4-inducible (CD4i) HIV-1 neutralizing antibodies (2, 12). Recently, two additional broadly neutralizing antibodies, PG9 and PG16, have been identified (22). Like E51, these antibodies include sulfotyrosines in their heavy-chain CDR3. However, unlike CD4i antibodies, their association with the HIV-1 envelope glycoprotein is not enhanced by CD4.
Sulfopeptides based on the sequence of the CCR5 amino terminus specifically bind gp120 and inhibit HIV-1 entry, but only at 50 to 100 μM concentrations (4, 10), precluding their use as therapeutics. A mimetic peptide derived from the heavy-chain CDR3 of E51, pΔE51 (CCR5mim1), associates with gp120 with higher affinity and more efficiently neutralizes HIV-1 entry (6, 14). CCR5mim1 retains the arrangement of CCR5 sulfotyrosines, but it outperforms CCR5-based peptides, likely because it is more flexible and soluble than CCR5-based peptides.
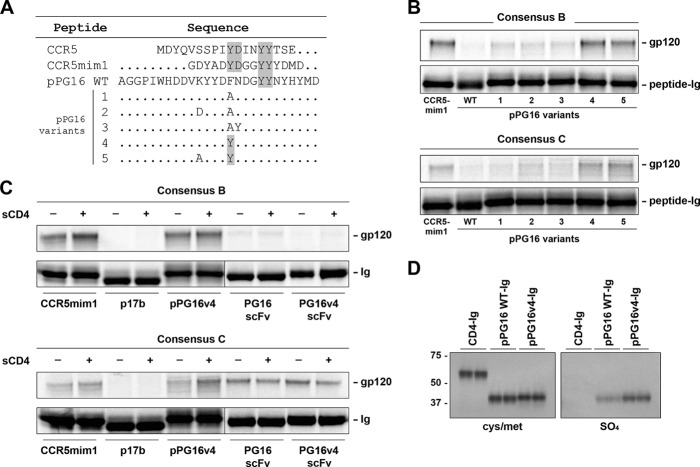
The example of CCR5mim1 suggested that additional antibody-derived peptides may bind gp120 more broadly and efficiently than either CCR5-based peptides or CCR5mim1. Accordingly, we investigated whether pPG16, a peptide based on the heavy-chain CDR3 region of PG16 (Fig. 1A), could precipitate gp120 using a previously described assay. We observed that pPG16-Ig, a fusion of this peptide with the human IgG1 Fc domain, bound metabolically labeled consensus B and C gp120 less efficiently than CCR5mim1-Ig (Fig. 1B). However, two pPG16-Ig variants precipitated similar amounts of consensus B gp120 and greater amounts of consensus C gp120 (Fig. 1B). Both variants included a phenylalanine-to-tyrosine substitution. We subsequently focused on variant 4 (pPG16v4-Ig) with this substitution alone (Fig. 1A). Unlike a single-chain form (scFv) of the PG16 antibody, binding of pPG16v4-Ig to consensus C gp120 was enhanced by soluble CD4 (sCD4) (Fig. 1C). Also, pPG16v4-Ig could incorporate more [35S]sulfate than wild-type pPG16-Ig, suggesting that the newly introduced tyrosine was modified by sulfate (Fig. 1D).
Fig 1.
pPG16v4-Ig, a variant of pPG16-Ig, precipitates HIV-1 gp120 more efficiently than the previously described CCR5-mimetic peptide-Fc fusion (CCR5mim1-Ig). (A) Amino acid sequence alignments of the CCR5 amino terminus, CCR5mim1, pPG16 wild type (WT), and pPG16 variants. Alterations to pPG16 are indicated. (B) [35S]cysteine and [35S]methionine-labeled consensus B or C gp120 molecules were precipitated with protein A-Sepharose and radiolabeled forms of the indicated peptide-Fc fusion and analyzed by SDS-PAGE. (C) An experiment similar to that shown in panel B, except the indicated peptide-Fc fusion or single-chain immunoadhesins were used to precipitate consensus B or C gp120 molecules in the presence or absence of soluble CD4. (D) The indicated Fc-fusion constructs were metabolically labeled with either [35S]cysteine and [35S]methionine (left panel) or [35S]sulfate (right panel) and analyzed as described for panel B. Note that all peptide-Fc fusions were generated in the presence of exogenous tyrosine-protein sulfotransferase 2.
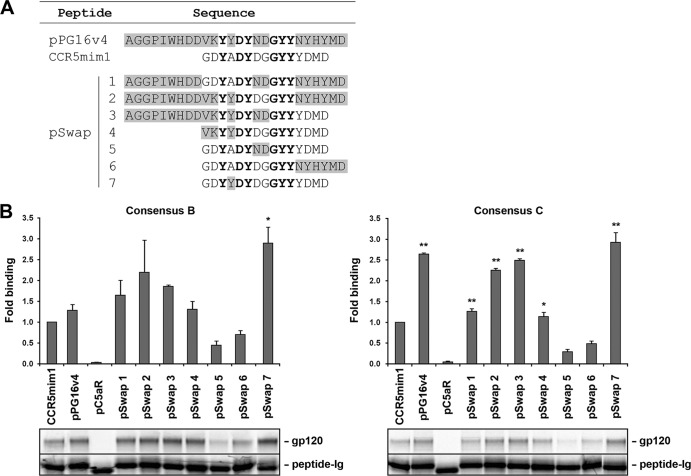
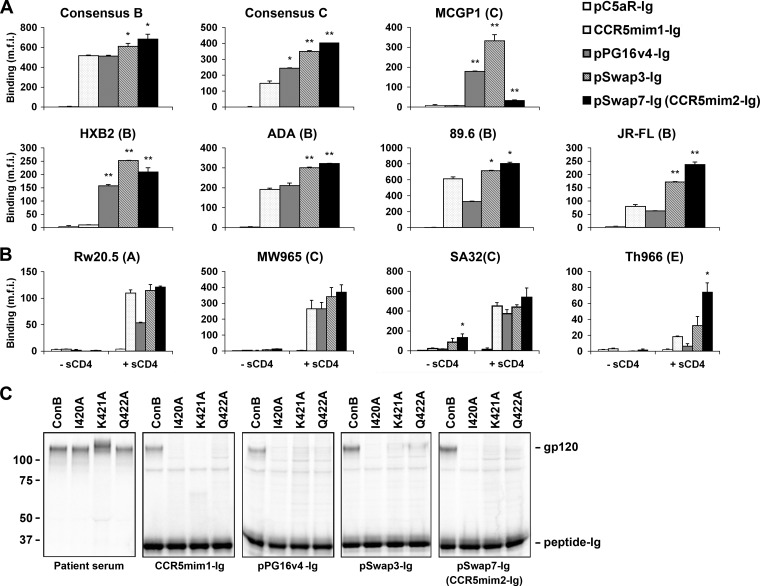
CCR5mim1, pPG16v4, and CCR5 share a arrangement of sulfotyrosines, facilitating their alignment. Based on this alignment, seven chimeras (pSwap1 to -7) between CCR5mim1 and pPG16v4 were made as Fc fusions (Fig. 2A). Several of these chimeras precipitated consensus B and C gp120 molecules with efficiency comparable to or better than that of either CCR5mim1-Ig or pPG16v4-Ig (Fig. 2B). We further compared two of these variants, pSwap3-Ig and pSwap7-Ig, with CCR5mim1-Ig and pPG16v4-Ig for their ability to bind a range of cell-surface-expressed HIV-1 envelope glycoproteins. In most cases, these variants bound envelope-glycoprotein-expressing cells more efficiently than CCR5mim1-Ig, pPG16v4-Ig, or a control Fc fusion with the sulfopeptide derived from the C5a receptor, pC5aR-Ig (Fig. 3A). In some cases, these peptides bound poorly, but their association was markedly enhanced by sCD4 (Fig. 3B). Also, pSwap3-Ig and pSwap7-Ig bound gp120 in the same region as both CCR5 and CCR5mim1, as indicated by their inability to bind consensus B (ConB) gp120 variants altered in a conserved CCR5-binding region (Fig. 3C) (6, 19). Given its close similarity to CCR5mim1 and its ability to bind envelope glycoprotein with higher affinity, we refer to pSwap7-Ig from this point on as “CCR5mim2-Ig.”
Fig 2.
Two chimeric peptides constructed from CCR5mim1 and pPG16v4 precipitate HIV-1 gp120 more efficiently than either original peptide. (A) Sequence alignment of pPG16v4, CCR5mim1, and chimeras thereof (pSwap1 to -7). Gray shading indicates pPG16v4 residues, unshaded letters indicate CCR5mim1 residues, and boldface lettering indicates common residues. (B) Experiments similar to that in Fig. 1B were performed. Bars indicate the ratio of the indicated gp120 to Fc fusion, measured by phosphorimaging and normalized to CCR5mim1-Ig. The bottom panel shows a representative experiment used to generate the figure. Single and double asterisks indicate significant differences (P < 0.05 and P < 0.005, respectively) from CCR5mim1.
Fig 3.
pSwap3-Ig and pSwap7-Ig (CCR5mim2-Ig) bind cell-surface-expressed HIV-1 gp120 envelope-glycoprotein more efficiently than CCR5mim1-Ig. (A) HEK239T cells were transfected to express the indicated envelope glycoproteins lacking most of their cytoplasmic domains. Cells were incubated with 50 nM the indicated peptide-Fc fusion proteins, washed, and analyzed by flow cytometry. m.f.i., mean fluorescence intensity. Single and double asterisks indicate significant differences (P < 0.05 and P < 0.005, respectively) from CCR5mim1. (B) An experiment similar to that in panel A, except that peptide-Fc fusions were incubated in the presence or absence of sCD4 (10 μg/ml), as indicated. (C) Experiments similar to that in Fig. 1B, except that consensus B gp120 I420A, K421A, and Q422A variants were precipitated with the patients' sera or the indicated peptide-Fc fusions.
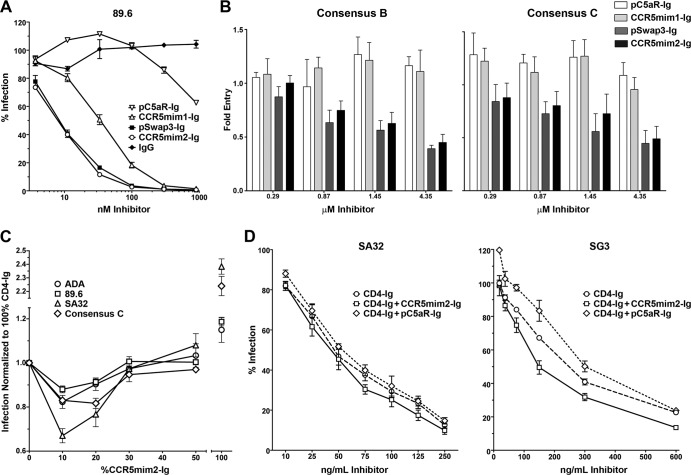
Next we analyzed pC5aR-Ig, CCR5mim1-Ig, pSwap3-Ig, and CCR5mim2-Ig using a previously described entry inhibition assay (6, 10). Both pSwap3-Ig and CCR5mim2-Ig neutralized the dualtropic, clade B isolate 89.6 more efficiently than CCR5mim1-Ig, with 50% inhibitory concentrations (IC50s) of approximately 10 nM (Fig. 4A). They better neutralized consensus B and C isolates, but with IC50s of approximately 4 μM (Fig. 4B). Using a previously described TZM-bl cell neutralization assay (16), we next investigated the ability of CCR5mim2-Ig to enhance CD4-Ig-mediated neutralization of HIV-1. When total protein was kept constant at the IC50 of CD4-Ig alone, a 9:1 ratio of CD4-Ig to CCR5mim2-Ig (4.5:1 on a molar basis) neutralized all pseudoviruses assayed more efficiently than CD4-Ig alone (Fig. 4C). This effect was evident at a 9:1 ratio for a range of total protein concentrations (Fig. 4D).
Fig 4.
CCR5mim2-Ig synergizes with CD4-Ig to neutralize HIV-1 infection. (A) The inhibitory activities of the indicated peptide-Fc fusions were measured with a TZM-bl neutralization assay using the envelope glycoprotein of the dualtropic clade B isolate 89.6. Infectivity is represented as a percentage of luciferase activity in the absence of inhibitor. (B) The indicated peptide-Fc fusions were assayed for their ability to limit infection of retroviruses pseudotyped with consensus B and C envelope glycoproteins in GHOST-CCR5 cells. Infection was measured as green fluorescent protein (GFP) activity by flow cytometry. (C) Relative infection of HIV-1 pseudotyped with the indicated envelope glycoproteins was measured with a TZM-bl neutralization assay in the presence of various ratios of CD4-Ig and CCR5mim2-Ig in which the total amount of protein was held constant. Horizontal axis indicates percentage of CCR5mim2-Ig. The total amount of protein for each isolate was chosen to be approximately that of the IC50 of CD4-Ig alone: 89.6 (clade B, R5X4), 50 ng/ml; ADA (clade B, R5), 40 ng/ml; SA32 (clade C, R5), 75 ng/ml; and consensus C (clade C, R5), 4 μg/ml. (D) A TZM-bl cell neutralization assay was performed at various concentrations of CD4-Ig and CCR5mim-Ig at a 9:1 ratio. The figure shows infection of cells with the SA32 (clade C, R5) and SG3 (clade B, X4) pseudoviruses in the presence of the CD4-Ig or CD4-Ig–peptide-Ig mixtures at various concentrations.
The synergy between CCR5mim2-Ig and CD4-Ig raises the possibility that a CCR5-mimetic peptide such a CCR5mim2 might increase the therapeutic utility of CD4-Ig. Despite its necessary breadth, CD4-Ig has been disappointing therapeutically because it has lower affinity for gp120 than effective neutralizing antibodies and because at low concentrations CD4-Ig enhances rather than inhibits HIV-1 entry (18, 20). This enhancement may occur because soluble forms of CD4 promote virion association with the coreceptor (21, 23). CCR5mim2-Ig can prevent coreceptor association with the virus and limit this enhancement. CCR5 mimetics like CCR5mim2-Ig have a second property that may also contribute to its potency as a partner for CD4-Ig. Unlike CD4-Ig, both arms of the dimeric CCR5 mimetic immunoadhesin can bind to gp120 monomers of an envelope glycoprotein trimer (14), perhaps more effectively preventing virus association with the cell. Finally, it is well established and consistent with our data here that sCD4 and CD4-Ig markedly enhance gp120 association with CCR5-mimetic peptides (4, 6, 10). We have also shown that in some cases, CCR5-mimetic peptides can decrease the off rate of CD4-mimetic peptides (14) and perhaps CD4-Ig from the envelope glycoprotein.
Combinations of CD4- and CCR5-mimetic peptides have one key advantage over most neutralizing antibodies: they associate with necessarily conserved regions of gp120 (15, 24). In contrast, antibody epitopes are larger than the CD4- and CCR5-binding sites and so must include variable residues that permit escape. Recent advances in gene therapy, such as self-complementary adeno-associated virus (scAAV), have enabled persistent expression of high levels of immunoadhesins but impose a size limit that precludes expression of full-length antibodies (13, 17). In contrast, both CD4-Ig and CCR5mim2-Ig can be easily expressed by scAAV vectors. Further study of their use in this context and further improvement of CCR5-mimetic peptides are therefore warranted. Finally, CCR5mim1, CCR5mim2, and pSwap3 may be useful in exploring the variation in the sulfotyrosine-binding pockets of gp120 or investigating the conformational transitions of the HIV-1 envelope glycoprotein.
Footnotes
Published ahead of print 29 August 2012
REFERENCES
- 1. Choe H, et al. 1996. The beta-chemokine receptors CCR3 and CCR5 facilitate infection by primary HIV-1 isolates. Cell 85:1135–1148 [DOI] [PubMed] [Google Scholar]
- 2. Choe H, et al. 2003. Tyrosine sulfation of human antibodies contributes to recognition of the CCR5 binding region of HIV-1 gp120. Cell 114:161–170 [DOI] [PubMed] [Google Scholar]
- 3. Cormier EG, Dragic T. 2002. The crown and stem of the V3 loop play distinct roles in human immunodeficiency virus type 1 envelope glycoprotein interactions with the CCR5 coreceptor. J. Virol. 76:8953–8957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cormier EG, et al. 2000. Specific interaction of CCR5 amino-terminal domain peptides containing sulfotyrosines with HIV-1 envelope glycoprotein gp120. Proc. Natl. Acad. Sci. U. S. A. 97:5762–5767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Dalgleish AG, et al. 1984. The CD4 (T4) antigen is an essential component of the receptor for the AIDS retrovirus. Nature 312:763–767 [DOI] [PubMed] [Google Scholar]
- 6. Dorfman T, Moore MJ, Guth AC, Choe H, Farzan M. 2006. A tyrosine-sulfated peptide derived from the heavy-chain CDR3 region of an HIV-1-neutralizing antibody binds gp120 and inhibits HIV-1 infection. J. Biol. Chem. 281:28529–28535 [DOI] [PubMed] [Google Scholar]
- 7. Farzan M, et al. 2002. The role of post-translational modifications of the CXCR4 amino-terminus in stromal-derived factor 1 alpha association and HIV-1 entry. J. Biol. Chem. 277:29484–29489 [DOI] [PubMed] [Google Scholar]
- 8. Farzan M, et al. 1998. A tyrosine-rich region in the N terminus of CCR5 is important for human immunodeficiency virus type 1 entry and mediates an association between gp120 and CCR5. J. Virol. 72:1160–1164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Farzan M, et al. 1999. Tyrosine sulfation of the amino terminus of CCR5 facilitates HIV-1 entry. Cell 96:667–676 [DOI] [PubMed] [Google Scholar]
- 10. Farzan M, et al. 2000. A tyrosine-sulfated peptide based on the N terminus of CCR5 interacts with a CD4-enhanced epitope of the HIV-1 gp120 envelope glycoprotein and inhibits HIV-1 entry. J. Biol. Chem. 275:33516–33521 [DOI] [PubMed] [Google Scholar]
- 11. Feng Y, Broder CC, Kennedy PE, Berger EA. 1996. HIV-1 entry cofactor: functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science 272:872–877 [DOI] [PubMed] [Google Scholar]
- 12. Huang CC, et al. 2007. Structures of the CCR5 N terminus and of a tyrosine-sulfated antibody with HIV-1 gp120 and CD4. Science 317:1930–1934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Johnson PR, et al. 2009. Vector-mediated gene transfer engenders long-lived neutralizing activity and protection against SIV infection in monkeys. Nat. Med. 15:901–906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kwong JA, et al. 2011. A tyrosine-sulfated CCR5-mimetic peptide promotes conformational transitions in the HIV-1 envelope glycoprotein. J. Virol. 85:7563–7571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kwong PD, et al. 1998. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature 393:648–659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Li M, et al. 2005. Human immunodeficiency virus type 1 env clones from acute and early subtype B infections for standardized assessments of vaccine-elicited neutralizing antibodies. J. Virol. 79:10108–10125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. McCarty DM. 2008. Self-complementary AAV vectors; advances and applications. Mol. Ther. 16:1648–1656 [DOI] [PubMed] [Google Scholar]
- 18. Moebius U, Clayton LK, Abraham S, Harrison SC, Reinherz EL. 1992. The human immunodeficiency virus gp120 binding site on CD4: delineation by quantitative equilibrium and kinetic binding studies of mutants in conjunction with a high-resolution CD4 atomic structure. J. Exp. Med. 176:507–517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Rizzuto CD, et al. 1998. A conserved HIV gp120 glycoprotein structure involved in chemokine receptor binding. Science 280:1949–1953 [DOI] [PubMed] [Google Scholar]
- 20. Sullivan N, Sun Y, Li J, Hofmann W, Sodroski J. 1995. Replicative function and neutralization sensitivity of envelope glycoproteins from primary and T-cell line-passaged human immunodeficiency virus type 1 isolates. J. Virol. 69:4413–4422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Trkola A, et al. 1996. CD4-dependent, antibody-sensitive interactions between HIV-1 and its co-receptor CCR-5. Nature 384:184–187 [DOI] [PubMed] [Google Scholar]
- 22. Walker LM, et al. 2009. Broad and potent neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Science 326:285–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wu L, et al. 1996. CD4-induced interactions of primary HIV-1 gp120 glycoproteins with the chemokine receptor CCR-5. Nature 184:179–183 [DOI] [PubMed] [Google Scholar]
- 24. Wyatt R, et al. 1998. The antigenic structure of the HIV gp120 envelope glycoprotein. Nature 393:705–711 [DOI] [PubMed] [Google Scholar]