Abstract
The mRNAs encoding the Rev and Env proteins of simian immunodeficiency virus (SIV) are unique because upstream translation start codons are present that may modulate the expression of these viral proteins. This is true for the regular mRNAs, but we also report novel mRNA splicing variants that encode up to five upstream AUG (uAUG) codons. Their influence on Rev and Env translation was measured by mutational inactivation in reporter constructs and in the SIVmac239 strain. An intricate regulatory mechanism was disclosed that allows the virus to express a balanced amount of these two proteins. This insight also allows the design of vector constructs that efficiently express these proteins.
INTRODUCTION
The RNA genome of the simian immunodeficiency virus (SIV) displays a complex splicing pattern with several splice donor (SD) and splice acceptor (SA) sites (Fig. 1) to allow the expression of all essential, regulatory, and accessory proteins (45, 51, 57). Translation initiation on eukaryotic mRNAs generally occurs via ribosomal scanning (34), in which translation initiation factors interact with the 40S ribosomal subunit at the mRNA 5′ cap structure. The ribosomal subunit then scans along the 5′ untranslated region (UTR) until it encounters an AUG translational start codon in a favorable sequence context. The optimal context of an AUG start codon in vertebrate cells is known as the Kozak consensus sequence: RCCAUGG, in which a purine at position −3, C at −1, and G at +4 have the strongest effects on the translation initiation efficiency (32). When an AUG is not in a favorable sequence context, translation initiation will be inefficient, and the ribosome may continue scanning until it encounters an AUG start codon further downstream (32). This mechanism of leaky scanning has been described for several viruses and enables these viral mRNAs to produce more than a single protein (30, 33, 48). In human immunodeficiency virus type 1 (HIV-1), the Vpu and Env proteins are encoded by the same mRNA because part of the ribosomes ignores the upstream Vpu start codon, which is in a weak Kozak context, and thus gains access to the downstream Env start codon (1, 35, 47). Other retroviruses, like Rous sarcoma virus (RSV), use upstream open reading frames (ORFs) to regulate the level of Gag translation (17).
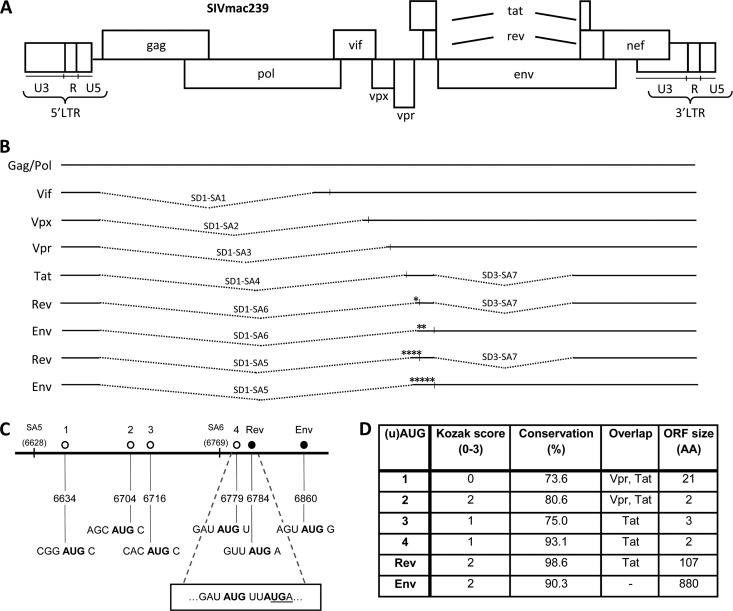
Fig 1.
SIV displays a complex splicing pattern, and upstream AUGs are present in the Rev and Env mRNAs. (A) The SIV DNA genome. (B) SIV mRNA splice variants with the encoded protein indicated on the left. The splice donor (SD) and splice acceptor (SA) sites used in SIV mRNA are indicated, along with the regular AUG start codons (vertical lines) and the upstream AUGs (*). (C) Start codons present between SA5 and the Env start codon in the SIV RNA genome. The uAUGs are depicted by an open circle, and the regular AUGs for Rev and Env by a filled circle with the name on top. The positions of SA5 and SA6 are marked. Below are the genomic positions and the sequence surrounding each AUG for comparison to the Kozak consensus sequence. The overlap between the uORF starting at uAUG4 and the Rev AUG is shown in a box with AUGs in bold and the uORF4 stop codon underlined. (D) Characteristics of each uAUG. Kozak score: a score was awarded based on the match with the Kozak consensus sequence (RCCAUGG) at positions −3 (A or G), −1 (C), and +4 (G), yielding a total score from 0 to 3. Conservation is the percentage of SIV isolates with the respective AUG based on all (n = 72) SIV sequences in the HIV Sequence Compendium 2011 (36). Overlap lists the viral ORFs with which each uAUG-ORF overlaps. ORF size is the length of the ORF served by each uAUG, indicated by the number of encoded amino acids (AA).
In SIVmac239, the singly spliced Env-encoding mRNA also encodes an upstream ORF, namely, Rev exon 1 (45, 51, 57). This situation is rather unique, as no other SIV transcripts have been reported to contain upstream AUGs (uAUGs) in front of the major ORF (Fig. 1B). SIV Rev is translated from its own doubly spliced mRNA. Surprisingly, we discovered that there is an additional uAUG codon present in these Rev and Env mRNAs. This uAUG is highly conserved among different SIV strains. Even more strikingly, we detected an alternative splicing event (switch from SA6 to upstream SA5) that puts three additional uAUG codons in front of the Rev and Env ORFs. We speculated that these multiple uAUGs may have a regulatory function in translation of the Rev and Env proteins. Here, we investigated the modulatory role of these uAUGs on Rev and Env translation using subgenomic reporter constructs and the impact on virus replication.
MATERIALS AND METHODS
Subgenomic reporter plasmids.
For the construction of the plasmids pSIV-SA5-rev-luc, pSIV-SA5-env-luc, and pSIV-SA6-env-luc, SIVmac239 sequences present in the pSIV-rtTA (15) template were PCR amplified with the forward primer (FP) SIV-mut-HindIII (GCGAAAGCTTGGCGGATGCATCCACTCCA; nucleotides matching SIV sequence in bold, HindIII site in italics) and the reverse primer (RP) SIV-mut-rev-NarI (AATGGCGCCGGGCCTTTCTTTATGTTTTTGGCGTCTTCGCTCATAACATATCCCCAAG; NarI site in italics), FP SIV-mut-HindIII and RP GV004 (AATGGCGCCGGGCCTTTCTTTATGTTTTTGGCGTCTTCTCCCATACTTACTTGTTTGA), and FP SIV-Rev-SA6-luc (CAAAAAGCTTGCTTGGGGATATGTTATG) and RP GV004, respectively. The PCR products were subsequently digested with HindIII and NarI and ligated into the corresponding sites of pGL3-control (Promega). For the construction of plasmid pSIV-SA6-rev-luc, the oligonucleotides SIV-rev-SA6-luc and SIV-mut-Rev-NarI were hybridized, and both strands were completed with Klenow DNA polymerase (Roche). The resulting DNA fragment was digested with HindIII and NarI and ligated into the corresponding sites of pGL3-control. The various uAUG mutations were subsequently introduced in these reporter plasmids by mutagenesis PCR (details available upon request). All nucleotide numbers refer to the positions in the SIVmac239 proviral DNA genome (GenBank accession number MM33262.1).
Construction of the SIVmac239 uAUG4 knockout mutant.
The plasmid pSIVmac239 encodes the wild-type (wt) SIVmac239 isolate (22). For the construction of SIVmac239-m4, the m4 mutation was first introduced in the SIV-rtTAopt-Tatstop construct (53). Separate PCRs were performed on SIV-rtTAopt-Tatstop with FP SIV-Tat-1 (GGTAGTGGAGGTTCTGGAAGA) and RP SIV-Tat-Splice-2 (GTTGGATATGGGTTTGTTTGATGCAGAAGATGTATT) and FP GV032 (AAAAAAGGCTTGGGGATTTGTTATGAGCAATCAC) and RP SIV-Env-8-GV (GTTGCTGCACTATCCCAGCC). The products were mixed and served as the template for a third PCR with FP SIV-Tat-1 and RP SIV-Env-8-GV. This product was digested with SphI and Kpn2I and ligated into the corresponding sites in SIV-rtTAopt-Tatstop to generate SIV-rtTAopt-Tatstop-m4. The latter vector was digested with SphI and Kpn2I, and the resulting fragment was ligated into the corresponding sites in SIVmac239 to create SIVmac239-m4.
Cells and virus cultures.
Human embryonic kidney (HEK) 293T cells were cultured in Dulbecco's modified Eagle medium (Gibco) supplemented with 10% fetal calf serum (Gibco), 40 units/ml penicillin, 40 μg/ml streptomycin, and 0.1 mM minimal essential medium nonessential amino acids (Gibco). 293T cells were cultured to 60% confluence in 2-cm2 wells and transfected with 1 μg of the SIVmac239 or SIVmac239-m4 plasmids to produce virus stocks as described previously (13). Cell-free culture supernatants were harvested after 48 h, and virus production was quantified by CA-p27 enzyme-linked immunosorbent assay (Advanced Bioscience Laboratories).
The PM1 (9, 37) and 174×CEM (25, 49) cell lines were cultured at 37°C and 5% CO2 in advanced RPMI 1640 (Gibco) containing 1% fetal bovine serum (Gibco), 2 mM l-glutamine (Gibco), 15 units/ml penicillin, and 15 μg/ml streptomycin. To assay virus replication, PM1 and 174×CEM cells were infected with equal amounts of virus (corresponding to 5 ng CA-p27) for 16 h at 37°C.
Peripheral blood mononuclear cells (PBMC) were isolated from cynomolgus macaques (41) and cultured as described previously (14). Cells were activated with 2 μg/ml phytohemagglutinin for 2 days and infected with equal amounts of virus (corresponding to 5 ng CA-p27) for 16 h at 37°C. Cells were maintained with 100 units/ml recombinant interleukin-2 following infection.
For all replication curves, the virus level in the culture medium was determined with a real-time PCR-based reverse transcriptase (RT) assay, in which avian myeloblastosis virus (AMV) RT was used as the standard (15, 39).
Luciferase assay.
To quantify protein production of each construct, 293T cells were cultured to 60% confluence in 2-cm2 wells and transfected with 100 ng of a luciferase reporter construct, 0.5 ng pRL-CMV, and 900 ng pBluescript (as carrier DNA) as previously described (13). The plasmid pRL-CMV (Promega), in which the expression of renilla luciferase is controlled by the cytomegalovirus (CMV) immediate-early enhancer/promoter, was cotransfected to allow correction for differences in transfection efficiency. The firefly and renilla luciferase production was measured after culturing the cells for 48 h. Protein production was calculated as the ratio between firefly and renilla luciferase activities and corrected for between-session variation (46).
Isolation of RNA.
For RNA analysis, 293T cells were cultured in 2-cm2 wells and transfected with 1 μg of the subgenomic reporter constructs by calcium phosphate precipitation (16). Cells were washed with phosphate-buffered saline (PBS) after 48 h, lysed in 350 μl RLT buffer (Qiagen), and homogenized with a QIAshredder column (Qiagen). Total cellular RNA was isolated with an RNeasy kit (Qiagen), and contaminating DNA was removed on the column with RNase-free DNase (Qiagen).
Northern blot analysis of RNA.
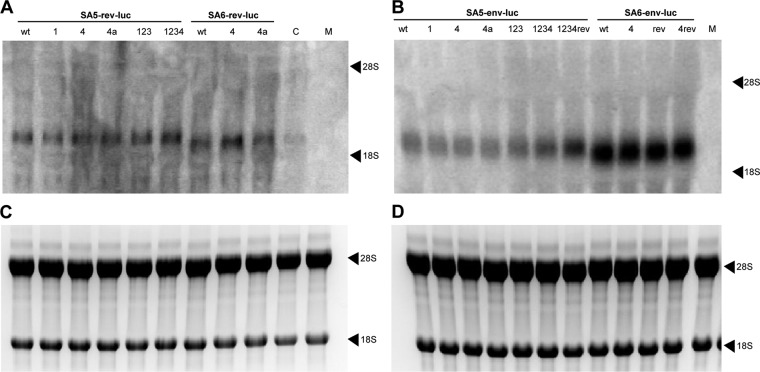
After electrophoresis of 5 μg RNA on a 1% agarose gel in 1× morpholinepropanesulfonic acid (MOPS) buffer (40 mM MOPS, 10 mM sodium acetate, pH 7.0) with 6.5% formaldehyde at 100 V for 4 h, RNAs were transferred onto a positively charged nylon membrane (Boehringer Mannheim) overnight by means of capillary force. RNAs were linked to the membrane in a UV cross-linker (Stratagene). The NcoI-XbaI luciferase fragment of the pGL3-control plasmid was labeled with [α-32P]dCTP with the High Prime DNA labeling kit (Roche). Prehybridization and hybridization of the membrane with the probe were done in ULTRAhyb buffer (Ambion) at 55°C for 1 and 16 h, respectively. The membrane was then washed twice for 15 min at room temperature in low-stringency buffer (2× SSC, 0.2% sodium dodecyl sulfate [SDS] [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate]) and twice for 30 min at 60°C in high-stringency buffer (0.1× SSC, 0.2% SDS). Images were obtained using a Storm 860 phosphorimager (Amersham Biosciences). The sizes of the luciferase transcripts were estimated using the ethidium bromide-stained 18S and 28S rRNAs as size markers. The amounts of ethidium bromide-stained 18S and 28S rRNAs were the same for all samples.
Slot blot analysis of RNA.
RNAs were dotted onto Zeta-probe blotting membrane (Bio-Rad) using a Bio-dot microfiltration apparatus (Bio-Rad). RNA (1 μg) was dissolved in glyoxal buffer (final concentration of 50% dimethyl sulfoxide [DMSO], 10 mM NaH2PO4, pH 7.2, and 1 M glyoxal) and incubated at 50°C for 1 h followed by cooling. The membrane was washed with TE (10 mM Tris-HCl, 1 mM EDTA, pH 8.0), and the samples were loaded into the slot and blotted onto the membrane by vacuum. The membrane was washed once with TE and once with 2× SSC-0.1% SDS. RNAs were linked to the membrane in a UV cross-linker (Stratagene), and the blot was hybridized, washed, and analyzed as described above for the Northern blot. Data analysis was performed with the ImageQuant software package. The background signal (mock transfection) was subtracted from all samples before the values were plotted.
Virus competition experiments, proviral DNA isolation, and sequencing.
Virus competition experiments were conducted in 174×CEM cells to determine the fitness of SIVmac239-m4 relative to the parental wild-type (wt) SIVmac239 virus. Pairwise competitions were started by transfecting equal amounts of each viral construct as previously described (27, 28, 55). The virus was passaged onto fresh cells at the peak of infection when massive syncytia were observed in order to continue the competition.
For proviral DNA analysis, infected cells were pelleted by centrifugation at 1,500 × g for 4 min and washed with phosphate-buffered saline. DNA was solubilized by resuspending the cells in TLE (10 mM Tris, 0.1 mM EDTA, pH 8.0) plus 0.5% Tween 20, followed by incubation with 200 μg/ml of proteinase K at 56°C for 60 min and subsequently at 95°C for 10 min. For the analysis of Rev-coding exon 1, proviral DNA sequences were PCR amplified from total cellular DNA with primers SIV-Tat-2 (GGGAACCATGGGATGAATG) and SIV-Env-4 (CCCTGTCATGTTGAATTTACAGCT). The PCR product was subsequently sequenced using the BigDye Terminator cycle sequencing kit (Applied Biosystems) and primer SIV-Tat-1 (GGTAGTGGAGGTTCTGGAAGA). The sequences were aligned using the CodonCode Aligner software package.
RESULTS
Multiple uAUGs in the SIV transcripts encoding Rev and Env proteins.
The SIV genome yields a complex array of mRNA transcripts to express all viral proteins (45, 51, 57). In principle, splicing ensures that the translational start codon of each protein-encoding ORF represents the first AUG on a dedicated mRNA. The situation seems to be more complex for the Rev ORF and in particular the Env ORF. Rev is translated from a doubly spliced mRNA, whereas Env is synthesized from a singly spliced mRNA. This Env mRNA has an upstream ORF (uORF) that encodes 23 Rev amino acids (exon 1) fused to another 50 unrelated amino acids before a stop codon is reached. This situation gets more complicated by two new findings. First, we discovered an additional AUG immediately upstream of the Rev start codon on the regular Rev and Env mRNAs. Second, we detected alternatively spliced RNAs that use SA5 instead of SA6 to create 5′-extended Rev and Env mRNAs. This alternative splicing event has been detected previously (45) and seems to represent a minor transcript. However, we observed that splicing to SA5 was increased upon the mutation of the Tat gene in a doxycycline-controlled SIVmac239 derivate (SIV-rtTA-TatY55A) (15) (unpublished results). The SA5 sequence as present in SIVmac239 is 89% conserved throughout the different SIV strains (36), and the 11% that differ in sequence may encode a similar splice acceptor site, as the SIVmac239 sequence does not match the classical splice signal (18). These SA5 transcripts do attach upstream sequences to the Rev and Env mRNAs with three additional uAUGs (Fig. 1). We therefore named them uAUGs 1 to 4 (Fig. 1C), the latter being the start codon immediately upstream of the Rev start, and decided to study their relevance for Rev and Env translation.
The sequence around the uAUGs was compared to the optimal Kozak consensus sequence, which includes the sequence of the second codon (21, 26, 29, 31). We gave the start codons a score from zero to three based on similarity to the consensus RCCAUGG sequence, where a point was awarded for positions −3, −1, and +4 if these nucleotides matched. The uAUGs have poor to moderately good scores, ranging from 0 for uAUG1 to 2 for uAUG2 (Fig. 1D). However, many of the regular HIV and SIV start codons are also in a suboptimal Kozak context (references 48 and 59 and our own analysis). We included the Rev and Env AUGs for comparison, and both are moderately good, with a Kozak score of 2 (Fig. 1D).
We subsequently scored the conservation of these uAUG codons in 72 SIV isolates (36) because important regulatory sequence motifs are expected to be maintained during virus evolution. uAUGs 1, 2, and 3 of the alternative SA5 transcript score moderately high at 73.6% to 80.6% conservation, but uAUG4 of the regular SA6 transcript is highly conserved in 93.1% of the viral isolates (Fig. 1D). Even the well-accepted start codons for Rev and Env do not yield 100% conservation (Fig. 1D). This analysis may be influenced by selection pressure imposed by overlapping ORFs, which are listed in Fig. 1D. uAUGs 1 and 2 overlap the Vpr and Tat ORFs, whereas uAUGs 3 and 4 overlap only with the Tat ORF. Thus, while uAUG4 is not unique in terms of overlap with viral ORFs, it is highly conserved.
We next analyzed whether these uAUGs have coding capacity in terms of the length of the corresponding ORFs (Fig. 1D). While uAUG1 encodes a 21-amino-acid peptide, all other uAUGs facilitate translation of maximally 2 to 3 amino acids before a termination codon is encountered. Blasting the 21-amino-acid peptide against the NCBI protein database produced only a hit with an N-terminally extended Rev protein from a particular SIV strain (isolate CPZ GAB1) that lacks the stop codon of the uAUG1-ORF (11). The potential for resumed scanning and reinitiation at the next AUG is linked to the length of the uORF (34, 38). Another important determinant of the reinitiation efficiency is the distance of the uORF stop codon to the start codon of the next ORF. The three-codon ORF initiated at uAUG4 terminates at a UGA codon that overlaps with the Rev AUG (Fig. 1C). It has been shown that suppression by uAUGs is most efficient when the uORF terminates downstream of the initiation site of the downstream ORF, most likely because ribosomes cannot scan backwards (34).
Taken together, Rev translation from the regular SA6 transcript could be regulated by uAUG4, and the alternative SA5 transcript has three additional uAUGs that may down-modulate Rev expression by absorbing the scanning ribosomes. Env translation seems even more complex as it faces the Rev start codon as an additional uAUG. We therefore set out to measure Rev and Env translation in the SA5 and SA6 contexts by mutation of the respective uAUG codons.
Design of uAUG mutants.
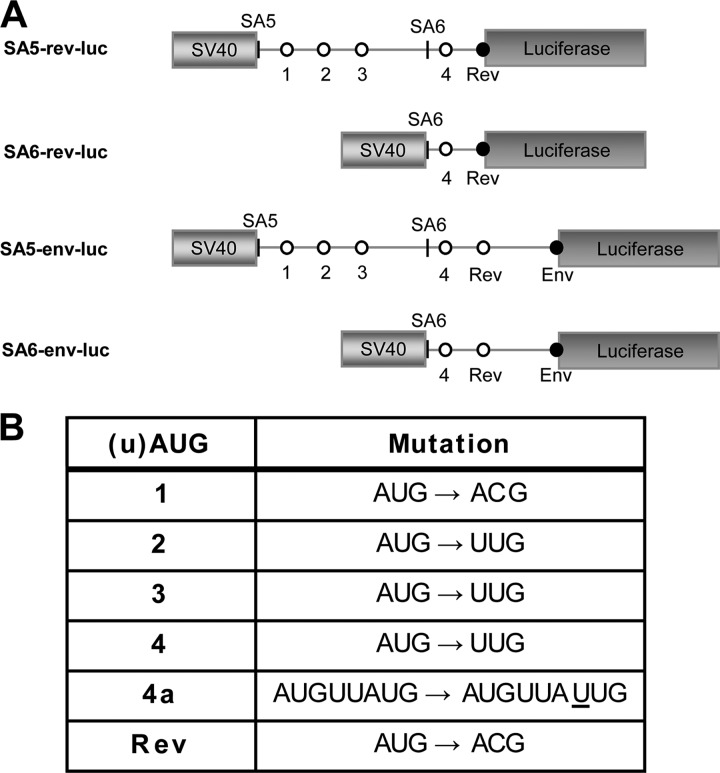
Subgenomic luciferase reporter constructs were made that are driven by the SV40 early promoter and that encode SIVmac239 sequences starting at SA5 (genomic position 6628) or SA6 (genomic position 6769). We purposely designed such a minimal expression cassette to avoid complexities present in the SIV genome, e.g., the Tat-LTR (long-terminal repeat) and Rev-RRE (Rev-responsive element) regulatory circuits. The luciferase reporter ORF was fused to the second codon of the Rev and Env ORFs, thus leaving the sequence context around the AUG codon intact (21, 26, 29, 31). This creates a basic set of four reporter constructs: SA5-rev-luc, SA6-rev-luc, SA5-env-luc, and SA6-env-luc (Fig. 2A). Inactivating mutations were introduced in individual or multiple uAUGs to study the effect on Rev and Env expression (Fig. 2B). The constructs were transfected into 293T cells, and the amount of luciferase protein was measured after 2 days as a measure of the translational efficiency.
Fig 2.
Design of subgenomic reporter constructs. (A) The subgenomic reporter constructs. SIV sequences starting at SA5 or SA6 were placed directly downstream of the SV40 promoter in the pGL3-control plasmid (Promega), and the second codon of the Rev or Env ORF was fused to the luciferase gene in order to preserve the Kozak motif. The uAUGs are depicted by white circles and the regular start codons by black circles. (B) The inactivating mutations for each AUG. These mutations were chosen to be silent in the overlapping Tat ORF for future testing in the full-length SIV genome. The 4a mutation is a single nucleotide insertion (underlined) that fuses uAUG4 to the Rev ORF and simultaneously inactivates the Rev AUG.
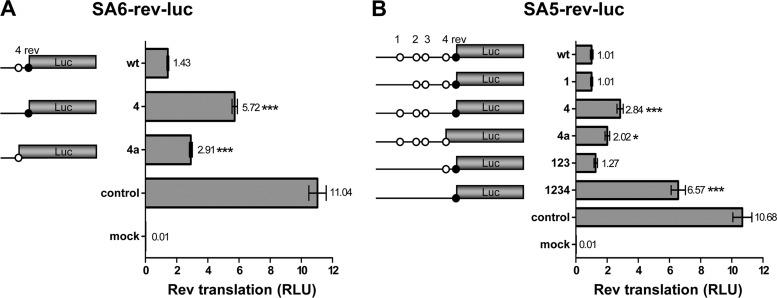
Impact of uAUGs on Rev translation.
The effect of uAUG mutation on Rev translation from SA5-rev-luc and SA6-rev-luc was measured. We will first discuss Rev expression from the regular SA6-rev-luc transcript (Fig. 3A). The wt Rev-luc construct, in which one uAUG is present (uAUG4), produces relatively little luciferase activity (13%) compared to the unrelated SV40-luciferase control construct. Interestingly, inactivation of uAUG4 stimulates Rev-luc expression approximately 4-fold, providing the first evidence that Rev translation is profoundly down-modulated by the capture of scanning ribosomes at uAUG4. To prove that uAUG4 is indeed recognized as a translational start codon, we made an additional construct in which the uAUG4 start was connected to the luciferase ORF by a single nucleotide insertion that simultaneously inactivates the Rev AUG (Fig. 2B). This 4a variant produced about twice as much luciferase as the wt Rev-luc fusion, confirming that uAUG4 absorbs a significant fraction of the scanning ribosomes to suppress Rev translation.
Fig 3.
Protein expression of subgenomic Rev reporters. 293T cells were transfected with the SA6-rev-luc reporter constructs (A) or with the SA5-rev-luc reporter constructs (B). Both constructs encode the firefly luciferase gene. Cells were cultured for 2 days, and the intracellular firefly luciferase level (relative light units [RLU]) was measured to quantify luciferase production, using renilla luciferase produced from the cotransfected pRL-CMV plasmid as an internal control. The ratio between firefly and renilla luciferase was calculated, and the results of 14 measurements (means ± standard errors of the means [SEM]) are shown. The mean value is shown next to each bar. Statistical analysis performed by one-way analysis of variance (ANOVA) demonstrated that luciferase production differed significantly between SA6-rev-luc (wt) and mutants 4 and 4a (A; ***, P < 0.0001); luciferase production differed significantly between SA5-rev-luc (wt) and mutants 4, 4a, and 1234 (B; *, P < 0.05). Control, cells transfected with pGL3-control; mock, cells transfected with pBluescript plasmid instead of firefly luciferase-encoding plasmid.
We next analyzed the impact of uAUG1 to uAUG4 inactivation in the context of the alternative SA5-rev-luc transcript (Fig. 3B). The wt SA5 transcript is an even weaker Rev-luc expresser than the wt SA6 construct, as demonstrated by the modest 9% expression level compared to the control SV40-luciferase construct. This could be due to the presence of four uAUG motifs. Inactivation of uAUG1 did not increase Rev translation, but the combined inactivation of uAUG1 to uAUG3 showed a small, although not significant, stimulatory effect. Inactivation of uAUG4 again demonstrated a profound increase of Rev translation. Removal of all uAUGs (construct 1234) caused an even greater 6.5-fold induction. The latter transcript expresses approximately 60% of the amount of luciferase produced by the SV40-luciferase control. Upon fusing uAUG4 to luciferase in variant 4a, we measured increased luciferase production (2-fold higher than with the wt construct), which again demonstrated that the upstream start codon is actively used by scanning ribosomes. These combined results indicate that uAUG4 and, to a lesser extent, uAUGs 1, 2, and 3 in the SA5 context play a regulatory role to modulate Rev translation.
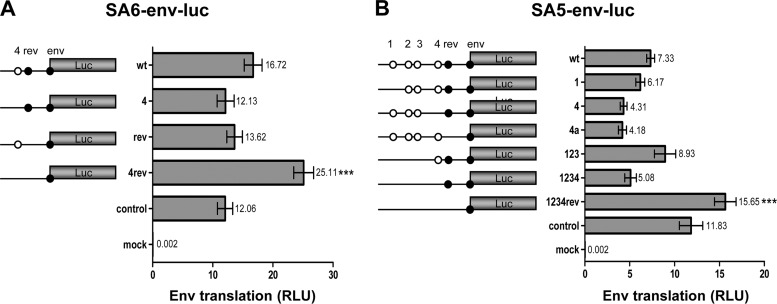
Impact of uAUGs on Env translation.
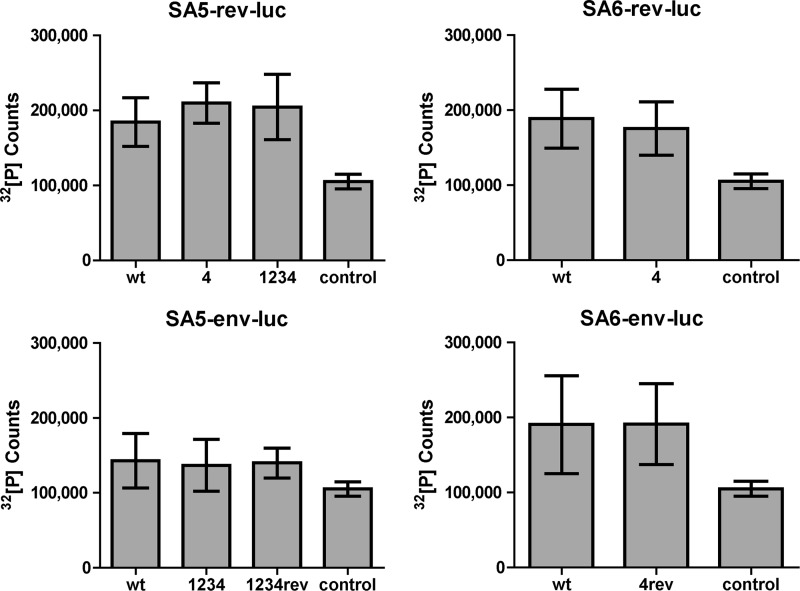
Next we tested the impact of uAUG motifs on Env translation. The regular SA6 transcript encodes two upstream start codons: uAUG4 and AUG-Rev (Fig. 4A). The wt Env-luciferase transcript clearly exhibits more luciferase activity than the control SV40-luciferase construct, which contrasts with the poorly translated Rev-luciferase transcript. In light of the scanning mechanism, it seems remarkable that the third start codon (Env) is used much more efficiently than the second start codon (Rev), in particular as both have a reasonably good Kozak signature (Fig. 1C). It is also remarkable that inactivation of uAUG4 and inactivation of AUG-Rev had a small negative impact on Env translation, while inactivation of both uAUGs increased Env expression nearly 2-fold. These seemingly complex results can be understood when involving a suppressive effect of uAUG4 on Rev translation (as observed in Fig. 3) and of AUG-Rev on Env translation. In the wt context, with both uAUG4 and AUG-Rev present, this results in an indirect positive effect of uAUG4 on Env translation, and Env translation is reduced upon uAUG4 inactivation (compare constructs wt and 4 in Fig. 4A). In the absence of AUG-Rev, uAUG4 has a suppressive effect on Env translation, and uAUG4 inactivation now increases Env translation (compare mutants rev and 4rev).
Fig 4.
Protein expression of subgenomic Env reporters. 293T cells were transfected with the SA6-env-luc reporter constructs (A) and with the SA5-env-luc reporter constructs (B). Luciferase production was analyzed as described for Fig. 3. Results of 14 measurements are shown (means ± SEM). The mean value is shown next to each bar. Statistical analysis indicated that luciferase production differed significantly between SA6-env-luc (wt) and mutant 4rev (A). There is also a statistically significant difference in luciferase production between SA5-env-luc (wt) and mutant 1234rev (B).
We tried to confirm some of these results in the context of the alternative SA5 transcript that has five uAUG codons (constructs 1234 and Rev) in front of the Env ORF (Fig. 4B). The impact on Env translation varied for the SA5-specific uAUGs: luciferase production was slightly reduced for mutant 1 and moderately increased for the combination mutant 123. Inactivation of uAUG4 resulted in a greater reduction of Env translation, both in the wt context (compare wt with mutant 4) and with multiple mutations (compare mutant 123 with mutant 1234). Removal of all uAUGs in mutant 1234rev boosted Env expression approximately 2-fold. The positive impact of uAUG4 and the negative impact of AUG-Rev on Env translation were thus confirmed in the SA5 context, but the effect of uAUGs 1, 2, and 3 was less pronounced. We also tested the 4a mutant that connects uAUG4 with the Rev ORF. As a result, uAUG4 will no longer interfere with translation of the Rev ORF, which potentially suppresses downstream Env translation. Indeed this manipulation enforced Env suppression, consistent with the idea that uAUG4 absorbs more ribosomes than AUG-Rev.
RNA stability.
To rule out any differences in transcription and/or mRNA stability of the different luciferase constructs, total cellular RNA was isolated from the transfected cells, and the luciferase transcripts were analyzed by Northern blotting (Fig. 5). Differences in size between the four sets of luciferase constructs correlate with the expected transcript length. All Rev-luc and Env-luc constructs showed a higher transcript level than the SV40-luciferase control that lacked virus-derived sequences. Moreover, a slightly higher level of transcript was observed for the SA6-env-luc set compared to the SA5-env-luc set. These results indicate that the viral sequences can influence transcript production or stability. Importantly, there were no visible differences in transcript level among constructs within a set. As a loading control, the rRNA was stained with ethidium bromide (Fig. 5C and D). The amount of rRNA was the same for all samples.
Fig 5.
Northern blot analysis of reporter transcripts. 293T cells were transfected with the wt and mutant Rev-luc (A, C) and Env-luc (B, D) plasmids. (A, B) After 48 h, total cellular RNA was isolated and size-separated on a denaturing gel, and the reporter RNA was visualized with a 32P-labeled luciferase probe by Northern blotting (control, pGL3-control; mock, pBluescript). (C, D) The 18S and 28S rRNA markers of the Rev-luc transfected cells (C) and the Env-luc transfected cells (D) were identified by ethidium bromide staining and used as a loading control.
To confirm these results, the luciferase mRNA level of the transfected cells was quantified by RNA slot blot analysis. We selected the mutants that exhibited the most deviant translation effect (Fig. 6). In each of the four transcript sets, the selected uAUG mutants yielded an amount of RNA comparable to that of the respective wt construct, indicating that there is no difference in RNA expression level within each set. These data confirm the results of the Northern blot analysis and indicate that the differences in luciferase expression of the mutant constructs (Fig. 3 and 4) are solely due to translational effects.
Fig 6.
RNA slot blot analysis of reporter transcripts. Total cellular RNA was isolated as described for Fig. 5, dotted onto a membrane, and hybridized with a 32P-labeled luciferase probe for quantification. The results of 5 measurements are shown (means ± SEM) after subtraction of background signal (mock transfection). Statistical analysis was performed and showed no significant differences between the wt and uAUG-mutated constructs for each group.
Virus replication and virus competition experiments.
To demonstrate the importance of the uAUGs in virus replication, uAUG4 was knocked out in the SIVmac239 molecular clone. This uAUG had the largest effect on translation in the luciferase reporter constructs, and it is the only uAUG present in the regular SA6 transcript. The uAUG in SIVmac239 was inactivated by introducing the same nucleotide substitution as that used in the luciferase construct (Fig. 2B), which was silent in the overlapping Tat ORF.
Replication of SIVmac239 and the mutant SIVmac239-m4 was tested in the PM1 and 174×CEM cell lines, which express the CD4 and CCR5 receptors (9, 25, 37, 49). Virus replication was monitored by measuring the virion-associated reverse transcriptase (RT) activity that accumulates in the culture supernatant. Surprisingly, SIVmac239-m4 replicated efficiently in both cell lines (data not shown). Therefore, we infected peripheral blood mononuclear cells (PBMC) isolated from cynomolgus macaques to study replication on primary cells. SIVmac239-m4 replicated efficiently on PBMC as well (Fig. 7A).
Fig 7.
Virus replication studies. (A) PBMC isolated from cynomolgus macaques were infected with equal amounts of 293T-produced SIVmac239 (wt) and SIVmac239-m4 (m4) virus (5 ng CA-p27). Replication was monitored by measuring the RT level in the culture supernatant. The results shown are representative of 2 independent experiments, and similar results were obtained with different amounts of virus input. (B) Virus competition experiments were conducted in 174×CEM cells. Pairwise competitions were started by transfecting an equimolar mixture of wt and mutant viral DNA constructs as previously described (27, 55, 56). The proviral DNA was analyzed by sequencing at several time points. The original input DNA (day 0) was included as a control, in which the double A/U signal (arrow) demonstrates the presence of a wt-m4 mixture.
The difference in replication between SIVmac239-m4 and the wt virus was too small to detect in our replication assays, but such assays are often not sensitive enough to detect minor fitness differences. We therefore performed a more sensitive competition experiment with the mutant and wt virus. An equimolar mixture of the wt and mutant SIVmac239 plasmids was transfected into 174×CEM cells. The virus was passaged onto fresh cells at the peak of infection when massive syncytia were observed in order to continue the competition. At the same time, cell samples were taken for DNA isolation, PCR amplification, and population-based sequence analysis. On 174×CEM cells, the wt virus outcompeted the uAUG4 knockout virus within the short time frame of 14 days (Fig. 7B). This indicates that there is a significant loss of fitness upon uAUG4 inactivation, which we approximate to be around 20% (27, 54, 56). Similar results were obtained in PM1 cells (data not shown). Thus, although the uAUG4 knockout variant replicates efficiently on several cell lines and PBMC, the presence of uAUG4 is required for optimal viral replication.
DISCUSSION
A survey of the 5′ UTR sequences of diverse HIV-1 isolates indicated the absence of uAUG codons (48, 59). It has been argued that uAUG codons are actively counterselected because they would interfere with the process of ribosomal scanning and translation of the viral proteins (3). Indeed, the introduction of such uAUG codons by mutation can severely hamper HIV-1 replication (12, 23), and scanning was recently confirmed as the major translation mechanism in HIV-1 (5). This does not mean that internal ribosome entry site (IRES)-mediated translation could not also occur under specific circumstances (7, 8, 20, 24, 43, 52, 58). In agreement with the scanning mode of translation, there are rarely any uAUGs present in the large variety of mRNAs produced by HIV and SIV. An exception in HIV is the bicistronic mRNA that encodes both Vpu and Env (1, 47), and this mRNA also contains a minimal uORF (35). We now describe a major exception for SIV.
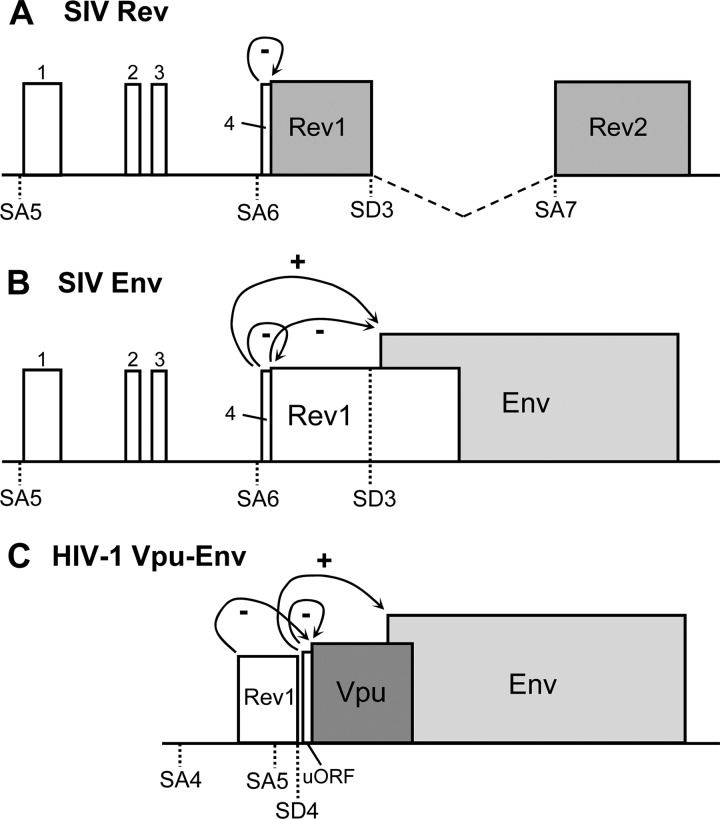
In SIV, we identified two uORFs in the Env mRNA: a minimal ORF directed by uAUG4 followed by the Rev ORF that runs into the Env ORF. This uAUG4 is highly conserved among SIV strains, implying that its presence has a purpose, in particular because it could be inactivated by a mutation that is silent in the overlapping Tat ORF. We constructed subgenomic luciferase reporters to quantify the effect of uAUG4 on Rev and Env translation. The results show that Rev is strongly downregulated by uAUG4 (Fig. 8A), while Env is upregulated (Fig. 8B). The latter effect can be explained by the negative effect of Rev translation on Env translation. uAUG4 may stimulate Env translation by blocking ribosomal access to the Rev AUG, which otherwise would suppress Env translation. These data suggest that uAUG4 actively recruits ribosomes that subsequently encounter a translational stop codon just downstream of the Rev AUG, thus bypassing the Rev ORF. We assume that these ribosomes can reinitiate scanning on the Rev-Env mRNA toward the Env AUG. Similar regulatory scenarios have been described previously for HIV-1 Vpu and Env translation (1, 35, 38) (Fig. 8C) but also in other viruses and eukaryotes (34, 40, 44, 50).
Fig 8.
Regulation of Rev and Env translation in HIV-1 and SIVmac239. Schematic representation of the SIV doubly spliced Rev mRNA (A), SIV singly spliced Env mRNA (B), and HIV-1 singly spliced Env mRNA (C). The SIV Rev and Env mRNAs mostly result from splicing at SA6, but transcripts spliced at SA5 have been observed. Similarly, the HIV-1 Vpu and Env mRNAs mostly result from splicing at SA5, but splicing at SA4 has been observed. The ORFs are boxed, with the dedicated ORFs in gray and the uORFs in white. (A) The presence of uAUG4 has a suppressive effect on Rev translation. Alternative splicing at SA5 adds three more uAUGs: 1, 2, and 3. (B) SIV Env mRNA has two uAUGs: uAUG4 and AUG-Rev. The presence of the Rev AUG has a suppressive effect on Env translation. uAUG4 has a downregulating effect on Rev translation and thus an indirect stimulatory effect on Env translation. Alternative splicing at SA5 adds three more uAUGs to the Env mRNA: uAUGs 1, 2, and 3. (C) HIV-1 Env mRNA has two uAUG codons: the Vpu AUG and a minimal uORF directly upstream of Vpu. The HIV-1 uORF has a suppressive effect on Vpu translation and thus an indirect stimulatory effect on Env translation (35). Alternative splicing at SA4 will result in an additional uORF for the Env mRNA: the Rev AUG, which has a suppressive effect on Vpu translation (1).
We also described an alternative mRNA for Rev and Env that is produced by splicing at SA5 instead of SA6, which adds three more uAUGs upstream of Rev and Env. This alternative splicing event has been described previously (45) but appears to occur at a low frequency (unpublished data). It is possible that the alternative mRNAs encode new ORFs and novel viral proteins, but their coding capacity is rather restricted (Fig. 1C). Colombini et al. (11) described that uAUG1 is the translation start codon for Rev in isolate CPZ GAB1. However, in SIVmac239 and most other isolates, a stop codon results in the translation of a 21-amino-acid polypeptide before the Rev ORF is reached. We tested the regulatory effect of the additional SA5-encoded start codons on Rev and Env translation but scored relatively minor effects compared to the impact of uAUG4.
uAUG4 had the greatest impact on translation of both Rev and Env in the subgenomic reporter constructs and is the only uAUG present in the regular SA6 transcript. Virus competition experiments showed that inactivation of this uAUG in SIVmac239 slightly reduced virus replication (Fig. 7B), which demonstrates that the presence of uAUG4 is required for optimal viral replication. The presence of regulatory loops such as the Tat-LTR and Rev-RRE circuits makes SIV a complex system to study the effects of uAUGs on the translation of specific proteins. The increased translation of Rev in the uAUG4 knockout mutant would theoretically result in an earlier switch from spliced to unspliced SIV mRNAs, resulting in reduced Tat expression and less transcriptional activation. The earlier switch to singly spliced Env and unspliced Gag-Pol mRNAs does not necessarily lead to increased Env protein levels, as its translation would be less efficient due to the absence of uAUG4 and an increase in ribosomal capture by the Rev AUG.
In general, we believe that the ORFs served by the many uAUG codons do not serve a protein-coding role but rather a regulatory role during the process of translation. This is most obvious for the ultrashort ORFs served by uAUGs 2, 3, and 4. We cannot formally exclude a role for the ORF1-encoded 21-amino-acid polypeptide and the 73-amino-acid protein that results from AUG-Rev usage on the singly spliced Env mRNA (Fig. 8B). The latter protein consists of a 23-amino-acid Rev domain (exon 1) that is connected to 50 unrelated amino acids. This exon 1 Rev domain does lack the critical motifs for RRE RNA binding and nuclear localization that are encoded by Rev exon 2 (2). There are other viral examples where the major role of significant parts of ORFs concerns the regulation of mRNA translation instead of an encoded protein function (4). Thus, we believe that these uORFs serve regulatory purposes to allow fine-tuning of viral gene expression. The SIV situation is particularly complex due to the presence of the many spliced mRNA variants. For instance, AUG-Rev is obviously critical for Rev translation on the Rev mRNA, and it simultaneously suppresses Env translation on the Env mRNA, which necessitates the presence of uAUG4 to suppress Rev translation. But this uAUG4-mediated suppression of Rev translation cannot be absolute in order to allow Rev expression from the Rev mRNA. We do not know whether the presence of uAUG4 is important to improve Env translation, reduce Rev expression, or both. Too much Rev expression may be detrimental because it could trigger an untimely switch from spliced to unspliced RNA, which causes reduced Tat expression and consequently suboptimal viral gene expression. Furthermore, Rev overexpression may be toxic for the cell (42). The efficacy of translation may also be influenced by other factors, such as the presence of RNA hairpins upstream of the AUG that inhibit ribosome scanning (19), the accessibility of the AUG within an RNA structure, and the binding of cellular proteins to the viral RNA (6, 10).
The information gained in this study helps us to design optimized SIV Rev expression vectors. If these vectors contain the sequence from SA6 onward and uAUG4 is inactivated, the expression level is increased 4-fold compared to that of the wt vector (Fig. 3). If the Rev expression vector contains the sequence from SA5 onward, removal of all uAUGs will increase the expression up to 6.5-fold. Env expression can also be optimized by inactivation of the uAUGs (uAUG4 and AUG-Rev) in the SA6 context.
Several HIV and SIV mRNAs do contain additional ORFs downstream of the dedicated ORF. For instance, the HIV-1 Tat mRNA can potentially also encode Rev and Nef from downstream sequences. However, early studies indicated that the scanning mechanism is operational such that only the dedicated 5′ ORF is served by ribosomes (38, 48). Nevertheless, some of the dedicated 5′ ORFs have a relatively weak AUG codon in a suboptimal Kozak context, allowing leaky scanning that can trigger translation of the downstream ORF. The Env mRNA for both HIV-1 and SIV is exceptional in that an ORF is situated upstream of the dedicated Env ORF. Early HIV-1 studies indicated that the upstream Vpu ORF has a weak AUG to allow sufficient leaky scanning and thus Env translation (38, 48). Two recent studies revealed even more regulatory aspects of HIV-1 Env translation. First, a uAUG was described immediately upstream of the Vpu ORF, which could thus affect Vpu expression and indirectly Env translation (Fig. 8C). Inactivation of this uAUG reduced Env expression, consistent with the idea that this uAUG augments access to the downstream Env ORF (35). Second, alternatively spliced HIV-1 RNAs with a 5′ extension (SA4 in Fig. 8C) were analyzed for the impact of the additional uAUG (AUG-Rev) on Vpu and Env translation (1). There is a rather surprising parallel in the HIV-1 and SIV scenarios for Env translation (Fig. 8). Both Env mRNAs have a uORF (Vpu in HIV-1, Rev in SIV) and a uAUG very close to the start of that uORF (uAUG4 in SIV), and alternative 5′-extended transcripts have been reported (using SA5 in SIV, SA4 in HIV-1) that add more uAUGs (uAUGs 1, 2, and 3 in SIV; AUG-Rev in HIV-1). Thus, the Env mRNAs seem unique among the many HIV and SIV transcripts in exhibiting a complex level of translational regulation.
ACKNOWLEDGMENTS
This work was supported by the AIDS Fonds (grant 2007028) and the Netherlands Organization for Scientific Research (Chemical Sciences Division; NWO-CW; Top grant).
We thank Alex Harwig for technical support. We thank Neil Almond and Mark Page (National Institute for Biological Standards and Control, United Kingdom) for providing macaque PBMC.
Footnotes
Published ahead of print 5 September 2012
REFERENCES
- 1. Anderson JL, Johnson AT, Howard JL, Purcell DFJ. 2007. Both linear and discontinuous ribosome scanning are used for translation initiation from bicistronic human immunodeficiency virus type 1 env mRNAs. J. Virol. 81:4664–4676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Berchtold S, Ries J, Hornung U, Aepinus C. 1994. Exchange of functional domains between Rev proteins of HIV-1 and SIVmac239 results in a dominant negative phenotype. Virology 204:436–441 [DOI] [PubMed] [Google Scholar]
- 3. Berkhout B. 1996. Structure and function of the human immunodeficiency virus leader RNA. Prog. Nucleic Acid Res. Mol. Biol. 54:1–34 [DOI] [PubMed] [Google Scholar]
- 4. Berkhout B, de Smit MH, Spanjaard RA, Blom T, van Duin J. 1985. The amino terminal half of the MS2-coded lysis protein is dispensable for function: implications for our understanding of coding region overlaps. EMBO J. 4:3315–3320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Berkhout B, Arts K, Abbink TEM. 2011. Ribosomal scanning on the 5′-untranslated region of the human immunodeficiency virus RNA genome. Nucleic Acids Res. 39:5232–5244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bolinger C, Boris-Lawrie K. 2009. Mechanisms employed by retroviruses to exploit host factors for translational control of a complicated proteome. Retrovirology 6:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Brasey A, et al. 2003. The leader of human immunodeficiency virus type 1 genomic RNA harbors an internal ribosome entry segment that is active during the G2/M phase of the cell cycle. J. Virol. 77:3939–3949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Buck CB, et al. 2001. The human immunodeficiency virus type 1 gag gene encodes an internal ribosome entry site. J. Virol. 75:181–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chen Z, Gettie A, Ho DD, Marx PA. 1998. Primary SIVsm isolates use the CCR5 coreceptor from sooty mangabeys naturally infected in West Africa: a comparison of coreceptor usage of primary SIVsm, HIV-2, and SIVmac. Virology 246:113–124 [DOI] [PubMed] [Google Scholar]
- 10. Clerzius G, Gélinas J-F, Gatignol A. 2011. Multiple levels of PKR inhibition during HIV-1 replication. Rev. Med. Virol. 21:42–53 [DOI] [PubMed] [Google Scholar]
- 11. Colombini S, et al. 1989. Structure of simian immunodeficiency virus regulatory genes. Proc. Natl. Acad. Sci. U. S. A. 86:4813–4817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Das AT, van Dam AP, Klaver B, Berkhout B. 1998. Improved envelope function selected by long-term cultivation of a translation-impaired HIV-1 mutant. Virology 244:552–562 [DOI] [PubMed] [Google Scholar]
- 13. Das AT, Harwig A, Vrolijk MM, Berkhout B. 2007. The TAR hairpin of human immunodeficiency virus type 1 can be deleted when not required for Tat-mediated activation of transcription. J. Virol. 81:7742–7748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Das AT, et al. 2008. Optimization of the doxycycline-dependent simian immunodeficiency virus through in vitro evolution. Retrovirology 5:44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Das AT, et al. 2007. Construction of a doxycycline-dependent simian immunodeficiency virus reveals a nontranscriptional function of Tat in viral replication. J. Virol. 81:11159–11169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Das AT, et al. 2004. Viral evolution as a tool to improve the tetracycline-regulated gene expression system. J. Biol. Chem. 279:18776–18782 [DOI] [PubMed] [Google Scholar]
- 17. Donzé O, Spahr PF. 1992. Role of the open reading frames of Rous sarcoma virus leader RNA in translation and genome packaging. EMBO J. 11:3747–3757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Faustino NA, Cooper Ta. 2003. Pre-mRNA splicing and human disease. Gen. Dev. 17:419–437 [DOI] [PubMed] [Google Scholar]
- 19. Firth AE, Brierley I. 2012. Non-canonical translation in RNA viruses. J. Gen. Virol. 93:1385–1409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gendron K, Ferbeyre G, Heveker N, Brakier-Gingras L. 2011. The activity of the HIV-1 IRES is stimulated by oxidative stress and controlled by a negative regulatory element. Nucleic Acids Res. 39:902–912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Grünert S, Jackson RJ. 1994. The immediate downstream codon strongly influences the efficiency of utilization of eukaryotic translation initiation codons. EMBO J. 13:3618–3630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Guan Y, Whitney JB, Diallo K, Wainberg MA. 2000. Leader sequences downstream of the primer binding site are important for efficient replication of simian immunodeficiency virus. J. Virol. 74:8854–8860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Haché G, Abbink TEM, Berkhout B, Harris RS. 2009. Optimal translation initiation enables Vif-deficient human immunodeficiency virus type 1 to escape restriction by APOBEC3G. J. Virol. 83:5956–5960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Herbreteau CH, et al. 2005. HIV-2 genomic RNA contains a novel type of IRES located downstream of its initiation codon. Nat. Struct. Mol. Biol. 12:1001–1007 [DOI] [PubMed] [Google Scholar]
- 25. Hoxie JA, et al. 1988. Biological characterization of a simian immunodeficiency virus-like retrovirus (HTLV-IV): evidence for CD4-associated molecules required for infection. J. Virol. 62:2557–2568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Jackson RJ, Hellen CUT, Pestova TV. 2010. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat. Rev. Mol. Cell Biol. 11:113–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Koken SE, van Wamel JL, Goudsmit J, Berkhout B, Geelen JL. 1992. Natural variants of the HIV-1 long terminal repeat: analysis of promoters with duplicated DNA regulatory motifs. Virology 191:968–972 [DOI] [PubMed] [Google Scholar]
- 28. Koken SE, et al. 1994. Intracellular analysis of in vitro modified HIV Tat protein. J. Biol. Chem. 269:8366–8375 [PubMed] [Google Scholar]
- 29. Kozak M. 1987. An analysis of 5′-noncoding sequences from 699 vertebrate messenger RNAs. Nucleic Acids Res. 15:8125–8148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kozak M. 1986. Bifunctional messenger RNAs in eukaryotes. Cell 47:481–483 [DOI] [PubMed] [Google Scholar]
- 31. Kozak M. 1986. Point mutations define a sequence flanking the AUG initiator codon that modulates translation by eukaryotic ribosomes. Cell 44:283–292 [DOI] [PubMed] [Google Scholar]
- 32. Kozak M. 1991. Structural features in eukaryotic mRNAs that modulate the initiation of translation. J. Biol. Chem. 266:19867–19870 [PubMed] [Google Scholar]
- 33. Kozak M. 1991. An analysis of vertebrate mRNA sequences: intimations of translational control. J. Cell Biol. 115:887–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kozak M. 2002. Pushing the limits of the scanning mechanism for initiation of translation. Gene 299:1–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Krummheuer J, et al. 2007. A minimal uORF within the HIV-1 vpu leader allows efficient translation initiation at the downstream env AUG. Virology 363:261–271 [DOI] [PubMed] [Google Scholar]
- 36. Kuiken C, et al. 2011. HIV sequence compendium 2011. Los Alamos National Laboratory, Theoretical Biology and Biophysics, Los Alamos, NM [Google Scholar]
- 37. Lusso P, et al. 1995. Growth of macrophage-tropic and primary human immunodeficiency virus type 1 (HIV-1) isolates in a unique CD4+ T-cell clone (PM1): failure to downregulate CD4 and to interfere with cell-line-tropic HIV-1. J. Virol. 69:3712–3720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Luukkonen BG, Tan W, Schwartz S. 1995. Efficiency of reinitiation of translation on human immunodeficiency virus type 1 mRNAs is determined by the length of the upstream open reading frame and by intercistronic distance. J. Virol. 69:4086–4094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Maudru T, Peden KW. 1998. Adaptation of the fluorogenic 5′-nuclease chemistry to a PCR-based reverse transcriptase assay. Biotechniques 25:972–975 [DOI] [PubMed] [Google Scholar]
- 40. Medenbach J, Seiler M, Hentze MW. 2011. Translational control via protein-regulated upstream open reading frames. Cell 145:902–913 [DOI] [PubMed] [Google Scholar]
- 41. Mills KH, et al. 1991. Vaccine-induced CD4+ T cells against the simian immunodeficiency virus Gag protein. Epitope specificity and relevance to protective immunity. J. Immunol. 147:3560–3567 [PubMed] [Google Scholar]
- 42. Miyazaki Y, et al. 1995. The cytotoxicity of human immunodeficiency virus type 1 Rev: implications for its interaction with the nucleolar protein B23. Exp. Cell Res. 219:93–101 [DOI] [PubMed] [Google Scholar]
- 43. Nicholson MG, Rue SM, Clements JE, Barber SA. 2006. An internal ribosome entry site promotes translation of a novel SIV Pr55(Gag) isoform. Virology 349:325–334 [DOI] [PubMed] [Google Scholar]
- 44. Oestreicher N, Scazzocchio C. 2009. Phenotypes of mutations in the 5′-UTR of a limiting transcription factor in Aspergillus nidulans can be accounted for by translational inhibition and leaky scanning. Genetics 181:1261–1272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Park IW, Steen R, Li Y. 1991. Characterization of multiple mRNA species of simian immunodeficiency virus from macaques in a CD4+ lymphoid cell line. J. Virol. 65:2987–2992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ruijter JM, et al. 2006. Factor correction as a tool to eliminate between-session variation in replicate experiments: application to molecular biology and retrovirology. Retrovirology 3:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Schwartz S, Felber BK, Fenyö EM, Pavlakis GN. 1990. Env and Vpu proteins of human immunodeficiency virus type 1 are produced from multiple bicistronic mRNAs. J. Virol. 64:5448–5456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Schwartz S, Felber BK, Pavlakis GN. 1992. Mechanism of translation of monocistronic and multicistronic human immunodeficiency virus type 1 mRNAs. Mol. Cell Biol. 12:207–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Sei Y, Inoue M, Yokoyama MM, Bekesi JG, Arora PK. 1990. Characterization of human B cell (DK) and promonocyte (U937) clones after HIV-1 exposure: accumulation of viral reverse transcriptase activity in cells and early syncytia induction against SupT1 cells. Cell Immunol. 125:1–13 [DOI] [PubMed] [Google Scholar]
- 50. Shung CY, Sunter G. 2009. Regulation of tomato golden mosaic virus AL2 and AL3 gene expression by a conserved upstream open reading frame. Virology 383:310–318 [DOI] [PubMed] [Google Scholar]
- 51. Unger RE, Stout MW, Luciw PA. 1991. Simian immunodeficiency virus (SIVmac) exhibits complex splicing for tat, rev, and env mRNA. Virology 182:177–185 [DOI] [PubMed] [Google Scholar]
- 52. Vallejos M, et al. 2011. Activity of the human immunodeficiency virus type 1 cell cycle-dependent internal ribosomal entry site is modulated by IRES trans-acting factors. Nucleic Acids Res. 39:6186–6200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. van der Velden GJ, Vink MA, Berkhout B, Das AT. 2012. Tat has a dual role in SIV transcription. J. Gen. Virol. 93:2279–2289 [DOI] [PubMed] [Google Scholar]
- 54. van Opijnen T, et al. 2004. Human immunodeficiency virus type 1 subtypes have a distinct long terminal repeat that determines the replication rate in a host-cell-specific manner. J. Virol. 78:3675–3683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. van Opijnen T, Kamoschinski J, Jeeninga RE, Berkhout B. 2004. The human immunodeficiency virus type 1 promoter contains a CATA box instead of a TATA box for optimal transcription and replication. J. Virol. 78:6883–6890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Verhoef K, Bauer M, Meyerhans A, Berkhout B. 1998. On the role of the second coding exon of the HIV-1 Tat protein in virus replication and MHC class I downregulation. AIDS Res. Hum. Retroviruses 14:1553–1559 [DOI] [PubMed] [Google Scholar]
- 57. Viglianti GA, Sharma PL, Mullins JI. 1990. Simian immunodeficiency virus displays complex patterns of RNA splicing. J. Virol. 64:4207–4216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Weill L, et al. 2010. A new type of IRES within gag coding region recruits three initiation complexes on HIV-2 genomic RNA. Nucleic Acids Res. 38:1367–1381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Yilmaz A, Bolinger C, Boris-Lawrie K. 2006. Retrovirus translation initiation: issues and hypotheses derived from study of HIV-1. Curr. HIV Res. 4:131–139 [DOI] [PMC free article] [PubMed] [Google Scholar]