Abstract
A protamine-like protein named P6.9 is thought to play a role in the condensation of genomes of the baculovirus Autographa californica multiple nucleopolyhedrovirus (AcMNPV) during an infection. Previous studies have shown that P6.9 is phosphorylated immediately upon synthesis and dephosphorylated upon the entry of the P6.9-DNA complex into the capsid. Here, we investigate the dynamic distribution of P6.9 in AcMNPV-infected Spodoptera frugiperda cells using an influenza virus hemagglutinin (HA)-tagged P6.9. Although a portion of P6.9-HA localized to the virogenic stroma, which is the center of viral DNA replication, transcription, and nucleocapsid assembly, the majority of P6.9-HA was distributed near the inner nuclear membrane throughout the course of infection. Antiserum against P6.9 detected specific phosphorylated forms of P6.9 at the edge of, but not within, the electron-dense matte regions of the virogenic stroma. Further analysis using immunoblotting revealed that at least 11 different phosphorylated forms of P6.9, as well as dephosphorylated P6.9, were present in association with occlusion-derived virions, although only dephosphorylated P6.9 was associated with budded virions.
INTRODUCTION
The family Baculoviridae comprises a diverse group of insect-specific viruses that are characterized by rod-shaped enveloped nucleocapsids containing large, double-stranded, circular, supercoiled DNA genomes (26). Baculovirus DNA replication, transcription, and nucleocapsid assembly occur within a subnuclear structure called the virogenic stroma (VS). Early in infection, newly formed nucleocapsids are transported from the nucleus through the cytoplasm and acquire an envelope to form budded virus (BV). At later stages of infection, nucleocapsids remain within the nucleus, where they become enveloped and occluded in a protein matrix to form occlusion-derived virus (ODV). BV spreads the infection from cell to cell, whereas ODV is responsible for virus transmission from insect to insect via oral infection (23). Although the two viral forms differ in the composition of their envelopes, their nucleocapsids appear to be similar in structure and are composed of a cylindrical capsid sheath and a nucleoprotein core.
It is well known that genomic DNA/RNA needs to be condensed and well compacted for the transmission of genetic information. Protamines are a diverse family of small, arginine-rich, positively charged proteins that are synthesized in the late-stage spermatids of many animals and plants to condense the spermatid genome into a genetically inactive state (2). A protamine-like protein named P6.9 is thought to bind and condense baculoviral DNA for packaging into capsids (27). When nucleocapsids enter nuclei through nuclear pores, it is thought that a capsid-associated kinase is involved in the release of the viral genome by phosphorylation of the nucleocore protein P6.9, which repels the negatively charged DNA (8, 32). Subsequently, a complex cascade of early to late gene expression takes place. The gene p6.9 is expressed as a late gene (25, 33), and evidence suggests that newly synthesized P6.9 is transiently phosphorylated in infected cells prior to nucleocapsid assembly (20). The switchover from early to late gene expression coincides with the start of viral DNA replication and intranuclear development of the VS (23). The VS consists of the electron-dense stromal regions, also referred to as stromal mattes, and electron-lucent intrastromal spaces (37). Nucleocapsid assembly occurs at the surface of the stromal mattes within the VS (5, 9, 37), at which point P6.9 is thought to be dephosphorylated and viral DNA is condensed and packaged with P6.9 to form a DNA-protein core within the capsid sheath (8, 12, 20, 27). Thus, the phosphorylation and dephosphorylation of P6.9 may be critical events in the life cycle of baculoviruses.
Because the VS is the center of viral DNA packaging and nucleocapsid assembly (5, 9, 37), it is thought that P6.9 would localize mainly in the VS. It was reported that newly synthesized viral DNA may initially associate with host histones and that P6.9 may serve to displace the histones from the viral genome during the encapsidation process (34). Such a protein exchange process has been reported to occur during spermiogenesis (17, 19). Although P6.9 was originally identified as a component of the nuclear matrix in infected cells (35), the distribution of de novo synthesized P6.9 and its possible relationship to nucleoprotein exchange have not yet been described. In this paper, we report that the majority of P6.9 is distributed near the inner nuclear membrane throughout the course of an infection, and a minority of P6.9 is present in the VS of Spodoptera frugiperda cells infected with Autographa californica multiple nucleopolyhedrovirus (AcMNPV). Moreover, we found that at least 11 different phosphorylated forms of P6.9 as well as dephosphorylated P6.9 are associated with ODV, whereas only dephosphorylated P6.9 was found in BV.
MATERIALS AND METHODS
Cells, viruses, and antibodies.
Sf9 cells, which were derived from the fall armyworm S. frugiperda (29), were grown in monolayer cultures at 27°C in TNM-FH medium (Invitrogen Life Technologies) supplemented with 10% heat-inactivated fetal bovine serum, penicillin (100 μg/ml), and streptomycin (30 μg/ml). BVs and ODVs of AcMNPV were propagated and purified as previously described (21). In all infection experiments, time zero was defined as the time the virus inocula were removed after a 1-h absorption period. In all transfection experiments, time zero was defined as the time when the viral DNA was removed after a 5-h incubation period.
Two peptides corresponding to amino acids (aa) 29 to 40 and 42 to 55 of AcMNPV P6.9 were synthesized by Abmart and were used to generate polyclonal antibodies in rabbits. The raised antibodies were named ab1002940 for aa 29 to 40 and ab1004255 for aa 42 to 55. The mouse monoclonal antibody against AcMNPV IE1 was a gift from L. A. Guarino (Texas A&M University). Rabbit polyclonal anti-hemagglutinin (HA) antibody was purchased from Abcam. Mouse monoclonal anti-actin antibody was purchased from Abmart.
Construction of viruses and a plasmid.
Recombinant viruses used in this study were constructed from the bacmid bMON14272, which contains an AcMNPV genome (15). Using λ Red homologous recombination in Escherichia coli as previously described (36), the internal 48-bp segment of p6.9 (AcMNPV nucleotides [nt] 86793 to 86840) of bMON14272 was replaced with a 1,038-bp chloramphenicol acetyltransferase (CAT) cassette to generate the p6.9-null recombinant bacmid bP6.9KO. The primers P6.9US1-Cm (5′-GCGTGTTCTGTAACTTCGGCGACCTGTCGATGAACGGCTCCTGGATCTTCTGTATGTGCGGGGTCTACCCGGAAGCTTCCCTTTCGTCTTCGAATAAA-3′; the HindIII site is underlined) and P6.9DS1-Cm (5′-AAATTACAGCTACATAAATTACACAATTTAAACATGGTTTATCGTCGCCGTCGCCGTTCTTCAACCGGTACCTAAACCAGCAATAGACATAAGCGGCT-3′) were used. The construct vAcp6.9KO-PH-GFP (vP6.9KO) was generated by the insertion of the polyhedrin (polh) and enhanced green fluorescence protein (egfp) genes into the polh locus of bP6.9KO via a site-specific transposition as previously described (36). Likewise, vAcp6.9:HA-PH-GFP (vP6.9:HA) was generated by the insertion of an influenza virus HA-tagged p6.9 (C-terminal fusion) as well as the polh and egfp genes into bP6.9KO. A wild-type control vAcPH-GFP (vAcWT) was generated by the insertion of the polh and egfp genes into bMON14272. All of these recombinants were verified by PCR analysis and isolated using a Qiagen Large-Construct kit (Qiagen). Cells were transfected with 1 μg of bacmid DNA using the Cellfectin liposome reagent (Invitrogen Life Technologies) to obtain BVs.
A plasmid expressing the S. frugiperda histone H4 gene was constructed in this study. Total Sf9 genomic DNA was extracted using a Universal Genomic DNA extraction kit (TaKaRa) and was used as a template. The open reading frame (ORF) of Sf-H4 was PCR amplified with primers Sf-H45K (5′-GGTACCATGACTGGTCGCGGCAAAGGC-3′; the KpnI site is underlined) and Sf-H43S (5′-GAGCTCTTACCCGCCAAAACCGTACAG-3′; the SacI site is underlined) under the following conditions: 94°C for 5 min and then 34 cycles of 94°C for 30 s, 60°C for 30 s, and 72°C for 1 min. The PCR product was digested with KpnI and SacI and then cloned into the pIZ/V5-His vector (Invitrogen Life Technologies) to generate pIZ-SfH4. An egfp-containing HindIII-KpnI fragment of pUC19egfp (donated by J. M. Vlak, Wageningen University, the Netherlands) was inserted into the HindIII-KpnI site of pIZ-SfH4 to construct the plasmid pIZ-SfH4-EGFP (H4G).
Virus growth curve.
Sf9 cells (1.0 × 106 cells/35-mm-diameter dish) were infected with vP6.9:HA or vAcWT in triplicate at a multiplicity of infection (MOI) of 10 times the 50% tissue culture infective doses (TCID50)/cell. The BV-enriched supernatant was harvested at 0, 12, 24, 36, and 48 h postinfection (h p.i.). The virus titer was determined in Sf9 cells using a TCID50 endpoint dilution assay (21).
Immunofluorescence microscopy.
A total of 2 × 105 Sf9 cells were seeded in a 35-mm glass-bottom culture dish (MatTek) and infected with vP6.9:HA or vAcWT at an MOI of 10 TCID50/cell. At the desired h p.i., cells were washed three times with phosphate-buffered saline (PBS) and fixed with 4% paraformaldehyde in PBS for 10 min at room temperature (RT). After washing three times with PBS, the fixed cells were permeabilized for 15 min at RT using 0.15% Triton X-100 in PBS containing 0.1% normal goat serum. Following three additional washes with PBS, the cells were blocked with 1% normal goat serum in PBS for 5 min, incubated with the primary antibody for 60 min, washed three times with blocking buffer, and then incubated with the secondary antibody for 60 min at RT. The secondary antibodies included goat anti-mouse IgG conjugated with Alexa Fluor 561 (Invitrogen Life Technologies) and goat anti-rabbit IgG conjugated with Alexa Fluor 647 (Invitrogen Life Technologies). Finally, the labeled cells were stained with 2 μg/ml 4,6-diamidino-2-phenylindole (DAPI) (Roche) for 3 min at RT prior to analysis. All images were collected using a Leica TCS SP5 confocal microscope according to the same parameters as those used for the mock-infected cells in each experiment.
Transmission electron microscopy.
A total of 1.0 × 106 Sf9 cells were transfected with 1 μg of bacmid DNA. At selected times posttransfection, cells were dislodged with a rubber policeman, pelleted by centrifugation at 200 × g for 5 min, and fixed in a solution of 4% glutaraldehyde and 1% paraformaldehyde in PBS at 4°C overnight. The fixed cells were washed, postfixed, dehydrated, and embedded with Spurr resin as previously described (13). Ultrathin sections were stained and subsequently observed using a JEM-1400 electron microscope at 120 kV.
Immunoelectron microscopy.
A total of 1.0 × 106 cells were infected with vP6.9:HA or vAcWT virus at an MOI of 5 TCID50/cell. For bromodeoxyuridine (BrdU) labeling experiments, the DNA precursor BrdU (Invitrogen Life Technologies) (1:100) was added to the culture medium at 12 h p.i. Because host DNA replication was halted by viral infection, BrdU was selectively incorporated into newly synthesized viral DNA (28). After 12 h of labeling, infected cells were pelleted by centrifugation at 200 × g for 5 min. The cell pellets were then fixed with 1% glutaraldehyde and 4% paraformaldehyde in PBS at 4°C for 2 h. Dehydration and embedding processes were performed as previously described with slight modifications (6, 24). Following three washes with 0.1 M PBS at 4°C, the fixed cells were dehydrated through a graded series of ethanol at 4°C, 30% ethanol for 30 min and 50% for 30 min, and then 50% ethanol at −35°C for 1 h, 70% ethanol for 1 h, 90% for 1 h, and 100% ethanol three times for 1 h each. The dehydrated cells then were infiltrated through a graded series of Lowicryl K4M resin in 30% ethanol at −35°C for 2 h, 70% ethanol for 2 h, and 100% ethanol three times for 2 h each. Polymerization was performed by UV irradiation (320 nm) for 72 h at −20°C, followed by irradiation at RT for 48 h. Ultrathin sections were immunolabeled and stained as previously described (36), although 0.1% NaN3 was used here.
Western blot analysis.
To separate the different phosphorylated forms of P6.9, proteins were electrophoresed on 15% acid urea polyacrylamide slab gels containing 6.25 M urea (AU-PAGE) as previously described (20, 22). For BV samples, supernatants collected from infected cells were centrifuged twice at 10,000 × g for 20 min to remove the cell debris. BVs were purified by gradient centrifugation and stored in 0.1 M Tris-EDTA (TE) until use as previously described (38). A total of 5.0 × 106 polyhedra from infected cells were lysed in 10 μl of alkaline carbonate buffer containing 0.1 M Na2CO3 and 0.01 M EDTA (pH 10.5) for 10 min at RT. After centrifugation at 10,000 × g for 5 min, the supernatants were acidified with a 1/20 volume of glacial acetic acid and used as the ODV samples. Denaturing of BV, ODV, and infected cell samples with AU gel sample buffer and the preparation of the separating gel and stacking gel were described by Oppenheimer and Volkman (20). The prepared gel was pre-electrophoresed in 0.9 M glacial acetic acid electrophoresis buffer at 250 V before the protein samples were loaded (1). After electrophoresis with the samples, AU gels were equilibrated and transferred to a 0.22-μm nitrocellulose membrane (Whatman) as previously described (4). The membranes were immunoblotted with either anti-P6.9 antibodies (1:1,000) or anti-HA antibody (1:10,000). A horseradish peroxidase-conjugated anti-rabbit IgG (1:5,000) secondary antibody was used with the enhanced chemiluminescence (ECL) system (GE Healthcare).
The specificity of anti-P6.9 antibodies was determined by SDS-PAGE Western blotting. Infected cells were lysed in 1× radioimmunoprecipitation assay (RIPA; Thermo Fisher Scientific) buffer on ice for 30 min and centrifuged at 14,000 × g for 10 min; the supernatants were then dissolved in 4× lithium dodecyl sulfate (LDS) buffer and heated for 10 min at 70°C prior to loading onto 4 to 12% NuPAGE Novex Bis-Tris gels (Invitrogen Life Technologies). After electrophoresis, proteins were transferred onto a 0.22-μm nitrocellulose membrane using an alkaline transfer buffer [25 mM 3-(cyclohexylamino)-1-propanesulfonic acid (CAPS), pH 11, with 10% methanol]. The remaining procedures were the same as that for the AU-PAGE Western blotting described above.
RESULTS
Construction of AcMNPV recombinants.
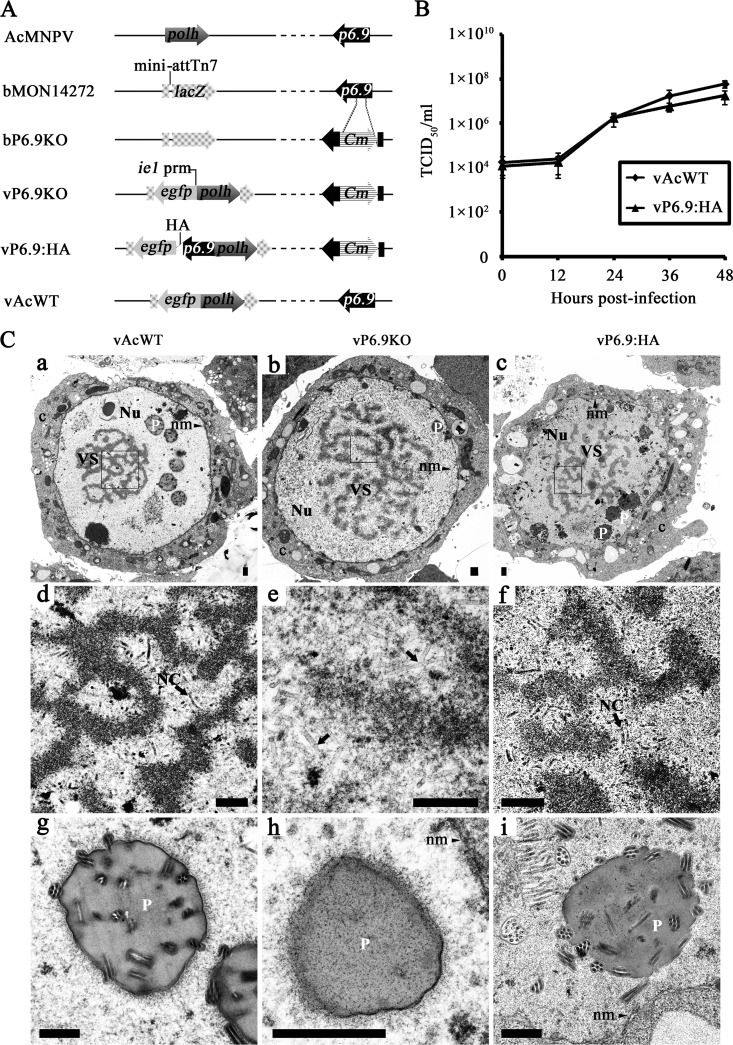
Although the potential role of P6.9 in the viral life cycle has been investigated (35) using immunoblot analyses and the results suggest that P6.9 localizes in the nuclear matrix, the expression profile and the migration of P6.9 from the cytoplasm to the VS in AcMNPV-infected Sf9 cells has not been determined in detail due to the lack of highly specific antibodies. In this study, a recombinant baculovirus, vP6.9:HA, was constructed to observe P6.9 localization during an infection. To investigate the biology of P6.9, we constructed a knockout bacmid in which a 48-bp segment near the center of the p6.9 gene was replaced with a chloramphenicol acetyltransferase (CAT) cassette. Subsequently, a fragment containing the AcMNPV p6.9 cassette fused to an HA tag as well as the egfp and polh genes were also inserted into the construct to create vP6.9:HA (Fig. 1A). The egfp and polh genes were also inserted into bP6.9KO to generate vP6.9KO and were inserted into bMON14272 to generate a “wild-type” AcMNPV control (vAcWT).
Fig 1.
Construction of the p6.9-HA-tagged virus and its fitness analysis. (A) The genes polyhedrin (polh) and p6.9 are indicated on the linear representation of the AcMNPV genome. Arrows denote protein-coding regions and the direction of transcription for each gene. bP6.9KO was generated by the replacement of a 48-bp fragment of p6.9 in the bMON14272 genome with a 1,038-bp CAT cassette. Foreign genes, including the enhanced green fluorescence protein gene (egfp) under the control of the ie1 promoter (prm) and polh, were inserted into the mini-attTn7 site of bP6.9KO by Tn7-mediated transposition. vP6.9:HA contains the p6.9 ORF tagged with an in-frame HA epitope sequence (indicated as a white triangle) under the control of its native promoter. vAcWT was constructed from bMON14272 as a positive control. (B) Comparison of the viral replication capacity of vP6.9:HA and vAcWT. Each data point represents the averages from three independent infections. Error bars indicate the means ± SD. (C) Transmission electron microscopy analysis. Sf9 cells were transfected with vAcWT (a, d, and g), vP6.9KO (b, e, and h), and vP6.9:HA (c, f, and i) and observed at 60 h posttransfection. Panels a to c display the enlarged nucleus (Nu), virogenic stroma (VS), nuclear membrane (nm), polyhedra (P), and cytoplasm (c). Panels d, e, and f are micrographs of the boxed regions in a, b, and c, respectively, with higher magnifications. Arrows indicate the rod-shaped nucleocapsids (NC) in the VS in panels d and f and electron-lucent tubular structures at the intrastromal spaces as shown in panel e. Panels g, h, and i show the polyhera (P) produced by vAcWT, vP6.9KO, and vP6.9:HA viruses, respectively. Scale bar, 500 nm.
To determine the effect of HA-tagged p6.9 on virus replication kinetics, the virus growth curve was determined using a TCID50 endpoint dilution assay. As shown in Fig. 1B, a steady increase in virus production occurred in Sf9 cells infected with either vP6.9:HA or vAcWT virus. At the same time points, the titers of vP6.9:HA and vAcWT viruses were almost equivalent. In contrast, there was no detectable increase in the number of infectious BVs in the vP6.9KO-transfected cells (data not shown), which is consistent with previous observations (30) that p6.9 is an essential gene for viral propagation. Ultrathin sections of Sf9 cells transfected with vP6.9:HA, vP6.9KO, or vAcWT constructs were examined by electron microscopy. Characteristic cytopathic effects, such as enlarged nuclei and the appearance of VS and polyhedra, were observed in cells that were individually transfected with the three viruses (Fig. 1C, a, b, and c). Mature nucleocapsids were present in the intrastromal spaces of the VS in the vP6.9:HA- or vAcWT-transfected cells and were morphologically indistinguishable (Fig. 1C, d and f). ODV-containing polyhedra were frequently observed within the ring zone of cells transfected with vP6.9:HA or vAcWT (Fig. 1C, g and i). However, many elongated and electron-translucent capsid-like structures were found in the intrastromal spaces of the vP6.9KO-transfected cells (Fig. 1C, e); virions were not observed in the polyhedra (Fig. 1C, h). The HA-tagged p6.9 did not appear to have an effect on viral replication relative to the vAcWT form, therefore we used the HA-tagged p6.9 for subsequent analyses.
Majority of P6.9 localizes near the inner nuclear membrane in AcMNPV-infected Sf9 cells.
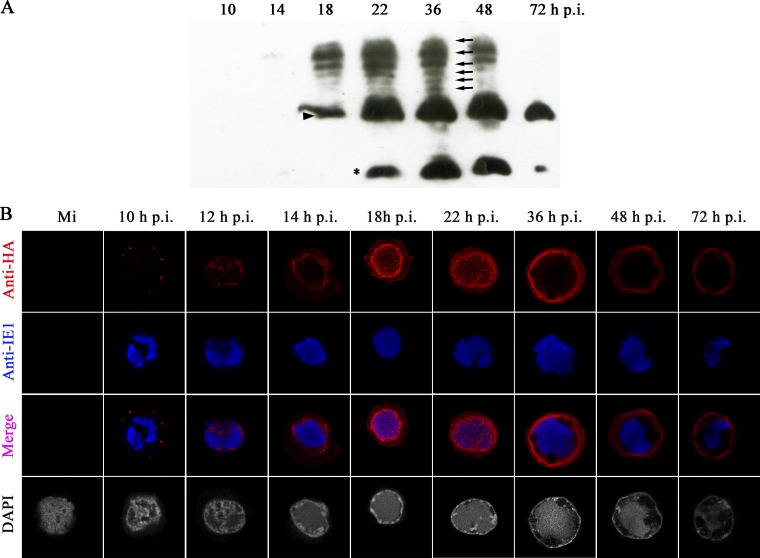
To determine the expression of P6.9, Sf9 cells were infected with vP6.9:HA and were subjected to AU gel electrophoresis and Western blot analysis using an anti-HA antibody. The AU gel system is able to separate highly similar basic proteins based on differences in size and effective charge (22). Using a longer exposure time, a band corresponding to P6.9 was first detected at 10 h p.i., which increased slightly at 14 h p.i. (data not shown). As shown in Fig. 2A, the level of P6.9 increased significantly by 18 h p.i. (indicated by a triangle). By 22 h p.i., P6.9 was expressed abundantly but decreased by 72 h p.i. After 14 h p.i., slower-migrating bands appeared as ladder-like structures (indicated by arrows). Since P6.9 has been previously shown to be phosphorylated (8, 20), this result suggested that the protein detected from 10 h p.i. on was unphosphorylated P6.9 and the ladder-like structures were different forms of phosphorylated P6.9. By 22 h p.i., a lower band was detected (indicated by an asterisk in Fig. 2A), thus suggesting that P6.9 is cleaved late in infection.
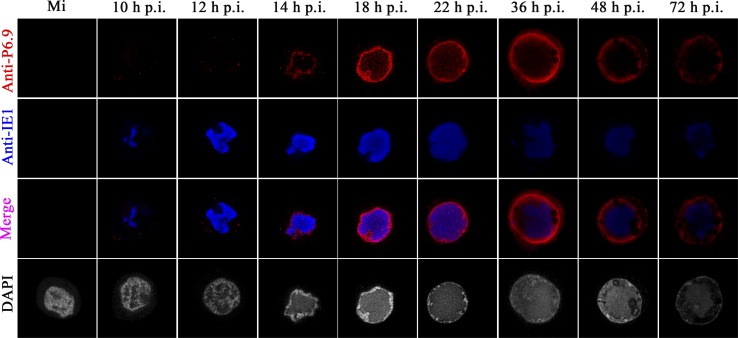
Fig 2.
Spatial-temporal expression of P6.9 in vP6.9:HA-infected Sf9 cells. Sf9 cells were infected with vP6.9:HA at an MOI of 10 TCID50/cell. (A) At the indicated h p.i., proteins from the infected cells were separated by 15% AU-PAGE and then immunoblotted with rabbit anti-HA antibody. The triangle indicates the unphosphorylated P6.9 band. The black arrows indicate possible phosphorylated P6.9 bands. The asterisk indicates a possible cleavage product of P6.9 produced in the late phase of infection. (B) Mock-infected (Mi) or infected Sf9 cells were subjected to indirect immunofluorescence microscopy to detect total P6.9 localization. Rabbit anti-HA and mouse anti-IE1 were used as primary antibodies. Goat anti-rabbit IgG conjugated with Alexa Fluor 647 antibody (red) and goat anti-mouse IgG conjugated with Alexa Fluor 561 antibody (pseudocolored blue) were used as secondary antibodies. Infected cells were stained with DAPI to directly visualize the nuclear DNA (pseudocolored gray).
To visualize the distribution of P6.9, vP6.9:HA-infected Sf9 cells were immunostained with anti-HA antibody and scanned using laser confocal microscopy (Fig. 2B). In addition, to investigate the location of P6.9 relative to the VS, the infected cells were immunostained with an anti-IE1 antibody, which serves as a marker for VS distribution (11, 18). Indirect immunofluorescence showed that P6.9 first appeared as discrete foci in the cytoplasm of vP6.9:HA-infected cells at 10 h p.i. Subsequently, at 12 h p.i., P6.9 was found as foci interspersed among the VS. As the VS matured from 14 to 22 h p.i., the majority of the P6.9 localized near the inner nuclear membrane, whereas a lower concentration of P6.9 was observed in the VS. The amount of P6.9 in the VS increased slightly at 18 h p.i. and became focused at 22 h p.i. During the late phase of infection the VS became condensed, and the majority of P6.9 remained located near the inner nuclear membrane.
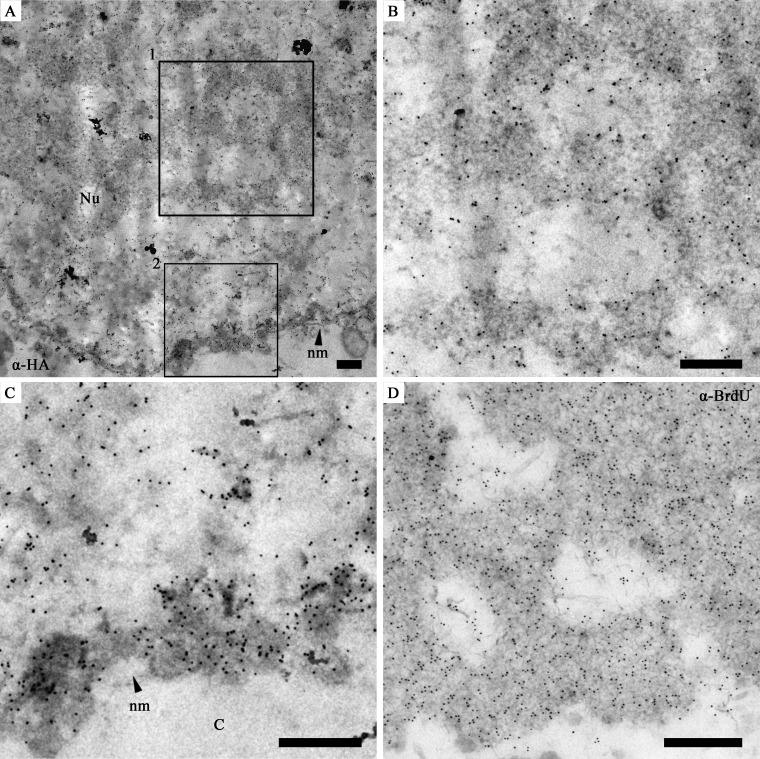
It was previously reported that P6.9 plays a role in viral DNA packaging and nucleocapsid assembly, which occurs within the VS. Moreover, a previous study suggested that P6.9 was a component of the VS (35). Thus, our observation of abundant P6.9 localized near the inner nuclear membrane of infected cells between 14 and 72 h p.i. was unexpected. To confirm this observation, Sf9 cells were infected with vP6.9:HA, and sample sections were subjected to electron microscopy. As expected, gold particle-labeled P6.9 was observed in the electron-dense stromal mattes of the VS (Fig. 3A) and associated with nucleocapsids at the periphery of the stromal mattes (Fig. 3B). Additionally, P6.9 was detected at the periphery of the inner nuclear membrane and was associated with the marginalized host chromatin (Fig. 3C). In addition, de novo synthesized viral DNA, labeled with an antibody against BrdU (Invitrogen Life Technologies), predominantly localized within the electron-dense regions of the VS (Fig. 3D) rather than at the periphery of the inner nuclear membrane (data not shown).
Fig 3.
Immunoelectron micrographs showing the distribution of P6.9 in vP6.9:HA-infected Sf9 cells. (A) A part of a vP6.9:HA-infected cell showing the heavily gold-labeled nucleus. Here, anti-HA was used as the primary antibody, and protein A conjugated with 10 nm gold was used at a 1:50 dilution. (B and C) Enlargement of the vP6.9:HA-infected cell highlighted in panel A. (B) Box 1 from panel A showing gold-labeled P6.9 in the VS. (C) Box 2 from panel A showing that gold-labeled P6.9 localized near the inner nuclear membrane of the nucleus (C). (D) The VS region of a vP6.9HA-infected Sf9 cell. The cell was labeled with BrdU and probed with mouse monoclonal anti-BrdU antibody. Goat anti-mouse secondary antibody conjugated with 10 nm gold was used at a 1:100 dilution. Nu, nucleus; nm, nuclear membrane. Scale bar, 500 nm.
Immunodetection of P6.9 in infected cells using the anti-P6.9 antibodies.
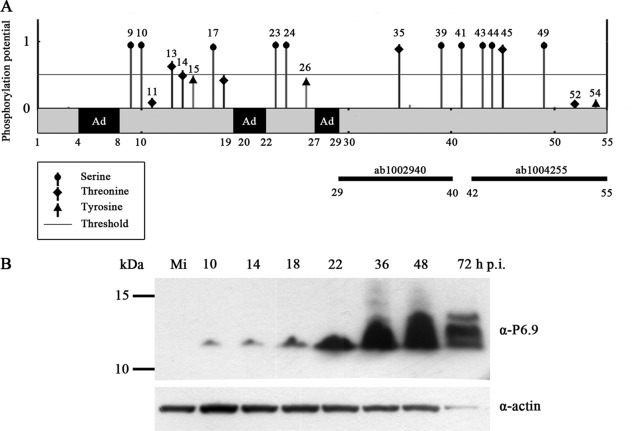
Because the total P6.9 detected using anti-HA antibody appeared to be composed of phosphorylated and unphosphorylated forms of P6.9:HA in vP6.9:HA-infected Sf9 cells, we examined the distribution of unphosphorylated P6.9 in infected cells. To this end, the anti-P6.9 antibodies were generated using two peptides corresponding to sequences of AcMNPV P6.9 (residues 29 to 40 and 42 to 55). The phosphorylation sites of P6.9 were predicted using the NetPhos 2.0 server (Fig. 4A). A total of 3 DNA-anchoring domains (Ad), which typically contain 3 to 7 arginine residues (3), were predicted at the N-terminal region of P6.9. The specificity of the antibodies was first investigated using SDS-PAGE Western blot analysis (Fig. 4B). By using a mixture of the two antibodies, a protein of approximately 13 kDa was first detectable at 10 h p.i. The protein level did not change at 14 h p.i. but increased at 18 h p.i. From 18 to 36 h p.i., the P6.9 expression level increased significantly. At 48 h p.i., P6.9 was abundantly expressed, and several higher bands appeared at 72 h p.i., which may represent phosphorylated forms of P6.9 (Fig. 4B).
Fig 4.
Schematic representation of AcMNPV P6.9 and Western blot analysis. (A) Predicted phosphorylation sites of P6.9 and the proposed DNA anchoring domains. The y axis indicates the predicted phosphorylation score obtained using NetPhos (http://www.cbs.dtu.dk/services/NetPhos/). If the score was greater than the threshold of 0.5, the site was predicted to be a potential phosphorylation site. Numbers denote the amino acid residues of P6.9; Ad, proposed DNA anchoring domains (regions containing 3 or more consecutive arginine residues). Two solid black bars indicate two peptides of P6.9 that were used as antigens to prepare the polyclonal antibodies against P6.9. (B) Time course analysis of P6.9 expression. Sf9 cells were mock infected (Mi) or infected with vAcWT at an MOI of 10 TCID50/cell. At the indicated h p.i., cells were collected and subjected to 4 to 12% SDS-PAGE (Invitrogen Life Technologies). Western blot analysis using anti-P6.9 antibodies (ab1002940 and ab1004255 mixture) and anti-actin antibody as shown. The molecular mass (kDa) standards are indicated on the left.
vAcWT-infected Sf9 cells were immunostained with the anti-P6.9 antibodies and scanned with laser confocal microscopy. No difference was observed in the distribution of P6.9 in the infected cells with the anti-HA antibody (Fig. 3) or the anti-P6.9 antibodies (Fig. 5), except that the P6.9 foci in the nuclei at 12 h p.i. were difficult to detect using the anti-P6.9 antibodies (Fig. 5).
Fig 5.
Spatial-temporal distribution of P6.9 in the vAcWT-infected Sf9 cells. Sf9 cells were mock infected (Mi) or infected with vAcWT virus at an MOI of 10 TCID50/cell. At different h p.i., cells were incubated with rabbit anti-P6.9 antibodies and mouse anti-IE1 antibody as the primary antibodies. The goat anti-rabbit IgG conjugated with Alexa Fluor 647 antibody (Red) and the goat anti-mouse IgG conjugated with Alexa Fluor 561 antibody (pseudocolored blue) were used as secondary antibodies. Infected cells were stained with DAPI to directly visualize the nuclear DNA (pseudocolored gray).
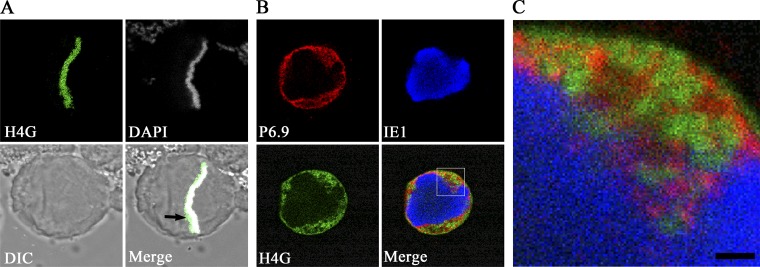
Because baculovirus infection causes host chromatin marginalization, we investigated whether P6.9 could colocalize with host chromatin. Therefore, Sf9 cells were transfected with the plasmid pIZ-SfH4-EGFP (named H4G), which expresses a GFP-tagged host chromatin marker, as previously described (18). In metaphase, the distribution of H4G was consistent with that of DAPI-labeled cellular DNA (Fig. 6A), thereby indicating that H4G can serve as a marker of cellular chromatin in Sf9 cells. At 24 h after transfection, cells were infected with AcMNPV. As shown in Fig. 6B, when the IE1-indicated VS occupied the central region of the nucleus at 12 h p.i. (Fig. 6B, IE1), the cellular chromatin localized to the margins of the nuclei (Fig. 6B, H4G). Although a portion of P6.9 was observed within the VS, P6.9 was enriched at the periphery of the inner nuclear membrane where the host chromatin also appeared to be located (Fig. 6B, merge and P6.9). However, P6.9 and the host chromatin did not completely colocalize, as shown in the enlarged photograph (Fig. 6C).
Fig 6.
P6.9 and the marginalized host chromatin localize to the same regions. (A) An H4G-transfected Sf9 cell in anaphase. (B) After transfection with H4-GFP (green), cells were infected with AcMNPV at an MOI of 10 TCID50/cell. At 12 h p.i., cells were probed with anti-P6.9 (red) and anti-IE1 (pseudocolored blue) antibodies. (C) A magnified image of the boxed region in panel B. Scale bar, 1 μm.
A specific form of P6.9 localizes to the edge of, but not within, the stromal mattes of the VS.
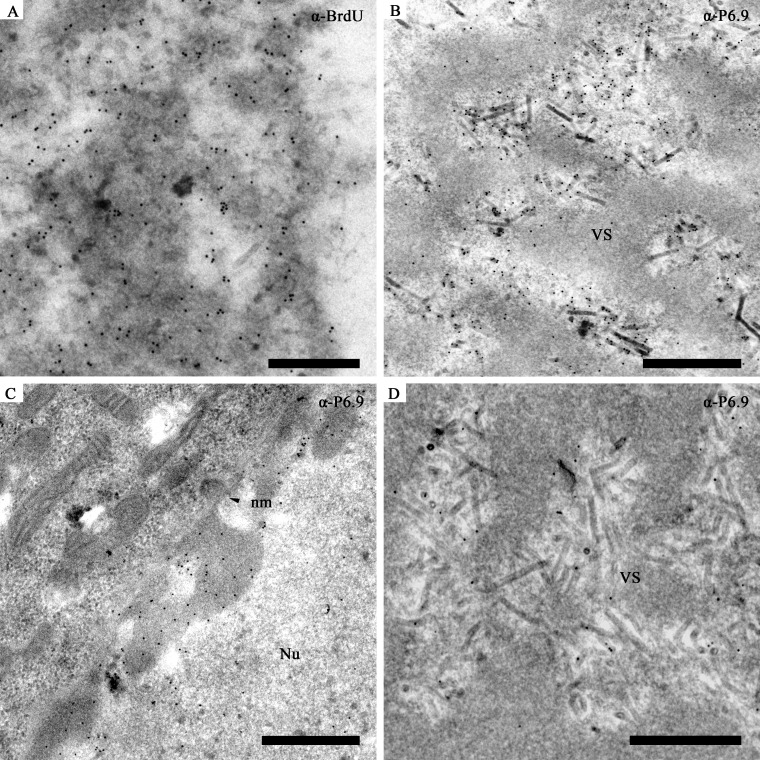
Immunoelectron microscopy was performed using the anti-P6.9 antibodies. As shown in Fig. 7A, the electron-dense regions of the VS in vAcWT-infected cells were densely labeled by the anti-BrdU antibody. Interestingly, the gold-labeled P6.9 was observed to localize near the edge of the electron-dense stromal mattes of the VS and to what appeared to be nucleocapsids undergoing assembly or being packaged (Fig. 7B). Few, if any, gold particles appeared within the electron-dense regions, thereby suggesting that P6.9 was not enriched in the electron-dense structures. Gold particles were also observed in the region of the marginalized chromatin near the inner nuclear membrane (Fig. 7C), which is consistent with the results of the indirect immunofluorescence assay. Gold particles were rarely detected in the cells in which the primary antibody was omitted (data not shown). Since anti-P6.9 is a mixture of ab1002940 and ab1004255, immunoelectron microscopy was performed using ab1002940 or ab1004255 alone to detect the distribution of P6.9 in vAcWT-infected Sf9 cells. The detected P6.9 still distributed at the edges of the stromal mattes, electron-lucent intrastromal spaces of the VS, and the marginalized inner nuclear membrane, similar to when both anti-P6.9 antibodies were used (data not shown).
Fig 7.
Electron microscopic analysis of P6.9 detected by the prepared anti-P6.9 antibodies. (A, B, and C) Images of vAcWT-infected Sf9 cells. (A) VS labeled with a mouse monoclonal anti-BrdU antibody. Goat anti-mouse secondary antibody conjugated with 10 nm gold was used at a 1:100 dilution. Gold particles were abundant in the electron-dense regions as well as at the nucleocapsids in the VS. (B and C) Infected cells were immunoreacted with the anti-P6.9 antibodies. P6.9 localized to the intrastromal spaces instead of the electron-dense regions. Protein A conjugated with gold (10 nm) was used at a 1:50 dilution. (B) The VS region of an infected cell. (C) The periphery region of a nucleus. (D) The VS region of an vP6.9:HA-infected Sf9 cell. The VS was labeled with anti-P6.9 antibodies. Protein A conjugated with gold (10 nm) was used at a 1:50 dilution. Scale bar, 500 nm.
To investigate whether the different results generated by anti-HA antibody and anti-P6.9 antibody were attributed to the HA tag or not, an immunoelectron microscopy experiment was performed using anti-P6.9 antibody to detect the HA-tagged P6.9 protein in vP6.9:HA-infected Sf9 cells (Fig. 7D). In the VS regions of vP6.9:HA-infected Sf9 cells, gold particles were only found at the edges of the stromal matte and the processing nucleocapsids (Fig. 7D). These results suggest that a specific form of P6.9 localized to the edge of the stromal mattes of the VS (Fig. 7D). In contrast, other form(s) of P6.9 (Fig. 3B) which are different from the specific one localized within the stromal mattes.
Phosphorylated and unphosphorylated P6.9 are associated with ODV.
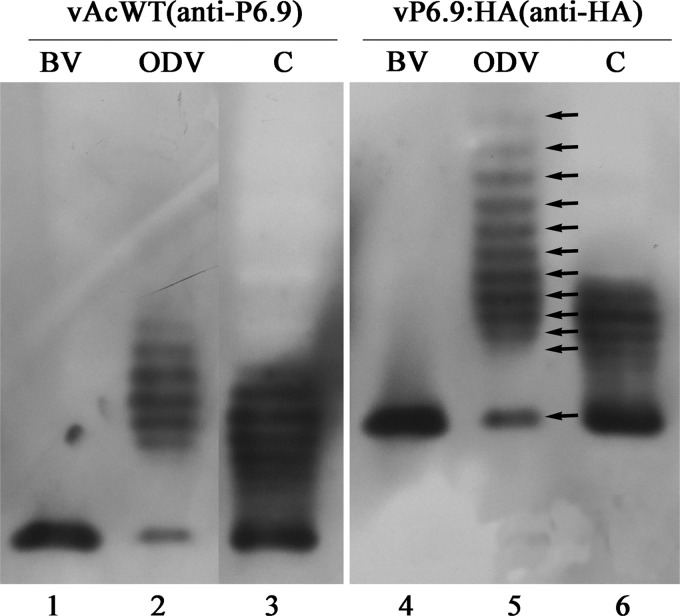
When protein samples were electrophoresed on AU gels and subjected to Western blot analysis using the anti-P6.9 antibodies or the anti-HA antibody, only one band, which represented unphosphorylated P6.9, was detected in the BVs derived from vAcWT- or vP6.9:HA-infected cells (Fig. 8, lanes 1 and 4). This result is consistent with the previous observations that P6.9 or its homologue, VP12, was present in an unphosphorylated form in mature nucleocapsids (20, 32). Similar forms of P6.9 were identified in the vAcWT- or vP6.9:HA-infected cells at 18 h p.i. In addition to the unphosphorylated P6.9, a ladder comprised of 6 forms of P6.9 which are likely to be phosphorylated was observed in infected cell extracts (Fig. 8, lanes 3 and 6). A similar ladder was also previously reported in AcMNPV-infected, radiolabeled cells (20). To our surprise, in addition to the unphosphorylated P6.9, at least 11 possible phosphorylated forms of P6.9 were detected by anti-HA antibodies, and at least 6 were detected using anti-P6.9 antibodies in ODV (Fig. 8, lanes 2 and 5). This result indicates that, in contrast to completely unphosphorylated P6.9 in BV, phosphorylated P6.9 is present in ODV.
Fig 8.
AU-PAGE Western blot analysis of P6.9 in BV, ODV, and infected cells. BVs and ODVs collected from vAcWT-infected cells were subjected to Western blotting with the anti-P6.9 antibodies (ab1002940 and ab1004255 mixture), whereas BVs and ODVs from vP6.9HA-infected cells were probed with the anti-HA antibody. Samples from cells at 18 h p.i. (C) are also shown.
DISCUSSION
P6.9 is a basic protamine-like protein and plays a role in the packaging of baculovirus DNA (5, 8, 20, 27). In AcMNPV-infected cells, P6.9 is phosphorylated immediately upon synthesis and is subsequently dephosphorylated. The half-life of phosphorylated P6.9 is less than 1 h (20). The dephosphorylation of P6.9 is thought to be important for the condensation of the nucleoprotein because it presumably modulates the affinity of P6.9 for DNA (8, 32). In this paper, a recombinant AcMNPV expressing HA-tagged P6.9 was constructed to investigate the distribution of P6.9 in infected cells. Indirect immunofluorescence microscopy demonstrated that large quantities of P6.9 localized near the periphery of the nucleus throughout the course of infection. Baculovirus infection can induce host chromatin marginalization, which is related to four virus genes (ie1, lef3, p143, and hr) (18). We observed that host chromatin was marginalized in the vP6.9KO-transfected cells (data not shown), thereby suggesting that the deletion of P6.9 had no effect on host chromatin marginalization. Using the GFP-tagged host histone H4 as a host chromatin marker, indirect immunofluorescence microscopy revealed that P6.9 was localized within the regions of the host chromatin; however, the regions where P6.9 localized did not precisely overlap those of H4 (Fig. 6). Immunoelectron microscopy confirmed that P6.9 was located within the region of marginalized host chromatin (Fig. 3C and 7C).
Protamine undergoes a phosphorylation-dephosphorylation cycle before being packaged into sperm chromatin during spermiogenesis (10, 14). A previous study also observed temporal perinuclear localization of protamine 1 during spermiogenesis and found that the binding of protamine to an inner nuclear protein lamin B receptor is required for the phosphorylation of protamine (17). Thus, the observation that abundant P6.9 localized near the inner nuclear membrane of the nucleus suggests that the docking of P6.9 is required for the phosphorylation of P6.9.
Immunoelectron microscopy using an anti-HA antibody showed that P6.9 forms were present in the electron-dense regions of stromal mattes, the surface of stromal mattes, and around the nucleocapsids in the pockets of stromal mattes of infected cells. However, when we used the P6.9 antiserum generated in this study, P6.9 was observed to localize primarily to the edge of the electron-dense stromal mattes of the VS and the processing nucleocapsids (Fig. 7B). Because the synthesized peptides used to generate anti-P6.9 antibodies had not been phosphorylated, they likely would be unable to detect P6.9 with phosphates in those regions. Therefore, anti-P6.9 antibodies are likely to identify C-terminal unphosphorylated forms of P6.9 but not total P6.9, as does anti-HA antibody. Our observations suggest for the first time that the dephosphorylation of P6.9, at least at the C terminus (aa 29 to 55) of P6.9, occurs at the surface of stromal mattes of the VS. Phosphoamino acid analysis of phosphorylated P6.9 obtained from the cells infected with AcMNPV indicated that serines and threonines, but not tyrosine, were phosphorylated (20). There are five serines (aa 39, 41, 43, 44, and 49) and two threonines (aa 35 and 45) present at the C-terminal region of AcMNPV P6.9. These residues have a high potential to be phosphorylated according to predictions using NetPhos (http://www.cbs.dtu.dk/services/NetPhos/) (Fig. 4A). By constructing AcMNPV genomes encoding either P6.9 proteins with C-terminal deletions or point mutations or chimeric P6.9 fusion proteins, a recently study has demonstrated that the C-terminal domain of P6.9 appears to be an essential genus-specific feature required for viral assembly (30).
Previous studies reported that only unphosphorylated P6.9 or its homologue, VP12, was present in mature nucleocapsids derived from BVs (20, 32). Only unphosphorylated P6.9 was consistently detected in BVs using the anti-P6.9 and anti-HA antibodies in the present study. However, several forms of phosphorylated P6.9 were found that were associated with ODV (Fig. 8). Anti-HA antibody could detect at least 5 more phosphorylated forms of P6.9 than did anti-P6.9 antibodies. It is probable that the phosphorylation of at least some of the P6.9 protein in ODV occurs after encapsidation, because baculoviruses apparently possess protein kinase activity associated with the nucleocapsids (16, 31). Another possibility is incomplete dephosphorylation of phosphorylated P6.9 during encapsidation (8). The nucleocapsids containing partially phosphorylated P6.9 might not be able to pass through the ring zone and therefore are destined to become ODV. In contrast, those nucleocapsids with completely unphosphorylated P6.9 could be readily transported to the cytoplasm to form BV.
P6.9 requires bivalent metal ions, such as Zn2+, to maintain the structural stability of the nucleocapsid, and the phosphorylation of P6.9 by the associated protein kinase may be triggered by the loss of metal ions (7, 8). The polyhedral lysis buffer in our experiment (Fig. 8) contained EDTA, which can chelate metal ions; thus, the observed phosphorylation of P6.9 in ODVs might be an artifact. To rule out this possibility, polyhedra from vP6.9:HA-infected cells were lysed in 0.1 M Na2CO3 without EDTA. Identical forms of phosphorylated P6.9 were observed, as previously detected (data not shown). Furthermore, prior to lysis, we heated the polyhedra at 95°C for 10 min to inactivate the enzymes packaged within the matrix of the polyhedra, and the 12 forms of P6.9 remained detectable (data not shown).
Western blot analysis of P6.9 temporal expression in virus-infected cells observed using AU-PAGE analysis showed that several forms of phosphorylated P6.9 create a ladder starting at 14 h p.i. A similar ladder of [H3]arginine-labeled phosphorylated P6.9 has been observed in AcMNPV-infected Sf cells (20). The multiphosphorylation of protamine was also found in trout testes cells (14). One P6.9 band that migrated faster than the unphosphorylated form of P6.9 appeared in both vP6.9:HA- and vAcWT-infected cells (data not shown) starting at 22 h p.i., which suggests that a cleavage event of P6.9 had occurred in the late phase of infection. Because the HA epitope was tagged at the C terminus of P6.9, the cleavage of P6.9 must have occurred at its N-terminal region. A previous study reported that it was difficult to detect [35S]methionine-labeled P6.9 (12), which may have been attributed to the cleavage of the methionine at the N terminus of P6.9. Oppenheimer and Volkman (20) found that cytochalasin D, a fungus-derived compound that interferes with actin polymerization, had no effect on the synthesis, phosphorylation, or dephosphorylation of P6.9 but found that it induced the proteolysis of P6.9. The proteolysis of P6.9 appeared to account for the production of BV. The biological significance of the cleavage of P6.9 in the infection cycle of AcMNPV remains unclear.
Based on the present observations and previous reports, a model for the role of P6.9 in baculoviruses is proposed. After synthesis in cytoplasm, the P6.9 proteins move into nuclei, initially appear as foci interspersed among the VS, and dock near the inner nuclear membrane for phosphorylation. Phosphorylated P6.9 is transported into the electron-dense stromal mattes of the VS and is dephosphorylated at the surface of stromal mattes to allow the nucleoprotein core to enter the preassembled capsid sheaths. In the early phase of infection, nucleocapsids with completely dephosphorylated P6.9 pass through the cytoplasm to form BV. In the late phase of infection, at least a fraction of the P6.9 proteins are phosphorylated by nucleocapsid-associated protein kinases, and the nucleocapsids remain in the ring zone to form ODV.
ACKNOWLEDGMENTS
We thank George F. Rohrmann (Oregon State University) for critically reading the manuscript and Just M. Vlak (Wageningen University and Research Center, Wageningen, the Netherlands) for valuable discussion. We are grateful to Linda A. Guarino (Texas A&M University) for the generous gift of the mouse monoclonal anti-AcMNPV IE1 antibody (IE1-4B7). We thank Dongwei Hu and Jian Hong (Zhejiang University, People's Republic of China) for their assistance with immunoelectron microscopy.
This research was supported by the National Basic Research Program of China (973 Program; no. 2009CB118903), the Hi-Tech Research and Development Program of China (863 Program; no. 2011AA10A204), the National Nature Science Foundation of China (no. 30900941), and the Fundamental Research Funds for the Central Universities (no. 09lgpy39).
Footnotes
Published ahead of print 5 September 2012
REFERENCES
- 1. Alfageme CR, Zweidler A, Mahowald A, Cohen LH. 1974. Histones of Drosophila embryos. Electrophoretic isolation and structural studies. J. Biol. Chem. 249:3729–3736 [PubMed] [Google Scholar]
- 2. Balhorn R. 2007. The protamine family of sperm nuclear proteins. Genome Biol. 8:227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Balhorn R, et al. 1999. The male gamete: from basic knowledge to clinical applications. Cache River Press, Vienna, IL [Google Scholar]
- 4. Delcuve GP, Davie JR. 1992. Western blotting and immunochemical detection of histones electrophoretically resolved on acid-urea-triton- and sodium dodecyl sulfate-polyacrylamide gels. Anal. Biochem. 200:339–341 [DOI] [PubMed] [Google Scholar]
- 5. Fraser MJ. 1986. Ultrastructural observations of virion maturation in Autographa californica nuclear polyhedrosis virus infected Spodoptera frugiperda cell cultures. J. Ultrastruct. Mol. Struct. Res. 95:189–195 [Google Scholar]
- 6. Fryer PR, Wells C, Ratcliffe A. 1983. Technical difficulties overcome in the use of Lowicryl 4KM EM embedding resin. Histochemistry 77:141–143 [DOI] [PubMed] [Google Scholar]
- 7. Funk CJ, Consigli RA. 1992. Evidence for zinc binding by two structural proteins of Plodia interpunctella granulosis virus. J. Virol. 66:3168–3171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Funk CJ, Consigli RA. 1993. Phosphate cycling on the basic protein of Plodia interpunctella granulosis virus. Virology 193:396–402 [DOI] [PubMed] [Google Scholar]
- 9. Harrap KA. 1972. The structure of nuclear polyhedrosis viruses. 3. Virus assembly. Virology 50:133–139 [DOI] [PubMed] [Google Scholar]
- 10. Ingles CJ, Dixon GH. 1967. Phosphorylation of protamine during spermatogenesis in trout testis. Proc. Natl. Acad. Sci. U. S. A. 58:1011–1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kawasaki Y, Matsumoto S, Nagamine T. 2004. Analysis of baculovirus IE1 in living cells: dynamics and spatial relationships to viral structural proteins. J. Gen. Virol. 85:3575–3583 [DOI] [PubMed] [Google Scholar]
- 12. Kelly DC, Lescott T. 1984. Baculovirus replication: phosphorylation of polypeptides synthesized in Trichoplusia ni nuclear polyhedrosis virus-infected cells. J. Gen. Virol. 65(Pt 7):1183–1191 [DOI] [PubMed] [Google Scholar]
- 13. Li Y, et al. 2005. vlf-1 deletion brought AcMNPV to defect in nucleocapsid formation. Virus Genes 31:275–284 [DOI] [PubMed] [Google Scholar]
- 14. Louie AJ, Dixon GH. 1972. Kinetics of enzymatic modification of the protamines and a proposal for their binding to chromatin. J. Biol. Chem. 247:7962–7968 [PubMed] [Google Scholar]
- 15. Luckow VA, Lee SC, Barry GF, Olins PO. 1993. Efficient generation of infectious recombinant baculoviruses by site-specific transposon-mediated insertion of foreign genes into a baculovirus genome propagated in Escherichia coli. J. Virol. 67:4566–4579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Miller LK, Adang MJ, Browne D. 1983. Protein kinase activity associated with the extracellular and occluded forms of the baculovirus Autographa californica nuclear polyhedrosis virus. J. Virol. 46:275–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Mylonis I, et al. 2004. Temporal association of protamine 1 with the inner nuclear membrane protein lamin B receptor during spermiogenesis. J. Biol. Chem. 279:11626–11631 [DOI] [PubMed] [Google Scholar]
- 18. Nagamine T, Kawasaki Y, Abe A, Matsumoto S. 2008. Nuclear marginalization of host cell chromatin associated with expansion of two discrete virus-induced subnuclear compartments during baculovirus infection. J. Virol. 82:6409–6418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Oliva R, Dixon GH. 1991. Vertebrate protamine genes and the histone-to-protamine replacement reaction. Prog. Nucleic Acid Res. Mol. Biol. 40:25–94 [DOI] [PubMed] [Google Scholar]
- 20. Oppenheimer DI, Volkman LE. 1995. Proteolysis of p6.9 induced by cytochalasin D in Autographa californica M nuclear polyhedrosis virus-infected cells. Virology 207:1–11 [DOI] [PubMed] [Google Scholar]
- 21. O'Reilly DR, Miller LK, Luckow VA. 1992. Baculovirus expression vectors: a laboratory manual. Oxford University Press, New York, New York [Google Scholar]
- 22. Panyim S, Chalkley R. 1969. High resolution acrylamide gel electrophoresis of histones. Arch. Biochem. Biophys. 130:337–346 [DOI] [PubMed] [Google Scholar]
- 23. Rohrmann GF. 2011. Baculovirus molecular biology, 2nd ed National Center for Biotechnology Information, Bethesda, MD: [PubMed] [Google Scholar]
- 24. Roth J, Bendayan M, Carlemalm E, Villiger W, Garavito M. 1981. Enhancement of structural preservation and immunocytochemical staining in low temperature embedded pancreatic tissue. J. Histochem. Cytochem. 29:663–671 [DOI] [PubMed] [Google Scholar]
- 25. Russell RL, Rohrmann GF. 1990. The p6.5 gene region of a nuclear polyhedrosis virus of Orgyia pseudotsugata: DNA sequence and transcriptional analysis of four late genes. J. Gen. Virol. 71(Pt 3):551–560 [DOI] [PubMed] [Google Scholar]
- 26. Theilmann DA, et al. 2005. Baculoviridae. Elsevier, London, United Kingdom [Google Scholar]
- 27. Tweeten KA, Bulla LA, Consigli RA. 1980. Characterization of an extremely basic protein derived from granulosis virus nucleocapsids. J. Virol. 33:866–876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Vanarsdall AL, Mikhailov VS, Rohrmann GF. 2007. Characterization of a baculovirus lacking the DBP (DNA-binding protein) gene. Virology 364:475–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Vaughn JL, Goodwin RH, Tompkins GJ, McCawley P. 1977. The establishment of two cell lines from the insect Spodoptera frugiperda (Lepidoptera; Noctuidae). In Vitro 13:213–217 [DOI] [PubMed] [Google Scholar]
- 30. Wang M, et al. 2010. Specificity of baculovirus P6.9 basic DNA-binding proteins and critical role of the C terminus in virion formation. J. Virol. 84:8821–8828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wang X. 1984. Early events in baculovirus replication. University of Oxford, Oxford, United Kingdom [Google Scholar]
- 32. Wilson ME, Consigli RA. 1985. Functions of a protein kinase activity associated with purified capsids of the granulosis virus infecting Plodia interpunctella. Virology 143:526–535 [DOI] [PubMed] [Google Scholar]
- 33. Wilson ME, Mainprize TH, Friesen PD, Miller LK. 1987. Location, transcription, and sequence of a baculovirus gene encoding a small arginine-rich polypeptide. J. Virol. 61:661–666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wilson ME, Miller LK. 1986. Changes in the nucleoprotein complexes of a baculovirus DNA during infection. Virology 151:315–328 [DOI] [PubMed] [Google Scholar]
- 35. Wilson ME, Price KH. 1988. Association of Autographa californica nuclear polyhedrosis virus (AcMNPV) with the nuclear matrix. Virology 167:233–241 [DOI] [PubMed] [Google Scholar]
- 36. Wu W, et al. 2006. Autographa californica multiple nucleopolyhedrovirus nucleocapsid assembly is interrupted upon deletion of the 38K gene. J. Virol. 80:11475–11485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Young JC, MacKinnon EA, Faulkner P. 1993. The architecture of the virogenic stroma in isolated nuclei of Spodoptera frugiperda cells in vitro infected by Autographa californica nuclear polyhedrosis virus. J. Struct. Biol. 110:141–153 [Google Scholar]
- 38. Yuan M, et al. 2011. Identification of Autographa californica nucleopolyhedrovirus ac93 as a core gene and its requirement for intranuclear microvesicle formation and nuclear egress of nucleocapsids. J. Virol. 85:11664–11674 [DOI] [PMC free article] [PubMed] [Google Scholar]