Abstract
Phosphorylation of the hepadnavirus core protein C-terminal domain (CTD) is important for viral RNA packaging, reverse transcription, and subcellular localization. Hepadnavirus capsids also package a cellular kinase. The identity of the host kinase that phosphorylates the core CTD or gets packaged remains to be resolved. In particular, both the human hepatitis B virus (HBV) and duck hepatitis B virus (DHBV) core CTDs harbor several conserved serine/threonine-proline (S/T-P) sites whose phosphorylation state is known to regulate CTD functions. We report here that the endogenous kinase in the HBV capsids was blocked by chemical inhibitors of the cyclin-dependent kinases (CDKs), in particular, CDK2 inhibitors. The kinase phosphorylated the HBV CTD at the serine-proline (S-P) sites. Furthermore, we were able to detect CDK2 in purified HBV capsids by immunoblotting. Purified CDK2 phosphorylated the S/T-P sites of the HBV and DHBV CTD in vitro. Inhibitors of CDKs, of CDK2 in particular, decreased both HBV and DHBV CTD phosphorylation in vivo. Moreover, CDK2 inhibitors blocked DHBV CTD phosphorylation, specifically at the S/T-P sites, in a mammalian cell lysate. These results indicate that cellular CDK2 phosphorylates the functionally critical S/T-P sites of the hepadnavirus core CTD and is incorporated into viral capsids.
INTRODUCTION
The human hepatitis B virus (HBV) continues to pose a significant health risk worldwide, causing more than one million deaths annually (52). Chronic HBV infection, estimated to affect 350 million people globally, dramatically elevates the risk for developing serious liver diseases, including cirrhosis and hepatocellular carcinoma. HBV is a member of the Hepadnaviridae family, which includes hepatotropic DNA viruses that consist of an enveloped icosahedral capsid enclosing an approximately 3-kb DNA genome in a partially double-stranded, relaxed circular (RC) form. These DNA viruses are also retroid viruses and encode a reverse transcriptase (RT) enzyme that converts a so-called pregenomic RNA (pgRNA) template to the RC DNA through reverse transcription within cytoplasmic capsids. Capsids are composed of multiple copies (180 or 240) of one virally encoded protein, the core or capsid protein (9, 63, 65, 71).
Phosphorylation of the hepadnavirus core protein is important for RNA packaging, DNA synthesis, and subcellular localization. The HBV core protein (HBc) contains three major serine-proline (S-P) phosphorylation sites in its C-terminal domain (CTD) (32). The duck hepatitis B virus (DHBV) core protein (DHBc) contains six known phosphorylation sites, four of which also have the serine/threonine-proline (S/T-P) motifs (43, 68). Mutational analyses indicate that phosphorylation of the core protein at these S/T-P sites is required for RNA packaging and DNA synthesis in HBV (29, 31). For DHBV, dynamic CTD phosphorylation at the S/T-P sites is required for complete DNA synthesis such that the S/T-P phosphorylation is needed for first-strand DNA synthesis and dephosphorylation is required for second-strand DNA synthesis and accumulation (4, 15, 51, 68). Phosphorylation at these sites has also been shown to regulate nuclear localization of HBc and DHBc (34, 62, 66).
Several kinases have been reported to phosphorylate the core protein in vitro, including protein kinase C (PKC) (23–26, 64), glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (12), a 46-kDa serine kinase (26), and serine-arginine protein kinases 1 and 2 (SRPK1 and -2) (10). In all these cases, the site(s) of core phosphorylation was never defined, except that SRPK1 and -2 were shown to phosphorylate the HBc CTD in vitro (10, 45) and in Escherichia coli (8). However, the SRPKs appear to have rather relaxed substrate specificity in these systems, phosphorylating mostly S176 and S178 in the HBc CTD and only weakly at the three S-P sites. Furthermore, SRPK1 and -2 do not appear to be responsible for phosphorylating HBc in human hepatic cells (70). Also, PKC is reported to disfavor proline at the P+1 position (10, 41) and is thus unlikely to be the kinase responsible for phosphorylating the CTD S/T-P sites. Indeed, previous studies have argued against a role for either PKC or protein kinase A (PKA) in phosphorylating HBc (10, 26). Therefore, the identity of the cellular kinase(s) that phosphorylates the core protein, in particular the functionally critical S/T-P sites in its CTD, remains to be resolved.
The HBV capsids were shown more than 30 years ago to display an endogenous protein kinase activity that can phosphorylate HBc (1). Since HBV encodes no proteins with kinase capability, it has long been presumed that the virus encapsidates a kinase of cellular origin. PKC has been reported to be incorporated into HBV capsids (23, 25, 48, 64). However, other reports have argued that neither PKC, PKA, nor casein kinase II (CKII) is the endogenous kinase (12, 25, 26). The aforementioned 46-kDa serine kinase was also proposed to be packaged in HBV capsids (26), but no further identification or characterization has since been reported. Thus, there is as yet no agreement on the identity of the endogenous kinase. In addition, although it was reported early on that an S residue(s) was phosphorylated by the endogenous kinase (18), the precise site(s) of HBc phosphorylated by the endogenous kinase has never been defined.
The S/T-P motifs in the HBc and DHBc CTD suggest that a proline-directed serine-threonine kinase(s) (35, 44) could be at least partially responsible for the phosphorylation of these proteins, specifically at these sites. We now report that cyclin-dependent kinase 2 (CDK2), a member of a major class of proline-directed kinases, can indeed phosphorylate the HBV and DHBV CTD in vitro and in vivo and also that CDK2, or a CDK2-like kinase, represents a major kinase packaged into HBV capsids.
MATERIALS AND METHODS
Plasmids, antibodies, and chemicals.
pCMV-HBV directs the expression of the wild-type (WT) HBV pgRNA under the control of the cytomegalovirus (CMV) promoter (13). pCMV-HBV/pol− was derived from pCMV-HBV by a frameshift mutation in the RT open reading frame after codon 108 and is defective in RT expression (14). HBc coding sequences, either full-length (amino acids [aa] 1 to 183) or CTD deleted (1 to 149), were PCR amplified and inserted into the pET11d vector for bacterial expression. HBc coding sequences (1 to 183), either WT or containing phosphorylation site mutations, were amplified by PCR and inserted into the pCI vector (Promega) to make pCI-HBc for expression in mammalian cells. HBc CTD (HCC) coding sequences (from 141 to 183, WT or phosphorylation site mutants) were amplified by PCR from pCMV-HBV and inserted into pGEX-KT (21) or pEBG (61), downstream of the glutathione S-transferase (GST) coding sequences, for expression of the GST-HBc CTD (GST-HCC) fusion proteins in bacteria and mammalian cells, respectively. pSP64pA-Dcore contains the WT DHBc coding sequence, derived from pCMV-DHBV (55), downstream of the SP6 promoter in the pSP64pA vector (Promega). The S-P motif S245A, P246G, S259A, and P260G point mutants were obtained from J. Summers (68) and subcloned into the pSP64pA-Dcore background. pcDNA-Dcore, either WT or containing phosphorylation site mutations, was described previously (4). DHBV CTD coding sequences (from 196 or 229 to 262 or from 196 to 228, either WT or containing phosphorylation site mutations) were subcloned from the pcDNA-Dcore background to pGEX-KT (21) or pEBG (61), downstream of the GST coding sequences, for expression of the GST-DHBc CTD (GST-DCC) fusion proteins in bacteria and mammalian cells, respectively.
Rabbit polyclonal anti-CDK2 and goat polyclonal anti-SRPK antibodies were obtained from Santa Cruz, a rabbit polyclonal antibody against HBc was purchased from Dako, a rabbit polyclonal antibody against DHBc was provided by W. Mason (22), and goat anti-rabbit IgG horseradish peroxidase (HRP)-conjugated antibody (Southern Biotech) or rabbit anti-goat IgG HRP-conjugated antibody (Sigma) was used as the secondary antibody. The protein kinase inhibitors CDK2 inhibitor III, CDK4/6 inhibitor IV, cdc7/CDK9 inhibitor, Gö6976, Gö6983, olomoucine, PKCβ inhibitor, and roscovitine were obtained from Calbiochem, bisindolylmaleimide I (Bisindo) was obtained from Calbiochem and Cell Signaling, RO-3306 (CDK1 inhibitor) was obtained from Enzo Life Sciences, 5,6-dichloro-1-β-d-ribofuranosylbenzimidazole (DRB) was obtained from Enzo Life Sciences and Cayman Chemical, H-7 was obtained from Calbiochem and Tocris Biosciences, H-8 was obtained from Cayman Chemical, and H-89 was purchased from Sigma. All protein kinase inhibitors were dissolved in dimethyl sulfoxide (DMSO).
Cell line maintenance and transient transfection.
HepG2 cells (human hepatoblastoma) and HEK293T cells (human embryonic kidney) were cultured in Dulbecco's modified Eagle's medium (DMEM)-F12 supplemented with 10% fetal bovine serum (FBS) (HyClone) (4, 39). HepG2 cells were transfected with FuGENE 6 (39), and HEK293T cells were transfected by calcium phosphate precipitation (4).
Purification of HBV capsids.
HBV capsids were prepared as previously described by sucrose gradient ultracentrifugation of HepG2 cell lysates that had been transfected with pCMV-HBV or pCMV-HBV/pol− (39, 43). Recombinant HBc was expressed in E. coli BL21-CodonPlus(DE3) cells. Bacterial induction and lysate preparation were carried out as described previously (21). Purification of the capsids was then performed similarly to capsid purification from HepG2 and LMH cells by sucrose gradient ultracentrifugation (40).
Alternatively, crude HBV capsids (WT or containing phosphorylation site mutations) were isolated for the endogenous kinase reaction. HepG2 cells were transfected with pCI-HBc, either WT or an AAA phosphorylation site mutant. Seven days posttransfection, the cells were lysed with lysis buffer (50 mM Tris, pH 8.0, 150 mM NaCl, 1 mM EDTA, 1% Nonidet P-40 [NP-40]) supplemented with complete protease inhibitor cocktail. Cell debris was removed after centrifugation of the lysate, and capsids were precipitated with polyethylene glycol (PEG) similarly to the core DNA isolation procedure described previously (4). The PEG pellet was collected by centrifugation and resuspended in TNE (10 mM Tris, pH 8.0, 100 mM NaCl, 1 mM EDTA). The crude capsid preparation was treated with 0.5 U micrococcal nuclease and CaCl2 (5 mM) for 30 min. The nuclease was inactivated by the addition of EDTA (10 mM). Proteinase K (final concentration, 4 mg/ml) was added to the crude capsids, and the mix was then incubated for 1 h at 37°C to further remove cellular proteins not protected by the capsids. Phenylmethylsulfonyl fluoride (PMSF) (final concentration, 1 mM) (Roche) and EDTA-free protease inhibitor cocktail (Roche) were added following the proteinase K digestion. Endogenous kinase assays were then performed as described below except that 15 mM MgCl2 was used.
Protease protection assays.
HepG2 HBV capsids in sucrose fractions were digested for 1.5 h with proteinase K-agarose beads (Sigma) at 37°C with gentle agitation. Following the digestion, the proteinase K resin was removed by centrifugation. The supernatant was collected and used for endogenous kinase reactions or sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and Western blot analysis. For capsid particle analysis, proteinase K-treated capsids were loaded onto a 1% agarose gel. When proteinase K-treated capsids were resolved by SDS-PAGE, the proteinase K digestion was stopped by adding proteinase K inhibitor (Calbiochem) to 1 mM (final concentration) and incubating the sample at room temperature for 10 min (27). The samples were then boiled in SDS sample buffer and loaded onto a 15% SDS-PAGE gel.
Endogenous kinase assays.
Purified HepG2 HBV capsids (25 to 50 ng) were used in 20-μl reaction mixtures that included kinase reaction buffer (10 mM sodium phosphate, pH 7.0, 10 mM MgCl2, and 0.01% Brij 35) supplemented with fresh EDTA-free protease inhibitor cocktail (Roche), 1 mM dithiothreitol (DTT), and 5 μCi [γ32P]ATP (3,000 Ci/mmol, 10 mCi/ml; Perkin Elmer). Endogenous kinase reactions were also carried out under conditions described below for PKCα with essentially the same results obtained. In reactions that included kinase inhibitors, the inhibitor was added to the reaction before the substrates. The final concentration of DMSO in each reaction was limited to 1%, as was that of the solvent control. Kinase reaction mixtures were incubated at 37°C for 1 h (1, 18). One half (10 μl) of each reaction mixture was loaded on a 1% agarose gel. After electrophoresis, the gel was washed in distilled water (dH2O) for several hours to remove free isotope from the gel. The gel was stained with Sypro ruby protein stain (Sigma, Lonza) and destained in 10% methanol, 7% glacial acetic acid. Capsid particles were visualized on a Kodak gel image station. Radiolabeled capsids were subsequently detected by autoradiography following vacuum drying of the gel. Some reactions were also resolved by SDS-PAGE on 15% acrylamide gels and visualized with Sypro ruby staining and autoradiography. Endogenous kinase activities were quantified by phosphorimaging of 32P capsids on a Bio-Rad FX Pro Plus molecular imager with the QuantityOne 1-D analysis software program.
Exogenous kinase assays.
Recombinant E. coli-derived HBV full-length or CTD-truncated (1 to 149) capsids were prepared as described above. Fifty to one hundred nanograms recombinant capsids were subjected to an exogenous kinase reaction under conditions similar to those of the endogenous reaction except that 1 ng CDK2-cyclin E1 (Millipore) or 5 ng SRPK1 (SignalChem) was also added. In experiments using PKCα, reactions included 0.01 U PKCα (Promega), PKC buffer (20 mM HEPES, pH 7.4, 1.67 mM CaCl2, 10 mM MgCl2) supplemented with fresh EDTA-free protease inhibitor cocktail (Roche), 1 mM DTT, and 5 μCi [γ32P]ATP (3,000 Ci/mmol, 10 mCi/ml; Perkin Elmer) and were incubated for 30 min at 30°C according to the manufacturer's recommendations. Essentially the same results were also obtained with PKCα using the endogenous kinase reaction conditions above. When GST-DCC fusion proteins were used as substrates, 1 μg of substrate was used in a 20-μl reaction mixture, and 5 μl of the reaction mixture was separated by 12.5% SDS-PAGE. In reactions that included kinase inhibitors, the inhibitor was added to the reaction before the substrates. The final concentration of DMSO in each reaction mixture was limited to 1%, as was the solvent control. Gels were stained with Sypro ruby as described above, and phosphorylated proteins were detected by autoradiography.
Western blotting.
HBV capsids purified from HepG2 cells via sucrose gradient centrifugation, recombinant GST-tagged CDK2 (Santa Cruz), and untransfected total HepG2 cell lysate were resolved on a 15% SDS-PAGE gel. Proteins were transferred to polyvinylidene difluoride (PVDF) membrane and detected by incubation with rabbit polyclonal antibody against human CDK2 (1:1,000 dilution), followed by the goat antirabbit secondary antibody (1:20,000 dilution) and Western Lightning enhanced chemiluminescence reagent (Perkin Elmer). Samples were also probed with goat polyclonal antibody against human SRPK (1:200 dilution), followed by anti-goat secondary antibody (1:10,000 dilution) and SuperSignal West Pico chemiluminescent substrate (Pierce).
Purification and lysis of HBV virions.
HBV virions were prepared as described previously (40, 43). Briefly, virions were precipitated from the culture medium of transfected HepG2 cells with PEG. The precipitated virions were resuspended at a 50-fold concentration in TNE with 0.05% β-mercaptoethanol and then digested with DNase I (1 mg/ml) for 1 h at 37°C. The digestion was terminated by the addition of EDTA, and the digested virions were subjected to cesium gradient ultracentrifugation for 96 h, 50,000 rpm, at 4°C. The virions from the virion core protein peak fraction were PEG precipitated overnight for further concentration. The concentrated virions were lysed by resuspension in TNE containing 1% NP-40, 10 mM DTT, 1 mM PMSF, and the complete protease inhibitor cocktail (Roche). The lysed virions were treated with 0.5 μg/μl proteinase K for 1 h at 37°C in order to remove the viral envelope proteins and other contaminating proteins. The virion-derived capsids were then subjected to the endogenous kinase reaction as described above.
GST fusion protein expression, metabolic labeling, and purification.
GST fusion proteins were purified from bacteria as previously described (21), with the exception that BL21-CodonPlus(DE3) cells were used here. GST fusion proteins were purified from HEK293T cells as described previously (61) with minor modifications. Briefly, HEK293T cells in 60-mm dishes were transfected with GST fusion construct expression plasmids. On the third day posttransfection, cells were lysed in 1 ml lysis buffer, supplemented with complete protease inhibitor cocktail (Roche) and phosphatase inhibitors (10 mM NaF, 10 mM sodium pyrophosphate, 2 mM Na3VO4, and 50 mM β-glycerophosphate). Cell debris was removed after centrifugation of the lysate, and the cleared supernatant was treated with RNase A (67 μg/ml) for 1.5 h at room temperature with gentle agitation. The supernatants were removed from the precipitates and used for purification with glutathione (GSH) affinity resin. Bound proteins were eluted in 20 mM GSH (Sigma) in 200 mM Tris, pH 8.0, with phosphatase and protease inhibitors, analyzed by SDS-PAGE, and detected by protein staining.
The [32P]orthophosphate metabolic labeling procedure was carried out as described previously (68). Briefly, 2 days posttransfection, 60-mm dishes of cells were washed twice in 4 ml phosphate-free DMEM (Invitrogen). Cells were phosphate starved for 6 h in phosphate-free DMEM supplemented with 1× sodium pyruvate (Invitrogen) and 10% dialyzed FBS (HyClone) and then incubated with 100 μCi [32P]orthophosphate (9,000 Ci/mmol, 150 mCi/ml; Perkin Elmer) for 16 h. For experiments including inhibitor treatment, 3 days posttransfection, 35-mm dishes of HEK293T cells were washed twice with phosphate-free DMEM and phosphate starved for 6 h. During the final hour of phosphate starvation, the indicated kinase inhibitor was added. The cells were then incubated with ∼80 μCi [32P]orthophosphate (9,000 Ci/mmol, 2 mCi/ml; Perkin Elmer) for an additional hour. GST-tagged proteins were purified as described above.
In vitro translation in RRL.
DHBc was translated and [35S]methionine labeled in rabbit reticulocyte lysate (RRL), using the pSP64pA-Dcore or pcDNA-Dcore constructs, according to the manufacturer's recommendations (Promega). Translation products were treated with 1 U calf intestinal alkaline phosphatase (CIAP) (New England BioLabs) per μl reaction mixture for 16 h at 37°C, mock treated, or left untreated. Where indicated, DHBc was translated in the presence of protein kinase inhibitors. To make the CTD-deleted DHBc (lacking aa 229 to 262) in RRL, pSP64pA-Dcore was linearized at the XmnI site (aa 228) before it was used for in vitro transcription and translation.
RESULTS
HBV capsids made in human hepatoma cells displayed endogenous protein kinase activity that could be blocked by inhibitors of CDKs and phosphorylated the S-P sites in the HBc CTD.
We observed phosphorylation of HBV capsids purified from HepG2 cells in an in vitro kinase reaction with no exogenous kinase added (Fig. 1), indicating the presence of a kinase associated with the purified capsids, as reported by others using HBV capsids from infected human liver and hepatoma cells (1, 12, 18, 25). Furthermore, the endogenous kinase activity was associated with the capsids independent of the viral polymerase or pgRNA, since HepG2 capsids made in the absence of polymerase (Pol−) and consequently unable to package pgRNA were also phosphorylated (Fig. 1D, lane 1).
Fig 1.
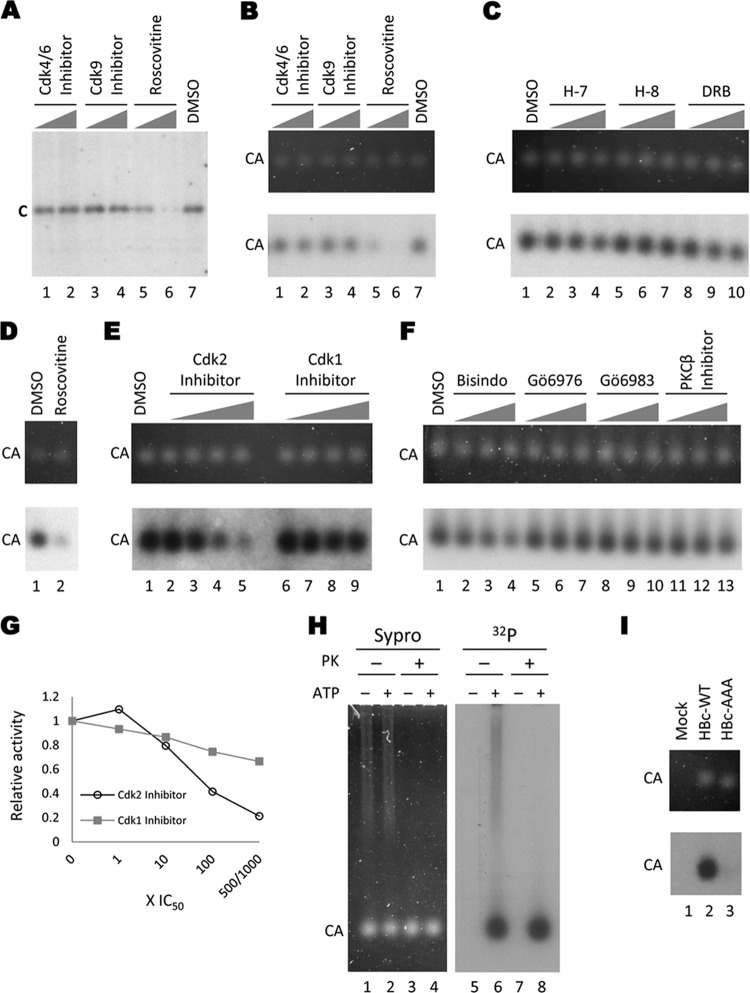
Endogenous kinase reactions with HepG2-derived HBV capsids. HBV capsids isolated from HepG2 cells transfected with pCMV-HBV (all lanes except in panels D and I) or pCMV-HBV/pol− (D) were phosphorylated by the encapsidated kinase in the in vitro endogenous kinase assay in the presence of the indicated kinase inhibitors. The reaction products were resolved by SDS-PAGE to visualize the total core protein (after disruption of the capsids by SDS) (A) or by agarose gel electrophoresis to visualize capsid particles (B to F). Core proteins were detected by autoradiography (A; B to F, bottom) or Sypro ruby staining (B to F, top). Kinase inhibitors used were CDK4/6 inhibitor (A and B [lane 1, 15 μM; lane 2, 150 μM]), CDK9 inhibitor (A and B [lane 3, 350 nM; lane 4, 3.5 μM]), roscovitine (A and B [lane 5, 10 μM; lane 6, 100 μM] and D [lane 2, 100 μM]), H-7 (C [lane 2, 37 μM; lane 3, 370 μM; lane 4, 750 μM]), H-8 (C [lane 5, 62 μM; lane 6, 470 μM; lane 7, 590 μM]), DRB (C [lane 8, 30 μM; lane 9, 200 μM; lane 10, 300 μM]), CDK2 inhibitor (E [lane 2, 0.5 μM; lane 3, 5 μM; lane 4, 50 μM; lane 5, 250 μM]), CDK1 inhibitor (E [lane 6, 35 nM; lane 7, 350 nM; lane 8, 3.5 μM; lane 9, 35 μM]), Bisindo (F [lane 2, 100 nM; lane 3, 1 μM; lane 4, 5 μM]), Gö6976 (F [lane 5, 62 nM; lane 6, 620 nM; lane 7, 3.1 μM]), Gö6983 (F [lane 8, 70 nM; lane 9, 700 nM; lane 10, 3.5 μM]), PKCβ inhibitor (F [lane 11, 210 nM; lane 12, 2.1 μM; lane 13, 10.5 μM]), or DMSO control (A and B [lane 7] and C to F [lane 1]). The graph in panel G shows the endogenous kinase activity in the presence of CDK2 inhibitor or CDK1 inhibitor at increasing concentrations relative to the DMSO control. Open circles, CDK2 inhibitor; filled squares, CDK1 inhibitor. (H) Protease protection. Sucrose gradient-purified HBV capsids from HepG2 cells were treated with proteinase K (lanes 3 and 4 and 7 and 8) or mock treated (lanes 1 and 2 and 5 and 6) and subjected to an endogenous kinase reaction (even-number lanes) or a control reaction with no ATP (odd-number lanes). The samples were resolved on an agarose gel and visualized by Sypro ruby staining (left panel) or autoradiography (right panel). (I) Endogenous kinase reactions with WT and mutant HBV capsids. WT (lane 2) or the phosphorylation site mutant (AAA) (lane 3) HBV capsids were harvested from transiently transfected HepG2 cells by PEG precipitation. The crude capsids were then treated with proteinase K before being subjected to the endogenous kinase reaction. A sample from mock-transfected cells was also included (lane 1). The reactions were resolved on an agarose gel; capsids were visualized by Sypro ruby staining (top panel), and labeling by the endogenous kinase was detected by autoradiography (bottom panel). C, core protein; CA, capsids; PK, proteinase K.
To identify the endogenous kinase(s), we attempted to block the endogenous kinase activity with chemical inhibitors specific for different kinases. We observed a dose-dependent decrease in phosphorylation of capsids, both WT and Pol−, treated with the broad-spectrum CDK inhibitor roscovitine (Fig. 1A and B, lanes 5 and 6, and D, lane 2; Table 1). Further, a CDK2-specific inhibitor also blocked endogenous kinase activity in a dose-dependent manner (Fig. 1E, lanes 2 to 5). In contrast, inhibitors against CDK4/6 and CDK9 showed little to no effect on endogenous phosphorylation of HepG2 capsids (Fig. 1A and B, lanes 1 to 4), nor did any of the multiple inhibitors of PKC tested (Fig. 1F, lanes 2 to 13).
Table 1.
Protein kinase inhibitors used in the endogenous kinase reaction and their known targets
| Inhibitor [reference] | Kinases | IC50 (Ki) | Effect (concn; %)a |
|---|---|---|---|
| Roscovitine (Seliciclib, CYC202) [36] | CDK1, CDK2, CDK5, CDK7, CDK9, CDK3 | 0.65 μM, 0.7 μM, 0.2 μM, 0.5–0.6 μM, 0.5 μM, 1.4 μM | 10 μM; 34 |
| Erk1 and -2 | 34 μM and 14 μM | 100 μM; 10 | |
| CDK4 and CDK6 | >100 μM | 250 μM; 5–7 | |
| Cdk2 inhibitor III (CVT-313) [7] | CDK2/A and -/E, CDK1 | 0.5 μM, 4.2 μM, | 5 μM; 80 |
| CDK4 | 215 μM | 50 μM; 42 | |
| MAPK, PKA, PKC | >1.25 mM | 250 μM; 21 | |
| Cdk1 inhibitor IV (RO-3306) [28, 58, 59] | CDK1/A, CDK1/B | 35 nM, 110 nM | 35 μM; 67 |
| CDK2 | 340 nM | ||
| CDK4 | >2 μM | ||
| Cdc7/Cdk9 Inhibitor (PHA-767491) [37] | cdc7, CDK9 | 10 nM, 34 nM | No inhibition at 100 μM |
| Cdk4/6 inhibitor IV | CDK4, CDK6, CDK5 | 1.5 μM, 5.6 μM, 25 μM | No inhibition at 200 μM |
| (CINK4) [54] | CDK1, CDK2/A or/E | >100 μM, >50 μM | |
| DRB [49, 69] | Casein kinase II | 6 μM | No inhibition at 300 μM |
| CDK9, CDK7, CDK8 | 3 μM, ∼20 μM, ∼20 μM | ||
| H-7 [20, 49] | PKA, PKC, PKG | (3 μM), (6 μM), (5.8 μM) | No inhibition at 750 μM |
| CDK8, CDK7 | 3.7 μM, 37 μM | ||
| H-8 [49] | CDK7 | 6.2 μM | No inhibition at 590 μM |
| CDK8 | 47 μM | ||
| Bisindo [57] | PKCα, -β, -γ | 10–20 nM | 5 μM; 69 |
| PKCδ, -ε | 100–200 nM | ||
| PKCζ | 6 μM | ||
| Gö6976 [47] | PKCα, PKCβI | 2.3 nM, 6.2 nM | No inhibition at 3.1 μM |
| PKCμ | 20 nM | ||
| Gö6983 [19] | PKCα, PKCβ | 7 nM | No inhibition at 3.5 μM |
| PKCγ, -δ, -ζ | 6 nM, 10 nM, 60 nM | ||
| PKCμ | 20 μM | ||
| PKCβ inhibitor [56] | PKCβII, -βI | 5 nM, 21 nM | No inhibition at 10.5 μM |
| PKCα, -γ, -ε | 331 nM, >1 μM, 2.8 μM |
Effect on the endogenous kinase reaction is expressed as the residual activity remaining in the presence of the indicated inhibitors as a percentage of the solvent (DMSO) control.
In order to further narrow down the possible CDK candidates, we tested additional inhibitors specific for CDK7 and CDK8: DRB, H-7, and H-8. We saw no inhibition of endogenous kinase activity by H-7 (Fig. 1C, lanes 2 to 4) even at concentrations 200-fold above the 50% inhibitory concentration (IC50) for CDK8 and more than 20-fold above the IC50 for CDK7. Similarly, concentrations of H-8 100-fold above the IC50 for CDK7 and more than 10-fold above the IC50 for CDK8 showed no effect on the endogenous kinase activity of HepG2 capsids (Fig. 1C, lanes 5 to 7). Furthermore, concentrations of DRB greater than 15-fold above the IC50 for CDK7 and CDK8 were also unable to inhibit the endogenous kinase activity of HepG2 capsids (Fig. 1C, lane 8 to 10). Taken together, these results suggested that CDK7 or CDK8 was unlikely to be the endogenous kinase. In addition, the lack of effect of H-7 suggested that PKA, PKC, or protein kinase G (PKG) was unlikely to be the endogenous kinase, and the lack of a DRB effect suggested that CKII and CDK9 were unlikely candidates (Table 1), in agreement with previous reports (12, 25, 26) and the results above obtained with specific PKC inhibitors and the CDK9 inhibitor.
Because relatively high concentrations of both roscovitine and the CDK2 inhibitor, which can inhibit CDK2 as well as CDK1, were necessary to efficiently block endogenous kinase activity, it remained possible that CDK1, in addition to or other than CDK2, was the packaged kinase. Therefore, we tested a specific inhibitor of CDK1, RO-3306, which has an approximately 10-fold greater potency against CDK1 than against CDK2. Increasing concentrations of the CDK1 inhibitor, up to 1,000-fold above the IC50 for CDK1, effected little decrease in endogenous kinase activity of HepG2 capsids (Fig. 1E, lanes 6 to 9, and G; Table 1). The slight inhibition of endogenous kinase activity at the highest concentration of CDK1 inhibitor likely reflected its inhibition of CDK2 at the high concentration. These results thus strongly suggest that CDK2 but not CDK1 was packaged in the capsids. The observed inhibitory effects of the CDK inhibitors (particularly the CDK2 inhibitor) on the endogenous kinase and the lack of inhibitory effects by a variety of other kinase inhibitors at similarly high or much higher concentrations (up to 10,000-fold) relative to their respective IC50s (Tables 1 and 2) indicated strongly that the observed inhibitory effects were specific. Although excess (100-fold above their IC50s or more) roscovitine and CDK2 inhibitor III were needed to show a strong effect on the endogenous kinase, both did start to show inhibitory effects at concentrations ca. 10-fold above their IC50s for CDK2. The relatively high concentrations of roscovitine and the CDK2 inhibitor needed to show a strong effect on the endogenous kinase were likely due to the fact that the kinase(s) packaged inside the capsids might not be as accessible as purified kinases, against which the IC50s of the various inhibitors are usually measured (Fig. 2).
Table 2.
Targeting one protein kinase with multiple inhibitors in the endogenous kinase reaction
| Kinase | Inhibitorsa |
|---|---|
| CDK2 | CDK2 inhibitor III |
| Roscovitine | |
| CDK1 inhibitor IV | |
| CDK4/6 inhibitor IV | |
| CDK1 | CDK1 inhibitor IV |
| Roscovitine | |
| CDK2 inhibitor III | |
| CDK4/6 inhibitor | |
| CDK7 | Roscovitine |
| DRB | |
| H-7 | |
| H-8 | |
| CDK8 | DRB |
| H-7 | |
| H-8 | |
| CDK9 | Roscovitine |
| cdc7/CDK9 inhibitor | |
| DRB | |
| PKC (multiple isoforms) | H-7 |
| Bisindo | |
| Gö6976 | |
| Gö6983 | |
| PKCβ inhibitor |
Only inhibitors denoted in boldface showed inhibitory effect on the endogenous kinase.
Fig 2.
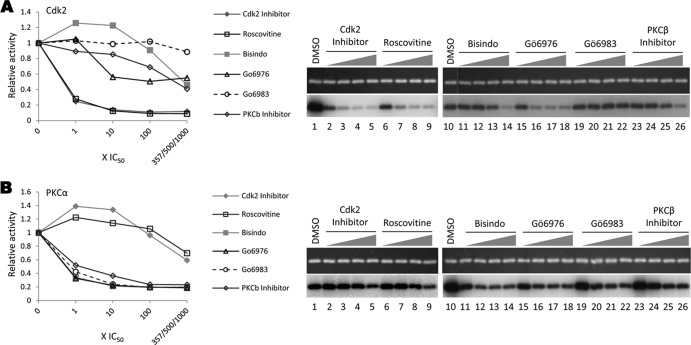
Activities of chemical inhibitors against purified kinases. In vitro kinase reactions were performed using GST-DCC3 as the substrate with CDK2 (A) or PKCα (B). Inhibitors tested were CDK2 inhibitor III (lanes 2 to 5) at concentrations 1×, 10×, 100×, and 500× IC50 for CDK2 (Table 1), roscovitine (lanes 6 to 9) at concentrations 1×, 10×, 100×, and 357× IC50 for CDK2, Bisindo (lanes 11 to 14) at concentrations 1×, 10×, 100×, and 1,000× IC50 for PKCα/β/γ, Gö6976 (lanes 15 to 18) at concentrations 1×, 10×, 100×, and 1,000× IC50 for PKCβ1, Gö6983 (lanes 19 to 22) at concentrations 1×, 10×, 100×, and 1,000× IC50 for PKCα/β, PKCβ inhibitor Gö6976 (lanes 23 to 26) at concentrations 1×, 10×, 100×, and 1,000× IC50 for PKCβ1, or DMSO control (lanes 1 and 10). The reactions were resolved by SDS-PAGE. Protein loading was visualized by Sypro ruby staining (top panels), and kinase activity was detected by autoradiography (bottom panels).
In addition, we employed multiple inhibitors against a given kinase (Tables 1 and 2) to help exclude off-target effects and multiple batches (and multiple suppliers in some cases; see Materials and Methods) of the same compound to ensure their functionality. To further verify the activities and specificities of the kinase inhibitors used, we performed exogenous kinase assays using DCC229 (see Fig. 8) as the substrate and tested the relevant inhibitors against purified CDK2 or PKC, the kinase most implicated as an endogenous kinase by previous reports (see the introduction). Both CDK2 inhibitor III and roscovitine inhibited CDK2 activity in a dose-dependent manner, as expected (Fig. 2A, lanes 2 to 9). Also as expected, none of the PKC inhibitors significantly blocked CDK2 activity (Fig. 2A, lanes 11 to 26). All the PKC inhibitors, none of which affected endogenous kinase activity, did result in dose-dependent inhibition of PKCα, whereas CDK2 inhibitor III and roscovitine, which potently suppressed the endogenous kinase, were ineffective against PKCα even at high concentrations (Fig. 2B). In addition, we also tested the CDK4/6 inhibitor, which was ineffective at inhibiting the endogenous kinase, against purified CDK4 and verified that the inhibitor was active (data not shown).
Fig 8.
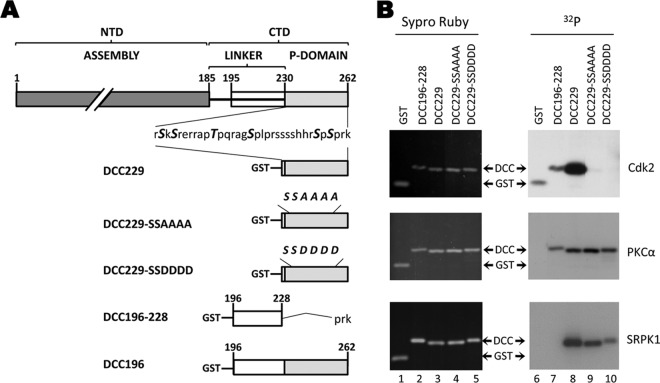
Phosphorylation of GST-DCC fusion proteins by purified kinases in vitro. (A) GST-DCC fusion constructs. Shown at the top is the domain structure of DHBc with the N-terminal assembly domain (NTD) and CTD indicated. The CTD phospho domain sequence (229 to 262) is shown below, with the 6 known phosphorylation sites highlighted in italics and bold. Below are diagrams of GST-DCC fusion proteins. DCC229 contains amino acids 229 to 262 (WT sequences) fused to GST. DCC229-SSAAAA contains the same portion of the DHBc CTD with alanine substitutions in the S/T sites (239, 245, 257, and 259). DCC229-SSDDDD contains aspartic acid substitutions at these same S/T sites. DCC196-228 contains the linker region (amino acids 196 to 228) with the phospho domain deleted, and DCC196 contains the linker region plus the phospho domain from 196 to 262 with WT sequences. Exogenous kinase reactions were performed using GST-DCC fusion proteins purified from bacteria as substrates with CDK2-cyclin E1, PKCα, or SRPK1, as indicated (B). DCC and GST-DCC fusion proteins are shown.
The endogenous kinase activity seen in our studies and by others (1, 18, 25, 30) could be due to a kinase that either cofractionates with the capsids in the sucrose gradient or is tightly associated with the exterior of the capsid. To test these possibilities, we performed proteinase K digestions of the capsid fractions followed by the endogenous kinase reaction. Proteinase K treatment of a capsid fraction (a less-pure fraction containing appreciable amounts of contaminating proteins was deliberately selected for this purpose) resulted in the loss of the contaminating proteins (the smear above the capsid band) (Fig. 1H, lanes 1 and 2 and 5 and 6 versus lanes 3 and 4 and 7 and 8), verifying the effectiveness of protease digestion. Following the endogenous kinase reaction, the labeled contaminating species were also eliminated by proteinase K, but neither the amount of capsids nor their labeling was affected. These results indicate that the endogenous kinase was indeed protected within the capsid.
We were next interested in determining if the endogenous kinase phosphorylated the S-P sites in the HBc CTD. We thus isolated from HepG2 cells HBV capsids, either the WT or the AAA mutant that had the three S residues in the S-P motifs replaced by A residues (see Fig. 5A), and conducted the endogenous kinase reactions. As shown in Fig. 1I, the AAA mutant capsid (lane 3) showed drastically reduced levels of endogenous kinase activity compared to the WT (lane 2), suggesting that the endogenous kinase phosphorylated mostly the S-P sites in the HBc CTD, although additional sites in HBc might also be weakly phosphorylated.
Fig 5.
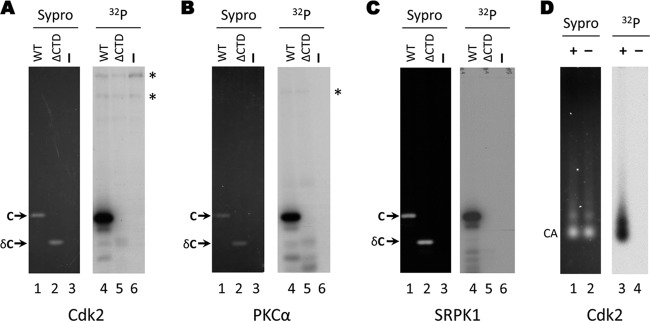
Phosphorylation of E. coli-derived HBV capsids by purified kinases in vitro. Exogenous kinase assays using CDK2-cyclin E1 (A), PKCα (B), or SRPK1 (C) were conducted with recombinant HBV capsids purified from bacteria. The products were resolved by SDS-PAGE (lanes 1 to 6) and visualized by autoradiography (lanes 1 to 3) or Sypro ruby staining (lanes 4 to 6). Lanes 1 and 4, full-length E. coli-derived capsid (WT; 1 to 183); lanes 2 and 5, E. coli-derived truncated capsid lacking the CTD phospho domain (ΔCTD; 1 to 149). Lanes 3 and 6, kinase assay containing no added substrate. (D) Exogenous CDK2-cyclin E1 kinase assays were conducted with E. coli-derived HBV capsids. The reaction products were resolved by agarose gel electrophoresis and visualized by Sypro ruby staining (lanes 1 and 2) or autoradiography (lanes 3 and 4). C, core protein; δC, ΔCTD, truncated core protein, 1 to 149; CA, capsids; *, background bands originating from the kinase preparation.
Detection of the endogenous protein kinase in HBV capsids by anti-CDK2 specific antibody.
To determine if a packaged kinase in HBV capsids was indeed CDK2, capsids purified from HepG2 cells by sucrose gradient centrifugation, with or without prior proteinase K digestion, were resolved by SDS-PAGE, and the presence of CDK2 in the capsids was detected by Western blotting using a specific antibody against CDK2 (Fig. 3). To verify the effectiveness of protease digestion, a capsid-negative fraction and a relatively impure capsid-positive fraction were selected. It was apparent that variable amounts of CDK2 migrated into the sucrose gradient and were detectable in the capsid-negative fraction (lane 5). However, the CDK2 signal in this fraction was completely removed by protease digestion (lane 6). Most of the CDK2 signal in the low-purity capsid fraction, which was evidenced by the cross-reactive bands above the CDK2 signal and total protein staining (not shown), was also eliminated by protease digestion (lane 8 versus lane 7), suggesting that most of the CDK2 in this relatively impure fraction was simply cofractionating with the capsids or perhaps was associated with the surface of the capsids. Importantly, some CDK2 remained clearly detectable in the fraction containing the capsids (lane 8), in contrast to the capsid-negative fraction (lane 6). Furthermore, with a highly purified capsid fraction (as evidenced by the lack of cross-reactive bands as well as total protein staining [data not shown]), it was clear that virtually all the CDK2 signal associated with the capsids was protected from protease digestion (lanes 9 and 10). Based on comparison with the CDK2 standards, we estimated that ca. 2 ng CDK2 (protected from protease digestion) was detected in ca. 500 ng capsids. Thus, protease-resistant CDK2 was clearly detectable by Western blotting in the HBV capsids, indicating that CDK2 was indeed packaged within HBV capsids, as further suggested by the kinase inhibitor results above.
Fig 3.
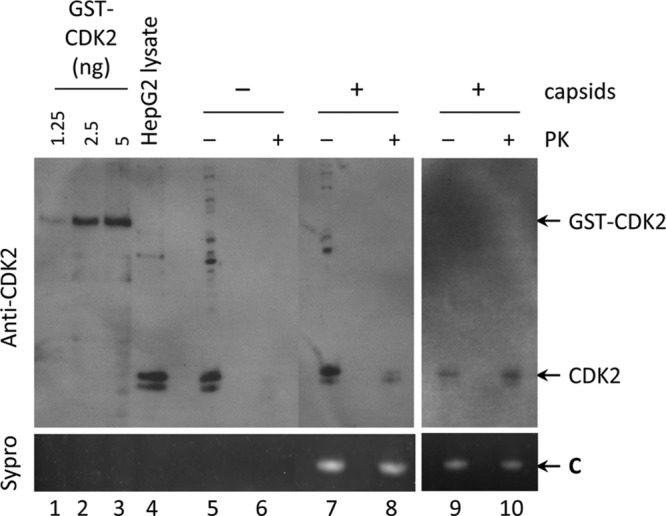
Detection of CDK2 in HBV capsids purified from HepG2 cells by Western blotting. HBV capsids (0.5 μg) were digested with proteinase K (PK) agarose beads as described in Materials and Methods. Proteinase K was then inactivated by the addition of the proteinase K inhibitor, and the sample was resolved by SDS-PAGE (lanes 8 and 10), along with the same amount of undigested capsids from the same fractions (lanes 7 and 9). Purified GST-CDK2 standards (lanes 1 to 3) and total lysate from HepG2 cells (lane 4) were loaded as controls for CDK2 detection. A sucrose fraction that did not contain HBV capsids was also treated with the proteinase K agarose beads to ensure that contaminating proteins were removed by the digestion (lane 6) or mock treated (lane 5). The capsids loaded in lanes 9 and 10 were from a capsid-positive sucrose fraction of higher purity than those loaded in lanes 7 and 8. CDK2 was detected by Western blotting using the anti-CDK2 antibody (top panel). HBc was visualized by Sypro ruby staining (bottom panel). C, core protein.
We also attempted to detect SRPK1 and -2 in the purified HBV capsids under the same conditions described for the detection of CDK2. We were unable to detect any SRPK from the proteinase K-treated capsids (data not shown). However, due to the relatively low sensitivity of the anti-SRPK antibody, a copy number of one kinase per capsid or lower would have escaped detection.
HBV capsids isolated from virions displayed an endogenous protein kinase activity that could also be blocked by CDK inhibitors.
To ascertain the relationship between the endogenous kinase activity detected in cytoplasmic HBV capsids and what was detected early on in secreted HBV virions, we purified HBV virions from transfected HepG2 cell culture medium, removed their envelope, and then either mock treated the released capsids or treated them with proteinase K to remove the viral envelope proteins and other contaminants. We then performed endogenous kinase reactions with these virion-derived capsids. Without the proteinase K treatment, the virion-derived capsids did display an endogenous kinase activity that phosphorylated HBc (Fig. 4, lane 1). However, it was also clear that additional proteins were also being phosphorylated in these reactions, as reported by others (1). These other proteins were mostly removed by the proteinase K digestion (Fig. 4, lane 2), suggesting that they were outside the capsids. Upon protease treatment, the major protein labeled by the endogenous kinase was the HBc protein (Fig. 4, lane 2). Notably, the endogenous kinase activity of the virion-derived capsids was also sensitive to the same inhibitors that inhibited the kinase activity detected in the cytoplasmic capsids, including the CDK2 inhibitor and roscovitine (Fig. 4, lane 2 versus lanes 3 and 4). In contrast, the endogenous kinase in the virions was relatively insensitive to Bisindo, a potent inhibitor of PKC, just as that in the cytoplasmic capsids was (Fig. 4, lane 5). In addition to HBc, there was another weakly labeled species (ca. 50 kDa) that appeared to be resistant to protease digestion and was not affected by the kinase inhibitors. This species likely represented a contaminating protein that was phosphorylated from outside the capsid but was not completely eliminated by proteinase K, since it disappeared upon more extensive proteinase K digestion (data not shown) and we never detected this species in cytoplasmic capsids. Together, these results suggest that the endogenous kinase activity observed in virion-derived capsid particles was similar if not identical to that seen in cytoplasmic capsids.
Fig 4.
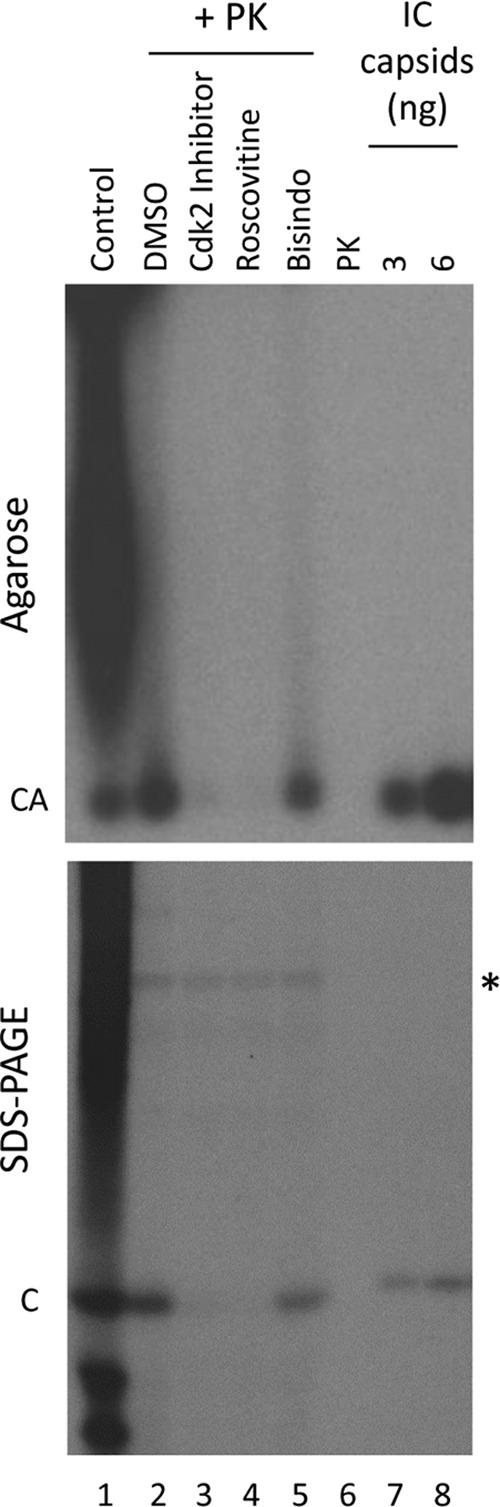
Endogenous kinase reactions with virion-derived HBV capsids. HBV capsids were released from virions purified from the medium of HBV-transfected HepG2 cells by NP-40 treatment. One control aliquot was removed, and the remaining virion-derived capsids were digested by proteinase K. The untreated (lane 1) or proteinase K-treated (lanes 2 to 5), virion-derived capsids (ca. 6 ng per reaction) were phosphorylated by the encapsidated kinase in endogenous kinase reactions in the presence of the indicated kinase inhibitors (lanes 3 to 5) or DMSO control (lane 2). Kinase inhibitors used included CDK2 inhibitor III (lane 3, 250 μM, 500× IC50 for CDK2), roscovitine (lane 4, 250 μM, 350× IC50 for CDK2), and Bisindo (lane 3, 5 μM, 500× IC50 for PKC). An equal amount of proteinase K alone was also loaded as an additional control (lane 6). In parallel, 6 ng (lane 7) or 12 ng (lane 8) of cytoplasmic HBV capsids from HepG2 cells (also treated with proteinase K; see Materials and Methods) were subjected to the endogenous kinase reaction. Half of each reaction product was resolved by agarose gel electrophoresis to visualize capsid particles (top), and proteinase K inhibitor was added to the other half to terminate the digestion before boiling in SDS sample buffer and resolving by SDS-PAGE to visualize the total core protein (bottom). The gels were dried, and labeled capsids or core proteins were detected by autoradiography. *, unknown labeled species. CA, capsids; C, core protein; PK, proteinase K; IC, intracellular.
Phosphorylation of HBV capsids purified from bacteria by CDK2 in vitro.
We next asked if CDK2 could phosphorylate HBV capsids purified from E. coli. CDK2-cyclin E1 was able to phosphorylate the HBV full-length capsids (Fig. 5A, lanes 1 and 4). Truncated HBV capsids (amino acids 1 to 149; ΔCTD) lacking the CTD were not phosphorylated by the CDK in vitro (Fig. 5A, lanes 2 and 5), indicating that phosphorylation occurred on the CTD of full-length HBc. Similarly, PKCα and SRPK1 also phosphorylated the full-length and not the truncated HBV capsids (Fig. 5B and C, lanes 1 and 4 versus lanes 2 and 5), again suggesting that phosphorylation occurred on the CTD. Furthermore, HBV capsids remained intact after the kinase reaction, migrating as a single distinct species on an agarose gel with the same mobility as unphosphorylated capsids (Fig. 5D, lanes 1 to 4, and data not shown). These results indicated that the HBc CTD of intact capsids assembled in bacteria was accessible to the exogenous kinases.
Phosphorylation of isolated HBc CTDs by purified CDK2 in vitro.
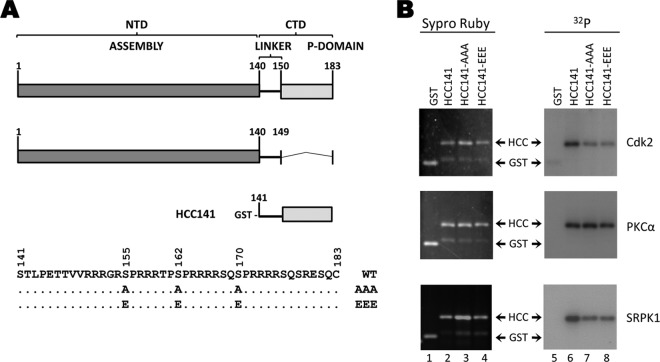
We then tested whether these kinases would preferentially phosphorylate the S-P motifs in the HBc CTD (Fig. 6A). Therefore, we used the GST-HBc CTD (GST-HCC) fusion proteins, WT and phosphorylation site mutants, that were purified from bacteria and tested the ability of CDK2-cyclin E1, PKCα, and SRPK1 to phosphorylate the fusion proteins. CDK2 phosphorylated the WT HCC fusion protein more efficiently than the S-P site mutants, although lower levels of phosphorylation were observed on both of the phosphorylation site mutants HCC141-AAA and -EEE (Fig. 6B, top row, lane 6 versus lanes 7 and 8). These results indicated that this proline-directed kinase could indeed phosphorylate the HBc CTD S-P sites, although it could also phosphorylate, to some extent, non-S-P sites under in vitro conditions. Similar to CDK2, SRPK1 also phosphorylated the WT HCC fusion protein to a greater extent than the phosphorylation site mutants (Fig. 6B, bottom row, lane 6 versus lanes 7 and 8), consistent with the previous report (10). On the other hand, PKCα phosphorylated all of the HCC constructs to a similar extent (Fig. 6B, middle row, lanes 6 to 8), indicating that, unlike CDK2, PKCα phosphorylated sites other than the S/T-P motifs.
Fig 6.
Phosphorylation of GST-HCC fusion proteins by purified kinases in vitro. (A) HBc constructs. Shown at the top is the domain structure of HBc with the N-terminal assembly domain (NTD) and CTD indicated. Below are diagrams of the HBcΔCTD protein, containing amino acids 1 to 149, and the GST-HCC fusion proteins, containing amino acids 141 to 183 fused to GST. The CTD sequence (aa 141 to 183) is shown below, with the 3 known phosphorylation sites indicated at amino acid positions S155, S162, and S170. HCC141 contains amino acids 141 to 183 (WT sequences) fused to GST. HCC141-AAA contains the same portion of the HBc CTD with alanine substitutions in the phosphorylation sites (155, 162, and 170). HCC141-EEE contains glutamic acid substitutions at these same sites. Exogenous kinase reactions were carried out using GST-HCC fusion proteins purified from bacteria as substrates with CDK2-cyclin E1, PKCα, or SRPK1, as indicated (B). The reactions were resolved by SDS-PAGE and visualized with Sypro ruby protein stain (lanes 1 to 4) and autoradiography (lanes 5 to 8). HCC, GST-HCC fusion proteins.
Inhibition of HBc and DHBc CTD phosphorylation in vivo by CDK inhibitors.
To obtain evidence for a role of CDK2 in CTD phosphorylation in vivo, we decided to treat HEK293T cells expressing the HBc or DHBc CTDs with roscovitine and the CDK2 inhibitor. HEK293T cells were chosen for their high transfection efficiency, which made it possible to readily visualize the purified CTDs both by total protein staining and by metabolic labeling. It is also important to note that HEK293T cells support efficient HBV and DHBV RNA packaging and DNA synthesis, and the DHBc CTD S/T-P site mutants display the same phenotypes in these cells as in LMH cells (33, 50, 61). Furthermore, we found that HBV capsids assembled in HEK293T cells also packaged a host kinase that was sensitive to the CDK2 inhibitor, as in HepG2 cells (data not shown). We thus expressed GST-HCC141, which contains the HBc CTD (141 to 183) fused to GST, or GST alone in HEK293T cells. The proteins were metabolically labeled for 2 h with [32P]orthophosphate, and kinase inhibitors were added during the second hour of labeling. We reasoned that this brief treatment would not cause excessive cytotoxicity or pleiotropic effects even with relatively high inhibitor concentrations but could increase the likelihood of revealing a role for CDK2 in CTD phosphorylation in cells. Indeed, we found that treatment of the cells with either roscovitine or the CDK2 inhibitor clearly decreased HCC phosphorylation (Fig. 7B, lanes 4 to 6) without affecting the HCC protein levels (Fig. 7A, lanes 4 to 6).
Fig 7.
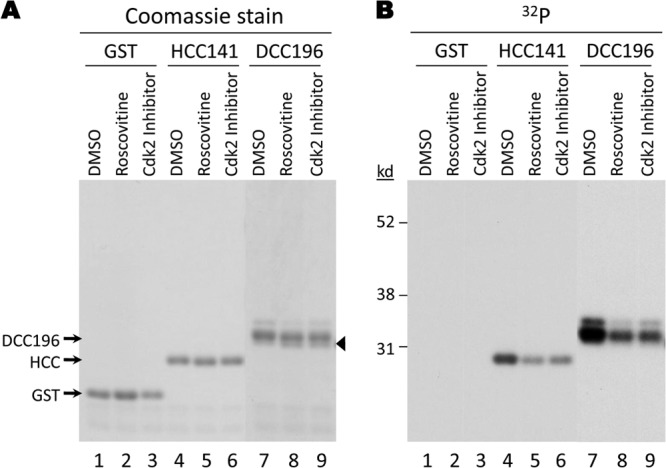
Effect of CDK inhibition on phosphorylation of GST-HCC and GST-DCC fusion proteins in HEK293T cells. HEK293T cells were transfected with plasmids to express GST-HCC141 (HBc CTD), GST-DCC196 (DHBc CTD), or GST. Three days posttransfection, the cells were labeled with [32P]orthophosphate in the absence (DMSO) (A and B, lanes 1, 4, and 7) or presence of the indicated kinase inhibitors (roscovitine, 200 μM [A and B, lanes 2, 5, and 8], or CDK2 inhibitor III, 200 μM [A and B, lanes 3, 6, and 9]). GST fusion proteins were purified with GSH affinity resin. The eluted 32P-labeled proteins were resolved by SDS-PAGE and visualized by Coomassie blue staining (A) or autoradiography (B). The fastest-migrating, presumably least-phosphorylated or nonphosphorylated, species enhanced by inhibitor treatment is indicated by the arrowhead.
Encouraged by these results, we also decided to test the effect of CDK2 inhibition on the phosphorylation in vivo of the DHBc CTD. Thus, we expressed GST-DCC196, which contains the DHBc CTD (196 to 262) fused to GST, in the HEK293T cells and metabolically labeled the protein using the same procedure as we did with the HCC protein above. An advantage of the DCC protein, relative to HCC, for these studies was the fact that DCC196 migrated as several bands (Fig. 7A and B, lane 7), indicative of several heterogeneously phosphorylated species at the S/T-P sites, as described earlier (33), which provided a simple means of monitoring in vivo phosphorylation of the DHBc CTD specifically at the S/T-P sites. Roscovitine and the CDK2 inhibitor affected the DCC196 fusion protein similarly in that both treatments caused a downward shift in the migrational heterogeneity, as well as an overall reduction of phosphate labeling. Clearly, the slower, hyperphosphorylated species were decreased after inhibitor treatment as judged by both protein staining and phosphate labeling (Fig. 7A and B, lanes 8 and 9). Further, both inhibitors increased the levels of the fastest-migrating, hypo- or unphosphorylated species (Fig. 7A, lanes 8 and 9). This downward mobility shift of DCC caused by the CDK inhibitors strongly indicated that the inhibitors in fact blocked CTD phosphorylation at the S/T-P sites. Importantly, as observed in the endogenous kinase reactions, the CDK2 inhibitor behaved similarly to roscovitine, suggesting that both inhibitors were targeting the same cellular kinase(s), likely CDK2.
Phosphorylation of isolated DHBc CTDs by purified CDK2 in vitro.
The above results indicated that the S/T-P sites in the DHBc CTD, like those in the HBc CTD, were subject to phosphorylation in vivo by the host CDK2. To determine if CDK2 could phosphorylate DHBc CTD in vitro, GST-DCC fusion proteins were expressed in and purified from bacteria and subjected to in vitro kinase assays with purified CDK2. These fusion proteins contained either WT sequences or S/T to -A or -D substitutions in the four S/T-P sites (Fig. 8A). When expressed in mammalian or avian cells or in the RRL, the isolated CTD was found to be phosphorylated (33), displaying the characteristic heterogeneity in migration similar to that of the full-length protein (4, 33). The phospho site mutants, DCC229-SSAAAA and DCC229-SSDDDD, retained the SK/SR phosphorylation sites at 230 and 232 and were also phosphorylated in cells but to a much lesser degree and migrated as a single fast species (33), which suggested that the CTD itself could serve as an appropriate substrate for identifying the kinase(s) that phosphorylates the S/T-P motifs. Therefore, we asked if CDK2 could phosphorylate the GST-DCC fusion proteins purified from bacteria, which are unphosphorylated. When the same GST-DCC constructs purified from bacteria were used in the in vitro reactions with purified kinases, we found that CDK2-cyclin E1 was indeed able to phosphorylate the WT CTDs (Fig. 8B, top row, lane 8). We observed only minimal labeling of GST itself or DCC196-228, containing the linker region (Fig. 8B, top row, lanes 6 and 7), both of which contain one S-P motif in GST itself. Interestingly, the in vitro phosphorylated WT fusion protein (Fig. 8B) did not display the characteristic up-shifted, multibanding pattern that was observed when the same fusion protein was expressed and phosphorylated in cells or in the RRL (Fig. 9) (33). Also, DCC229-SSAAAA and DCC229-SSDDDD showed little to no phosphorylation, indicating that the S/T-P sites but not the upstream SK/SR sites were phosphorylated by CDK2 in vitro (Fig. 8B, top row, lanes 9 and 10), indicating a high degree of selectivity of CDK2 for the S/T-P sites in the DHBc CTD in vitro. Conversely, PKCα phosphorylated all of the CTD-containing constructs, with the S/T-P site mutants phosphorylated as efficiently as the WT CTD (Fig. 8B, middle row, lanes 8 to 10), reminiscent of the results seen with the HCC fusion proteins. Also, the upstream linker region (DCC196-228), containing multiple T residues but no S/T-P motifs, was phosphorylated by PKCα to almost the same degree as the downstream phospho domain containing the four S/T-P sites (Fig. 8B, middle row, lane 7). Thus, although PKCα could phosphorylate the DHBc CTD, it did not phosphorylate the S/T-P sites, consistent with the known bias of PKC against S/T-P sites (10, 41). In addition, we showed that SRPK1 could phosphorylate the DHBc CTD (Fig. 8B, bottom row, lane 8), as it did the HBc CTD. However, SRPK1 also phosphorylated the DHBc CTD when its S/T-P sites were mutated (Fig. 8B, bottom row, lanes 9 and 10), indicating that SRPK1 did not display the same level of selectivity as CDK2 for the S/T-P sites.
Fig 9.
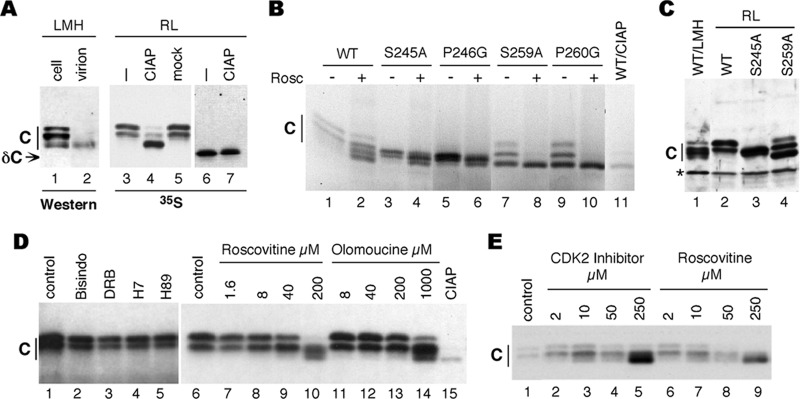
Phosphorylation of DHBc in RRL. (A) Left panel, Western blot analysis of DHBc, using the anti-DHBc antibody, from transfected LMH cell lysate (lane 1) or isolated from virions (lane 2). Middle and right panels, DHBc, full length (lanes 3 to 5) or CTD truncated (1 to 228; lanes 6 and7), was translated and 35S labeled in RRL (RL), treated with calf intestinal alkaline phosphatase (CIAP) (lanes 4 and 7), mock treated (lane 5), or left untreated (lanes 3 and 6), resolved by SDS-PAGE, and detected by autoradiography. (B) Individual alanine substitutions of DHBc at the indicated serine residues or glycine substitutions at the adjacent proline residues were translated along with the WT core protein. Roscovitine (200 μM) was added to the even-number reactions. CIAP-treated WT translation served as a control of dephosphorylated core (lane 11). (C) Western blot analysis of DHBc expressed in LMH cells versus expression in RRL. WT DHBc expressed in LMH cells (lane 1) or in RRL (lane 2) and the S245A and S259A mutants expressed in RRL were resolved by SDS-PAGE and detected by Western blotting using the anti-DHBc antibody. (D) 35S-labeled DHBc was translated in the absence or presence of the indicated protein kinase inhibitors. Lanes 1 and 6, solvent control (DMSO); lane 2, Bisindo (10 μM, 1,000× IC50 for PKC) (57); lane 3, DRB (500 μM, 83× IC50 for CKII) (69); lane 4, H-7 (1,000 μM, over 300× Ki for PKA and over 150× Ki for PKC) (20); lane 5, H-89 (100 μM, 740× IC50 for PKA) (11); lanes 7 to 10, roscovitine (2× to 286× IC50 for CDK1 and CDK2) (36); lanes 11 to 14, olomoucine (1× to 143× IC50 for CDK1 and CDK2) (2, 60). A CIAP-treated reaction mixture served as a control for the dephosphorylated core protein (lane 15). (E) 35S-labeled DHBc was translated in the absence or presence of the indicated protein kinase inhibitors. Lane 1, solvent control (DMSO); lanes 2 to 5, CDK2 inhibitor III (4× to 500× IC50 for CDK2) (7); lanes 6 to 9, roscovitine (3× to 357× IC50 for CDK1 and CDK2). C, DHBc; δC, CTD-truncated DHBc; *, cross-reacting cellular protein.
Phosphorylation of DHBc at the CTD in cell lysate that was blocked by CDK inhibitors.
In our attempts to develop a cell-free system to analyze core protein phosphorylation, we noticed that DHBc, when translated in RRL, migrated as two major species, comigrating with the hyperphosphorylated core species expressed in LMH cells (Fig. 9A, lanes 1 and 3, and C, lanes 1 and 2). The core protein isolated from DHBV virions is known to be completely unphosphorylated (43, 46) and migrates as a single band, faster than those made in RRL (Fig. 9A, lane 2). The core protein species made in RRL were converted to one major, faster-migrating species, comigrating with the virion core protein, upon CIAP treatment (Fig. 9A, lane 4). These results thus indicated that a kinase(s) in RRL was able to phosphorylate DHBc. The migration of a CTD-truncated construct was unaffected by CIAP treatment (Fig. 9A, lanes 6 and 7), suggesting that phosphorylation (and CIAP dephosphorylation) of the full-length core was occurring at the CTD.
Replacement of S245 and S259 in the S-P motifs with alanine resulted in a change in mobility similar to what was previously shown in LMH cells (68), indicating that phosphorylation in RRL occurred on these particular sites as in vivo (Fig. 9B, lane 1 versus lanes 3 and 7, and C, lane 2 versus lanes 3 and 4). The migration pattern of the S259 mutant was not exactly the same in RRL as in LMH cells. In LMH cells, this mutant produced one fast-migrating species comigrating with the unphosphorylated DHBc. In RRL, it produced three species, but the predominant species also comigrated with unphosphorylated DHBc. This could suggest that S259 phosphorylation may be somewhat different in RRL versus LMH cells. However, the lack of the weak, slower-migrating species of the S259 mutant in LMH cells could also be a reflection of dynamic core dephosphorylation or phosphorylation-associated core conformational changes in the cell that were not completely reproduced in the lysate. Nevertheless, it was clear that S259 was phosphorylated in RRL as in cells. Moreover, substitutions of the adjacent prolines (P246 and P260) with glycines resulted in the same phenotype as the serine (S245 and S259) substitutions, again as reported in vivo (Fig. 9B, lanes 5 and 9) (68), indicating the importance of the proline in the kinase target motif. Furthermore, as we have recently reported (33), replacement of all four S/T-P sites or all six phosphorylation sites in DHBc resulted in a mobility shift to a single fast-migrating band in the RRL, and the DHBc CTD by itself was also phosphorylated in the RRL at the S/T-P sites, similar to what we reported in LMH cells (4).
Additionally, treatment with roscovitine caused a downward mobility shift, suggesting that DHBc phosphorylation in RRL might be mediated by a CDK(s) (Fig. 9D, lanes 7 to 10, and B, lanes 2, 4, 6, 8, and 10), consistent with the results above obtained in cells and with purified kinases. To help further identify the kinase(s) in RRL that mediated core phosphorylation, we tested the ability of a panel of different kinase inhibitors to block the phosphorylation of DHBc. Other than roscovitine, only olomoucine (Fig. 9B, lanes 11 to 14), another CDK inhibitor, was able to induce a dose-dependent, downward shift in core protein mobility, signifying a decrease in the hyperphosphorylated species and a corresponding increase in the hypophosphorylated or nonphosphorylated species. We also tested the CDK2-specific inhibitor (as suggested by results obtained above in the cells, with purified kinases, and with the HBV endogenous kinase reaction) and observed a dose-dependent, downward shift in core protein mobility (Fig. 9E, lanes 2 to 5), similar to what was observed with roscovitine (Fig. 9E, lanes 6 to 9). It was not surprising that high concentrations of roscovitine, olomoucine, or CDK2 inhibitor III were needed to show a strong effect on DHBc phosphorylation in the RRL, since these inhibitors are competitive with respect to ATP, which is present in the RRL translation reaction at 5 mM to facilitate translation (Promega). None of the other inhibitors tested, including inhibitors of PKC, PKA, and CKII, showed any effect on the mobility of DHBc at concentrations 100- to 1,000-fold above the respective IC50 or Ki (Fig. 9D).
DISCUSSION
In this study, we have provided multiple lines of evidence, employing purified kinases, cell lysate, and living cells as sources of host kinases, a large panel of kinase inhibitors, and specific CTD phospho site mutants, that cellular CDK2 can carry out the phosphorylation of the HBc and DHBc proteins in vitro and in vivo, specifically at the S/T-P sites within the CTD that are critical for viral replication. First, a broad-spectrum CDK inhibitor and a specific CDK2 inhibitor blocked the endogenous kinase activity of HBV capsids purified from human cells, which we showed to phosphorylate the S-P sites in the HBc CTD. Second, we have obtained direct evidence that CDK2 could be detected within purified capsids by immunoblot analysis. Third, we have shown here that full-length but not CTD-deleted HBc made in E. coli and fusion proteins containing the HBc and DHBc CTD could be phosphorylated by purified CDK2. Fourth, the sites of CDK2 phosphorylation in the purified system were shown to be the S/T-P motifs in the CTD. Finally, chemical inhibitors against CDK2 inhibited HBc and DHBc CTD phosphorylation in living cells and could block the phosphorylation of full-length DHBc at the S/T-P sites of its CTD in mammalian cell lysate.
CDK1 (CDC2) has been suggested, based on the sequence motif, to phosphorylate the HBc CTD (67) and a T-P site in the N-terminal assembly domain of DHBc (3). On the other hand, one previous report argued against a role of CDK1 or CDK2 because depletion of CDK1 and CDK2 from cell lysate did not affect HBc phosphorylation in vitro (10). However, it is clear that sufficient amounts of CDK activity remained in the depleted lysate to detectably phosphorylate histone H1, and therefore it was possible that the residual CDK1 or CDK2 activity remaining after depletion might have been sufficient for the HBc phosphorylation observed. Alternatively, a CDK(s) other than CDK1 or CDK2 in the cell lysate might have substituted for CDK1 or CDK2 in phosphorylating HBc. The consistent findings reported here using the different in vitro and in vivo systems in support of a role for CDK, particularly CDK2, in phosphorylating the CTD in the cell are also in accord with reported evidence suggesting the HBV replication requires the host hepatocytes to exit from quiescence (G0) but then stall in the G1 phase (with no entry into S) of the cell cycle (5, 16, 17, 42) and the fact that CDK2 is selectively active in G1 and may be activated during HBV replication (5, 6, 38, 53).
Consistent with the findings obtained using purified CDK2, cell lysate, and living cells in combination with CDK inhibitors, our results obtained using a large panel of kinase inhibitors in the context of the endogenous kinase assay also strongly suggest that a CDK(s), particularly CDK2, is a major capsid-associated kinase. The lack of effect by inhibitors of other CDKs, including CDK4/6, -7, -8, and -9, and of PKC, PKA, and CKII made the dose-dependent inhibition of the endogenous kinase by the CDK2 inhibitor all the more remarkable. The specificity of the endogenous kinase for the S-P sites in the HBc CTD, as demonstrated for the first time here, is also consistent with the identification of the endogenous kinase as a proline-directed kinase. Again, the incorporation of CDK2 into capsids is consistent with the notion that HBV replication requires the host cells to be in G1 and may be associated with CDK2 activation, as discussed above. All these results, along with the detection of CDK2 in the HBV capsids directly by the anti-CDK2 antibody, indicate that CDK2 represents at least one of the major endogenous kinases in the capsids.
We note that our evidence for a role of CDK2 in phosphorylating HBc and DHBc CTDs at the S/T-P sites in vitro and in vivo and for its incorporation into HBV capsids assembled in human cells does not rule out the involvement of other cellular kinases in CTD or core protein phosphorylation or their incorporation into the capsids. Indeed, the SK/SR motif of the DHBc CTD (43) likely requires a kinase other than CDKs given the strong preference of CDKs for a P downstream of the S/T sites. Also, we have found that Erk2, a member of the mitogen-activated protein kinase (MAPK) family, the other major group of proline-directed kinases (35, 44) besides CDKs, could phosphorylate the S/T-P sites in the CTD in vitro (L. Ludgate and J. Hu, unpublished results), although its role in CTD phosphorylation in vivo or relationship to the endogenous kinase remains to be determined. On the other hand, although we could confirm that purified PKCα is able to phosphorylate the HBc (and DHBc) CTD, our results indicate that PKC is unlikely to be responsible for phosphorylating the S/T-P sites or to represent a major endogenous kinase packaged in the HBV capsids. In addition, although we and others (10) could show that SRPK can phosphorylate HBc in vitro and we have also shown here that SRPK could phosphorylate the DHBc CTD in vitro, there has been no evidence to date to indicate that SRPK is packaged into HBV capsids or is responsible for phosphorylating the CTD in vivo. However, the lack of well-characterized and commercially available SRPK-specific inhibitors and the relatively low sensitivity of the currently available SRPK antibodies do not allow a clear resolution of these issues at present.
Interestingly, in vitro phosphorylation of the GST-DCC fusion proteins at the S/T-P sites by purified CDK2 did not produce the migrational heterogeneity as detected by SDS-PAGE, which is characteristic of the same fusion constructs phosphorylated in cells or in the cell lysate and of the full-length core protein phosphorylated at the CTD S/T-P motifs in cell lysate or in cells (4, 33, 68). Thus, phosphorylation of the S/T-P sites per se is not sufficient to induce the conformational changes in the CTD that are thought to be responsible for the distinct mobility changes associated with CTD phosphorylation. The possibility thus arises that another host function, distinct from the kinase(s), is required to mediate these conformational changes, which may in turn be necessary for phosphorylation-dependent roles of the core protein in viral replication. Alternatively, we cannot exclude the possibility that the combination of the CTD sites phosphorylated by the purified kinases was not entirely the same as that in the cell or the cell lysate.
We have shown here that unphosphorylated HBV capsids made in E. coli, which package RNA nonspecifically, could be phosphorylated at their CTDs by exogenous kinases, indicating that their CTDs were at least partially accessible on the outside of these capsids. On the other hand, it has been reported recently that empty, but not RNA-containing, HBV capsids expose their CTDs to allow the binding of SRPK, as visualized by cryo-electron microscopy (8). Together, these results suggest that the CTD of the RNA-containing HBV capsids may also be exposed but only transiently or at low frequency such that it can be detected by 32P labeling in the exogenous kinase reaction (highly sensitive) but not by methods that require stable or high-frequency CTD exposure.
It remains to be determined how CDK2 or any other host kinase gets packaged into the HBV capsids. Our results here indicate that neither the viral RT, pgRNA, nor any hepatocyte-specific host factor is required for kinase packaging and fit well with previous reports of endogenous kinase activity observed in recombinant HBV capsids made in insect cells and in empty core particles from infected livers (1, 18, 30). These results thus suggest that the core protein alone may be able to capture and package the host kinase(s). The most straightforward explanation would be that the assembling capsid captures the kinase that is specifically interacting with, and phosphorylating, the core subunits, perhaps reflecting rapid capsid assembly while core phosphorylation is taking place. Our finding that the endogenous kinase may be the same as or related to the kinase that phosphorylates the CTD is consistent with this notion. The estimated one-copy-CDK2-per-capsid stoichiometry further suggests that the packaging of one CDK2 molecule may prevent further CDK2 packaging. However, other possibilities exist and remain to be explored.
Due to the long interval (many hours to days) between core phosphorylation and any measurable levels of viral RNA packaging and DNA synthesis, it would be difficult to directly test the effects of kinase inhibitors on viral replication in cells. Not only does kinase redundancy have to be considered, but also cytotoxicity and potentially pleiotropic effects of kinase inhibition over an extended period of time in vivo could obscure its specific effects on viral replication. Indeed, our efforts so far to demonstrate directly a role of CDK2 in HBV replication in vivo have been hampered by these complications. However, the known importance of the CTD S/T-P site phosphorylation in HBV and DHBV RNA packaging and DNA synthesis strongly suggests that a CDK (particularly CDK2) is an important host factor that is required for viral replication. Establishment of cell-free systems that can recapitulate aspects of hepadnavirus RNA packaging and/or DNA synthesis that are regulated by CTD phosphorylation would facilitate future efforts to determine directly if CDKs or other cellular kinases play a functional role in viral replication.
ACKNOWLEDGMENTS
We thank Jesse Summers for plasmids and William Mason for the anti-DHBc antibody.
This work was supported by Public Health Service grant R01AI43453 from the National Institutes of Health.
Footnotes
Published ahead of print 5 September 2012
REFERENCES
- 1. Albin C, Robinson WS. 1980. Protein kinase activity in hepatitis B virus. J. Virol. 34:297–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Alessi F, et al. 1998. The cyclin-dependent kinase inhibitors olomoucine and roscovitine arrest human fibroblasts in G1 phase by specific inhibition of CDK2 kinase activity. Exp. Cell Res. 245:8–18 [DOI] [PubMed] [Google Scholar]
- 3. Barrasa MI, Guo JT, Saputelli J, Mason WS, Seeger C. 2001. Does a cdc2 kinase-like recognition motif on the core protein of hepadnaviruses regulate assembly and disintegration of capsids? J. Virol. 75:2024–2028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Basagoudanavar SH, Perlman DH, Hu J. 2007. Regulation of hepadnavirus reverse transcription by dynamic nucleocapsid phosphorylation. J. Virol. 81:1641–1649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Benn J, Schneider RJ. 1995. Hepatitis B virus HBx protein deregulates cell cycle checkpoint controls. Proc. Natl. Acad. Sci. U. S. A. 92:11215–11219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bouchard M, Giannakopoulos S, Wang EH, Tanese N, Schneider RJ. 2001. Hepatitis B virus HBx protein activation of cyclin A-cyclin-dependent kinase 2 complexes and G1 transit via a Src kinase pathway. J. Virol. 75:4247–4257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Brooks EE, et al. 1997. CVT-313, a specific and potent inhibitor of CDK2 that prevents neointimal proliferation. J. Biol. Chem. 272:29207–29211 [DOI] [PubMed] [Google Scholar]
- 8. Chen C, Wang JC, Zlotnick A. 2011. A kinase chaperones hepatitis B virus capsid assembly and captures capsid dynamics in vitro. PLoS Pathog. 7:e1002388 doi:10.1371/journal.ppat.1002388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Crowther RA, et al. 1994. Three-dimensional structure of hepatitis B virus core particles determined by electron cryomicroscopy. Cell 77:943–950 [DOI] [PubMed] [Google Scholar]
- 10. Daub H, et al. 2002. Identification of SRPK1 and SRPK2 as the major cellular protein kinases phosphorylating hepatitis B virus core protein. J. Virol. 76:8124–8137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Davies SP, Reddy H, Caivano M, Cohen P. 2000. Specificity and mechanism of action of some commonly used protein kinase inhibitors. Biochem. J. 351:95–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Duclos-Vallee JC, Capel F, Mabit H, Petit MA. 1998. Phosphorylation of the hepatitis B virus core protein by glyceraldehyde-3-phosphate dehydrogenase protein kinase activity. J. Gen. Virol. 79(Pt 7):1665–1670 [DOI] [PubMed] [Google Scholar]
- 13. Fallows DA, Goff SP. 1995. Mutations in the epsilon sequences of human hepatitis B virus affect both RNA encapsidation and reverse transcription. J. Virol. 69:3067–3073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gao W, Hu J. 2007. Formation of hepatitis B virus covalently closed circular DNA: removal of genome-linked protein. J. Virol. 81:6164–6174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gazina EV, Fielding JE, Lin B, Anderson DA. 2000. Core protein phosphorylation modulates pregenomic RNA encapsidation to different extents in human and duck hepatitis B viruses. J. Virol. 74:4721–4728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gearhart TL, Bouchard MJ. 2010. Replication of the hepatitis B virus requires a calcium-dependent HBx-induced G1 phase arrest of hepatocytes. Virology 407:14–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gearhart TL, Bouchard MJ. 2010. The hepatitis B virus X protein modulates hepatocyte proliferation pathways to stimulate viral replication. J. Virol. 84:2675–2686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gerlich WH, Goldmann U, Muller R, Stibbe W, Wolff W. 1982. Specificity and localization of the hepatitis B virus-associated protein kinase. J. Virol. 42:761–766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gschwendt M, et al. 1996. Inhibition of protein kinase C mu by various inhibitors. Differentiation from protein kinase c isoenzymes. FEBS Lett. 392:77–80 [DOI] [PubMed] [Google Scholar]
- 20. Hidaka H, Inagaki M, Kawamoto S, Sasaki Y. 1984. Isoquinolinesulfonamides, novel and potent inhibitors of cyclic nucleotide dependent protein kinase and protein kinase C. Biochemistry 23:5036–5041 [DOI] [PubMed] [Google Scholar]
- 21. Hu J, Anselmo D. 2000. In vitro reconstitution of a functional duck hepatitis B virus reverse transcriptase: posttranslational activation by Hsp90. J. Virol. 74:11447–11455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jilbert AR, et al. 1992. Rapid resolution of duck hepatitis B virus infections occurs after massive hepatocellular involvement. J. Virol. 66:1377–1388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kann M, Gerlich WH. 1994. Effect of core protein phosphorylation by protein kinase C on encapsidation of RNA within core particles of hepatitis B virus. J. Virol. 68:7993–8000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kann M, Sodeik B, Vlachou A, Gerlich WH, Helenius A. 1999. Phosphorylation-dependent binding of hepatitis B virus core particles to the nuclear pore complex. J. Cell Biol. 145:45–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kann M, Thomssen R, Kochel HG, Gerlich WH. 1993. Characterization of the endogenous protein kinase activity of the hepatitis B virus. Arch. Virol. Suppl. 8:53–62 [DOI] [PubMed] [Google Scholar]
- 26. Kau JH, Ting LP. 1998. Phosphorylation of the core protein of hepatitis B virus by a 46-kilodalton serine kinase. J. Virol. 72:3796–3803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kore AR, et al. 2009. Synthesis and application of MeOSuc-Ala-Ala-Pro-Phe-CH2Cl as potent proteinase K inhibitor. Bioorg. Med. Chem. Lett. 19:1296–1300 [DOI] [PubMed] [Google Scholar]
- 28. Krasinska L, Cot E, Fisher D. 2008. Selective chemical inhibition as a tool to study Cdk1 and Cdk2 functions in the cell cycle. Cell Cycle 7:1702–1708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lan YT, Li J, Liao W, Ou J. 1999. Roles of the three major phosphorylation sites of hepatitis B virus core protein in viral replication. Virology 259:342–348 [DOI] [PubMed] [Google Scholar]
- 30. Lanford RE, Notvall L. 1990. Expression of hepatitis B virus core and precore antigens in insect cells and characterization of a core-associated kinase activity. Virology 176:222–233 [DOI] [PubMed] [Google Scholar]
- 31. Lewellyn EB, Loeb DD. 2011. Serine phosphoacceptor sites within the core protein of hepatitis B virus contribute to genome replication pleiotropically. PLoS One 6:e17202 doi:10.1371/journal.pone.0017202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Liao W, Ou JH. 1995. Phosphorylation and nuclear localization of the hepatitis B virus core protein: significance of serine in the three repeated SPRRR motifs. J. Virol. 69:1025–1029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ludgate L, Adams C, Hu J. 2011. Phosphorylation state-dependent interactions of hepadnavirus core protein with host factors. PLoS One 6:e29566 doi:10.1371/journal.pone.0029566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mabit H, Breiner KM, Knaust A, Zachmann-Brand B, Schaller H. 2001. Signals for bidirectional nucleocytoplasmic transport in the duck hepatitis B virus capsid protein. J. Virol. 75:1968–1977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. 2002. The protein kinase complement of the human genome. Science 298:1912–1934 [DOI] [PubMed] [Google Scholar]
- 36. Meijer L, Bettayeb K, Galons H. 2007. (R)-Roscovitine (CYC202, Seliciclib), p 187–226 In Smith PJ, Yue EW. (ed), Inhibitors of cyclin-dependent kinases as anti-tumor agents. CRC Press, Boca Raton, FL [Google Scholar]
- 37. Montagnoli A, et al. 2008. A Cdc7 kinase inhibitor restricts initiation of DNA replication and has antitumor activity. Nat. Chem. Biol. 4:357–365 [DOI] [PubMed] [Google Scholar]
- 38. Mukherji A, Janbandhu VC, Kumar V. 2007. HBx-dependent cell cycle deregulation involves interaction with cyclin E/A-cdk2 complex and destabilization of p27Kip1. Biochem. J. 401:247–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Nguyen DH, Hu J. 2008. Reverse transcriptase- and RNA packaging signal-dependent incorporation of APOBEC3G into hepatitis B virus nucleocapsids. J. Virol. 82:6852–6861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ning X, et al. 2011. Secretion of genome-free hepatitis B virus—single strand blocking model for virion morphogenesis of para-retrovirus. PLoS Pathog. 7:e1002255 doi:10.1371/journal.ppat.1002255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Nishikawa K, Toker A, Johannes FJ, Songyang Z, Cantley LC. 1997. Determination of the specific substrate sequence motifs of protein kinase C isozymes. J. Biol. Chem. 272:952–960 [DOI] [PubMed] [Google Scholar]
- 42. Ozer A, et al. 1996. Effect of hepatocyte proliferation and cellular DNA synthesis on hepatitis B virus replication. Gastroenterology 110:1519–1528 [DOI] [PubMed] [Google Scholar]
- 43. Perlman DH, Berg EA, O'Connor BP, Costello CE, Hu J. 2005. Reverse transcription-associated dephosphorylation of hepadnavirus nucleocapsids. Proc. Natl. Acad. Sci. U. S. A. 102:9020–9025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Pinna LA, Ruzzene M. 1996. How do protein kinases recognize their substrates? Biochim. Biophys. Acta 1314:191–225 [DOI] [PubMed] [Google Scholar]
- 45. Porterfield JZ, et al. 2010. Full-length hepatitis B virus core protein packages viral and heterologous RNA with similarly high levels of cooperativity. J. Virol. 84:7174–7184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Pugh J, Zweidler A, Summers J. 1989. Characterization of the major duck hepatitis B virus core particle protein. J. Virol. 63:1371–1376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Qatsha KA, Rudolph C, Marme D, Schachtele C, May WS. 1993. Go 6976, a selective inhibitor of protein kinase C, is a potent antagonist of human immunodeficiency virus 1 induction from latent/low-level-producing reservoir cells in vitro. Proc. Natl. Acad. Sci. U. S. A. 90:4674–4678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Rabe B, et al. 2009. Nuclear entry of hepatitis B virus capsids involves disintegration to protein dimers followed by nuclear reassociation to capsids. PLoS Pathog. 5:e1000563 doi:10.1371/journal.ppat.1000563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Rickert P, Corden JL, Lees E. 1999. Cyclin C/CDK8 and cyclin H/CDK7/p36 are biochemically distinct CTD kinases. Oncogene 18:1093–1102 [DOI] [PubMed] [Google Scholar]
- 50. Scaglioni PP, Melegari M, Wands JR. 1997. Posttranscriptional regulation of hepatitis B virus replication by the precore protein. J. Virol. 71:345–353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Schlicht HJ, Bartenschlager R, Schaller H. 1989. The duck hepatitis B virus core protein contains a highly phosphorylated C terminus that is essential for replication but not for RNA packaging. J. Virol. 63:2995–3000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Seeger C, Mason WS, Zoulim F. 2007. Hepadnaviruses, p 2977–3029 In Knipe DM, Howley PM. (ed), Fields virology, 5th ed Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 53. Singh AK, Swarnalatha M, Kumar V. 2011. c-ETS1 facilitates G1/S-phase transition by up-regulating cyclin E and CDK2 genes and cooperates with hepatitis B virus X protein for their deregulation. J. Biol. Chem. 286:21961–21970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Soni R, et al. 2001. Selective in vivo and in vitro effects of a small molecule inhibitor of cyclin-dependent kinase 4. J. Natl. Cancer Inst. 93:436–446 [DOI] [PubMed] [Google Scholar]
- 55. Summers J, Smith PM, Horwich AL. 1990. Hepadnavirus envelope proteins regulate covalently closed circular DNA amplification. J. Virol. 64:2819–2824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Tanaka M, et al. 2004. Synthesis of anilino-monoindolylmaleimides as potent and selective PKCbeta inhibitors. Bioorg. Med. Chem. Lett. 14:5171–5174 [DOI] [PubMed] [Google Scholar]
- 57. Toullec D, et al. 1991. The bisindolylmaleimide GF 109203X is a potent and selective inhibitor of protein kinase C. J. Biol. Chem. 266:15771–15781 [PubMed] [Google Scholar]
- 58. Vassilev LT. 2006. Cell cycle synchronization at the G2/M phase border by reversible inhibition of CDK1. Cell Cycle 5:2555–2556 [DOI] [PubMed] [Google Scholar]
- 59. Vassilev LT, et al. 2006. Selective small-molecule inhibitor reveals critical mitotic functions of human CDK1. Proc. Natl. Acad. Sci. U. S. A. 103:10660–10665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Vesely J, et al. 1994. Inhibition of cyclin-dependent kinases by purine analogues. Eur. J. Biochem. 224:771–786 [DOI] [PubMed] [Google Scholar]
- 61. Wang X, Grammatikakis N, Hu J. 2002. Role of p50/CDC37 in hepadnavirus assembly and replication. J. Biol. Chem. 277:24361–24367 [DOI] [PubMed] [Google Scholar]
- 62. Weigand K, Knaust A, Schaller H. 2010. Assembly and export determine the intracellular distribution of hepatitis B virus core protein subunits. J. Gen. Virol. 91:59–67 [DOI] [PubMed] [Google Scholar]
- 63. Wingfield PT, Stahl SJ, Williams RW, Steven AC. 1995. Hepatitis core antigen produced in Escherichia coli: subunit composition, conformational analysis, and in vitro capsid assembly. Biochemistry 34:4919–4932 [DOI] [PubMed] [Google Scholar]
- 64. Wittkop L, et al. 2010. Inhibition of protein kinase C phosphorylation of hepatitis B virus capsids inhibits virion formation and causes intracellular capsid accumulation. Cell Microbiol. 12:962–975 [DOI] [PubMed] [Google Scholar]
- 65. Wynne SA, Crowther RA, Leslie AG. 1999. The crystal structure of the human hepatitis B virus capsid. Mol. Cell 3:771–780 [DOI] [PubMed] [Google Scholar]
- 66. Yeh CT, Liaw YF, Ou JH. 1990. The arginine-rich domain of hepatitis B virus precore and core proteins contains a signal for nuclear transport. J. Virol. 64:6141–6147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Yeh CT, Wong SW, Fung YK, Ou JH. 1993. Cell cycle regulation of nuclear localization of hepatitis B virus core protein. Proc. Natl. Acad. Sci. U. S. A. 90:6459–6463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Yu M, Summers J. 1994. Phosphorylation of the duck hepatitis B virus capsid protein associated with conformational changes in the C terminus. J. Virol. 68:2965–2969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Zandomeni RO. 1989. Kinetics of inhibition by 5,6-dichloro-1-beta-d-ribofuranosylbenzimidazole on calf thymus casein kinase II. Biochem. J. 262:469–473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Zheng Y, Fu XD, Ou JH. 2005. Suppression of hepatitis B virus replication by SRPK1 and SRPK2 via a pathway independent of the phosphorylation of the viral core protein. Virology 342:150–158 [DOI] [PubMed] [Google Scholar]
- 71. Zlotnick A, et al. 1996. Dimorphism of hepatitis B virus capsids is strongly influenced by the C-terminus of the capsid protein. Biochemistry 35:7412–7421 [DOI] [PubMed] [Google Scholar]