Abstract
Genetic variability is a hallmark of RNA virus populations. However, transmission to a new host often results in a marked decrease in population diversity. This genetic bottlenecking is observed during hepatitis C virus (HCV) transmission and can arise via a selective sweep or through the founder effect. To model HCV transmission, we utilized chimeric SCID/Alb-uPA mice with transplanted human hepatocytes and infected them with a human serum HCV inoculum. E1E2 glycoprotein gene sequences in the donor inoculum and recipient mice were determined following single-genome amplification (SGA). In independent experiments, using mice with liver cells grafted from different sources, an E1E2 variant undetectable in the source inoculum was selected for during transmission. Bayesian coalescent analyses indicated that this variant arose in the inoculum pretransmission. Transmitted variants that established initial infection harbored key substitutions in E1E2 outside HVR1. Notably, all posttransmission E1E2s had lost a potential N-linked glycosylation site (PNGS) in E2. In lentiviral pseudoparticle assays, the major posttransmission E1E2 variant conferred an increased capacity for entry compared to the major variant present in the inoculum. Together, these data demonstrate that increased envelope glycoprotein fitness can drive selective outgrowth of minor variants posttransmission and that loss of a PNGS is integral to this improved phenotype. Mathematical modeling of the dynamics of competing HCV variants indicated that relatively modest differences in glycoprotein fitness can result in marked shifts in virus population composition. Overall, these data provide important insights into the dynamics and selection of HCV populations during transmission.
INTRODUCTION
Hepatitis C virus (HCV) is a positive-sense RNA enveloped virus belonging to the genus Hepacivirus within the family Flaviviridae. Globally, an estimated 170 million people are infected with HCV. Only about 15 to 20% of the population resolve acute HCV infection; the vast majority go on to develop a chronic infection, which is implicated in liver cirrhosis and hepatocellular carcinoma (65, 66). HCV continues to represent a significant global health burden, and vaccines to prevent new infections are much needed.
HCV circulates within an infected host as a swarm of genetically distinct but closely related variants (7). This genetic variability is unevenly distributed throughout the viral genome. The most variable regions are the E1E2 genes encoding the envelope glycoproteins (30). Both E1 and E2 are heavily glycosylated, and these glycans have been shown to be integral in protein folding, cell entry, and antibody shielding (23, 24). E1E2 evolution is driven by a combination of positive and purifying selection and neutral sequence drift (5, 59). Functional requirements for receptor binding, membrane fusion, and entry (17) exert a strong purifying effect, but it is set against the need for the virus to escape host antibody responses (27), which often target epitopes that overlap or are proximal to these functionally important regions (26, 32, 50, 52, 55, 63). Therefore, E1E2 evolution is driven by a subtle interplay between functional constraint and immune escape.
HCV is primarily transmitted via the transcutaneous route. Prior to the discovery of HCV and establishment of robust blood-screening techniques, a major route of infection was via transfusion of contaminated blood or blood products. Currently, the major risk factor in countries with well-developed screening procedures is intravenous drug use (1). Additionally, health care worker needle-stick injury (22, 46), transmission between sexual partners (6), horizontal and vertical intrafamilial transmission (9), and mother-to-child transmission (68) have all been reported. Although nosocomial and iatrogenic HCV transmission is rare in Western nations (64), these routes of transmission are still a major source of new infections in developing countries; for example, in Egypt, there are an estimated 500,000 new infections annually (44).
Development of an effective vaccine will need to elicit immunity that targets those variants that are successfully transmitted. Heterosexual transmission of HIV-1 often results in a significant virus population bottleneck due to the founder effect, where a single virus establishes the initial infection (14, 56). Characterization of the envelope glycoproteins from these founder viruses has shown that they have an increased sensitivity to antibody-mediated neutralization (14). In contrast, in HIV-1 transmission in men who have sex with men and in intravenous drug users (IDUs), there is wide variation in the number of viral strains that initiate the new infection (3, 33).
Studying HCV transmission is challenging because most cases of acute infection are asymptomatic and therefore go unnoticed. Most studies of acute infection have utilized samples obtained after seroconversion, when the virus has already been exposed, and responded, to the early immune response. A number of studies have been performed using samples obtained during acute infection of humans and chimpanzees, and the restricted diversity observed posttransmission indicates that productive infection is initiated by a single or a limited number of viral variants (8, 34, 36, 54, 60, 67). However, there are few studies that have tracked the entire transmission process from donor to recipient, and those that have, have focused on analysis of the first hypervariable region (HVR1) of E2 in the chimpanzee model (54, 60). Other regions of the envelope glycoproteins contain important functional and neutralizing determinants (16, 26, 27, 32, 43, 48–50, 52, 63), and the natural histories of HCV infection in chimpanzees and humans differ. Very limited studies on liver transplant cohorts have shown that reinfection of grafts is characterized by E1E2 variants that permit efficient entry and exhibit neutralization-resistant phenotypes (20). However, the presence of naturally occurring neutralizing antibodies in transplant patients and posttransplant therapy undoubtedly influences reinfection.
Envelope glycoprotein determinants that define the ability of HCV to establish infection in a new host have not been identified. Such insight will be critical for the rational design of entry-targeted strategies that prevent HCV infection. To address this significant shortfall, we have utilized single-genome amplification (SGA) of E1E2 genes to study transmission of serum-derived HCV to SCID/Alb-uPA chimeric mice with grafted human liver cells (29, 41). This small-animal model is ideal for these studies, as infection is established in grafted human liver cells, and the lack of an adaptive immune response provides a good surrogate for the acute phase of infection. Comparative analyses of donor and recipient sequences were performed, in conjunction with a range of phylogenetic tests. Importantly, recovery of full-length E1E2s also enabled us to investigate the phenotypic consequences of E1E2 selection upon transmission. Together, our data reveal important E1E2 determinants facilitating successful HCV transmission and establishment of initial infection in a new host.
MATERIALS AND METHODS
Source of samples.
HCV RNA samples were obtained as described in a previously published study (32). Briefly, 11 human liver-chimeric SCID/Alb-uPA mice, transplanted with human hepatocytes from different donors, were inoculated intrajugularly with 100 μl genotype 1a HCV-infected serum KP (2.3 × 106 IU/ml). The infections were monitored over time (Table 1) by tail bleed sampling. HCV RNA in mouse serum was quantified by a real-time 24 TaqMan PCR assay (32). RNA was recovered from serum aliquots using a commercially available RNA extraction kit (Qiagen) and resuspended in 20 μl of H2O. Donor serum (KP) was obtained from an HCV+ patient in the chronic phase of infection, with time since initial infection estimated at 5 years.
Table 1.
Summary of sample detailsa
| Patient/mouse ID | Population sampling | Serum HCV (IU/ml) | Hepatocyte donora | Time postinoculation (days) | No. of SGA ampliconsb |
|---|---|---|---|---|---|
| KP | KP | 1 × 106 | 36 | ||
| A594 | 594_1 | 8.09 × 105 | II | 32 | 13 + bulk |
| 594_2 | 1.01 × 106 | II | 59 | 17 | |
| N666c | 666_1 | 2.69 × 106 | I | 14 | 22 |
| 666_2 | 2.49 × 106 | I | 28 | 20 | |
| N714c | 714_1 | 1.16 × 106 | I | 14 | 21 |
| 714_2 | 1.54 × 106 | I | 28 | 19 | |
| A931 | 931_1 | 8.61 × 107 | III | 21 | 11 |
| 931_2 | 2.94 × 107 | III | 42 | 13 | |
| A931 | 8.04 × 107 | III | 28 | NA, bulk | |
| A594 | A594 | 8.09 × 105 | II | 32 | NA, bulk |
| A583 | A583 | 8.40 × 105 | II | 32 | NA, bulk |
| A596 | A596 | 1.73 × 105 | II | 32 | NA, bulk |
| A585 | A585 | 2.99 × 105 | II | 32 | NA, bulk |
| A587 | A587 | 9.79 × 106 | II | 32 | NA, bulk |
| A902 | A902 | 4.46 × 107 | III | 28 | NA, bulk |
| A909 | A909 | 1.96 × 107 | III | 28 | NA, bulk |
| A965 | A965 | 7.73 × 106 | III | 28 | NA, bulk |
I, 36-year-old female; II, 45-year-old female, 92% viability polycystic tissue; III, 4-year-old male, 92% viability normal tissue.
Bulk indicates samples for which majority consensus sequences were generated on undiluted cDNA samples. NA, not applicable.
Mouse N666 and mouse N714 were control mice in the HCV monoclonal antibody protection study previously reported (32).
All mice were housed and treated according to Canadian Council on Animal Care guidelines. Experimental approval came from the University of Alberta Animal Welfare Committee, and human sera and hepatocytes were obtained following informed written consent of all donors with ethics approval from the University of Alberta, Faculty of Medicine Research Ethics Board.
SGA of HCV E1E2.
To accurately assess the dynamics of HCV transmission, an SGA approach was employed, followed by direct sequencing (12). Full-length E1E2 sequences (amino acids 170 to 746, corresponding to the polyprotein sequence of the reference strain H77 [accession no. NC_004102]) from sequential samples (2 time points) derived from 4 HCV-infected chimeric mice (666, 714, 594, and 931) and donor inoculum (KP) were amplified, using previously described primers (31). Nine microliters of HCV RNA was used in a 20-μl reverse transcription reaction with 10 pmol primer OAS 1a using a commercially available Thermoscript cDNA synthesis kit (Invitrogen) according to the manufacturer's protocol. The resulting viral cDNAs were serially diluted, and aliquots of 2-fold dilutions (1 μl) were used as templates in the first round of a nested full-length E1E2 PCR. Amplification reactions were set up in 20-μl volumes containing 4 pmol of primer OAS 1a and primer EOS (31), 200 mM deoxynucleoside triphosphates (dNTPs), 0.5 U of Platinum Taq High Fidelity polymerase (Invitrogen), 1× High Fidelity polymerase buffer, and 2 mM MgSO4. The PCR cycling parameters were as follows: initial denaturation at 94°C for 2 min, followed by 35 cycles of 94°C for 15 s, 50°C for 30 s, and 68°C for 3 min, with a final extension step at 68°C for 10 min. Two microliters of first-round product was subsequently used as the template in second-round reactions with primers 1ASGT1a and 170gt1, using amplification and cycling conditions identical to the first round but increasing the cycle number to 45. Amplification of a 2-fold dilution series of cDNA titration PCRs revealed the dilution at which the concentration of viral cDNA was <1 molecule per μl. Subsequently, multiple E1E2 amplicons for each sample were generated at this endpoint dilution (to give a frequency of ≤3/10 PCR-positive reactions). The amplification products were directly sequenced using BigDye v1.1 (Applied Biosystems). Chromatographs were manually inspected, and amplicons exhibiting dual peaks at a single nucleotide position, resulting from amplification from >1 starting template molecule, were excluded from further analysis.
Sequence analysis and phylogenetic reconstruction.
Nucleotide sequences were aligned according to overlying amino acid translations. Alignments were performed using Mega 4.0 (62), with manual adjustment to ensure maintenance of the open reading frame. Substitutions occurring across viral populations were visualized using the Highlighter Tool (http://hcv.lanl.gov/content/sequence/HIGHLIGHT/highlighter.html), where the KP consensus sequence was used as a master sequence. Consensus sequences were generated using the Consensus Maker tool (http://hcv.lanl.gov/content/sequence/CONSENSUS/consensus.html). Phylogenetic relationships between generated E1E2 sequences were calculated utilizing the maximum-likelihood (ML) criterion implemented by PAUP version 4.0b10 (61) using the best-fit substitution model for the data calculated in Modeltest version 3.7 (53). ML trees were rooted on the consensus sequence from the donor inoculum (KP_con).
Assessment of the number of transmitting variants and evolutionary rates.
To define the number of variants that established initial infection following mouse inoculation and to estimate substitution rates, time-scaled phylogenies of each data set were generated using a Bayesian Monte Carlo Markov Chain (MCMC) method implemented in BEAST (version 1.6.0; available from http://beast.bio.ed.ac.uk/) (18, 19, 58). Evolutionary rate estimates and phylogenies were obtained using the SRD06 model and two relaxed clock models: uncorrelated lognormal and uncorrelated exponential. The MCMC search was set to at least 10,000,000 iterations, so that the effective sampling size for the parameters under study reached more than 200. Trees were sampled every 1,000th generation with a 10% burn-in and then summarized using Tree Annotator v.1.6.0 (also available from http://beast.bio.ed.ac.uk/). The most appropriate model was identified by calculating the Bayes factor (BF) using the program Tracer v1.5 (http://beast.bio.ed.ac.uk/). Phylogenies were visualized using FigTree v1.3.1. Branches that had a time to most recent common ancestor (tMRCA) estimated to have occurred after the known time of inoculation were collapsed to enable identification of the number of effective transmitting variants that establish initial infection.
Patient HLA typing and epitope prediction.
Donor KP's HLA class I and II genotype was determined by the histocompatibility laboratory at the University of Alberta Hospital (Edmonton, AL, Canada) using a low-resolution PCR method with sequence-specific primer mixes. E1E2 epitopes previously reported to be presented by the donor's HLA class I alleles were identified with the help of the Immune Epitope Database (http://www.immuneepitope.org/). Peptides that were likely to be presented by the donor HLA class I alleles were predicted using the SYFPEITHI (http://www.syfpeithi.de/home.htm) and BIMAS (http://www-bimas.cit.nih.gov/molbio/hla_bind/) websites.
HCV pseudovirus infection.
HCV E1E2 sequences, bearing one of 4 possible key residue combinations (SNHV, SDHV, TDYD, and TDYV) were ligated into a pcDNA3.1 V5 D-TOPO mammalian expression vector (Invitrogen). The rare transitory variant (SDYV) and the minority posttransmission variant TDYA were not tested (V/A have very similar physiochemical properties). Huh-7.5 cells were propagated in Dulbecco's Modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS)-1% nonessential amino acids. Primary human hepatocytes were isolated according to a previously published protocol (45) and were maintained in Williams E medium supplemented with 10% FBS-5 mM HEPES-insulin-dexamethasone. The cells were plated at 4 × 104 cells/cm2 and infected the following day. Pseudoviruses were generated by transfecting 293T cells with plasmids encoding human immunodeficiency virus gag/pol, luciferase, and either HCV E1E2 glycoproteins, vesicular stomatitis virus glycoprotein (VSV-G), or no-envelope (ΔE1E2) control as previously reported (25). Supernatants were harvested 48 h posttransfection, clarified, and filtered through a 0.45-μm membrane. Virus-containing medium was incubated with target cells for 8 h in the presence or absence of anti-CD81 monoclonal antibody 2s131 (40) (5 μg/ml) overnight, unbound virus was removed, and the medium was replaced with DMEM containing 3% FBS. At 72 h postinfection, the medium was removed and the cells were lysed with cell lysis buffer (Promega, Madison, WI). Luciferase activity was determined by the addition of luciferase substrate and measured for 10 s in a luminometer (Lumat LB 9507).
Mathematical modeling.
An ordinary differential equation model was employed to analyze the dynamics of two competing HCV variants. The model is an extension of the basic model used to represent viral dynamics (51), incorporating two HCV variants and neglecting the effects of treatment in order to reflect the conditions of our study. Certain assumptions are required to formulate the model: target cells (T) are produced at rate λ, die at rate d, and are infected by variant 1 (V1) at rate k and by variant 2 (V2) at rate γk, where γ is >1 (reflecting a fitness advantage for variant 2); infected cells (I1 and I2) die at rate δ. We assume equal production and clearance rates for both variants: p and c, respectively. The equations are thus dT/dt = λ − dT − kV1T − γkV2T, dI1/dt = kV1T − δI1, dI2/dt = γkV2T − δI2, dV1/d2 = pI1 − cV1, and dV2/d2 = pI2 − cV2.
For simplicity, only two variants are considered (though we can assume V1 accounts for all variants except TDYV/A, which is given by V2), but the model could be easily extended to consider any number. To match our data, we set I1 (0) equal to I2 (0) equal to 0 cells ml−1 to reflect all host cells initially being healthy, and we assume that variant 1 formed 99% of our initial inoculum, with variant 2 making up the remaining 1%; thus, V1 (0) is equal to 2.277 × 106 and V2 (0) is equal to 2.3 × 104 virions ml−1. From Dahari et al. (13), we take the initial concentration of target liver cells to be 1.87 × 107 cells ml−1 and scale this by 0.7 to reflect our chimeric livers consisting of 70% human cells, making T (0) equal to 1.309 × 107 cells ml−1. Neumann et al. (47) estimated viral reproduction and infection rates to be as follows: P = 100 virions cell−1 day−1 and k = 3 × 10−7 ml virion−1 day−1. In addition, the viral clearance rate and infected-cell death rate are assumed to be as follows: c = 5 days−1 and δ = 0.5 day−1 (corresponding to half-lives of 2.7 h and 12 h, respectively). However, given that our mouse model is immunodeficient, we investigated a range of c and δ values decreasing from those stated above. Half-life estimates of target cells ranged from 50 to 500 days (13). We investigated both extremes of this interval, considering the following: 0.013 ≤ d ≤ 0.0014 day−1. Finally, assuming that, in the absence of a viral load the target cell concentration is at quasi-steady state (dT/dt = 0), we take λ to be equal to dT (0). The equations were solved numerically in Matlab 7.12.0 (R2011a).
RESULTS
Experimental transmissions.
Defining the genotypic and phenotypic envelope glycoprotein determinants underlying successful HCV transmission is challenging, as natural acute infection is often asymptomatic. To overcome this limitation, we used the well-defined and robust human liver-chimeric SCID/Alb-uPA mouse model to study HCV envelope glycoprotein evolution at transmission. Eleven chimeric mice, transplanted with human hepatocytes from different donors, were inoculated via the intrajugular route with 100 μl genotype 1a HCV-infected serum KP (2.3 × 106 IU/ml) to simulate natural exposure to virus. In total, we obtained 36 SGA E1E2 sequences from the KP inoculum and 136 temporally sampled SGA E1E2 sequences derived from viruses circulating in four recipient chimeric mice (24 to 44 sequences for each chimeric mouse; 11 to 22 sequences for each time point). For the purposes of comparison, bulk E1E2 amplifications were also performed for the seven additional experimentally infected chimeric mice (Table 1). As an internal experimental control, bulk E1E2 amplicons were also generated and sequenced for chimeric mice 594 and 931. Importantly, SGA of full-length E1E2 enabled us to investigate the phenotypic consequences of E1E2 upon transmission.
Molecular determinants underlying experimental HCV transmission.
To define E1E2 sequence evolution during these experimental HCV transmission events, nucleotide sequences derived from the donor inoculum and recipient mice were combined and subjected to phylogenetic analyses (Fig. 1). There were two major clades evident, one containing the majority of the donor inoculum sequences and a second containing the mouse-derived sequences and two donor sequences (KP7 and KP22). The sequences derived from the recipient mice were interspersed with one another, and there was no evidence of clustering according to individual recipient mice.
Fig 1.
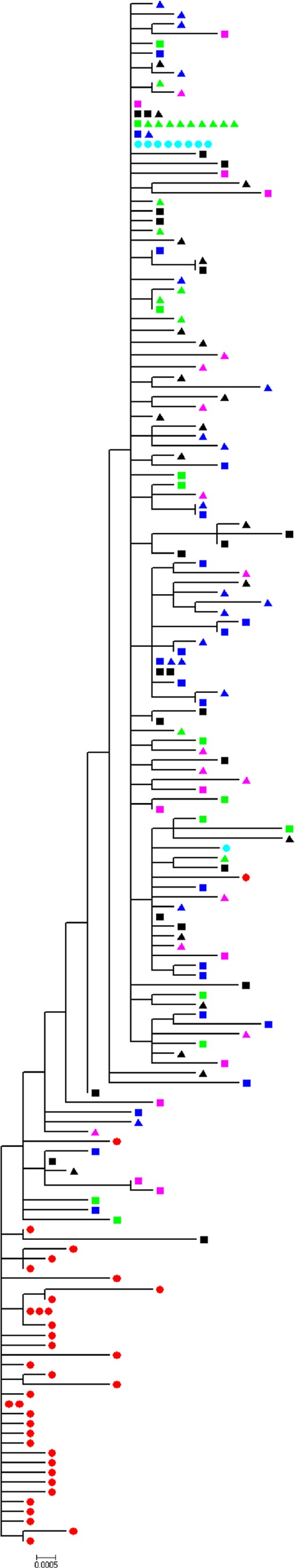
Combined phylogenetic reconstruction of donor and recipient viral E1E2 populations. Shown is a maximum-likelihood tree (HKY + Γ) derived using SGA and bulk E1E2 sequences present in the KP donor inoculum and in chimeric mice posttransmission. Red circles, KP inoculum; green squares, mouse 594 time point 1; green triangles, mouse 594 time point 2; black squares, mouse 666 time point 1; black triangles, mouse 666 time point 2; blue squares, mouse 714 time point 1; blue triangles, mouse 714 time point 2; pink squares, mouse 931 time point 1; pink triangles, mouse 931 time point 2; turquoise circles, bulk amplified sequences. The scale bar is proportional to the genetic distance and represents 0.0005 nucleotide substitution per site.
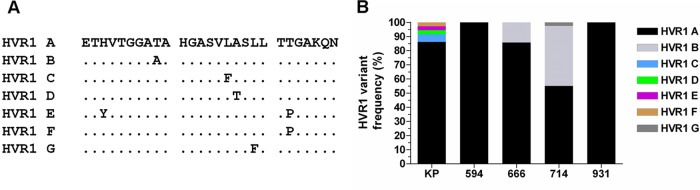
To determine whether HVR1 sequences altered following transmission, we determined the relative frequency of each HVR1 deduced amino acid sequence in the inoculum and recipient mice. A total of seven HVR1 amino acid variants were identified (HVR1 A to G) (Fig. 2A), and the frequency distribution of these seven variants revealed that the major HVR1 sequence in donor plasma was also the majority variant in all recipient chimeric mice (Fig. 2B). The majority sequence (HVR1 A) occurred in 83% of all analyzed sequences. This HVR1 variant represented over 86% of donor sequences and constituted 100% of HVR1 sequence populations circulating in mice 594 and 931. None of the minor HVR1 variants present in the KP inoculum (HVR1 C to F) were transmitted to recipient chimeric mice. Chimeric mice 714 and 666 also yielded another sequence variant (HVR1 B) that was not detected in the donor. This variant constituted over 42% of the HVR1 sequence population in chimeric mouse 714. This mouse also contained an additional minor variant (HVR1 G). All of the new HVR1 variants arose through a single amino acid substitution. These data show that there are no major differences in the HVR1 regions of viruses obtained pre- and posttransmission and indicate that determinants associated with successful transmission reside outside HVR1.
Fig 2.
Stability of HVR1 sequences upon transmission. (A) Alignment of HVR1 amino acid sequence variants (A to G) present in donor/recipient populations. (B) Frequencies of HVR1 variants circulating within donor/recipient hosts.
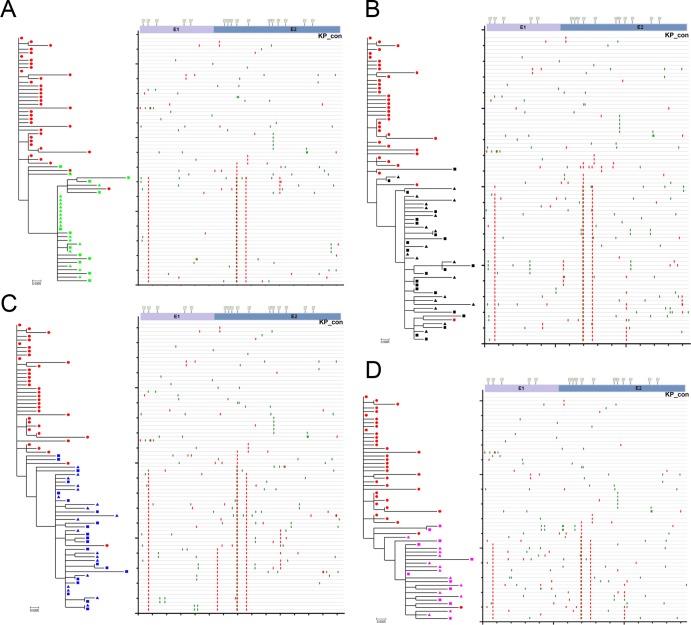
To investigate this further, full-length E1E2 sequences were analyzed using a combination of phylogenetic analyses and Highlighter plots (Fig. 3A to D). Although the overall intrahost sequence diversity was low, multiple distinct variants were identified in the donor and in each recipient. Highlighter plots and phylogenetic trees revealed mutational patterns associated with pre- and posttransmission populations. These data revealed a consistent pattern of nonsynonymous (amino acid- changing) substitutions associated with transmission, irrespective of the chimeric mouse recipient, which were distributed throughout the E1E2 genes. Analysis of the deduced amino acid sequence alignments identified four E1E2 amino acid substitutions at positions 198, 448, 474, and 570 that were associated with productive infection in the recipient mice (Fig. 4A). The residues at positions 198, 448, 474, and 570 appeared in one of 5 possible combinations: SNHV, SDHV, TDYD, SDYV, and TDYV/A. The frequencies of these motifs in the SGA sequence data set obtained for the inoculum and the recipient chimeric mice are summarized in Fig. 4B. The major E1E2 variant in the inoculum population possessed the SNHV motif. This combination was evident in approximately 90% of all the donor E1E2 amplicons. However, this sequence motif was undetectable in all recipient chimeric mice. Without exception, all of the mouse-derived sequences contained the N448D substitution. This residue is part of an NXS/T potential N-linked glycosylation site (PNGS) that is highly conserved in the donor inoculum and in the majority of globally sampled HCV strains. Minor circulating E1E2 variants containing the amino acid motifs SDHV and TDYD were detected in both the donor inoculum and all recipient mice. Variants containing the TDYV/A motif, which was undetectable in the donor population, were the dominant variant posttransmission, irrespective of the recipient chimeric-mouse viral population analyzed. An additional minority variant (SDYV) was detected at the first sampling time point from the chimeric mice 666 and 931. This rare transitory variant was undetectable in the donor population and at the second sampling time point in chimeric mice 666 and 931. Together, these data demonstrate that, while HVR1 sequences remain stable during HCV transmission, key amino acid substitutions elsewhere in E1E2 are selected for during transmission or during the initial phase of infection in the new host.
Fig 3.
Phylogenetic trees and patterns of substitution in sequences derived during experimental HCV transmission from the KP inoculum to chimeric mice 594, 666, 714, and 931. Chimeric mouse 594 (A), 666 (B), 714 (C), and 931 (D)-derived sequences were analyzed via phylogenetic reconstructions (left panels) and Highlighter plots (right panels) with pretransmission KP donor sequences included. The ML trees are rooted on the KP consensus (KP_con) sequence, which is the master sequence in corresponding Highlighter plots. The Highlighter plots depict the relative locations of synonymous (green vertical bars) and nonsynonymous (red vertical bars) substitutions in each E1E2 amplicon compared to the pretransmission master sequence. A schematic representation of the E1E2 gene, depicting the locations of PNG sites and the E1/E2 boundary, is provided above the Highlighter plots for the purposes of positional referencing. The scale bar is proportional to the genetic distance and represents 0.0005 nucleotide substitution per site.
Fig 4.
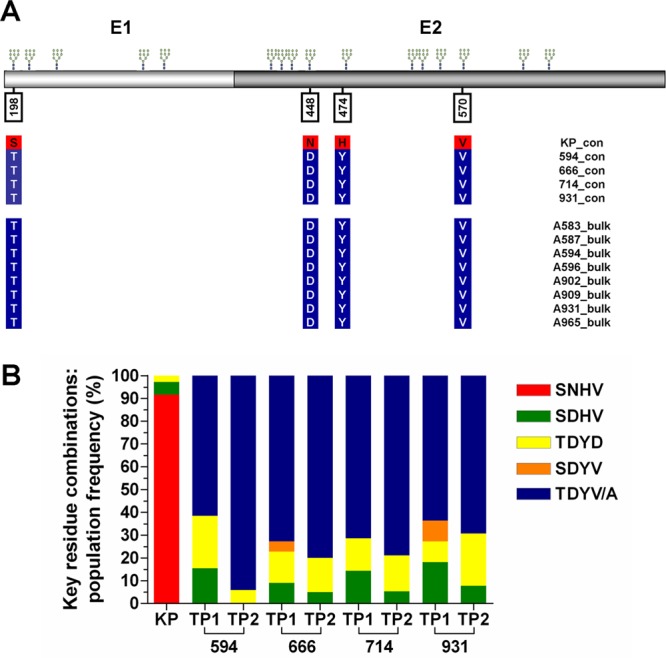
Identification of key residues involved in HCV transmission and the frequencies of key residue combinations in donor and recipient viral E1E2 populations. (A) Schematic E1E2 diagram depicting the locations of four key residues involved in transmission. The positions of conserved potential N-glycosylation sites are indicated above the E1E2 protein. The coordinates given are relative to the homologous positions in the H77 reference strain polyprotein (accession no. NC_004102). The colored vertical columns located below key residues indicate consensus amino acids present in the E1E2 SGA populations of donor and four recipient chimeric (top), as well as consensus, amino acid sequences derived from bulk amplified RNA obtained from eight other transmission experiments using the same KP inoculum (bottom). (B) Frequencies of key residue combinations circulating within donor/recipient hosts. TP1, time point 1; TP2, time point 2.
Identification of viral lineages that establish initial infection.
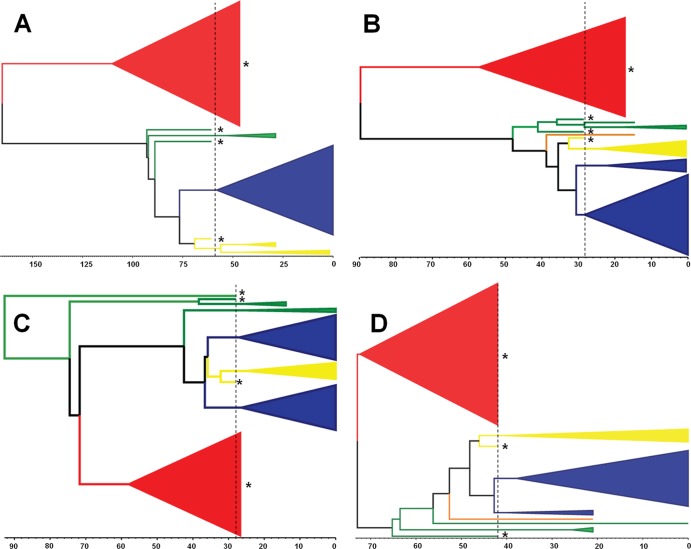
Individual donor/recipient transmission events (KP/594, KP/666, KP/914, and KP/714) were further interrogated via BEAST analyses to estimate the rates of evolution and the number of transmitted viral lineages (defined as variants present in the inoculum that became detectable, established lineages within the new host) (Fig. 5A to D). The estimated mean substitution rates ranged from 5 × 10−5 to 1 × 10−4 nucleotide substitutions per day. These analyses also showed that HCV infection was established by multiple lineages derived from the inoculum virus population, and in each mouse, one or more lineages representing viruses containing the TDYV/A motif became dominant. Also, in each mouse, there were one or more lineages associated with minor variants containing the sequence motif TDYD or SDHV. These motifs were also present in the predicted sequence of the most recent common ancestor for each of these lineages. The estimated occurrence time of these predicted ancestral sequences predated the time of the inoculation, indicating that these variants were present at low frequency in the original inoculum, rather than arising independently in each mouse postinoculation. In summary, these data demonstrate that multiple variants established infection in each chimeric mouse and point to a selective sweep occurring following transmission, which resulted in selective outgrowth of lineages possessing the motif TDYV/A.
Fig 5.
Identification of the number of transmitted lineages for each experimental infection event. Shown are donor/recipient pairs KP/594 (A), KP/666 (B), KP/714 (C), and KP/931 (D). The lineages are color coded according to key residue combinations: red, SNHV; green, SDHV; yellow, TDYD; orange, SDHV; blue, TDYV/A. The scale bar located below each tree is proportional to the time. The units correspond to days, with 0 representing the final sampling date for each individual chimeric mouse and the dotted line representing the time of experimental infection. Lineages/variants present in the KP inoculum are indicated by asterisks.
HLA typing and epitope prediction.
To investigate the possibility that the observed substitutions at these key residues might be due to reversion of escape mutations selected for by T-cell responses in the KP donor, patient HLA typing and mapping of epitopes known or predicted to be presented by the donor HLA class I alleles against the mouse consensus sequence were conducted. Details of the donor HLA type and a map of the locations of HCV E1E2 epitopes to which CD8 T-cell responses restricted by HLA-A*02, A*11, B*51, and Cw7 have been reported in the literature. None of the previously reported epitopes presented by these HLA alleles overlapped with residue 198, 448, 474, or 570 are presented in Fig. 6. Furthermore, when we used the SYFPEITHI (http://www.syfpeithi.de/home.htm) and BIMAS (http://www-bimas.cit.nih.gov/molbio/hla_bind/) websites to predict peptides containing or adjacent to residues 198, 448, 474, and 570 that were likely to be presented by the donor HLA class I alleles A*02, A*11, B*40, B*51, and Cw7, both analyses failed to identify any peptides at these sites with predicted binding to donor class I alleles comparable to that of the known T-cell epitope sequences we tested in parallel. Therefore, we have no evidence that the amino acid substitutions that were selected for in the immunocompromised mice represented T-cell escape reversions posttransmission.
Fig 6.
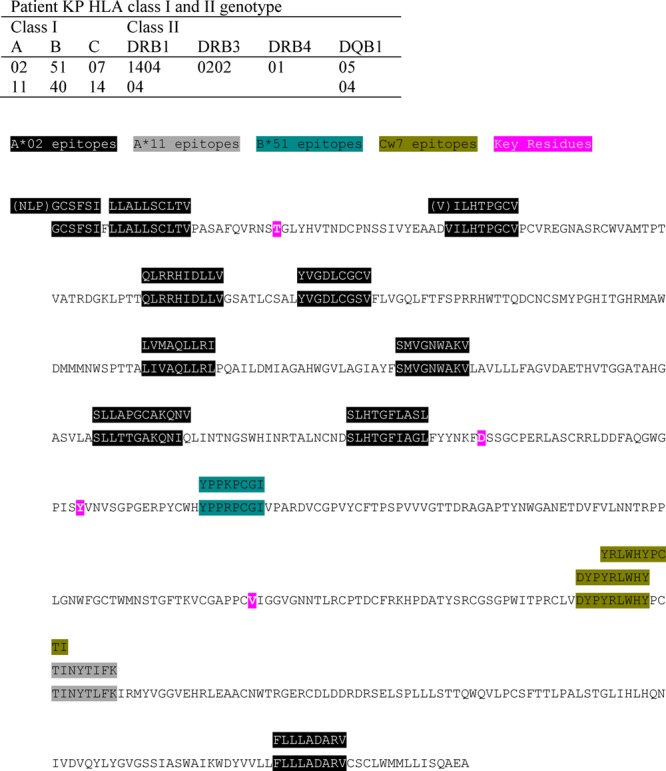
Locations of reported T-cell epitopes restricted by the donor KP's HLA class I alleles in the E1E2 sequence. The mouse consensus E1E2 amino acid sequence is shown, with the four amino acid substitutions associated with establishment of productive infection following HCV transmission highlighted in pink. Sequences corresponding to the locations of reported T-cell epitopes restricted by the donor KP's HLA class I alleles are highlighted in different colors, with the peptide sequence(s) to which T-cell responses were demonstrated shown above them.
The N448D substitution confers enhanced viral entry.
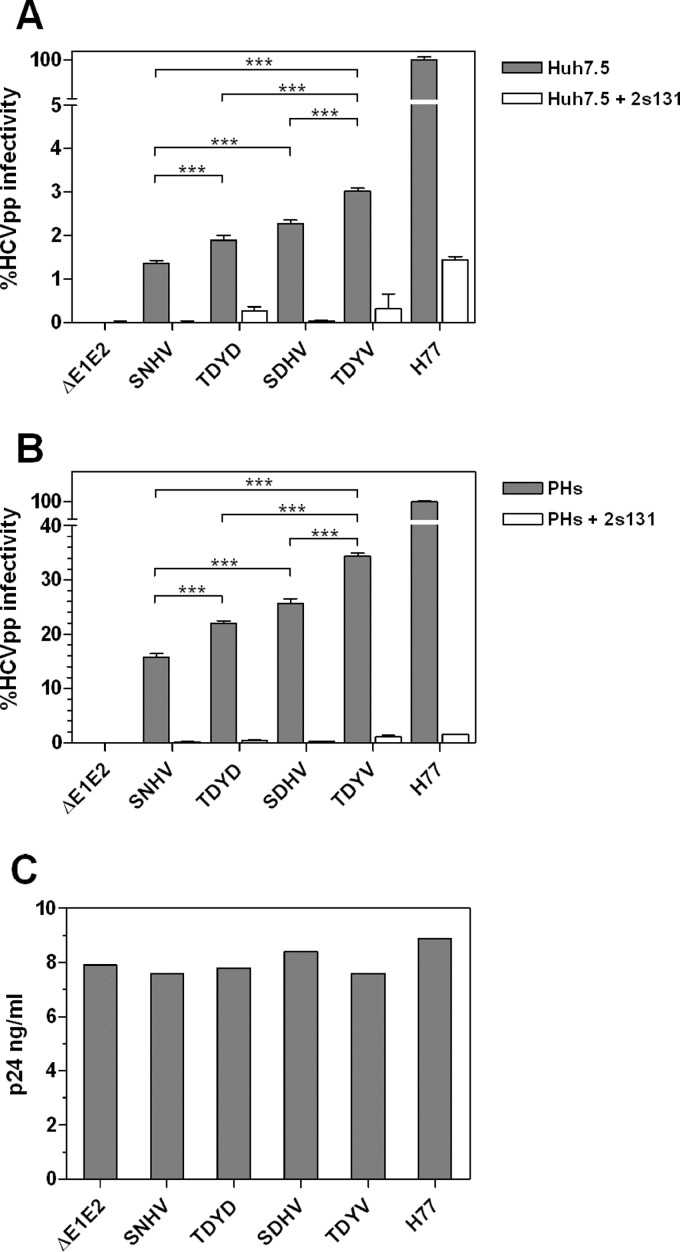
To determine whether the selective sweep observed in the mice was due to increased entry fitness, we determined the relative entry efficiency conferred by selected E1E2 variants present in the inoculum and in the mice. Lentiviral pseudotypes carrying the various E1E2s were generated and tested for the ability to infect both Huh7.5 cells and primary hepatocytes. In Huh7.5 cells, the overall level of infectivity conferred by the KP- and mouse-derived E1E2s was consistently lower than (<5%) the control H77c E1E2 (Fig. 7A). Therefore, we repeated the HCVpp assays using primary hepatocytes (PHs) as the target cells, which improved the relative infectivity of the KP- and mouse-derived E1E2 psuedoparticles (Fig. 7B). In both PHs and Huh7.5 cells, the TDYV variant (mouse major variant) was significantly more infectious (P < 0.001) than the SNHV variant (KP donor majority sequence). The minor variants recovered from both the mice and the KP inoculum conferred intermediate levels of infectivity. All N448D PNGS knockouts containing E1E2s had significantly increased capacity for cellular entry compared to the N448-containing major variant present in the inoculum. At the antibody concentration used, no variants were demonstrably more sensitive to neutralization by the anti-CD81 MAb 2s131. The significant increase in infectivity demonstrated between the majority circulating pre- and posttransmission E1E2s provides a likely mechanism for the selective sweep that occurred following HCV transmission.
Fig 7.
Infectivities of HCVpp bearing donor and recipient variant E1E2s in hepatoma cells and primary hepatocytes. (A and B) HCVpp infectivities conferred by the major inoculum E1E2 variant (SNHV), the major recipient E1E2 variant (TDYV), and two minor variants present in both the inoculum and recipient mice (SDHV and TDYD). Mean infectivity values are expressed as percentages of the infectivity conferred by H77c E1E2 in Huh7.5 cells (A) and PHs (B). Infectivity assays were performed in the presence or absence of anti-CD81 monoclonal antibody (MAb) 2s131, and the values presented are means for six replicates from two independent experiments with associated error bars (standard deviations). The associated significance values indicate that the TDYV variant confers more efficient entry than the SDHV, TDYD, and SNHV variants. Additionally, both the SDHV and TDYD variants confer improved capacity for entry compared to the SNHV variant. Differences in the mean infectivities conferred by each HCV E1E2 were assessed using analysis of variance (ANOVA) with infectivity: ***, P < 0.001. (C) p24 concentrations for each of the variant HCVpp preparations used to infect Huh7.5 cells or PHs.
Mathematical modeling.
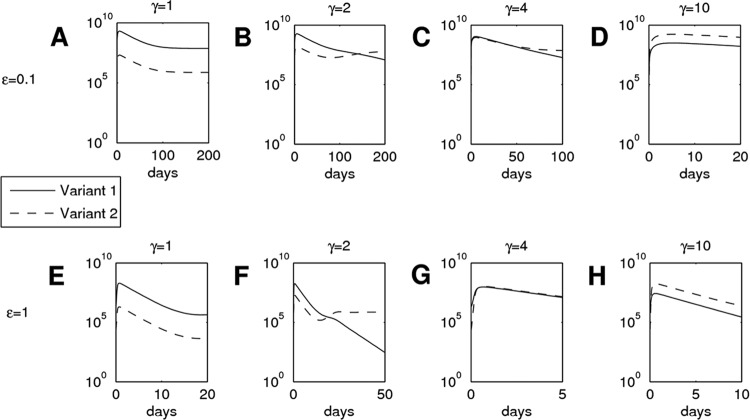
To confirm whether the increased capacity for entry in vitro could result in the observed selective outgrowth of the major mouse variant (TDYV/A) in vivo, the dynamics of competing HCV variants were mathematically modeled. The majority of parameters required for the mathematical analysis were determined from our own experimental data or from previously published literature. However, some of those derived from the literature are applicable only to mice with a functioning immune system. To account for the difference between these and our model, we scaled the relevant parameters to facilitate easy comparison between mouse models with functioning and nonfunctioning immune systems. In this analysis, we treated the population as two distinct variants: viruses containing key residues combinations other than TDYV/A (variant 1, with decreased entry fitness) and viruses that possessed TDYV/A (variant 2, with increased entry fitness). Our model indicates that for a γ value of >1, variant 2 will always outgrow variant 1, but the time at which this occurs depends upon the parameters employed (Fig. 8). Decreasing the initial concentration of target cells delays the time to outgrowth (the virions having fewer cells in which to replicate), although increasing it makes little difference, suggesting a saturation effect. Similarly, lowering the rate of target cell death also slows down the process.
Fig 8.
Mathematical modeling of HCV population expansion following transmission. The virus populations were modeled for two hypothetical variants, 1 and 2. If γ is equal to 1, the two variants have identical rates of infection of target cells; increasing γ gives variant 2 a fitness advantage. “ε = 0.1” is used to represent immunodeficient mice (A to D), while “ε = 1” represents mice with a functioning immune system (E to H). The y axes are plotted on a logarithmic scale.
When each variant is given equal entry fitness, variant 1 (due to its higher initial concentration) will remain dominant throughout the time course, while increasing the relative fitness of variant 2 provides sufficient advantage to ensure that it becomes the dominant population. For example, if the rate of infection of variant 2 is four times that of variant 1 (i.e., 1.2 × 106 ml virion−1 day−1), it will comprise 60% of the total viral load after only 20 days, despite starting at a drastically lower initial concentration. Larger increases in this rate can result in variant 2 becoming dominant almost immediately. Given the quantitative sensitivity of the model to the parameter choice, it would be unwise to make detailed predictions about the time taken to achieve dominance, but it is clear that a relatively subtle difference in the ability of a variant to infect a target cell can result in its overtaking all other variants in a matter of days.
DISCUSSION
In this study, the SCID/Alb-uPA chimeric mouse model was utilized to study HCV population dynamics following transmission. Such studies in natural infection have been hampered due to the difficulty in obtaining donor-recipient blood samples. Our data also highlight the fact that studying full-length E1E2, rather than gene fragments such as HVR1, yields important insight into determinants that shape HCV entry and transmission. Indeed, this study demonstrates that key E1E2 amino acid substitutions that predominate in viral populations following transmission are located outside HVR1.
Importantly, we used SGA to recover viral envelope genes from donor and recipient mice, followed by direct sequencing. This is preferable to standard bulk amplification and cloning, as it excludes polymerase-induced nucleotide misincorporation, amplicon resampling, selective amplification of specific isolates, cloning bias, and the generation of in vitro recombinants via polymerase template switching (56). Furthermore, SGA is not affected by the limitations of ultradeep-sequencing (UDS) technologies, where assigning linkage of the various substitutions remains challenging. The SGA approach has become the gold standard for sequence-based studies for a range of infectious agents (2, 4, 28, 33, 35, 56, 57) yet is still widely ignored within the HCV field.
Analysis of the E1E2 sequences derived from the inoculum and recipient mice revealed that the sequences formed two main clades, one containing sequences derived wholly from the inoculum and another containing mainly mouse-derived sequences. Surprisingly, there was no evidence of sequence clustering according to the recipient mouse. This contrasts with studies of common-source transmissions of HCV, where there is clear evidence of recipient/patient-specific clades (38). Factors that might shape patient-specific lineages include differential donor selection pressures (e.g., host-specific humoral and cellular immunity) or simply a founder effect arising from inoculation with low titers of genetically distinct variants. The genetic signature of the founder effect (establishment of infection from a single strain) is indistinguishable from a selective sweep (a selectively advantageous genetic variant is swept to fixation in the recipient) in contemporaneously sampled populations. However, knowledge of the titer and genetic composition of the inoculum allows us to assess which of these two competing evolutionary scenarios is most likely to have given rise to the change in variant frequencies observed in the recipient chimeric mice. We believe our data indicate that a selective sweep occurred, whereby variants that were present at undetectable frequencies in the donor inoculum became the major variant circulating in all experimentally infected chimeric mice. Coalescent analysis of individual transmission events indicated that one or more variants harboring the advantageous E1E2 motif were likely present in the inoculum rather than arising independently in each chimeric mouse, posttransmission.
While the KP inoculum used to infect each chimeric mouse possessed limited E1E2 genetic diversity, around 86% of recovered E1E2s from the donor serum were unique variants. Although 2.3 × 105 virions were used to seed each new infection, Bayesian coalescent analyses indicate that only a restricted number of variants established each initial infection. All E1E2s recovered posttransmission contained the N448D PNGS knockout, with selective amplification of variants bearing the TDYV/A motif occurring in all recipient chimeric mice. Indeed, increasing frequency of TDYV/A-bearing variants between the two sampling points was observed in all four animals (Fig. 4B), suggesting this variant is moving toward fixation in each population and pointing to a selective sweep occurring upon experimental HCV transmission. However, we were unable to detect any variants transmitted possessing the SNHV motif, which was the majority circulating strain in the inoculum. Therefore, we are unable to discount the possibility that the marked shifts observed in population composition posttransmission may be due to strong purifying selection against SNHV-bearing variants. In this scenario, the predominant variant in the inoculum possesses a deleterious phenotype in the chimeric mice and is removed from circulating populations via purifying selection.
In our transmission experiments, the major HVR1 amino acid variants pre- and posttransmission were identical, which is in agreement with other transmission studies (34, 37, 54, 60). HVR1 is often used as a surrogate marker for entire E1E2 genetic variation, and the HVR1 clonotype has been used as a basis for choosing E1E2 clones for downstream analyses (20). However, our data indicate that this approach can be flawed. In all of the separate transmission experiments, the major HVR1 variants were identical, and differences in entry fitness were associated with 4 amino acid residues (198, 448, 474, and 570) outside HVR1. All posttransmission E1E2s harbored an N→D PNGS knockout, which presumably conferred a selective advantage in each chimeric mouse host. A PNGS knockout at position 448 (corresponding to glycan E2N4) has previously been shown to abrogate H77 E1E2 pseudoparticle infectivity (21), although a similar knockout does not abrogate HCVcc infectivity (24). In the HCVcc system, the removal of a glycan at position 448 has been shown to render JFH-1 E1E2 more susceptible to antibody-mediated neutralization (24). Therefore, the absence of neutralizing antibodies and the increased fitness of the TDYV variant provide a likely mechanism for the selective amplification of this strain in all recipient chimeric mice. Unfortunately, we were unable to obtain any of the original KP serum to perform neutralization tests to validate this hypothesis.
In our hands, the KP- and mouse-derived E1E2 clones conferred very low-level infectivity in HCVpp when used to infect Huh7.5 cells. However, infectivity relative to H77 E1E2 was markedly increased when primary hepatocytes were used as the target cell. This contrasts with a previous study of HCVpp entry using E1E2 derived from a liver transplant setting, where primary hepatocytes were less efficient at supporting entry than Huh7.5 cells (20). It is unclear what underlies this discrepancy although hepatocyte donor-specific genetic differences may play a role. Importantly the infectivity conferred by the majority mouse-derived E1E2 was significantly greater than that conferred by the major variant present in the KP inoculum in both cell types. In the liver transplant setting, variants with increased capacity for cellular entry and also increased resistance to autologous neutralization were shown to preferentially reinfect the grafted liver (20). This report also showed that one of these variants also demonstrated increased fitness in a chimeric mouse. Given their neutralization-resistant phenotype, it is intriguing that these variants did not dominate pretransplantation. It is important to note that there are significant differences between acute HCV infection and liver graft reinfection following transplantation, not least the presence of host immunity, which will help shape the evolution of the reinfecting virus. Also, corticosteroids given post-liver transplantation have been shown to increase the infectivity of HCV by altering the expression levels of entry factors (10, 11), which may also help to select for particular viral variants in this unique setting.
Selective outgrowth of specific viral variants could also be driven by host factors, including genetic polymorphisms. To minimize this effect, we used mice that had hepatocyte grafts from a number of distinct donors. HCV infection of recipient chimeric mice is dependent on a high percentage of human grafts in each SCID/Alb-uPA mouse (42). It has previously been demonstrated that HCV does not replicate in mouse hepatocytes (15, 39). Thus, the productive viral replication and infectious-particle production observed in recipient chimeric mice can only be sustained by engrafted human hepatocytes. The observed E1E2 adaptations in recipient chimeric-mouse virions will therefore have been driven to fixation by their ability to enter and replicate in human hepatocytes, indicating that the chimeric-mouse system is a useful surrogate model to enable interrogation of HCV transmission to an HCV-naïve host and establishment of initial infection prior to seroconversion.
In summary, we have applied a robust methodological approach to enable interrogation of the dynamics of HCV transmission in the SCID/Alb-uPA chimeric-mouse model. Our data show that the environment of the new host dramatically alters the virus population following transmission. Identification of the molecular determinants of transmitted E1E2 glycoproteins that establish initial infection will be relevant to the design of vaccines and other preventative strategies.
ACKNOWLEDGMENTS
We thank Francois-Loic Cosset for provision of the HCVpp packaging and reporter plasmids, Jens Bukh for the H77c E1E2 plasmid, and Charles Rice for Huh7.5 cells.
This work was supported by the Medical Research Council (G0801169), the European Union (MRTN-CT-2006-035599), and the Nottingham Digestive Diseases Centre Biomedical Research Unit. P.B. is a Jenner Institute Investigator. M.L. is supported by NIH grants R21AI080916 and R01AI079031. D.L.T. and N.K. hold Canadian Institute of Health Research grants.
Footnotes
Published ahead of print 1 August 2012
REFERENCES
- 1. Alter MJ. 2002. Prevention of spread of hepatitis C. Hepatology 36:S93–98 [DOI] [PubMed] [Google Scholar]
- 2. Anderson JA, et al. 2010. HIV-1 populations in semen arise through multiple mechanisms. PLoS Pathog. 6:e1001053 doi:10.1371/journal.ppat.1001053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bar KJ, et al. 2009. Wide variation in the multiplicity of HIV-1 infection among injection drug users. J. Virol. 84:6241–6247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Brown RJ, et al. 2011. Intercompartmental recombination of HIV-1 contributes to env intrahost diversity and modulates viral tropism and sensitivity to entry inhibitors. J. Virol. 85:6024–6037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Brown RJP, et al. 2007. Cross-genotype characterization of genetic diversity and molecular adaptation in hepatitis C virus envelope glycoprotein genes. J. Gen. Virol. 88:458–469 [DOI] [PubMed] [Google Scholar]
- 6. Buchbinder SP, et al. 1994. Hepatitis C virus infection in sexually active homosexual men. J. Infect. 29:263–269 [DOI] [PubMed] [Google Scholar]
- 7. Bukh J, Miller RH, Purcell RH. 1995. Genetic heterogeneity of hepatitis C virus: quasispecies and genotypes. Semin. Liver Dis. 15:41–63 [DOI] [PubMed] [Google Scholar]
- 8. Bull RA, et al. 2011. Sequential bottlenecks drive viral evolution in early acute hepatitis C virus infection. PLoS Pathog. 7:e1002243 doi:10.1371/journal.ppat.1002243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cavalheiro NP, De La Rosa A, Elagin S, Tengan FM, Araújo ES, Barone AA. 2009. Hepatitis C: sexual or intrafamilial transmission? Epidemiological and phylogenetic analysis of hepatitis C virus in 24 infected couples. Rev. Soc. Bras. Med. Trop. 42:239–244 [DOI] [PubMed] [Google Scholar]
- 10. Ciesek S, et al. 2010. Glucocorticosteroids increase cell entry by hepatitis C virus. Gastroenterology 138:1875–1884 [DOI] [PubMed] [Google Scholar]
- 11. Ciesek S, Wedemeyer H. 2012. Immunosuppression, liver injury and post-transplant HCV recurrence. J. Viral Hepat. 19:1–8 [DOI] [PubMed] [Google Scholar]
- 12. Curran R, et al. 2002. Evolutionary trends of the first hypervariable region of the hepatitis C virus E2 protein in individuals with differing liver disease severity. J. Gen. Virol. 83:11–23 [DOI] [PubMed] [Google Scholar]
- 13. Dahari H, et al. 2005. Mathematical modeling of primary hepatitis C infection: noncytolytic clearance and early blockage of virion production. Gastroenterology 128:1056–1066 [DOI] [PubMed] [Google Scholar]
- 14. Derdeyn CA, et al. 2004. Envelope-constrained neutralization-sensitive HIV-1 after heterosexual transmission. Science 303:2019–2022 [DOI] [PubMed] [Google Scholar]
- 15. Dorner M, et al. 2011. A genetically humanized mouse model for hepatitis C virus infection. Nature 474:208–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Drummer HE, Boo I, Maerz AL, Poumbourios P. 2006. A conserved Gly436-Trp-Leu-Ala-Gly-Leu-Phe-Tyr motif in hepatitis C virus glycoprotein E2 is a determinant of CD81 binding and viral entry. J. Virol. 80:7844–7853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Drummer HE, Poumbourios P. 2004. Hepatitis C virus glycoprotein E2 contains a membrane-proximal heptad repeat sequence that is essential for E1E2 glycoprotein heterodimerization and viral entry. J. Biol. Chem. 279:30066–30072 [DOI] [PubMed] [Google Scholar]
- 18. Drummond A, Ho S, Phillips M, Rambaut A. 2006. Relaxed phylogenetics and dating with confidence. PLoS Biol. 4:e88 doi:10.1371/journal.pbio.0040088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Drummond AJ, Rambaut A. 2007. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 7:214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Fafi-Kremer S, et al. 2010. Viral entry and escape from antibody-mediated neutralization influence hepatitis C virus reinfection in liver transplantation. J. Exp. Med. 207:2019–2031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Goffard A, et al. 2005. Role of N-linked glycans in the functions of hepatitis C virus envelope glycoproteins. J. Virol. 79:8400–8409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Goldberg D, Anderson E. 2004. Hepatitis C: who is at risk and how do we identify them? J. Viral Hepat. 11:12–18 [DOI] [PubMed] [Google Scholar]
- 23. Helle F, et al. 2007. The neutralizing activity of anti-hepatitis C virus antibodies is modulated by specific glycans on the E2 envelope protein. J. Virol. 81:8101–8111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Helle F, et al. 2010. Role of N-linked glycans in the functions of hepatitis C virus envelope proteins incorporated into infectious virions. J. Virol. 84:11905–11915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hsu M, et al. 2003. Hepatitis C virus glycoproteins mediate pH-dependent cell entry of pseudotyped retroviral particles. Proc. Natl. Acad. Sci. U. S. A. 100:7271–7276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Johansson DX, et al. 2007. Human combinatorial libraries yield rare antibodies that broadly neutralize hepatitis C virus. Proc. Natl. Acad. Sci. U. S. A. 104:16269–16274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Keck ZY, et al. 2004. Hepatitis C virus E2 has three immunogenic domains containing conformational epitopes with distinct properties and biological functions. J. Virol. 78:9224–9232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Keele BF, et al. 2008. Identification and characterization of transmitted and early founder virus envelopes in primary HIV-1 infection. Proc. Natl. Acad. Sci. U. S. A. 105:7552–7557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kneteman NM, Toso C. 2009. In vivo study of HCV in mice with chimeric human livers. Methods Mol. Biol. 510:383–399 [DOI] [PubMed] [Google Scholar]
- 30. Kumar U, Brown J, Monjardino J, Thomas HC. 1993. Sequence variation in the large envelope glycoprotein (E2/NS1) of hepatitis C virus during chronic infection. J. Infect. Dis. 167:726–730 [DOI] [PubMed] [Google Scholar]
- 31. Lavillette D, et al. 2005. Characterization of host-range and cell entry properties of the major genotypes and subtypes of hepatitis C virus. Hepatology 41:265–274 [DOI] [PubMed] [Google Scholar]
- 32. Law M, et al. 2008. Broadly neutralizing antibodies protect against hepatitis C virus quasispecies challenge. Nat. Med. 14:25–27 [DOI] [PubMed] [Google Scholar]
- 33. Li H, et al. 2010. High multiplicity infection by HIV-1 in men who have sex with men. PLoS Pathog. 6:e1000890 doi:10.1371/journal.ppat.1000890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lin HJ, Seeff LB, Barbosa L, Hollinger FB. 2001. Occurrence of identical hypervariable region 1 sequences of hepatitis C virus in transfusion recipients and their respective blood donors: divergence over time. Hepatology 34:424–429 [DOI] [PubMed] [Google Scholar]
- 35. Liu W, et al. 2010. Origin of the human malaria parasite Plasmodium falciparum in gorillas. Nature 467:420–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Manzin A, et al. 2000. Dominant role of host selective pressure in driving hepatitis C virus evolution in perinatal infection. J. Virol. 74:4327–4334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Manzin A, et al. 1998. Evolution of hypervariable region 1 of hepatitis C virus in primary infection. J. Virol. 72:6271–6276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. McAllister J, et al. 1998. Long-term evolution of the hypervariable region of hepatitis C virus in a common-source-infected cohort. J. Virol. 72:4893–4905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. McCaffrey AP, et al. 2002. Determinants of hepatitis C translational initiation in vitro, in cultured cells and mice. Mol. Ther. 5:676–684 [DOI] [PubMed] [Google Scholar]
- 40. Mee CJ, et al. 2010. Hepatitis C virus infection reduces hepatocellular polarity in a vascular endothelial growth factor-dependent manner. Gastroenterology 138:1134–1142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Mercer DF, et al. 2001. Hepatitis C virus replication in mice with chimeric human livers. Nat. Med. 7:927–933 [DOI] [PubMed] [Google Scholar]
- 42. Meuleman P, Leroux-Roels G. 2008. The human liver-uPA-SCID mouse: a model for the evaluation of antiviral compounds against HBV and HCV. Antiviral Res. 80:231–238 [DOI] [PubMed] [Google Scholar]
- 43. Meunier J-C, et al. 2008. Isolation and characterization of broadly neutralizing human monoclonal antibodies to the E1 glycoprotein of hepatitis C virus. J. Virol. 82:966–973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Miller FD, Abu-Raddad LJ. 2010. Evidence of intense ongoing endemic transmission of hepatitis C virus in Egypt. Proc. Natl. Acad. Sci. U. S. A. 107:14757–14762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Mitry RR. 2009. Isolation of human hepatocytes. Methods Mol. Biol. 481:17–23 [DOI] [PubMed] [Google Scholar]
- 46. Mizuno Y, et al. 1997. Study of needlestick accidents and hepatitis C virus infection in healthcare workers by molecular evolutionary analysis. J. Hosp. Infect. 35:149–154 [DOI] [PubMed] [Google Scholar]
- 47. Neumann AU, et al. 1998. Hepatitis C viral dynamics in vivo and the antiviral efficacy of interferon-alpha therapy. Science 282:103–107 [DOI] [PubMed] [Google Scholar]
- 48. Owsianka A, et al. 2005. Monoclonal antibody AP33 defines a broadly neutralizing epitope on the hepatitis C virus E2 envelope glycoprotein. J. Virol. 79:11095–11104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Owsianka AM, et al. 2008. Broadly neutralizing human monoclonal antibodies to the hepatitis C virus E2 glycoprotein. J. Gen. Virol. 89:653–659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Owsianka AM, et al. 2006. Identification of conserved residues in the E2 envelope glycoprotein of the hepatitis C virus that are critical for CD81 binding. J. Virol. 80:8695–8704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Perelson AS. 2002. Modelling viral and immune system dynamics. Nat. Rev. Immunol. 2:28–36 [DOI] [PubMed] [Google Scholar]
- 52. Perotti M, et al. 2008. Identification of a broadly cross-reacting and neutralizing human monoclonal antibody directed against the hepatitis C virus E2 protein. J. Virol. 82:1047–1052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Posada D, Crandall KA. 1998. MODELTEST: testing the model of DNA substitution. Bioinformatics 14:817–818 [DOI] [PubMed] [Google Scholar]
- 54. Ray SC, et al. 2000. Hypervariable region 1 sequence stability during hepatitis C virus replication in chimpanzees. J. Virol. 74:3058–3066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Sabo MC, et al. 2011. Neutralizing monoclonal antibodies against hepatitis C virus E2 protein bind discontinuous epitopes and inhibit infection at a postattachment step. J. Virol. 85:7005–7019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Salazar-Gonzalez JF, et al. 2008. Deciphering human immunodeficiency virus type 1 transmission and early envelope diversification by single-genome amplification and sequencing. J. Virol. 82:3952–3970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Schnell G, Joseph S, Spudich S, Price RW, Swanstrom R. 2011. HIV-1 replication in the central nervous system occurs in two distinct cell types. PLoS Pathog. 7:e1002286 doi:10.1371/journal.ppat.1002286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Shapiro B, Rambaut A, Drummond AJ. 2006. Choosing appropriate substitution models for the phylogenetic analysis of protein-coding sequences. Mol. Biol. Evol. 23:7–9 [DOI] [PubMed] [Google Scholar]
- 59. Simmonds P. 2004. Genetic diversity and evolution of hepatitis C virus—15 years on. J. Gen. Virol. 85:3173–3188 [DOI] [PubMed] [Google Scholar]
- 60. Sugitani M, Shikata T. 1998. Comparison of amino acid sequences in hypervariable region-1 of hepatitis C virus clones between human inocula and the infected chimpanzee sera. Virus Res. 56:177–182 [DOI] [PubMed] [Google Scholar]
- 61. Swofford DL. 1998. PAUP*: Phylogenetic Analysis Using Parsimony (*and other methods). Sinauer Associates, Sunderland, MA [Google Scholar]
- 62. Tamura K, Nei DJM, Kumar S. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596–1599 [DOI] [PubMed] [Google Scholar]
- 63. Tarr AW, et al. 2006. Characterization of the hepatitis C virus E2 epitope defined by the broadly neutralizing monoclonal antibody AP33. Hepatology 43:592–601 [DOI] [PubMed] [Google Scholar]
- 64. Thomson PC, et al. 2011. A case of hepatitis C virus transmission acquired through sharing a haemodialysis machine. Nephrol. Dial. Transplant. Plus 4:33–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Tong MJ, El-Farra NS, Reikes AR, Co RL. 1995. Clinical outcomes after transfusion-associated hepatitis C. N. Engl. J. Med. 332:1463–1466 [DOI] [PubMed] [Google Scholar]
- 66. Villano AS, Vlahov D, Nelson EK, Cohn S, Thomas LD. 1999. Persistence of viremia and the importance of long-term follow-up after acute hepatitis C infection. Hepatology 29:908–914 [DOI] [PubMed] [Google Scholar]
- 67. Wang GP, Sherrill-Mix SA, Chang K-M, Quince C, Bushman FD. 2010. Hepatitis C virus transmission bottlenecks analyzed by deep sequencing. J. Virol. 84:6218–6228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Weiner AJ, et al. 1993. A unique, predominant hepatitis C virus variant found in an infant born to a mother with multiple variants. J. Virol. 67:4365–4368 [DOI] [PMC free article] [PubMed] [Google Scholar]